Abstract
We have isolated the human GRAF gene (for GTPase regulator associated with the focal adhesion kinase pp125FAK). This gene was fused with MLL in a unique t(5;11)(q31;q23) that occurred in an infant with juvenile myelomonocytic leukemia. GRAF encodes a member of the Rho family of the GTPase-activating protein (GAP) family. On the protein level, it is 90% homologous to the recently described chicken GRAF gene that functions as a GAP of RhoA in vivo and is thus a critical component of the integrin signaling transduction pathway. The particular position of the human GRAF gene at 5q31 and the proposed antiproliferative and tumor suppressor properties of its avian homologue suggest that it also might be pathogenetically relevant for hematologic malignancies with deletions of 5q. To investigate this possibility, we sequenced 4–5 individual cDNA clones from 13 cases in which one allele of GRAF was deleted. We found point mutations within the GAP domain of the second GRAF allele in one patient. In two additional patients we found an insertion of 52 or 74 bp within the GRAF cDNA that generates a reading frame shift followed by a premature stop codon. GRAF maps outside the previously defined commonly deleted 5q31 region. Nevertheless, inactivation of both alleles in at least some cases suggests that deletions and mutations of the GRAF gene may be instrumental in the development and progression of hematopoeitic disorders with a del(5q).
Chromosome abnormalities associated with hematologic malignancies alter the normal structure and function of genes that control cell proliferation and differentiation either in a positive or negative fashion (1). Normal cell division is positively regulated or activated through signal transduction pathways composed of extracellular signals, receptor G proteins, protein kinases, and transcription factors (1, 2). The genes encoding such proteins also are known as protooncogenes, because mutations turn them into dominant oncogenes with altered properties. Thus, disruption of one allele generally is sufficient to disturb normal cell division and initiate a neoplastic phenotype. Such protooncogenes frequently are affected by chromosome translocations (3). As a result, fusion of the participating genes leads to either abnormal activation or to the generation of novel chimeric oncogenes with new functions (3, 4). The majority of translocations that occur at 11q23 in acute leukemias disrupt the mixed-lineage leukemia (MLL) gene within a small region of 8.3 kb and fuse it to a variety of different partner genes. Twenty-three of those MLL-partner genes were cloned and sequenced, which make the detection of the leukemia-specific chimeric mRNA possible by reverse transcription–PCR (RT-PCR) (5–9).
The products of many so-called tumor suppressor genes, on the other hand, inhibit the same signal transduction pathway and are negative regulators of the cell cycle (10). Because mutations in these genes act recessively and result in a loss of function, uncontrolled cell growth takes place only after inactivation or elimination of both alleles. Most often, one allele is deleted, whereas the second one may be functionally compromised by various other mutations. The most common structural aberrations encountered in myelodysplastic syndromes (MDSs) and acute myeloid leukemias (AMLs) are deletions of the long arm of chromosome 5 (11–17). Although the size and position of the deleted segments may vary considerably, band 5q31.1 is consistently lost in 90% of cases (11, 13, 15, 18–20). This finding led to the notion that this critical region must harbor a tumor suppressor gene whose loss or inactivation is critical for the development of these malignant myeloid disorders. However, the variability of the deletions, the paucity of highly polymorphic markers, and the large number of attractive candidate genes within this region has hampered the identification and isolation of such a gene considerably (11, 13, 15–19).
The detection of a unique t(5;11)(q31;q23) in an infant with juvenile myelomonocytic leukemia and a MLL gene rearrangement provided an opportunity to clone another MLL fusion partner gene. We recovered a member of the GTPase-activating protein (GAP) family, which we identified as the human homologue of the described avian GRAF gene (21). Its particular chromosomal location and its potential growth inhibitory and antioncogenic properties prompted us to investigate its potential involvement in hematological malignancies with 5q abnormalities further.
Materials and Methods
Case History.
Juvenile myelomonocytic leukemia was diagnosed in a 4-month-old boy accordingly to the criteria of the European Working Group on Childhood Myelodysplastic Syndromes (22). The infant was admitted to the hospital because he had developed a striking livid color of the skin with small nodular infiltrations. Physical examination revealed an slightly enlarged liver and spleen (2 cm below costal margin). Blood count was hemoglobin 9.8 g/dl, platelets 286,000/μ, and leukocytes 19,000 μl. Bone marrow was infiltrated with 3% myelomonocytic blasts. Subsequently, the amount of blast cells raised up to 40% during a period of 2 months. Then, the boy was treated according to the German AML-BFM 93 chemotherapy protocol and received a bone marrow transplantation (BMT) from an HLA-identical sibling. He currently is in complete remission 3 years after BMT. At diagnosis of juvenile myelomonocytic leukemia, cytogenetic analysis of G-banded preparations detected a clonal t(5;11)(q31;q23) as the sole karyotype abnormality (Fig. 1a).
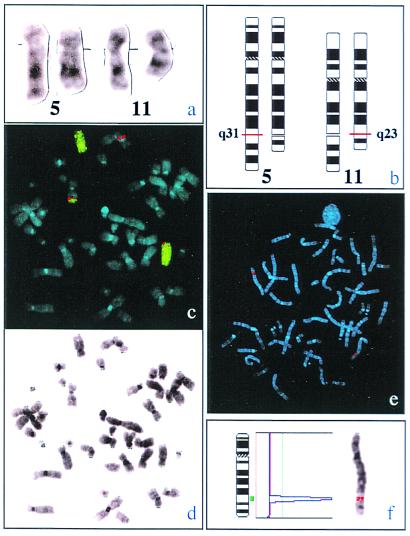
Figure 1.
(a) Partial karyotype showing the normal (outside) and translocated (inside) homologues of chromosomes 5 and 11. (b) Schematic presentation of the translocation t(5;11)(q31;q23). (c) As a result of the translocation the MLL-yeast artificial chromosome clone (red) is split and partly translocated to chromosome 5 (green painting probe), thus generating signals on both chromosomes 11 as well as on the affected chromosome 5. (d) G-banding of a representative metaphase from the leukemic clone. (e and f) Hybridization of a 1,176-bp clone of the human GRAF gene onto a representative normal metaphase chromosome confirms its location at 5(q31).
5′ and 3′ Rapid Amplification of cDNA Ends (RACE)-PCR and Nucleotide Sequencing.
We performed RACE-PCR with the Marathon cDNA amplification kit and nested PCR (CLONTECH) according to the manufacturer's instructions. For the 3′ RACE PCR, we used the two MLL-specific sense primers (MLL 5′-TCATCCCGCCTCAGCCACCTACTACAGGACCGC-3′; MLL 5′-CAAGAAAAGAAGTTCCCAAAACCACTCCTAGTGAGCC-3′) that were located in exon 5 of MLL (23). The first 250 bp of the human GRAF sequence were used to construct two GRAF antisense primers (GRAF 5′-TGCAGGACGGGGGCTTGGAGTCACTGCT-3′, GRAF 5′-AGCTGGGCATTGGTGAGAGGCATATCGGGCACGGTGTT-3′), which were necessary for the 5′ RACE-PCR.
RT-PCR and Long-Range PCR.
RNA and DNA were either isolated with the guanidium-isothiocyanat method or with ion exchange chromatography, respectively. For RT-PCR we denatured 5 μg of total RNA at 70°C for 5 min. cDNA synthesis was carried out at 42°C for 60 min with 100 pmol of random nucleotide hexamers in 20 μl. For sequence analysis of GRAF in patients with MDS/AML and del(5q) the entire coding sequence of GRAF was amplified in five overlapping fragments (C0–C4) by using the oligonucleotides shown in the supplementary material and Table 1, which are published on the PNAS web site, www.pnas.org. To increase specificity we used a seminested PCR approach. The concentration of each primer was 4 pmol in the first round or 20 pmol in the second PCR round. Amplification was carried out for 35 cycles with denaturation at 94°C for 10 sec, annealing at 65°C for 30 sec, and strand extension for 30 sec at 72°C. We analyzed 4–5 clones of each patient, except for patients #2 and #12 in whom 20 clones were sequenced because of the low amount of blast cells in their bone marrow. Samples that had sequence variations of GRAF were amplified and cloned a second time to exclude misincorportaion of nucleotides by Taq polymerase. For the long-range amplification of the genomic DNA we applied a nested long-range PCR protocol (24). The primers used for long-range amplification of the normal MLL allele have been described (25).
Determination of the Genomic MLL/GRAF Fusion by Fingerprinting.
After amplification of both the genomic unrearranged MLL and the MLL/GRAF fusion by long-range PCR we digested the PCR products by DdeI or Tru91 (Boehringer Mannheim). The digests generate protruding 5′ ends containing adenosine residues. The restriction fragments were end-labeled with fluorescein-11-dUTP (Amersham-Buchler, Braunschweig, Germany). After removal of unincorporated dUTP with the help of a Nucleotide Removal Kit (Qiagen, Düsseldorf, Germany) the digested fragments were subjected to genescan analysis (genescan 672 software, Perkin–Elmer). By comparing the restriction pattern of the normal unrearranged MLL and the rearranged MLL/GRAF allele we were able to localize the chromosomal breakpoints on the genomic DNA level with a range of 200 bp. The exact fusion sites were determined by DNA sequencing. The detailed protocol of the fingerprint procedure has been described (25).
RT-PCR, fluorescence in situ hybridization (FISH), Northern and Southern blot, as well as the immunohistochemical studies were performed by standard protocols. For detailed description and PCR primers used for amplification of fragments C0–C4, see Table 1.
Results
Molecular Analysis of the t(5;11)(q31;q23).
Southern blot and FISH analysis confirmed that the cytogenetically detected t(5;11)(q31;q23) disrupts the MLL gene (Fig. 1 a–d and see Fig. 9, which is published as supplemental material). RACE-PCR revealed four clones in which the downstream MLL sequences were replaced by 250 bp of non-MLL cDNA. By 5′ and 3′ RACE-PCR we recovered a full-length cDNA of 3,163 nt with an ORF of 759 aa (GenBank accession no. Y10388). Homology scans indicated that we identified a member of the Rho family of small GTPases whose predicted protein is 90% homologous to the product of a cDNA clone obtained from a chicken library (21). Because the Src homology 3 (SH3) domain of this particular avian protein mediates binding to the pp125 focal adhesion kinase (pp125FAK) protein, this clone was termed GRAF (for GTPase regulator associated with pp125FAK). The predicted fusion protein consists of the N-terminal part of MLL and the C-terminal part of GRAF. The ORF of the chimeric mRNA is maintained and enables the formation of a chimeric oncoprotein. The MLL part retains the putative “AT-hook” DNA binding domain and the DNA methyltransferase motifs present in the MLL amino-terminal region (26, 27). The GRAF part of the fused gene retains the SH3 domain, but not the GAP domain. To analyze the second allele of GRAF in the patient with translocation t(5;11), we sequenced DNA of five individual clones and found no mutation of the cDNA.
RT-PCR revealed that the fusion mRNA of the MLL/GRAF is expressed, whereas the reciprocal GRAF/MLL is not (Fig. 2). We cannot exclude that this negative result on expression of the der(5) GRAF/MLL fusion transcript is caused by alternative splicing of the mRNA. With long-range PCR, we were able to amplify an approximately 13-kb fragment of the MLL/GRAF sequence on the genomic DNA level in the case with the t(5;11), but in none of four leukemic cell lines or three healthy individuals. We amplified an approximately 11-kb long intron of the GRAF gene in which the break and fusion with MLL took place (Fig. 2B). At the genomic DNA level, we determined the structure of the MLL/GRAF gene fusion. At nucleotide 3234 of the breakpoint cluster region (BCR) of MLL (GenBank accession no. U04737) (23), we found a 1,215-bp fragment encompassing nucleotide numbers 3205–4420 of the BCR region of MLL that was inversely inserted between MLL and nucleotide 4460 of intron 12 of GRAF (Fig. 2C). The DNA sequence of the genomic MLL/GRAF fusion has been deposited to the GenBank as well (accession no. AF260130). In contrast to MLL, there was no duplication or deletion of GRAF intronic sequences at the MLL/GRAF genomic breakpoint.
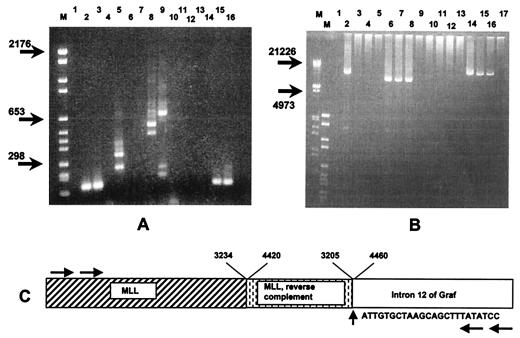
Figure 2.
(A) Results of RT-PCR analyses. Lane M contains a size marker VI (Boehringer Mannheim). Lanes 1, 4, 7, and 12 are negative controls in which the cDNA was replaced by sterile water. Normal fragments are obtained from the patient's unaffected GRAF allele (lane 2) and from the cell line Mono-Mac6 (lane 3). A MLL/GRAF fusion mRNA is detected in the sample with a t(5;11)(q31;q23) (lane 5) but not in the cell line lacking this translocation (lane 6). Normal fragments are obtained from the patient's unaffected MLL allele (lane 8) and the cell line Mono-Mac6 (lane 9). The additional fragments in lanes 5, 8, and 9 are generated by MLL splice variants. Further analysis reveals that the reciprocal GRAF/MLL fragment is neither present in the patient's sample (lane 10) nor in the cell line (lane 11). Control amplifications with primers specific for the ABL gene are shown in lanes 15 (patient sample) and 16 (cell line). (B) Long-range PCR results of genomic DNA. Lanes M contain the size markers III and VI (Boehringer Mannheim). Lane 1 is a negative control. A MLL/GRAF fusion product is detected in the patient with the t(5;11)(q31;23) (lane 2) but not in the control cell line Mono-Mac6 (lane 3). Lanes 4, 5, 9, and 17 are negative controls. A normal 8-kb fragment that covers the breakpoint cluster region of the unaffected MLL alleles in the patient with the t(5;11)(q31;q23) (lane 6), in a healthy individual (lane 7), and in the Mono-Mac cell line (lane 8) is seen. No reciprocal GRAF/MLL gene fragment is detected in any of these samples (lanes 10–13), whereas in all of them an approximately 13-kb long intron of GRAF becomes evident (lanes 14–16). (C) Sequence and schematic representation of the inverted duplication of MLL within the genomic MLL/GRAF fusion. Numbering of nucleotides within the breakpoint region of MLL according to ref. 23. The horizontal arrows indicate the positions of the primers used for amplification of the genomic MLL/GRAF fusion seen in lane 2 of B.
Motifs of GRAF and its mRNA Expression in Human Tissues.
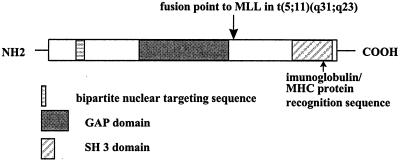
The search of the prosite database revealed a GAP domain (amino acids 390–538) and an SH3 domain (amino acids 701–759) at the carboxyl terminus of the protein (Fig. 3). Other homologous members of the Rho family include the Rho-related GAP proteins, β-chimerin and BCR (28). Analysis of the human GRAF protein sequence with the motif software (Oxford Molecular, Oxford, U.K.) revealed a bipartite nuclear targeting sequence (amino acids 120–137), 10 potential protein kinase C, and four cAMP/cGMP-dependent protein kinase phosphorylation sites.
Figure 3.
Schematic diagram of partial human GRAF protein.
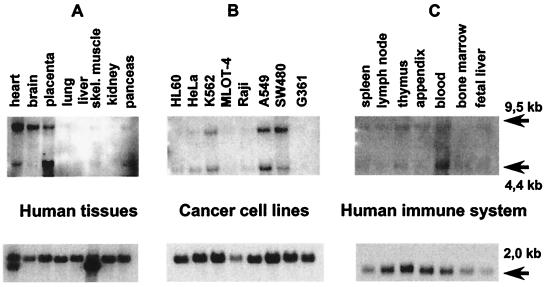
Northern blot analysis of the human GRAF disclosed a major and a minor transcript with 9.5 and 4.4 kb, respectively (Fig. 4A). The expression patterns in various normal tissues and cell lines are shown in Fig. 4. A Zoo blot that was performed with a 1,167-bp clone showed that this gene is conserved in all of the examined mammalian tissues including monkey, cow, and dog. The probe did not hybridize to yeast DNA (data not shown).
Figure 4.
(A) Northern blot analysis of poly(A)+ mRNAs derived from various normal human tissues and cancer cell lines (B). (C) The GRAF gene is only weakly expressed in various hematopoietic and immune system tissues. A control hybridization with β-actin is shown in the bottom row.
Immunohistochemical Findings.
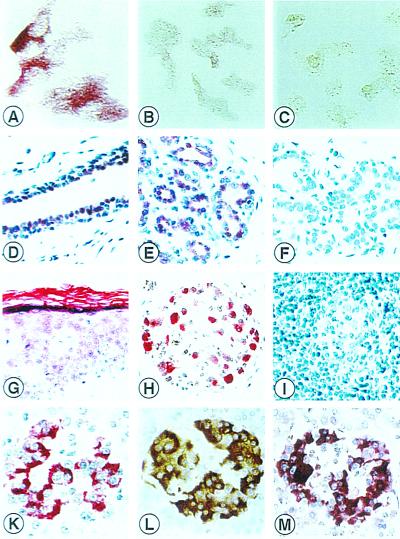
We tested the specificity of the polyclonal anti-GRAF antisera ED98015 and ED98016 in frozen and paraffin-embedded specimens of expressing and nonexpressing cell lines and tissues. We observed a strong cytoplasmatic labeling in the transfected cell line KMST6 that was transfected with an expression vector to overproduce GRAF (Fig. 5A), in the cells of the islets of Langerhans (Fig. 5K) and in tumor cells from a clear cell carcinoma of the kidney. Frozen sections and paraffin-embedded specimens of the mammary gland were only moderately labeled (Fig. 5 D and E). Negative controls consisted of incubating the above described cell preparations with both preimmune sera or with mouse anti-rabbit Ig (Fig. 5 B, C, and F).
Figure 5.
Human GRAF protein in a cell line and paraffin-embedded specimens. Specificity of the anti-GRAF antiserum tested with KMST6 cells. Strong labeling of transfected KMST6 cells with ED98016 (A). No labeling was observed with preimmune serum (B) and in untransfected KMST6 cells (C). Human GRAF protein in epithelial cells. Nuclear and cytoplasmatic immunoreactivity in ductal and acinary cells of the mammary (D and E), negative control (F). Weak GRAF staining in squamous epithelial cells of the skin and strong staining in the stratum corneum of the skin (G). Strong GRAF protein expression in ganglion cells (H). No staining of GRAF in lymphocytes and germinal center cells of a lymph node (I). Large amounts of GRAF protein were detected in insulin-producing β cells within the islets of Langerhans as detected by immunohistohemical single and double-staining: GRAF-staining APAAP red (K), insulin-staining LSAB brown (L), and red-brown staining product indicating strong GRAF (red) expression in insulin (brown) containing B cells (M). Original magnifications: ×100 A–C; ×40 D–M.
GRAF Protein Expression in Human Tissues.
The distribution patterns of GRAF protein in normal human tissues are summarized in Table 2, which is published as supplemental material. In total, more than 1,100 histological structures were analyzed by immunostaining. GRAF protein was widely expressed in epithelial tissues of the vast majority of organs. It was not detected in nonvascular supporting and connective tissue of mesenchymal origin, white adipose tissue, the stromal cells of many organs, hyaline cartilage, and, surprisingly, the myelo- and lymphopoeitic cell system. In contrast, we noticed a cytoplasmatic expression in the erythropoeitic cell lineage. For further details see the supplemental material.
Chromosomal Assignment of the GRAF Gene to 5(q31) and FISH Analysis of Patients with 5q Abnormality.
Hybridization of a 1,167-bp cDNA clone of the human GRAF gene onto normal metaphase chromosomes confirmed its position at 5(q31) (Fig. 1 e and f). Previously, two contig maps of the smallest commonly deleted region of 5q31 in malignant myeloid disorders (20) as well as the distinct classical 5q syndrome (14) were published. We were unable to find GRAF sequences in any of the following P1 artificial chromosome PAC/P1 and yeast artificial chromosome clones that have been used to delineate this particular deleted regions: P299F9, P244J5, P161J9, P14O2, 186K10, P86A12, P103F16, P13P5, P235N5, P273E22, P36N17, P38I10, P133 M3, 939F12, 816D6, 745D10, 936H1, 939A5, 632E12, 888E12, 743H4, 848D1, 370A8, 176D1, 335D9, 187D16, 204O11, 267F5, 11N14, and 54D23.
To assess the loss of GRAF in hematologic neoplasms with a cytogenetically proven 5q deletion, we performed FISH on bone marrow cells of 10 patients with either MDS (five cases) or AML (five cases) (for cytogenetic details see Table 3, which is published as supplemental material). Three patients had a deletion as the sole karyotype abnormality, and seven patients had complex karyotype changes. For FISH on patient's material we used the P1 clone L0649Q8 that, as confirmed by Southern blot and PCR analyses, contains the GRAF gene and maps to 5q31 (data not shown). We found that in these 10 patients one allele of GRAF was consistently lost in more than 75% of cells. In addition, we found three patients with AML in whom the 5q deletion was not apparent cytogenetically (patients 11–13, Table 3), because it was either too small or obscured by the complex karyotype changes. However, the fact that only one GRAF signal was detected in patients 11–13 by FISH indicates that this gene region was definitely deleted.
Disruption of the Second GRAF Allele in Three Patients with 5q Deletions.
To search for possible mutations in the residual GRAF allele, we sequenced the entire cDNA of bone marrow samples of all 13 patients. In consideration of the possibility that such mutations might be restricted to the comparatively small blast cell population, we cloned the GRAF cDNA in five overlapping fragments and sequenced 4–5 independent clones.
The point mutations resulted in amino acid changes of the GAP domain. In patient #5 it was an A to G exchange at nucleotide 1255 that substitutes a serine for an asparagine at amino acid 417. Because we did not detect this point mutation in 15 healthy controls, it is most likely not an polymorphism.
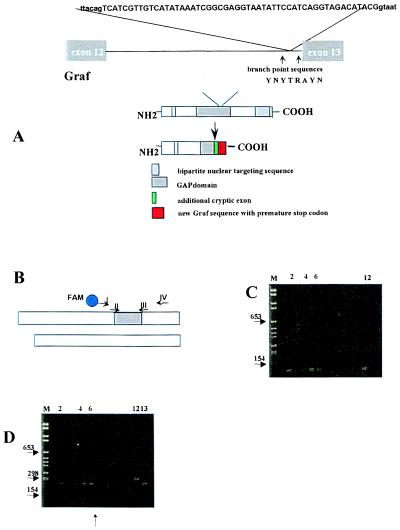
In the leukemic cells of two patients, the cDNA of the residual GRAF allele was compromised by a 52-bp or 74-bp insertion (Table 3). The insertions occurred at the splice junction of exons 12 and 13 (52 bp, patient #7) and exons 15 and 16 (74 bp, patient #3) (Fig. 6A). The respective exon/intron boundaries are located at nucleotides 1290/91 and 1993/94. In both cases, the insertion leads to a reading frame shift that generates a stop codon. The predicted truncated GRAF protein of patient #7 lacks almost the entire GAP domain and therefore is most likely not functional. The 52-bp insertion originates from a 524-bp long sequence that is upstream of the 3′ splice site of the adjacent nearly 13-kb long intron (Fig. 2B, lanes 14–16, and Fig. 6A). We sequenced the entire intron and found that its sequence (reverse complement) corresponds to nucleotides 32086–44631 of the bacterial artificial chromosome clone 118L13 (GenBank accession no. 005348). The 52-bp insertion (for sequence see Fig. 6A) derives from nucleotides 32560–32611 of 118L13. Because it is embedded in perfect splice donor and acceptor sites, it most likely results from aberrant splicing. Using chimeric RT-PCR primers (II and III in Fig. 6B) that specifically amplify the aberrantly spliced fragment, we found a faint band in the blood DNA from healthy donors with a nested PCR only (Fig. 6C). We tried to semiquantitatively assess the relative amount of inserted versus normal RNA in 15 healthy volunteers. For this purpose, we applied a competitive RT-PCR assay with fluorescence-labeled universal primers (I, IV) that share primer-binding sites of both fragments. In this competitive RT-PCR, the smaller, regularly spliced normal GRAF mRNA that does not contain the 52-bp insertion is preferably amplified and results in an overestimation of the normal GRAF mRNA product. As expected, we found only the regularly spliced fragment in the healthy volunteers. In contrast, both the normal and aberrantly spliced fragments were present in an approximately equal amount in patient #7 (lane 6 in Fig. 6D and lane 2 in Fig. 7). In support of this finding, two of four sequenced GRAF cDNA-clones harbored the 52-bp insertion. The more abundant generation of the mRNA with the 52-bp insertion in this particular patient might result from a sequence variation at the intronic branch point or the 3′ polymidine tract. Because sequence analysis of 400 bp of the respective intron did not reveal any alteration in 10 individual clones from the patient as well as in 12 healthy controls, the pathogenetic mechanism leading to this aberrantly spliced mRNA remains unexplained. The 74-bp insertion of patient #3 is located in the 60-kb long intron between exons 15 and 16. Databank searches revealed that the 74-bp sequence corresponds to nucleotides 128142–128215 of the human bacterial artificial chromosome clone 205e20. In accordance with the other patient, we identified the splice donor and acceptor sites at the insertion boundaries as well as potential lariat branch-point sequences. Three of five sequenced clones carried this insertion that presumably generates a stop codon. The predicted protein lacks the SH3 domain of GRAF that was shown to be necessary for the interaction with the focal adhesion kinase (21).
Figure 6.
(A) Insertion of 52-bp (capital letters) derived from intron 13 into the final cDNA found in patient #7. The surrounding intronic sequences are shown in lowercase letters. This leads to a reading frame shift followed by a premature stop codon. The GAP domain of Graf is substantially shortened. The intronic regions that were sequenced in patient #7 and 12 healthy controls are indicated by arrows. The splice branch site consensus sequence is shown as follows: Y represents T or C, R either A or G. (B) Schematic representation of both cDNA fragments that were coamplified by universal primers I and IV for assessment of their relative amount. Primers II and III amplify the aberrantly spliced fragment only. (C) Nested PCR analysis using the first-round primers I and IV and the second-round primers II and III. M, molecular weight marker. Lane 1, negative control. Four of 15 healthy blood donors expressed the aberrantly spliced fragment in their mononuclear cells (lanes 2 and 4–6) because a faint PCR product was seen. Lane 12, positive control. (D) Single-round PCR analysis using primers I and IV. Lane 6, two differently sized PCR products are seen from the cDNA of patient #7 even after only one round of PCR. Positive plasmid controls containing the 52-bp insertion (lane 12) or not (lane 13). In each RT-PCR, 2 μg of total RNA was subjected to cDNA synthesis and processed in parallel.
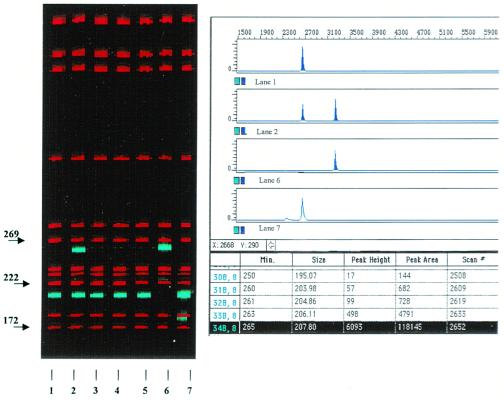
Figure 7.
genescan analysis of single-round PCR products using universal primers I and IV. Lane 2, two PCR products of 207 bp and 259 bp were obtained from patient #7. Lanes 1 and 3–5, PCR products derived from healthy volunteers showing only the 207-bp fragment. Lanes 6 and 7, positive controls.
Discussion
We have isolated and characterized a MLL fusion partner, GRAF, by cloning the breakpoints of a t(5;11)(q31;q23) that had occurred in an infant with juvenile myelomonocytic leukemia. Similar translocations were previously reported in only five patients with leukemia (29–32), whereas the involvement of MLL was analyzed by FISH in only one of them (29). GRAF is a GAP-encoding gene. It is highly homologous to the only other GAP-encoding gene currently known, BCR, which also is involved in a leukemia-associated chromosomal translocation. BCR is fused to ABL in the t(9,22)(q34;q11) that typically occurs in chronic myeloid and acute lymphoblastic leukemias. Interestingly, the functionally important GAP domain is lost in both the predicted hybrid proteins BCR/ABL and MLL/GRAF.
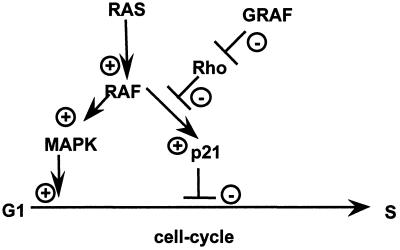
The GRAF gene is the human homologue of a recently isolated avian cDNA. It encodes the first known regulator of the Rho family of small GTPases that binds to a tyrosine kinase (21, 28). The Rho family belongs to the RAS superfamily and consists of five distinct types of GTP-binding proteins: RhoA, RhoB, RhoC, and RhoD; Rac1 and Rac2; Cdc42 and G25K; TC10, and RhoG (28, 33, 34). Rho, Rac, and Cdc42 regulate the organization of the actin cytoskeleton. They control the assembly of actin stress fibers and focal adhesion complexes that anchor the actin cytoskeleton to the plasma membrane. Rho family GTPases also play a role in the regulation of growth control, because each of the three GTPases (Rho, Rac, and Cdc42) can induce Swiss 3T3 cells to progress through G1 and to enter the S phase (35). Conversely, inhibitors of Rho, Rac, and Cdc42 block serum-induced DNA synthesis. The avian GRAF protein binds to the C-terminal domain of pp125FAK, one of the tyrosine kinases predicted to be a critical component of the integrin signaling transduction pathway, in a SH3 domain-dependent manner and stimulate the GTPase activity of the GTP-binding protein RhoA. Thus, GRAF acts as a negative regulator of RhoA. Olson and colleagues found recently that RhoA suppresses p21 (36), a well-known inhibitor of the cell cycle, when cells are transformed by oncogenic RAS (37–39). This observation leads to the simple but attractive hypothesis that the loss of function of GRAF prevents the physiologic down-regulation of RhoA. Thus, the mitigation or elimination of the negative regulatory function of the GRAF may lead to the repression of p21. If so, the GRAF-defective cell will be driven into the S phase (Fig. 8.). The first preliminary evidence as to how GRAF itself is regulated was provided by Taylor et al. (40), who found that GRAF is an in vivo target for mitogen-activated protein kinase.
Figure 8.
Hypothetical model about the role of GRAF in RAS-mediated signaling. MAPK, mitogen-activated protein kinase.
GRAF maps telomeric to the previously delineated commonly deleted 5(q31) region and therefore cannot be considered to be the long searched tumor suppressor gene whose functional elimination supposedly triggers the development of MDS and AML. However, the rather heterogeneous extensions of such 5q deletions probably delete several different genes that might be pathogenetically relevant for the initiation and progression of these diseases. In support of the potential tumor suppressor function of GRAF, we detected functionally relevant sequence alterations in the residual allele in three of 13 samples in which the other allele was deleted. In patient #5, the asparagine at position 417 corresponds to position 241 of the partial clone of the chicken GRAF (asparagine) and position 90 of p50RhoGAP (threonine), the GAP that interacts with Rho family proteins. Analysis of the crystal structure of RhoA and p50RhoGAP revealed that Thr-90 is involved in hydrogen-boding interaction between RhoGAP and RhoA (41).
We found a “cryptic out of frame” splicing that prematurely truncates the translation of the GRAF protein in patients #3 and #7. The detection of the truncated GRAF protein was not possible with the use of our polyclonal anti-GRAF antisera ED98015 or ED98016. They only bind to the C terminus of GRAF that is not translated in the leukemic cells of both patients. Although traces of these aberrantly spliced transcripts also were detected in the peripheral blood of healthy controls with nested PCR and insertion-specific primers, their levels were significantly higher in the leukemic samples. This shift in the relative amount of transcripts must result from an alteration that promotes such cryptic splicing in the neoplastic cells. However, the molecular basis for this phenomenon remains unclear, because the motifs that are responsible for correct splicing (3′ polymidine tract, branch point lariat sequence, and donor and acceptor sites) were preserved in the samples.
Further experiments are needed to clarify the relationship between RhoA, p21, and GRAF in the context of development of leukemias and probably other tumors. However, based on the fact that GRAF does not lie within the previously defined commonly deleted region of chromosome 5q31 and that the majority of our cases displayed the GRAF wild-type sequences within their second GRAF allele we conclude that the loss of genes other than GRAF are of importance in the development of leukemias and del(5q) as well.
Supplementary Material
Acknowledgments
The MLL yeast artificial chromosome-clone 13HH4 was kindly provided by Dr. Bryan Young (St. Bartholomew's Hospital, London). This work was supported by grants from the Deutsche Forschungsgemeinschaft (Bo1549/2–1) and Forschungshilfe “Station Peiper.”
Abbreviations
- GRAF
GTPase regulator associated with the focal adhesion kinase pp125FAK
- MLL
mixed-lineage leukemia
- RT-PCR
reverse transcription–PCR
- MDS
myelodysplastic syndrome
- AML
acute myeloid leukemia
- GAP
GTPase-activating protein
- RACE
rapid amplification of cDNA ends
- FISH
fluorescence in situ hybridization
- SH3
Src homology 3
- BCR
breakpoint cluster region
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. Y10388).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.150079597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.150079597
References
- 1.Varmus H E, Lowell C A. Blood. 1994;83:5–9. [PubMed] [Google Scholar]
- 2.Levine A J. Sci Med. 1995;1:28–37. [Google Scholar]
- 3.Rabbitts T H. Nature (London) 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 4.Hunger S P, Cleary M L. Semin Cancer Biol. 1993;4:387–399. [PubMed] [Google Scholar]
- 5.Repp R, Borkhardt A, Haupt E, Kreuder J, Brettreich S, Hammermann J, Nishida K, Harbott J, Lampert F. Leukemia. 1995;9:210–215. [PubMed] [Google Scholar]
- 6.Bernard O A, Berger R. Genes Chromosomes Cancer. 1995;13:75–85. doi: 10.1002/gcc.2870130202. [DOI] [PubMed] [Google Scholar]
- 7.Rowley J D. Semin Hematol. 1999;36:59–72. [PubMed] [Google Scholar]
- 8.DiMartino J F, Cleary M L. Br J Haematol. 1999;106:614–626. doi: 10.1046/j.1365-2141.1999.01439.x. [DOI] [PubMed] [Google Scholar]
- 9.Kourlas P J, Strout M P, Becknell B, Veronese M L, Croce C M, Theil K S, Krahe R, Ruutu T, Knuutila S, Bloomfield C D, Caligiuri M A. Proc Natl Acad Sci USA. 2000;97:2145–2150. doi: 10.1073/pnas.040569197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skuse G R, Ludlow J W. Lancet. 1995;345:902–906. doi: 10.1016/s0140-6736(95)90015-2. [DOI] [PubMed] [Google Scholar]
- 11.Boultwood J, Fidler C, Lewis S, Kelly S, Sheridan H, Littlewood T J, Buckle V J, Wainscoat J S. Genomics. 1994;19:425–432. doi: 10.1006/geno.1994.1090. [DOI] [PubMed] [Google Scholar]
- 12.LeBeau M M. Cancer Surv. 1997;15:143–159. [Google Scholar]
- 13.Willman C L, Sever C E, Pallavicini M G, Harada H, Tanaka N, Slovak M L, Yamamoto H, Harada K, Meeker T C, List A F, Taniguchi T. Science. 1993;259:968–971. doi: 10.1126/science.8438156. [DOI] [PubMed] [Google Scholar]
- 14.Boultwood J, Fidler C, Soularue P, Strickson A J, Kostrzewa M, Jaju R J, Cotter F E, Fairweather N, Monaco A P, Müller U, et al. Genomics. 1997;45:88–96. doi: 10.1006/geno.1997.4899. [DOI] [PubMed] [Google Scholar]
- 15.Boultwood J, Fidler C, Lewis S, MacCarthy A, Sheridan H, Kelly S, Oscier D, Buckle V J, Wainscoat J S. Blood. 1993;82:2611–2616. [PubMed] [Google Scholar]
- 16.Pedersen B. Anticancer Res. 1993;13:1913–1916. [PubMed] [Google Scholar]
- 17.Van den Berghe H, Michaux L. Cancer Genet Cytogenet. 1997;94:1–7. doi: 10.1016/s0165-4608(96)00350-0. [DOI] [PubMed] [Google Scholar]
- 18.Boultwood J, Lewis S, Wainscoat J S. Blood. 1994;84:3253–3260. [PubMed] [Google Scholar]
- 19.Pedersen B. Leukemia. 1996;10:1883–1890. [PubMed] [Google Scholar]
- 20.Zhao N D, Stoffel A, Wang P W, Eisenbart J D, Espinosa R, III, Larson R A, Le Beau M M. Proc Natl Acad Sci USA. 1997;94:6948–6953. doi: 10.1073/pnas.94.13.6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hildebrand J D, Taylor J M, Parsons J T. Mol Cell Biol. 1996;16:3169–3178. doi: 10.1128/mcb.16.6.3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niemeyer C M, Arico M, Basso G, Biondi A, Rajnoldi A C, Creutzig U, Haas O, Harbott J, Hasle H, Kerndrup G, et al. Blood. 1997;89:3534–3543. [PubMed] [Google Scholar]
- 23.Gu Y, Alder H, Nakamura T, Schichman S A, Prasad R, Canaani O, Saito H, Croce C M, Canaani E. Cancer Res. 1994;54:2327–2330. [PubMed] [Google Scholar]
- 24.Cheng S, Chang S-Y, Gravitt P, Respess R. Nature (London) 1994;369:684–685. doi: 10.1038/369684a0. [DOI] [PubMed] [Google Scholar]
- 25.Leis T, Repp R, Borkhardt A, Metzler M, Schläger F, Harbott J, Lampert F. Leukemia. 1998;12:758–763. doi: 10.1038/sj.leu.2401006. [DOI] [PubMed] [Google Scholar]
- 26.Alder H, Buchberg A M, Canaani E, Chatterjee D, Croce C M, Gu Y, Ma Q, Nakamura T, Nelson K K, Siracusa L D. Proc Natl Acad Sci USA. 1993;90:6350–6354. doi: 10.1073/pnas.90.13.6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeleznik-Le N J, Harden A M, Rowley J D. Proc Natl Acad Sci USA. 1994;91:10610–10614. doi: 10.1073/pnas.91.22.10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hotchin N A, Hall A. Cancer Surv. 1996;27:311–322. [PubMed] [Google Scholar]
- 29.Kearney L, Bower M, Gibbons B, Das S, Chaplin T, Nacheva E, Chessells J M, Reeves B, Riley J H, Lister T A, Young B D. Blood. 1992;80:1659–1665. [PubMed] [Google Scholar]
- 30.Harrison C J, Cuneo A, Clark R, Johansson B, Lafage-Pochitaloff M, Mugneret F, Moorman A V, Secker-Walker L M European 11q23 Workshop Participants. Leukemia. 1998;12:811–822. doi: 10.1038/sj.leu.2401017. [DOI] [PubMed] [Google Scholar]
- 31.Itoh M, Okazaki T, Tashima M, Sawada H, Uchiyama T. Leuk Res. 1999;23:677–680. doi: 10.1016/s0145-2126(99)00033-8. [DOI] [PubMed] [Google Scholar]
- 32.Bower M, Chaplin T, Das S, Kearney L, Gibbons B, Riley J H, Lister T A, Young B D. Leukemia. 1993;7:S34–S39. [PubMed] [Google Scholar]
- 33.Macara I G, Lounsbury K M, Richards S A, McKiernan C, Bar-Sagi D. FASEB J. 1996;10:625–630. doi: 10.1096/fasebj.10.5.8621061. [DOI] [PubMed] [Google Scholar]
- 34.Murphy G A, Solski P A, Jillian S A, De la Ossa P P, D'Eustachio P, Der C J, Rush M G. Oncogene. 1999;18:3831–3845. doi: 10.1038/sj.onc.1202758. [DOI] [PubMed] [Google Scholar]
- 35.Olson M F, Ashworth A, Hall A. Science. 1995;269:1270–1272. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- 36.Olson M F, Paterson H F, Marshall C J. Nature (London) 1998;394:295–299. doi: 10.1038/28425. [DOI] [PubMed] [Google Scholar]
- 37.Yang Z Y, Perkins N D, Ohno T, Nabel E G, Nabel G J. Nat Med. 1995;1:1052–1056. doi: 10.1038/nm1095-1052. [DOI] [PubMed] [Google Scholar]
- 38.Mercer, W. E. (1998) J. Cell. Biochem.50–54. [PubMed]
- 39.Cai K, Dynlacht B D. Proc Natl Acad Sci USA. 1998;95:12254–12259. doi: 10.1073/pnas.95.21.12254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor J M, Macklem M M, Parsons J T. J Cell Sci. 1999;112:231–242. doi: 10.1242/jcs.112.2.231. [DOI] [PubMed] [Google Scholar]
- 41.Rittinger K, Walker P A, Eccleston J F, Nurmahomed K, Owen D, Laue E, Gamblin S J, Smerdon S J. Nature (London) 1997;388:693–697. doi: 10.1038/41805. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.