Abstract
We performed a large-scale cDNA analysis to explore the transcriptome of the budding yeast Saccharomyces cerevisiae. We sequenced two cDNA libraries, one from the cells exponentially growing in a minimal medium and the other from meiotic cells. Both libraries were generated by using a vector-capping method that allows the accurate mapping of transcription start sites (TSSs). Consequently, we identified 11,575 TSSs associated with 3,638 annotated genomic features, including 3,599 ORFs, to suggest that most yeast genes have two or more TSSs. In addition, we identified 45 previously undescribed introns, including those affecting current ORF annotations and those spliced alternatively. Furthermore, the analysis revealed 667 transcription units in the intergenic regions and transcripts derived from antisense strands of 367 known features. We also found that 348 ORFs carry TSSs in their 3′-halves to generate sense transcripts starting from inside the ORFs. These results indicate that the budding yeast transcriptome is considerably more complex than previously thought, and it shares many recently revealed characteristics with the transcriptomes of mammals and other higher eukaryotes. Thus, the genome-wide active transcription that generates novel classes of transcripts appears to be an intrinsic feature of the eukaryotic cells. The budding yeast will serve as a versatile model for the studies on these aspects of transcriptome, and the full-length cDNA clones can function as an invaluable resource in such studies.
Keywords: alternative splicing, antisense transcript, noncoding RNA, transcription start site
Remarkable progress has been witnessed in the field of functional genomics in the first decade after the yeast genome sequencing (1). The systematic gene deletion project has revealed 1,105 essential genes (2) and has provided researchers with a set of strains deleted for individual genes, which facilitates a truly comprehensive phenotypic analysis as well as a systematic screening of synthetic lethal interactions (3). For the transcriptome analysis, DNA microarray has been extensively used to examine the expression of ≈6,000 genes under a plethora of conditions. This led to the functional discovery of novel genes based on their coexpression patterns shared with other known genes (4). More recently, chromatin immunoprecipitation integrated with microarray technology has enabled the genome-wide localization of transcription factors to accelerate analysis of gene regulatory networks (5). For proteome analysis, the expression, localization, and interactions of each protein have been extensively analyzed by systematic epitope tagging, fluorescent protein tagging, two-hybrid system, and affinity capture mass spectrometry (6–11).
Despite these remarkable achievements, we have been unable to determine the exact number of genes in this simple organism. Comparative sequencing has extensively revised genome annotations, leading to substantial reduction in gene numbers (12, 13). On the other hand, various approaches have been used to detect many novel genes (14–16). Although evolutionary conservation is one of the strongest lines of evidence, a more direct proof for the DNA segment being a gene would be provided by transcription into RNA. However, most yeast transcriptome studies have used microarrays to examine the expression of the selected genes. Yeast transcripts have not been extensively analyzed in an open-ended manner, except for the pioneering work using the serial analysis of gene expression (SAGE) technique (17). This is in good contrast with studies on higher eukaryotes, where extensive cDNA analysis is indispensable for genome annotation. Because of the lack of cDNA analysis, transcription start sites (TSSs), promoters, and 5′-UTRs have remained elusive for most yeast genes.
To complement this least-explored field of yeast functional genomics, we performed a large-scale full-length cDNA analysis. This analysis has not only provided the largest TSS data set but has also revealed a large number of previously overlooked RNAs transcribed from both strands of intergenic and intragenic regions. These results provide concrete evidence for the unexpected complexity of budding yeast transcriptome, which was also suggested by recent studies using 5′-SAGE (18) and tiling array hybridization (19). The yeast likely shares many of the novel features of the transcriptome with mammals and other higher eukaryotes, underscoring its importance as a model organism for studies on novel classes of RNAs.
Results
A Large-Scale Sequencing of Vector-Capped cDNA Clones.
We performed a large-scale sequencing of cDNA clones that were constructed by using a “vector-capping” method (20, 21). This method converts a full-length (i.e., 5′-capped) mRNA into a cDNA clone that has an additional dG at its 5′-end. In contrast, no nucleotides are added to the clones derived from a truncated (i.e., uncapped) mRNA and those generated by incomplete reverse transcription. Accordingly, if a cDNA clone has at its 5′-end an additional dG that is not encoded by the genome, we can assume that the dG is the product of cap-dependent nucleotide addition, and that the clone is a full-length one (for example, see Fig. 6, which is published as supporting information on the PNAS web site). A sizable fraction of the full-length clones was reported to have successive dT and/or dG residue(s) upstream of the cap-dependent dG (21). Thus, in this study, we considered a clone to be full-length if it started with a dT0∼ndG1∼m sequence not encoded by the genome (i.e., typical G-cap).
We prepared two cDNA libraries, S288C-SD and SK1-Spo. The S288C-SD library was generated from the S288C strain used for genome sequencing. The S288C cells were cultivated in a minimal medium to induce many genes involved in various biosynthetic pathways. The SK1-Spo library was generated from SK1, a strain that sporulates at high efficiency. The SK1 cells were cultivated in an acetate medium to induce many meiotic genes.
We subjected these clones to single-pass sequencing from their 5′-ends and obtained 51,026 sequences (Table 1, which is published as supporting information on the PNAS web site). Data on these sequences can be obtained from UT Genome Browser at http://yeast.utgenome.org (for example, see Fig. 1A).
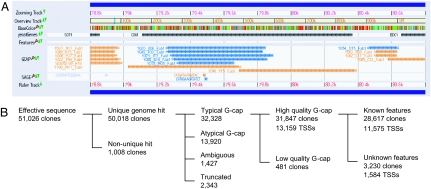
Fig. 1.
Data of the large-scale cDNA analysis. (A) A screenshot of the UT Genome Browser depicting a region including SDT1, COG1, and EDC1 (coordinate 78,801–80,800 of chromosome 7). Each bar in the GCAP track indicates the 5′-single-pass sequence of each cDNA clone, whose ID is shown at the left side of the bar. Blue and orange indicate Watson and Crick strands, respectively. The sharp (#) at the end of line indicates that the clone is a full-length one with a G-cap-derived nucleotide addition. This screen shows four and one full-length clones for SDT1 and COG1, respectively. In addition, it displays five, one, and one full-length clones for antisense transcripts of COG1, a transcript starting within the COG1 ORF, and a transcription unit lying between COG1 and EDC1 (TU #257 in Table 8), respectively. (B) Breakdown of the cDNA data. Starting from 51,026 clones, 13,159 TSSs were identified (see text for detail).
TSSs.
From the 51,026 sequences, we selected 31,847 for TSS analysis, because each of them hit a single unique genomic locus in BLAST search and started with the typical G-cap structure (i.e., dT0∼ndG1∼m with average Phred score ≥20; Fig. 1B, Table 2, which is published as supporting information on the PNAS web site). These 31,847 sequences correspond to 13,159 independent TSSs.
These TSSs were subsequently assigned to the annotated features on the yeast genome according to the following algorithm. If a sequence is mapped to an annotated feature or its 300-bp upstream region, it is assigned to that feature. However, if the sequence has a poly(A) tail starting from the region upstream of the feature, it is not assigned to that feature but is regarded as an independent transcript. In the case of sequences assigned to multiple features, we manually assigned each of them to a single feature. Consequently, we assigned 28,617 sequences (11,575 TSSs) to 3,638 annotated features including 3,599 known ORFs, thereby providing the largest yeast TSS data set (Table 3, which is published as supporting information on the PNAS web site). Although 3,303 of the 3,638 features have at least one TSS in the region upstream of the features, the other 335 have TSSs only inside the features: this suggests errors in the annotation of initiation codons and/or internal promoter activities (see below). The remaining 3,230 sequences (1,584 TSSs) include transcripts derived from intergenic regions, those transcribed from antisense strands of known features, and those spanning two or more consecutive genomic features or potential bicistronic RNAs.
We compared our TSS data with those recently obtained by 5′-SAGE (18). Of the 1,693 genes that had hits in both studies, 702 (≈41%) shared at least one TSS (data not shown). Because many genes have two or more TSSs (see below), and most TSSs in both data sets were hit only once, the rate of overlap would increase upon enhanced sampling.
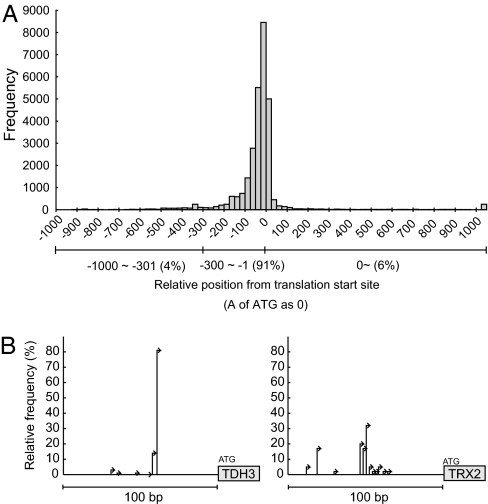
The relative position of each TSS from the initiation codon is shown in Fig. 2A. Most TSSs are mapped within the 100-bp region upstream of the initiation codon in accordance with previous notions as well as the results of recent 5′-SAGE and tiling array hybridization studies (18, 19). We examined the correlation between TSS position and gene category by using the ART-EX tool (ref. 22; http://itolab.cb.k.u-tokyo.ac.jp/BIRD/GATC-PCR/cgi-bin/ART.pl). The results indicated that the genes encoding plasma membrane proteins and involved in cell cycle regulation and/or protein phosphorylation tend to have longer 5′-UTRs (data not shown), as was reported in the tiling array study (19).
Fig. 2.
Transcription start sites. (A) Distribution of TSSs around the presumptive initiation codon. (B) Two typical patterns of TSS distribution.
It should be noted that 6% of the TSSs are mapped within the ORFs (Fig. 2A). Although those close to the 5′-end of the ORF may indicate incorrect annotation of the initiation codons, a sizable fraction is mapped considerably deeper inside the ORFs; 348 ORFs have at least one TSS in their 3′-halves (for example, see Figs. 1A and 5A). It is likely that these TSSs represent independent transcription units driven by promoters within the ORFs. In this context, it is intriguing to note that the prevalence of such “exonic” promoters in the protein-coding genes was recently revealed in mammals (23).
Fig. 5.
Antisense transcription. (A) A screenshot of the UT Genome Browser depicting GAL1-GAL10 locus. Two cDNA clones (blue lines) were isolated for the antisense transcript of GAL10, which had been hit by a SAGE tag (17). Note that GAL10 also has an internally primed sense transcript (orange lines), which was also detected by the tiling array study (segment ID 1028; ref. 19). (B) The expression of GAL10 is suppressed or induced in the presence of dextrose (Dex) or galactose (Gal), respectively. Coinduction of the sense and antisense transcripts was observed upon addition of Gal.
The TSS data also indicate that the yeast genes have generally two or more TSSs. Of the 1,130 annotated features to which at least five full-length cDNA clones were assigned, only eight (0.7%) have a single unique TSS (Table 3). The distance between the most distal and most proximal TSSs was examined in the regions upstream of the 1,040 features, to each of which at least five full-length cDNA clones were mapped. The distance is 91.4 bp on average and ≥10 bp in 94.5% of the cases (Table 3). Notably, the distribution of TSSs can be classified into two classes. Some genes use a single dominant TSS, whereas others use TSSs more evenly, occasionally having several modest peaks (Fig. 2B). Similar findings were also reported for mammalian genes in a large-scale analysis of TSSs (23).
5′-Untranslated Regions.
We examined whether each 5′-UTR contains any ORFs, because the upstream ORFs (uORFs) may play a pivotal role in translation regulation (24). Although 2,415 5′-UTRs were found to have at least one uORF (Table 4, which is published as supporting information on the PNAS web site), most were not evolutionarily conserved as reported (25), and some were excluded from the shorter forms of the transcripts. This information may help in identifying functional uORFs.
We found four transcripts spanning two adjacent ORFs that may function as bicistronic transcripts (Fig. 7, which is published as supporting information on the PNAS web site). Although two of these carry overlapping ORFs (i.e., YBR126W-A/YBR126W-B and YDR133C/YDR134C), the other two have two nonoverlapping ORFs. The transcript of PMP1 contains a downstream ORF YCR024C-B, which was identified by expression profiling and mass spectrometry (16). The transcript of URA6 contains a uORF YKL023C-A, which was identified by comparative sequencing of six Saccharomyces species (12, 13). It is intriguing to examine whether these two ORFs are translated either coordinately or independently from a single mRNA. Although bicistronic transcripts were reported for YMR181C-RGM1 and GIM3-YCK2 (19, 26), our full-length cDNA clones contained only RGM1 and YCK2, indicating that at least a fraction of these two genes is transcribed as conventional monocistronic mRNAs. In contrast, we have no evidence to date for the monocistronic expression of YCR024C-B and URA6.
The present cDNA analysis revealed the presence of in-frame AUG codons upstream of the annotated ORFs in the transcripts of 15 genes (Table 5, which is published as supporting information on the PNAS web site). Although most of these codons are not evolutionarily conserved, it is likely that the case for LAP3 is biologically relevant (Fig. 8, which is published as supporting information on the PNAS web site). One of the seven LAP3 cDNA clones is capable of encoding an isoform with a 29-aa N-terminal extension. Intriguingly, the 29-aa sequence is conserved among other Saccharomyces species and is predicted to function as a mitochondrial targeting signal by the iPSORT program (27). Indeed, the systematic GFP-tagging experiment revealed that LAP3 localizes to both the cytoplasm and the mitochondria (7). Thus, LAP3 likely uses these two TSSs to generate mitochondrial and cytoplasmic isoforms.
The analysis also identified 32 genes carrying introns in their 5′-UTRs. Of these, ORF annotation should be corrected in 10 genes (Tables 6 and 7, which are published as supporting information on the PNAS web site). For example, ATG annotated as the initiation codon of GIM4, PRP5, and YPR153W was found to be intronic. We also found that the splice donor site is inaccurately annotated in three intron-carrying ribosomal protein genes RPL20A, RPL20B, and RPL26B, wherein an intronic ATG was regarded as the initiation codon. In IWR1, YFR045W, YJR005C-A, and YKR005C, the presumptive upstream in-frame stop codon was shown to be intronic, leading to an N-terminal extension of each ORF. For example, the N-terminal end of IWR1 can be extended to include 113 amino acids (Fig. 9, which is published as supporting information on the PNAS web site). The extended form of IWR1 can be better aligned throughout its ORF with its orthologs in Kluyveromyces lactis, Ashbya gossypii, Candida albicans, and Debaryomyces hansenii, and it has a molecular weight consistent with that observed in gel electrophoresis (28).
Introns.
The present cDNA analysis detected 258 introns in 243 annotated genes, including eight genes with two introns, and seven previously undescribed transcription units (Table 6). These introns comprise 256 GT-AG introns and two noncanonical GC-AG introns (COX5B and SRC1). Of these 258 introns, 45 are previously undescribed (Table 7). In addition to the three introns in the ribosomal protein genes described above, the acceptor site of the intron in LSM7 had been erroneously annotated (Table 7). Its correction led to an insertion of eight amino acids into the sequence of LSM7, which eliminated a gap in the alignment between LSM7 and its fungal orthologs. We also detected many rare splicing variants of known introns that often disrupted the ORFs (Table 7). However, the three cases described below leading to significant alteration in ORFs are of interest.
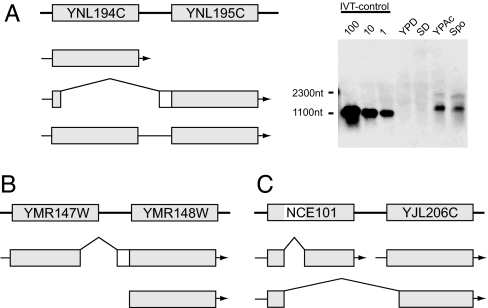
YNL194C encodes a protein of unknown function, which shows homology to YDL222C, a component of the newly identified organelle eisosome (29). The detailed analysis of the cDNA clones assigned to this locus revealed three different forms (Fig. 3A). One form contains the entire ORF of YNL194C and terminates between YNL194C and YNL195C, thereby being assigned to YNL194C. The second form encodes a protein composed of N-terminal five amino acids of YNL194C followed by 26 residues derived from the intergenic region and 251 residues of YNL195C. In addition, we found a third form carrying both ORFs. Although it may represent the unspliced precursor of the second form, Northern blot analysis using a YNL195C-specific probe detected a ≈2,000-nt band corresponding to this form, in addition to a ≈1,000-nt band corresponding to the second form (Fig. 3A). Thus, the third form may function as a bicistronic mRNA. Notably, we have failed to obtain any full-length clone carrying only YNL195C.
Fig. 3.
Alternative splicing events. The bold line with squares with ORF/gene names shown at the top of each image indicates the genome map, and the arrows shown below the map indicate the transcripts. (A) YNL194C and YNL195C share the same promoter to generate three mRNAs (Left), including a potential bicistronic one detected as a ≈2,000-nt band in Northern blot analysis of SK1 cells (Right). IVT, in vitro transcription. (B) YMR148W has two distinct promoters, and YMR147W may be an upstream exon of YMR148W used in the transcript starting from the distal promoter. (C) A case of transcription-induced chimera between NCE101 and YJL206C.
Based on these findings and the results of RT-PCR assays, we assume that YNL194C and YNL195C comprise a single transcription unit. If the transcription terminates between the two ORFs, the transcript encodes YNL194C. However, if it proceeds downstream of YNL195C, the primary transcript is either spliced to an mRNA encoding YNL195C with a 31-aa N-terminal extension (i.e., the second form) or exported to the cytoplasm as a potential bicistronic mRNA (i.e., the third form). Of interest, the clones of the first form were recovered from mitotic cells (i.e., the S288C-SD library), whereas those of the second and third forms were derived from meiotic cells (i.e., the SK1-Spo library), suggestive of their regulated expression. Indeed, Northern blot analysis of SK1 cells indicated the induced expression of the second and third forms in sporulation and acetate media (Fig. 3A). Because the induction was observed to a lesser extent in S288C cells (data not shown), we assume that it is not an SK1-specific event.
The transcript of YMR147W was found to be spliced to its downstream ORF YMR148W (Fig. 3B). Intriguingly, the transcript encodes a hybrid protein between YMR147W and YMR148W. Although we isolated two YMR147W clones, both were spliced to exclude the 29-aa sequence encoded by its 3′-end portion. Of interest, this sequence was less conserved among its orthologs, except for the first three amino acids that correspond to the splicing junction. Thus, we assume that YMR147W is not an independent ORF but an upstream exon of YMR148W. Because we isolated many full-length cDNA clones containing only YMR148W, it appears to have two distinct promoters, the upstream promoter generating the spliced transcript with the upstream exon (i.e., YMR147W) and the downstream one generating the unspliced transcript, presumably producing functionally diverged protein isoforms.
The NCE101 gene is involved in protein secretion and has an intron. We found a cDNA clone in which the first exon of NCE101 is not spliced to its second exon but to its downstream neighbor YJL206C (Fig. 3C). Intriguingly, the clone retains an ORF capable of encoding a hybrid protein between NCE101 and YJL206C, although the splicing disrupts the Zn2Cys6-DNA-binding motif of the latter. Of note, both NCE101 and YJL206C have distinct TSSs of their own. Thus, the clone represents a so-called transcription-induced chimera or transcription-mediated gene fusion, which was recently shown to occur at a substantial frequency in mammals as a potential means to increase protein complexity (30, 31).
Previously Undescribed Transcription Units.
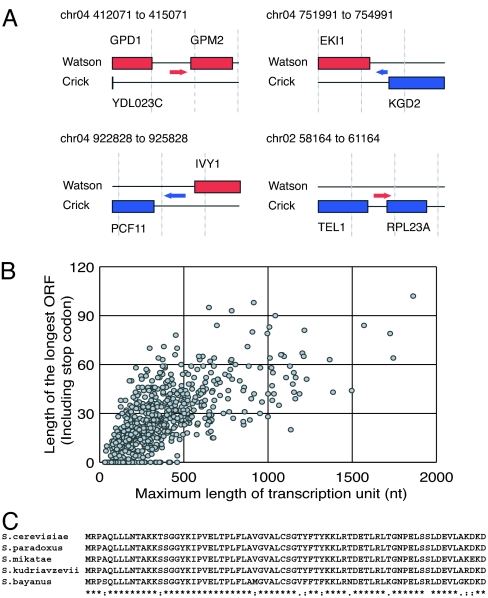
As described earlier, we identified sequences that are not assigned to any of the current annotated features. From these sequences, we identified 667 isolated transcripts expressed from the regions currently regarded as “intergenic.” To elucidate their structures, we determined the 3′-ends of these cDNA clones. Consequently, we succeeded in revealing the full-length structure of 504 transcription units (for example, see Figs. 1A and 4A; for detail, see Table 8, which is published as supporting information on the PNAS web site).
Fig. 4.
Transcription units in intergenic regions. (A) Examples of isolated transcription unit. Boxes and arrows indicate annotated ORFs and transcripts, respectively. Red and blue indicate features on Watson and Crick strands, respectively. (B) The longest ORF size was plotted against the size of each transcription unit. (C) Conservation of the ORF encoded by the transcription unit no. 633. Identical (∗), conservative (:), and semiconservative (.) residues are indicated.
These transcripts have ORFs with length ranging from 0 to 102 aa (Fig. 4B). Our initial data set included six additional transcripts carrying an evolutionarily conserved ORF longer than 50 aa. However, they were designated as YDR374W-A, YDL007C-A, YCL048W-A, YPR154C-A, YKL065W-A, and YLR146W-A during the course of this work, based solely on their evolutionary conservation. Our data provide evidence for the expression of these six newly annotated ORFs, consolidating their identities as bona fide genes. We found a transcription unit termed no. 633 capable of encoding a 72-aa protein that is highly conserved among various Saccharomyces species (Fig. 4C) and other fungi (data not shown). This gene is split by an intron, which probably caused the gene to escape detection by comparative sequence analysis. On the other hand, we found 79 transcripts lacking ORFs of any length (Fig. 4B). Nevertheless, some of these transcripts are conserved at the nucleotide sequence level and may function as noncoding RNAs (Table 8).
One may argue that most of these transcripts are cryptic unstable transcripts (CUTs), which are rapidly degraded by the coordinated action of a poly(A) polymerase TRF4 and the nuclear exosome (32). Because these CUTs were shown to accumulate upon loss of the nuclear exosome subunit RRP6 (32), it would be intriguing to examine whether the transcripts we identified increase in their amounts in the rrp6Δ mutant. A recent whole-genome microarray study compared wild-type yeast and rrp6Δ mutant to reveal 374 probes that displayed a ≥2-fold increase in mutant/wild-type signal ratio, corresponding to potential CUTs (33). We examined the microarray used in the study to find that 682 probes showed any overlap with the 667 transcription units. Remarkably, the 374 and 682 probes share only 41 in common, which corresponds to 39 transcription units (Table 9, which is published as supporting information on the PNAS web site). These results indicate that most, if not all, of the intergenic transcripts identified in this study from the wild-type strains are not CUTs. In accordance with our results, the tiling array study also detected a number of intergenic transcripts from a wild-type strain (19).
In addition to these isolated transcription units, we also identified 582 full-length cDNA clones that are transcribed from the antisense strands of 367 known genomic features (Table 10, which is published as supporting information on the PNAS web site). The inclusion of nonfull-length clones can result in as many as 1,092 potential antisense clones for 717 known genomic features. Although some pairs of sense and antisense transcripts almost entirely overlap with each other, most appear to overlap only at their 5′- or 3′-end portions (for example, see Figs. 1A and 5A).
Even in the well characterized GAL1-GAL10 locus, we identified transcripts antisense to GAL10 (Fig. 5A). Intriguingly, the antisense transcription of GAL10 was suggested by SAGE analysis (17) and was detected in the tiling array study (segment ID 474; ref. 19). Because the expression of GAL10 can be induced by galactose, this sense-antisense pair provides a good model to examine how the induction of sense-strand transcription affects its antisense transcript. We used real-time PCR to quantify GAL10 and its antisense transcripts in the presence of dextrose and galactose, which suppresses and induces the expression of GAL10, respectively. As shown in Fig. 5B, galactose caused not only a drastic induction of the sense transcript but also a significant (i.e., ≈5-fold) induction of the antisense transcript. Because such concordant regulation of sense/antisense pairs is frequently observed in mammals (34), it would be intriguing to examine the expression of more pairs in the yeast.
Discussion
This study demonstrated that, even in an intensively analyzed simple organism such as S. cerevisiae, an in-depth analysis of transcriptome can further improve the genome annotation by identification of TSSs and introns. Furthermore, it revealed unexpected complexity of the yeast transcriptome. Most, if not all, yeast genes likely have two or more TSSs. TSSs are widely distributed on both strands of intergenic and intragenic regions to generate thousands of novel transcripts, including isolated small transcripts, antisense transcripts to known genes, and sense transcripts that start within the known genes. This study also revealed examples of alternative splicing, including a case of transcription-mediated gene fusion event. These features are essentially similar to those recently observed in the mammalian transcriptome (23, 34, 35).
A recent study also revealed the complexity of budding yeast transcriptome by using the tiling array hybridization (19). Remarkably, this study included almost every gene and successfully defined both the 5′- and 3′-UTR boundaries for 2,223 genes. Tiling-array hybridization, by its nature, provides an averaged view of various TSSs at a resolution limited by the spacing of the probes. In contrast, cDNA analysis revealed a variety of transcripts to define each TSS at single-nucleotide resolution. It also provides definitive data on introns. For example, the tiling array study detected an unannotated, isolated transcript (segment ID 1950) at nucleotide positions 216,037–216,157 on chromosome 4. Our analysis revealed that this segment is not an isolated transcript but an upstream exon of ARF2. The two methods show good agreement with regard to the detection of novel transcripts, as exemplified by the GAL1-GAL10 locus. Although the presence of these novel transcripts was suggested by the SAGE experiments (17, 18), these two studies unequivocally revealed their prevalence throughout the genome.
Genome-wide active transcription was reported for higher eukaryotes, and it has resulted in an intense debate on its potential roles in gene regulation and evolution (36–38). The results on budding yeast may indicate that genome-wide transcription is an intrinsic feature of the eukaryotic cell. The yeast can serve as a versatile model to learn the biology of these novel transcripts, and the full-length cDNA clones can function as an invaluable resource in such studies.
Materials and Methods
Yeast Strains, Cultivation, and RNA Extraction.
The strain S288C was obtained from the Biological Resource Center at the National Institute of Technology and Evaluation (no. NBRC1136, MATα SUC2 mal mel gal2 CUP1 [cir+]). A strain with SK1 background was obtained from the American Type Culture Collection (no. 204722, MATa/MATα HO can1 gal2 cup1). Each strain was precultured in YPD medium (39) at 30°C overnight. The S288C were further cultivated in SD medium (39). The SK1 cells were cultivated in YPAc [1% (wt/vol) yeast extract/2% (wt/vol) peptone/2% sodium acetate] for 40 h and then transferred to a sporulation medium [1% (wt/vol) sodium acetate] to induce sporulation for 2.5 h. Total RNAs were extracted by using a hot-phenol method (40) with some modifications.
Library Construction and DNA Sequencing.
Two vector-capped cDNA libraries were constructed by Hitachi (Tokyo) from the S288C and SK1 RNAs using pGCap1 and pGCapzf3 as cloning vectors, respectively. Single-pass sequencing from the 5′-end of each insert was performed by using appropriate vector primers.
Data Processing.
The sequence of each clone was analyzed with BLAST against the vector and yeast genome sequence. We wrote a program to process the BLAST outputs, which recognizes the insert, checks the presence of introns, assigns the sequence to a genomic feature, and examines the G-cap and poly(A) sequence. For detail, see legend to Table 1.
Northern Blot Hybridization.
Total yeast RNA (5 μg) was denatured in 1.5 M glyoxal/15 mM tetramethylammonium phosphate (pH 7.0)/75% (vol/vol) DMSO at 55°C for 25 min and separated on 1% agarose gel. A positive control RNA was prepared by in vitro transcription of an appropriate plasmid template by using the T7 RiboMax Express Large Scale RNA production system (Promega, Madison, WI). After the electrophoresis, RNAs were electroblotted onto Hybond N+ membrane (Amersham, Piscataway, NJ) and fixed with baking at 80°C for 2 h. Probe labeling, hybridization, and detection were performed by using the ECL Direct Labeling and Detection System (Amersham).
Determination of poly(A) Site.
We sequenced the 3′-terminal portion of the clones for transcripts derived from intergenic regions to determine their poly(A) sites as follows. At first, the insert of each clone was amplified with biotinylated reverse primer (M13-RV: 5′-CAG GAA ACA GCT ATG AC-3′) and unlabeled forward primer (M13-F: 5′-GTA AAA CGA CGG CCA G-3′), purified with AMpure (Agencourt Bioscience, Beverly, MA) and digested either with MboI or Tsp509I. The 3′-end fragment was trapped onto DynaBeads Streptavidin M280 (Dynal, Carlsbad, CA) and ligated with an adaptor prepared by annealing HsM (5′-ACA ATT CAC AGA CAT TCC GCT CAC AAT AAG ATC TCT GCA CTG CGC TCA CAT CG-3′) with C (5′-(Phosphate) GAT CCG ATG TGA GCG CCA-3′ and 5′-(Phosphate) AAT TCG ATG TGA GCG CCA-3′ for MboI and Tsp509I digested fragments, respectively). Then the adaptor-ligated 3′-end fragment was amplified with M13-RV primer and an adaptor-specific primer (HsASP: 5′-GAC ATT CCG CTC ACA ATA AGA TCT C-3′) and subjected to direct sequencing with the HsASP primer. The clones refractory to this protocol were sequenced by using gene-specific primers.
Real-Time PCR Assay.
cDNA was prepared from 1 μg of total RNAs by using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA). The primers GAL10_F (5′-TTT TTG GGC AAC GTT CAC AGT-3′) and GAL10_R (5′-TAA ACC AGA TAG GGC CAA ACG-3′) were used in reverse transcription of the antisense and sense transcripts, respectively. Real-time PCR was performed by using both primers and Platinum SYBR green qPCR SuperMix-UDG w/ROX (Invitrogen). A series of diluted genomic DNAs in carrier RNA solution was used as standards to approximate the copy number of each transcript per cell. Amplification and detection were performed on ABI7000 SDS system (Applied Biosystems, Foster City, CA) by using the following thermal profile: prefacing incubation at 95°C for 5 min followed by 40 cycles of three-step amplification at 95°C for 30 sec, 55°C for 1 min, and 72°C for 1 min.
Data Availability.
All of our cDNA data can be browsed at http://yeast.utgenome.org and downloaded from http://itolab.cb.k.u-tokyo.ac.jp/GCap.
Supplementary Material
Acknowledgments
We thank Seishi Kato and Satoshi Okada for advice on vector-capping clones and SK1 cells, respectively. This work was partly supported by a grant-in-aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and a grant-in-aid for Scientific Research (B) from the Japanese Society of Promotion of the Science.
Abbreviations
- TSS
transcription start site
- SAGE
serial analysis of gene expression
- uORF
upstream ORF
- CUT
cryptic unstable transcript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The 5′-end sequence data for the 31,847 full-length cDNA clones have been deposited in the DNA Data Bank of Japan, www.ddbj.nig.ac.jp (DDBJ ID codes DB636784–DB668630).
References
- 1.Dolinski C, Botstein D. Genome Res. 2005;15:1611–1619. doi: 10.1101/gr.3727505. [DOI] [PubMed] [Google Scholar]
- 2.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 3.Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 4.Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, et al. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 5.Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 6.Ghaemmaghami S, Huh WK, Bower, Howson RW, Belle A, Dephoure N, O′Shea EK, Weissman JS. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 7.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 8.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 9.Ito T, Chiba T, Ozawa R, Yosida M, Hattori M, Sakaki Y. Proc Natl Acad Sci USA. 2001;98:4569–7574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Shultz J, Rick JM, Michon AM, Cruciat CM, et al. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 11.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 12.Cliften P, Sudarsanama P, Desikan A, Fulton L, Fulton B, Majors J, Waterston R, Cohen BA, Johnston M. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- 13.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 14.Olivas WM, Muhlrad D, Parker R. Nucleic Acids Res. 1997;25:4619–4625. doi: 10.1093/nar/25.22.4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar A, Harrison PM, Cheung KH, Lan N, Echols N, Bertone P, Miller P, Gerstein MB, Snyder M. Nat Biotechnol. 2002;20:58–63. doi: 10.1038/nbt0102-58. [DOI] [PubMed] [Google Scholar]
- 16.Oshiro G, Wodicka LM, Washburn MP, Yates JR, III, Lockhart DJ, Winzeler EA. Genome Res. 2002;12:1210–1220. doi: 10.1101/gr.226802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Velculescu VE, Zhang I, Zhou W, Vogelstein J, Basrai MA, Bassett DE, Jr, Hieter P, Vogelstein B, Kinzler KW. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z, Dietrich FS. Nucleic Acids Res. 2005;33:2838–2851. doi: 10.1093/nar/gki583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. Proc Natl Acad Sci USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohtake H, Ohtoko K, Ishimaru Y, Kato S. DNA Res. 2004;11:305–309. doi: 10.1093/dnares/11.4.305. [DOI] [PubMed] [Google Scholar]
- 21.Kato S, Ohtoko K, Ohtake H, Kimura T. DNA Res. 2005;12:53–62. doi: 10.1093/dnares/12.1.53. [DOI] [PubMed] [Google Scholar]
- 22.Oyama T, Yoshida M, Kamegai S, Kitano K, Miura F, Kawaguchi N, Onda M, Satou K, Ito T. Genome Informatics. 2003;14:312–313. [Google Scholar]
- 23.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. Nat Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 24.Vilela C, McCarthy JE. Mol Microbiol. 2003;49:859–867. doi: 10.1046/j.1365-2958.2003.03622.x. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z, Dietrich FS. Curr Genet. 2005;48:77–87. doi: 10.1007/s00294-005-0001-x. [DOI] [PubMed] [Google Scholar]
- 26.He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A. Mol Cell. 2003;12:1439–1452. doi: 10.1016/s1097-2765(03)00446-5. [DOI] [PubMed] [Google Scholar]
- 27.Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S. Bioinformatics. 2002;18:298–305. doi: 10.1093/bioinformatics/18.2.298. [DOI] [PubMed] [Google Scholar]
- 28.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 29.Walther TC, Brickner JH, Aguilar PS, Bernales S, Pantoja C, Walter P. Nature. 2006;439:998–1003. doi: 10.1038/nature04472. [DOI] [PubMed] [Google Scholar]
- 30.Akiva P, Toporik A, Edelheit S, Peretz Y, Diber A, Shemesh R, Novik A, Sorek R. Genome Res. 2006;16:30–36. doi: 10.1101/gr.4137606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parra G, Reymond A, Dabbouseh N, Dermitzakis ET, Castelo R, Thomson TM, Antonarakis SE, Guigo R. Genome Res. 2006;16:37–44. doi: 10.1101/gr.4145906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 33.Davis CA, Ares M., Jr Proc Natl Acad Sci USA. 2006;103:3262–3267. doi: 10.1073/pnas.0507783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 35.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 36.Mattick JS, Makunin IV. Hum Mol Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 37.Mendes Soares LM, Valcarcel J. EMBO J. 2006;25:923–931. doi: 10.1038/sj.emboj.7601023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bickel KS, Morris DR. Mol Cell. 2006;22:309–316. doi: 10.1016/j.molcel.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 39.Adams A, Gottschling DF, Kaiser CA, Stearns T. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1997. [Google Scholar]
- 40.Iyer V, Struhl K. Proc Natl Acad Sci USA. 1996;93:5208–5212. doi: 10.1073/pnas.93.11.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of our cDNA data can be browsed at http://yeast.utgenome.org and downloaded from http://itolab.cb.k.u-tokyo.ac.jp/GCap.