Abstract
Novel systems of inducible gene expression are presented in which CRE-M, an altered form of cre recombinase (cre), is fused to and activated by ligand binding to two forms of the androgen receptor (AR) ligand binding domain (LBD). Selective activation or inactivation of gene transcription is induced upon the addition of appropriate ligand. The coupling of this cre-LBD system with our previously reported highly sensitive assay to measure cre activity in vitro using a dual fluorescent gene switch reporter provides a novel, high-throughput assay system for identifying compounds that bind to and activate various forms of the LBD of androgen receptor. This method can similarly be applied to screen compounds for their activating properties on other steroid hormone LBDs. Three different forms of the AR-LBD were fused to CRE-M, including the wild-type AR-LBD (wt), a non-ligand binding truncated form, LBD (T), and a mutated form (Thr→Ala substitution) identified in the LNCaP prostate cancer cell line, LBD (LNCaP). We demonstrate a 10-fold induction of cre activity by the addition of androgen agonists to the CRE-M-AR-LBD(wt) fusion protein, but not in the presence of the anti-androgen, flutamide. However, cre activity can be induced by flutamide with the CRE-M-AR-LBD(LNCaP) fusion protein. Similar activation properties were obtained when these fusion proteins were expressed using adenoviral vectors. When combined with our previously reported cre-lox gene switch system, the CRE-M-AR-LBD system can be utilized in gene therapy systems in which a therapeutic product may be initially expressed, replaced by a second product, or turned-off following exposure to ligand. This provides an important, additional level of regulation to gene therapy sytems.
INTRODUCTION
The inducible alteration of gene expression is a powerful tool to study gene function in vitro and in vivo and to induce biologic activities for gene therapy (1–4). The use of cre recombinase to activate or inactivate gene expression selectively has been widely used for this purpose (5,6). We have previously demonstrated that our gene switch technology using our novel CRE-M adaptation of the cre-lox system provides the ability to initiate, amplify, switch or extinguish tissue-specific gene expression in a manner which can be applied to both in vitro and in vivo systems (7).
A primary advantage of the CRE-M modification is that it allows for the placement of cre recombinase sequences and loxP sites within the same plasmid (7). This was achieved by introducing an intron with splice sites into the single exon which codes for cre recombinase. The CRE-M modification is applicable to the development of improved transgenic animal methodologies and for gene therapy applications where one vector instead of two can be used to express cre leading to a recombination event and altered expression of a gene product. This approach, however, is still limited by the tissue-specificity and developmental timing determined by the particular promoter that is used. By coupling a steroid hormone ligand binding domain to cre, an additional level of regulation of cre activity can be introduced.
Previous studies have demonstrated that the fusion of steroid hormone ligand binding domains to cre makes the cre activity responsive to the binding of an appropriate ligand. This has been demonstrated for various forms of the estrogen receptor (ER) and progesterone receptor ligand binding domains (LBD) (8–13). These approaches have proved generally successful, but may be limited by relatively high background levels of cre activity in the absence of ligand.
In this study, we have utilized a fusion-protein approach using the LBD of the androgen receptor (AR) to add an additional level of regulation to our CRE-M-gene switch system and to develop additional cre-regulatory systems which may offer particular advantages over existing systems. We have fused the ligand binding domain of the wild-type androgen receptor and two mutant forms of the androgen receptor to our modified cre-recombinase (CRE-M). We demonstrate that cre-activity is minimized unless it is induced by the addition of an appropriate ligand for the AR-LBD in which case cre recombinase becomes functional. This system has also been adapted to adenoviral delivery systems.
When utilized in the context of our gene switch approach (7), the expression of one gene can be replaced by the expression of a second gene, or expression of a gene can simply be turned off when no longer needed. The latter case may be particularly useful when the prolonged expression of a gene therapy product may be potentially toxic or serious side effects have occurred.
The system we have developed can also be applied for high-throughput screening of compounds with ligand-binding activities to various steroid hormone receptors. Cre activity is induced by the addition of appropriate ligands to the CRE-M-AR-LBD constructs depending upon the specificity of the LBD being utilized. When utilized in conjuction with our highly sensitive fluorescent gene switch reporter assay, the CRE-M-AR-LBD system can readily be applied to high throughput screening of compounds for their affinities to various forms of the AR-LBD. This technology can be utilized to identify compounds that may potentially activate or inhibit the AR signaling pathway, through the binding to wild-type or mutant LBDs. Compounds identified using this screening approach may be further validated for their therapeutic or toxicologic potentials.
MATERIALS AND METHODS
Construction of plasmids
Development of reporter plasmids pCMVe-βAc-STOP-luc and pCMV-EGFP/RFP has been described previously (7).
Generation of pCMV-CRE-M(BglII). The 3′ coding sequence for modified cre recombinase CRE-M contained in plasmid p218 (7) was PCR amplified. PCR amplification using the primer set OLN 200T (5′-CTT-CAG-GCG-CGC-GGT-CTG-GCA-GTA-AAA-ACT-ATC-CA-3′) and OLN 201 B (5′-TAG-ACG-CGT-CAT-AGA-TCT-CCA-TCT-TCC-AGC-AGG-CGC-ACC-ATT-GCC-C-3′). The 812 bp amplification product was digested with BamHI and MluI, gel purified and ligated together with both the BamHI and MscI fragment from p218 containing the 5′ end of CRE-M and the MscI–MluI fragment from pBS 185 (Life Technologies, Rockville, MD) containing a polyadenylation sequence resulting in the generation of the plasmid pCMV-CRE-M(BglII) (p259).
Generation of of pSP72hAR. The plasmid containing the cDNA of human (h)AR was used as a template for the PCR amplification of the AR-LBD using the primers pairs OLN202 T (5′-ATT-CTA-GAT-CTG-ATG-ACT-CTG-GGA-GCC-CGG-AAG-CTG-AAG-AAA-CTT-GG-3′) and OLN 203 B (5′-GTA-GGA-TCC-ACG-CGT-TCA-TTG-GGT-GTG-GAA-ATA-GAT-GGG-CTT-GAC-3′). The 903 bp amplification product was digested with XbaI and BamHI enzymes and ligated into the XbaI and BglII of pSP-72 (Promega, Madison, WI) resulting in pSP72hAR (p258).
Generation of pCMV-CRE-M-AR(wt). To generate a fusion gene consisting of cre recombinase and the AR LBD, plasmid p258 was digested with BglII and MluI and the hAR fragment coding for LBD was isolated. This fragment was cloned into pCMV-CRE-M(BglII) by digesting two separate aliquots of pCMV-CRE-M(BglII) with ScaI/BglII or with ScaI/MluI. The ligation of these three fragments resulted in the generation of pCMV-CRE-M-AR(wt).
Generation of AR-LBD mutants. To generate plasmids containing mutant LBDs fused to hAR, plasmid p258 was digested with BglI and BsgI and the 1.8 kb fragment was isolated. A second aliquot of p258 was digested with BglI and DraIII and the 1.3 kb fragment was isolated. Both of these fragments were ligated to a double-stranded oligonucleotide OLN 204T/OLN 205B (5′-TGC-ATC-AGT-TCG-CTT-TTG-ACC-TGC-TAA-TCA-AGT-CAC-ACA-TG-3′ and 5′-GTG-TGA-CTT-GAT-TAG-CAG-GTC-AAA-AGC-GAA-CTG-ATG-CAG-C-3′, respectively) resulting in p264 containing the hAR-LBD(LNCaP) mutation, or to OLN 206/207 (5′-TGT-AGC-AGT-TCA-CTT-TTG-ACC-TGC-TAA-TCA-AGT-CAC-ACA-TG-3′ and 5′-GTG-TGA-CTT-GAT-TAG-CAG-GTC-AAA-AGT-GAA-CTG-CTA-CAG-C-3′, respectively) resulting in p267 containing the hAR-LBD(T) early termination mutation. The hAR-LBD(LNCaP) mutant contains the identical Thr to Ala mutation in the hAR-LBD identified in AR codon 868 (14).
Generation of pCMV-CRE-M-AR(T) and pCMV-CRE-M-AR(LNCaP). To generate expression plasmids containing CRE-M fused to the mutated forms of AR-LBD, the MluI–BglII fragment containing hAR-LBD (LNCaP) from p264 was cloned into the MluI and BglII sites of pCMV-CRE-M(BglII) resulting in the generation of pCMV-CRE-M-AR(LNCaP). Similarly, the MluI–BglII fragment from p267 containing hAR-LBD(T) was similarly cloned into plasmid pCMV-CRE-M(BglII) resulting in the generation of pCMV-Cre-AR(T).
Generation of adenovirus expressing cre or CRE-M-AR(LNCaP)
The XbaI fragment containing cre recombinase from plasmid pCMV-cre-K (p209) described previously (7) or the CRE-M-AR(LNCaP) fragment from pCMV-CRE-M-AR(LNCaP) was blunt ended and individually ligated into the pAdenoVator-CMV5 transfer vector (Qbiogene, Carlsbad, CA) which was previously linearized with PmeI. In order to obtain infectious adenovirus containing cre recombinase, the transfer plasmids were processed according to manufacturer’s instructions resulting in the generation of replication-defective Adeno-cre and Adeno-CRE-M-AR(LNCaP).
PCR conditions
DNA plasmid template 10 ng, 300 µM dNTP, 1 mM MgSO4, 300 nM primers set, 2.5 U of Pfx polymerase in total 50 µl reaction volume. Thermocycler settings were 94°C for 2 min for one cycle, followed by 94°C for 30 s, 68°C for 30 s and 72°C for 1 min (30 cycles), followed by 72°C for 7 min.
Tissue culture
CHO-KI and PC-3 cells (ATCC, Rockville, MD) were propagated in RPMI-1640 medium supplemented with 10% fetal calf serum (FCS) (Life Technologies, Rockville, MD), 1 mM sodium pyruvate, 10 mM HEPES buffer, 2 mM glutamine, 50 U/ml of penicillin G sodium and 50 µg/ml of streptomycin sulfate. Charcoal stripped FCS (HyCLONE, Logan, UT) was used for all studies that required hormone-ligand induction.
Cell transfection
For ligand induction studies, cells were trypsinized (Life Technologies, Rockville, MD), washed in culture media and centrifuged at 190× g for 5 min at room temperature. Cell pellets were resuspended in 1× PBS, centrifuged and resuspended in RPMI-1640 without phenol red and supplemented with charcoal/dextran-treated FCS (HyClONE, Logan, UT). Cells were plated at a density of 1.5 × 105 cells/well in 3 ml of culture media 24 h prior to transfection using 6-well plates (Costar, Corning, NY). On the morning of transfection, 2 ml of fresh media supplemented with or without hormone-ligand were added to plates. Transfections with luciferase reporter plasmids were performed 2 h later in which each well received 2 µg of total plasmid DNA in 6 µl of FuGENE™ 6 (Roche, Indianapolis, IN) in a total of 100 µl of serum-free transfection solution. Studies using fluorescent reporters were similarly performed with the amount of transfected DNA indicated in figure legends. Culture media with ligand was replaced 24 h later. Luciferase assays or microscopy was performed 48 h post-transfection. Trans fections were performed in triplicate and experiments were repeated at least three times. Cells were visualized using a Zeiss ICM 405 fluorescent microscope. GFP was visualized using a 436 nm excitation wavelength and a 510 nm emission filter. RFP was visualized using a 546 nm excitation wavelength and a 580 nm emission filter. Dual fluorescence was observed using 460–490 nm excitation wavelengths and a 510 emission filter.
Luciferase assay
The Dual-Luciferase™ Reporter assay system (Promega, Madison, WI) was used to determine the activity of firefly luciferase (Photinus pyralisus) and sea pansy renilla luciferase (Renilla reniformis) according to the manufacturer’s instructions. Cells were washed in 1× PBS and 500 µl passive lysis solution was added per well. Lystate was collected after 15 min and centrifuged at 13 000 r.p.m. for 30 s. 20 µl of supernatant was used for luciferase assays.
Infection of cells with Adeno-cre and Adeno-CRE-M-AR(LNCaP)
CHO-K1 or QBI293A cells were plated 24 h before transfection and infection in 6-well plates as described above. Two hours before transfection-infection, 2 ml of culture media supplemented with charcoal-striped serum was replaced with fresh media with or without ligand at the indicated concentrations. Cells were subsequently transfected with 0.95 µg of reporter plasmid pCMV-EGFP/RFP (7) using FuGene-6 according to the manufacturer’s instructions (Roche, Indianapolis, IN). Two hours post-transfection, cells were infected with 500 µl of supernatant containing replication defective adenovirus produced by the adenoviral packaging cell line as indicated previously. Cells received fresh ligand supplementations every 24 h.
RESULTS
Construction of inducible CRE-M using androgen-receptor ligand-binding domain variants
Construction of pCMV-CRE-M(BglII) was performed as described in Materials and Methods and is depicted in Figure 1. In order to test the functionality of this altered form of CRE-M, co-transfection experiments were performed in which pCMV-EGFP/RFP (p212) was co-transfected with or without pCMV-CRE-M(BglII). No lox recombination occurred in the absence of pCMV-CRE-M(BglII) since cells only expressed EGFP and not RFP. However, the addition of pCMV-CRE-M(BglII) did lead to lox recombination as evidenced by the expression of RFP (data not shown). This demonstrated that the modification to CRE-M by the addition of the BglII site did not inhibit cre recombinase activity.
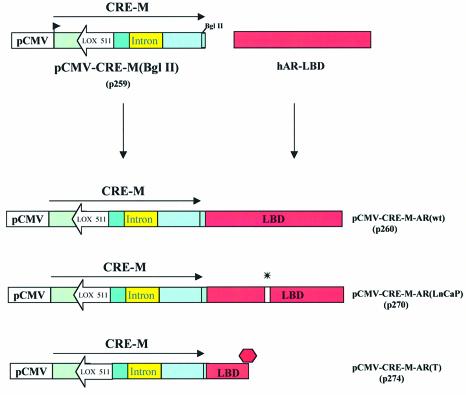
Figure 1.
Generation of ligand inducible cre recombinase. Strategy to generate variants of ligand inducible cre recombinase by fusing various forms of the AR-LBD to CRE-M (see Materials and Methods).
Having established that the BglII site in CRE-M was not deleterious to CRE-M function, three forms of the ligand-binding domain of the human androgen receptor were added separately to pCMV-CRE-M(BglII) (Fig. 1). One construct had the wild-type AR-LBD fused to CRE-M resulting in the construct designated pCMV-CRE-M-AR(wt) (p260). The construct pCMV-CRE-M-AR(LNCaP) contained CRE-M fused to the AR LBD with the mutation identified in the AR-LBD in LnCaP cells (14). The third construct generated contained CRE-M fused to a mutated AR-LBD in which a stop codon was introduced early in the 5′ coding sequence of the LBD [pCMV-CRE-M-AR(T)]. This construct was designed to have no LBD function and would essentially serve as CRE-M without LBD regulation.
Induction of cre activity with androgens or antiandrogen by CRE-M fused to variant AR-LBD
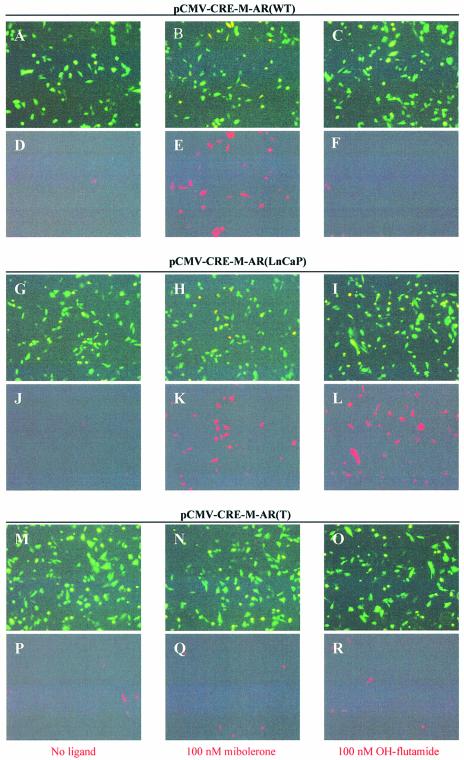
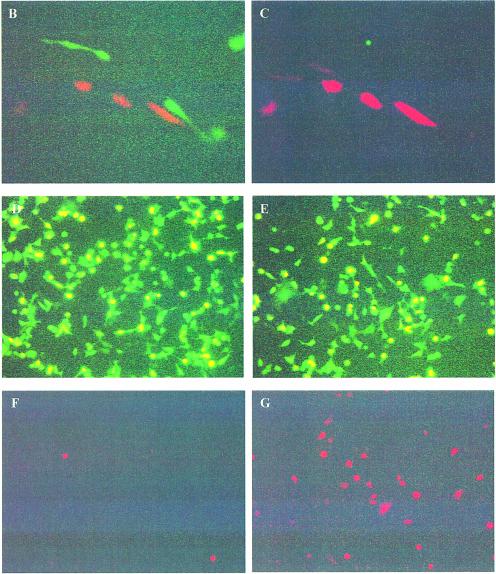
We wished to test the specificity of cre recombinase induction using various androgen analogues. As can be seen in Figure 2, when pCMV-CRE-M-AR(wt) was co-transfected with pCMV-EGFP/RFP (p212) (7), cre activity was only induced by the addition of the androgen agonist mibolerone, but not by the androgen antagonist, OH-flutamide (Fig. 2D–F). However, when pCMV-CRE-M-AR(LNCaP) containing the mutant LBD was co-transfected with pCMV-EGFP/RFP (p212), both mibolerone and OH-flutamide induced cre activity (Fig. 2K–L). However, when pCMV-CRE-M-AR(T) was co-transfected with pCMV-EGFP/RFP, neither mibolerone nor OH-flutamide induced cre activity (Fig. 2Q–R). These results demonstrate that different analogues of androgens could activate CRE-M, depending upon the form of LBD used. We found that the basal level of cre activity using the cre-AR(wt) construct was slightly lower compared with that observed using a previously reported cre-ER construct (data not shown) (15).
Figure 2.
Functional assay of inducible cre recombinase activity induced by ligand. CHO-K1 cells were transiently transfected with reporter plasmid pCMV-EGFP/RFP (0.95 µg) and co-transfected with 0.75 µg of pCMV-CRE-M-AR(WT) (A–F), pCMV-CRE-M-AR(LNCaP) (G–L) or pCMV-CRE-M-AR(T) (M–R) in absence of ligand (A, D, G, J, M, P), in presence of 100 nM mibolerone (B, E, H, K, N, Q) or 100 nM OH-flutamide (C, F, I, L, O, R). Fluorescence microscopy using GFP filter (A, B, C, G, H, I, M, N, O), RFP filter (D, E, F, J, K, L, P, Q, R). Assay was performed 48 h after exposure to ligand (magnification 100×).
Quantification of cre activity induced by ligands
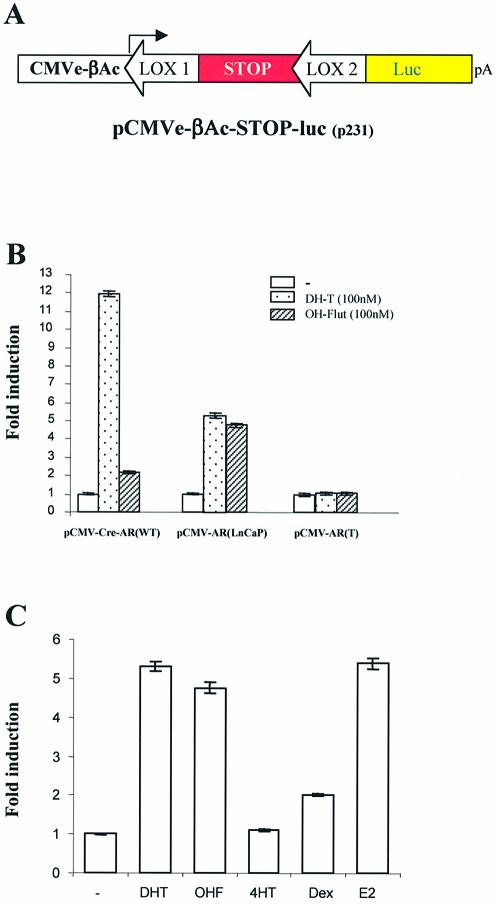
In order to quantify the induction of cre activity by various ligands, the pCMV-CRE-M-AR variants were co-transfected with our reporter construct pCMVe-βAc-STOP-luc (7) (Fig. 3A). The addition of 100 nM DHT resulted in a 12-fold induction of luciferase activity when pCMV-CRE-M-AR(wt) was used, whereas 100 nM OH-flutamide had a minimal effect on luciferase activity (Fig. 3B). However, both DHT and OH-flutamide were able to induce a 5-fold increase in luciferase activity when pCMV-CRE-M-AR(LNCaP) was used (Fig. 3B). As expected, no induction in luciferase was observed when pCMV-CRE-M-AR(T) was used.
Figure 3.
Relative induction of cre recombinase activity by different ligands. (A) Representation of luciferase reporter construct to assay cre recombinase activity. (B) PC-3 cells were co-transfected with reporter plasmid pCMVe-βAc-STOP-luc (0.1 pmol) and plasmids (0.1 pmol) coding for modified inducible cre recombinase in the absence or presence of ligand for a period of 48 h. Transfection efficiency was normalized with pTK-Rl. (C) PC-3 cells were co-transfected with reporter plasmid pCMVe-βAc-STOP-luc and pCMV-CRE-M-AR(LNCaP) (0.001 pmol) in presence of selected ligands at indicated ligand concentration (48 h exposure to ligand). Transfection efficiency was normalized with pTK-Rl which expresses renilla luciferase.
We further tested the ability of additional ligands to induce cre activity using pCMV-CRE-M-AR(LNCaP). DHT, OH-flutamide, 17-OH-estradiol (Fig. 3C) and the progesterone analogue RU-486 (data not shown) induced similar levels of cre activity. The addition of the anti-estrogen, tamoxifen, did not lead to a significant induction of luciferase activity, although there was a modest response to dexamethasone (Fig. 4B).
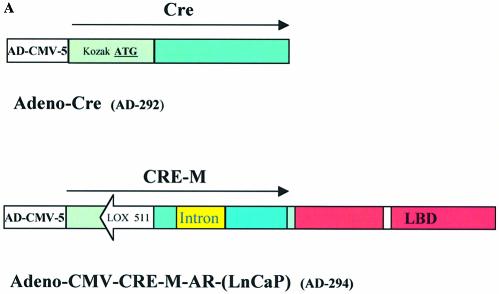
Figure 4.
Functional assay for inducible cre recombinase activity generated by adenovirus. (A) Schematic representation of Adeno-Cre and Adeno-CMV-CRE-M-AR(LNCaP). (B and C) CHO-K1 cells transfected with 0.95 µg reporter plasmid pCMV-EGFP/RFP and infected with Adeno-Cre. Fluorescence microscopy using GFP filter (B) or RFP filter (C). Assay performed 48 h post-infection (magnification 400×). (D–G) Cell line QBI-293A was transfected with 0.95 µg of reporter plasmid pCMV-EGFP/RFP and 2 h later was infected with Adeno-CMV-CRE-M-AR(LNCaP) (E–G) in the absence of ligand (D, F) or in the presence of 100 nM OH- flutamide (E, G). Assay was performed 48 h post-infection. Fluorescence microscopy using GFP filter (D, E) or using RFP filter (F, G) (magnification 100×).
Inducible expression of cre activity by adenoviral delivery
The coding sequence for cre recombinase, with modifications for improved translation as previously described (7), and the CRE-M-AR(LNCaP) coding sequence were individually inserted into the adenoviral vector (Fig. 4A). Infectious viral particles were produced as described in Materials and Methods. CHO-K1 cells were transfected with pCMV-EGFP/RFP (p212) and subsequently infected with Adeno-CRE-M. As seen in Figure 4B and C, lox recombination occurred as expression of RFP was observed demonstrating cre activity. When pCMV-EGFP/RFP (p212) was transfected into cells and subsequently infected with adeno-CRE-M-AR(LNCaP), cre recombinase activity could be induced by OH-flutamide (Fig. 4D–G). No recombination with RFP expression was observed without infection of adeno-CRE-M-AR(LNCaP) (data not shown). These results demonstrate that cre-activity can be induced by the addition of an appropriate ligand when delivered in an adenoviral vector system and that this method can turn gene expression on or off.
DISCUSSION
The controlled induction of gene expression is a powerful tool for studying aspects of many molecular processes including development and oncogenesis (16–20). Several systems have been developed where cre recombinase is induced to elicit recombination of lox sites leading to genomic rearrangements (21–23) alterations in gene expression (7,24,25) or the introduction of particular mutations (26,27). This technology has been particularly useful for conditional alterations in gene expression in genetically engineered mice (1,5,28–31).
We have previously described systems in which a modified cre recombinase (CRE-M) and a pair of lox sites can be assembled into a single vector (7). This system provides a means to use a single vector to conduct cre-lox based experiments instead of using two separate vectors with cre and lox sites in trans. Using this single vector approach, we have shown that gene expression from a weak, but tissue-specific promoter can be amplified.
In the series of experiments reported here, an additional level of regulation of cre activity was achieved through the fusion of variant forms of the AR-LBD to CRE-M. We have demonstrated that the induction of cre activity is dependent upon the ligand binding properties of the particular AR-LBD variant. When the wild-type AR-LBD is fused to CRE-M, cre recombinase activity is induced by the androgen agonists testosterone, dehydrotestosterone and mibolerone (data not shown), but not by the androgen antagonist OH-flutamide. However, when the ligand specificity of the AR-LBD has been altered through a mutation, the ligand inducibility of cre activity is similarly altered.
In this study, the mutant AR-LBD, which has been identified in the LNCaP cell line (14), was fused to CRE-M. In this case, cre activity could be induced by both androgen agonists and the anti-androgen OH-flutamide. This is consistent with previous reports where altered ligand binding and activation properties of the AR LNCaP mutant has been previously characterized (14,32–34). We have demonstrated that cre activity can be induced using CRE-M-AR-LBD(LNCaP) with high (non-physiological) concentrations of progesterone and E2, but not by the antiestrogen, tamoxifen or dexamethasone.
Our results demonstrate that the AR-LBD can be used to activate cre activity when used with ligands that activate the particular form of the AR-LBD. The application of the Cre-ER-LBD systems have previously been shown to activate cre activity when appropriate ligands are added (15). We compared the induction of our CRE-M-AR-LBD construct with the original cre-ER-LBD and found the CRE-M-AR-LBD construct to have lower background activity. Similar approaches have previously been reported in which variants of the GR-LBD (35) and PR LBDs have been fused to cre recombinase (13). However, such systems may not be ideal for activating cre activity in studies utilizing female mice, particularly those involving the study of mammary biology or cancer since such ligands may interfere with normal hormone signaling in the target tissue or may alter expression of the commonly used MMTV LTR. Experiments which study the therapeutic effects of antiestrogens such as tamoxifen in mice could be performed in combination with the cre-AR-LBD (wt)-lox-based system to additionally alter the expression of a gene of interest uncoupled to the exposure of tamoxifen. Such an experiment could not be performed using tamoxifen and the existing cre-ER system without simultaneously activating cre. Further in vivo studies will address this point to determine the potential advantages of the cre-AR-LBD (wt) system.
In addition, we have demonstrated that the CRE-M-LBD system can be delivered to cells using adenoviral vectors. A major concern of gene therapy approaches relates to potential toxicity of compounds produced by the gene therapy vector. Using our gene-switch approach, we demonstrate that an inducible form of cre recombinase may be particularly useful to inactivate gene expression from gene therapy vectors designed to contain lox P sites flanking the therapeutic gene. Expression of the therapeutic gene can be extinguished using the inducible cre-lox gene switch system to terminate the expression once adequate therapy has been administered or if toxicity develops from the gene-therapy vector.
The system we have developed may also be employed as a unique and novel assay for high throughput screening of compounds that may affect steroid hormone signaling, either as an activator or inhibitor, based upon their ability to bind various forms of the AR-LBD. The high sensitivity of our system in conjunction with high specificity required to activate Cre-AR will allow for the detection of relatively weak AR ligands which offers a significant advantage over other systems to screen for ligand binding activities. Using the gene switch reporter system, which is activated by cre recombinase, we can assess whether a particular compound binds to the LBD and activates cre. This assay can be adapted to screen numerous compounds for binding to the AR-LBD. Compounds which result in the expression of the reporter gene products (such as luciferase or RFP) would be identified as activating a particular LBD and can be studied further. Similar reporter systems can be developed to test ligand binding to other steroid hormone LBDs. This approach could rapidly identify compounds within the environment that may have steroid hormone activities and be related to environmental risks for disease, including hormone-related cancers.
REFERENCES
- 1.Le Y. and Sauer,B. (2000) Conditional gene knockout using cre recombinase. Methods Mol. Biol., 136, 477–485. [DOI] [PubMed] [Google Scholar]
- 2.Fuhrmann-Benzakein E., Garcia-Gabay,I., Pepper,M.S., Vassalli,J.D. and Herrera,P.L. (2000) Inducible and irreversible control of gene expression using a single transgene. Nucleic Acids Res., 28, e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaisser F. (2000) Inducible gene expression and gene modification in transgenic mice. J. Am. Soc. Nephrol., 11, suppl. 16, S95–S100. [PubMed] [Google Scholar]
- 4.Komatsu K., Suzuki,S., Shimosegawa,T., Miyazaki,J.I. and Toyota,T. (2000) Cre-loxP-mediated bax gene activation reduces growth rate and increases sensitivity to chemotherapeutic agents in human gastric cancer cells. Cancer Gene Ther., 7, 885–892. [DOI] [PubMed] [Google Scholar]
- 5.Reichardt H.M., Kellendonk,C., Tronche,F. and Schutz,G. (1999) The Cre/loxP system–a versatile tool to study glucocorticoid signalling in mice. Biochem. Soc. Trans, 27, 78–83. [DOI] [PubMed] [Google Scholar]
- 6.Gorman C. and Bullock,C. (2000) Site-specific gene targeting for gene expression in eukaryotes. Curr. Opin. Biotechnol., 11, 455–460. [DOI] [PubMed] [Google Scholar]
- 7.Kaczmarczyk S.J. and Green,J.E. (2001) A single vector containing modified cre recombinase and LOX recombination sequences for inducible tissue-specific amplification of gene expression. Nucleic Acids Res., 29, e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Metzger D. and Chambon,P. (2001) Site- and time-specific gene targeting in the mouse. Methods, 24, 71–80. [DOI] [PubMed] [Google Scholar]
- 9.Vallier L., Mancip,J., Markossian,S., Lukaszewicz,A., Dehay,C., Metzger,D., Chambon,P., Samarut,J. and Savatier,P. (2001) An efficient system for conditional gene expression in embryonic stem cells and in their in vitro and in vivo differentiated derivatives. Proc. Natl Acad. Sci. USA, 98, 2467–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiba H., Chambon,P. and Metzger,D. (2000) F9 embryonal carcinoma cells engineered for tamoxifen-dependent Cre-mediated site-directed mutagenesis and doxycycline-inducible gene expression. Exp. Cell Res., 260, 334–339. [DOI] [PubMed] [Google Scholar]
- 11.Kellendonk C., Tronche,F., Monaghan,A.P., Angrand,P.O., Stewart,F. and Schultz,G. (1996) Regulation of Cre recombinase activity by the synthetic steroid RU 486. Nucleic Acids Res., 24, 1404–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minamino T., Gaussin,V., DeMayo,F.J. and Schneider,M.D. (2001) Inducible gene targeting in postnatal myocardium by cardiac-specific expression of a hormone-activated Cre fusion protein. Circ. Res., 88, 587–592. [DOI] [PubMed] [Google Scholar]
- 13.Wunderlich F.T., Wildner,H., Rajewsky,K. and Edenhofer,F. (2001) New variants of inducible Cre recombinase: a novel mutant of Cre-PR fusion protein exhibits enhanced sensitivity and an expanded range of inducibility. Nucleic Acids Res., 29, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veldscholte J., Ris-Stalpers,C., Kuiper,G.G., Jenster,G., Berrevoets,C., Classen,E., van Rooij,H.C., Trapman,J., Brinkmann,A.O. and Mulder,E. (1990) A mutation in the ligand binding domain of the androgen receptor of human LNCaP cells affects steroid binding characteristics and response to anti-androgens. Biochem. Biophys. Res. Commun., 173, 534–540. [DOI] [PubMed] [Google Scholar]
- 15.Metzger D., Clifford,J., Chiba,H. and Chambon,P. (1995) Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc. Natl Acad. Sci. USA, 92, 6991–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo J.L., Yang,Q., Tong,W.M., Hergenhahm,M., Wang,Z.Q. and Hollstein,M. (2001) Knock-in mice with a chimeric human/murine p53 gene develop normally and show wild-type p53 responses to DNA damaging agents: a new biomedical research tool. Oncogene, 20, 320–328. [DOI] [PubMed] [Google Scholar]
- 17.Houghton L. and Rosenthal,N. (1999) Regulation of a muscle-specific transgene by persistent expression of Hox genes in postnatal murine limb muscle. Dev. Dyn., 216, 385–397. [DOI] [PubMed] [Google Scholar]
- 18.Prall O.W., Rogan,E.M. and Sutherland,R.L. (1998) Estrogen regulation of cell cycle progression in breast cancer cells. J. Steroid Biochem. Mol. Biol., 65, 169–174. [DOI] [PubMed] [Google Scholar]
- 19.Jackson E.L., Willis,N., Mercer,K., Bronson,R.T., Crowley,D., Montoya,R., Jacks,T. and Tuvenson,D.A. (2001) Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev., 15, 3243–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manickan E., Satoi,J., Wang,T.C. and Liang,T.J. (2001) Conditional liver-specific expression of simian virus 40 T antigen leads to regulatable development of hepatic neoplasm in transgenic mice. J. Biol. Chem., 276, 13989–13994. [DOI] [PubMed] [Google Scholar]
- 21.Osborne B.I., Wirtz,U. and Baker,B. (1995) A system for insertional mutagenesis and chromosomal rearrangement using the Ds transposon and Cre-lox. Plant J., 7, 687–701. [DOI] [PubMed] [Google Scholar]
- 22.VanDeursen J., Fornerod,M., Van Rees,B. and Grosveld,G. (1995) Cre-mediated site-specific translocation between nonhomologous mouse chromosomes. Proc. Natl Acad. Sci. USA, 92, 7376–7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlake T. et al. (1999) Predetermined chromosomal deletion encompassing the Nf-1 gene. Oncogene, 18, 6078–6082. [DOI] [PubMed] [Google Scholar]
- 24.Brooks A.I., Muhkerjee,B., Panahian,N., Cory-Slechta,D. and Federoff,H.J. (1997) Nerve growth factor somatic mosaicism produced by herpes virus-directed expression of cre recombinase. Nat. Biotechnol., 15, 57–62. [DOI] [PubMed] [Google Scholar]
- 25.Nagy A., Mones,C., Ivanyi,E., Pawling,J., Gertsenstein,M., Hadjantonakis,A.K., Pirity,M. and Rossant,J. (1998) Dissecting the role of N-myc in development using a single targeting vector to generate a series of alleles. Curr. Biol., 8, 661–664. [DOI] [PubMed] [Google Scholar]
- 26.Vazquez J.C., Nogues,C., Rucker,E.B. and Piedrahita,J.A. (1998) Factors affecting the efficiency of introducing precise genetic changes in ES cells by homologous recombination: tag-and-exchange versus the Cre-loxp system. Transgen. Res., 7, 181–193. [DOI] [PubMed] [Google Scholar]
- 27.Arin M.J., Longley,M.A., Wang,X.J. and Roop,D.R. (2001) Focal activation of a mutant allele defines the role of stem cells in mosaic skin disorders. J. Cell Biol., 152, 645–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu Y. and Bradley,A. (2001) Engineering chromosomal rearrangements in mice. Nature Rev. Genet., 2, 780–790. [DOI] [PubMed] [Google Scholar]
- 29.Lewandoski M. (2001) Conditional control of gene expression in the mouse. Nature Rev. Genet., 2, 743–755. [DOI] [PubMed] [Google Scholar]
- 30.Metzger D. and Feil,R. (1999) Engineering the mouse genome by site-specific recombination. Curr. Opin. Biotechnol., 10, 470–476. [DOI] [PubMed] [Google Scholar]
- 31.Hadjantonakis A.K., Pirity,M. and Nagy,A. (1999) Cre recombinase mediated alterations of the mouse genome using embryonic stem cells. Methods Mol. Biol., 97, 101–122. [DOI] [PubMed] [Google Scholar]
- 32.Veldscholte J., Berrevoets,C.A., Brinkmann,A.O., Grootegoed,J.A. and Mulder,E. (1992) Anti-androgens and the mutated androgen receptor of LNCaP cells: differential effects on binding affinity, heat-shock protein interaction and transcription activation. Biochemistry, 31, 2393–2399. [DOI] [PubMed] [Google Scholar]
- 33.Wilding G., Chen,M. and Gelmann,E.P. (1989) Aberrant response in vitro of hormone-responsive prostate cancer cells to antiandrogens. Prostate, 14, 103–115. [DOI] [PubMed] [Google Scholar]
- 34.Veldscholte J., Voorhorst-Ogink,M.M., Bolt-de Vries,J., van Rooij,H.C. and Trapman,J.E. (1990) Unusual specificity of the androgen receptor in the human prostate tumor cell line LNCaP: high affinity for progestagenic and estrogenic steroids. Biochim. Biophys. Acta, 1052, 187–194. [DOI] [PubMed] [Google Scholar]
- 35.Brocard J., Feil,R., Chambon,P. and Metzger,D. (1989) A chimeric Cre recombinase inducible by synthetic, but not by natural ligands of the glucocorticoid receptor. Nucleic Acids Res., 26, 4086–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]