Abstract
A great diversity of pheromone structures are used by moth species (Insecta: Lepidoptera) for long-distance mating signals. The signal/response channel seems to be narrow for each species, and a major conundrum is how signal divergence has occurred in the face of strong selection pressures against small changes in the signal. Observations of various closely related and morphologically similar species that use pheromone components biosynthesized by different enzymes and biosynthetic routes underscore the question as to how major jumps in the biosynthetic routes could have evolved with a mate recognition system that is based on responses to a specific blend of chemicals. Research on the desaturases used in the pheromone biosynthetic pathway for various moth species has revealed that one way to make a major shift in the pheromone blend is by activation of a different desaturase from mRNA that already exists in the pheromone gland. Data will be presented to support the hypothesis that this process was used in the evolution of the Asian corn borer, Ostrinia furnacalis species. In that context, moth sex-pheromone desaturase genes seem to be evolving under a birth-and-death process. According to this model of multigene family evolution, some genes are maintained in the genome for long periods of time, whereas others become deleted or lose their functionality, and new genes are created through gene duplication. This mode of evolution seems to play a role in moth speciation, as exemplified by the case of the Asian corn borer and European corn borer, Ostrinia nubilalis species.
Chemical communication systems in insects have provided exciting challenges to researchers in chemistry, biochemistry, physiology, ecology, genetics, and behavior for over four decades. Much of this research has been focused on moths in the order Lepidoptera, which is the second largest insect order with well over a hundred thousand described species. Most of the hundreds of species studied have been found to use a long-distance chemical communication system for attracting mates (www.nysaes.cornell.edu/fst/faculty/acree/pheronet/index.html). Initially, pheromone components were characterized, and behavior mediated by these chemical cues was studied. However, increased knowledge of the precise blends used by different species only raised more questions on many aspects of the communication system. Underlying these questions was the fundamental curiosity about the extensive radiation seen in moths and what role pheromones played in the speciation process. How did sex pheromones evolve and could changes in this mating system give rise to isolated populations that become new species? Studies on pheromone biosynthetic pathways provided basic information on this issue, but the use of molecular techniques in the postgenomics era has become essential in addressing these questions.
Pheromone Biosynthetic Pathways
By the 1980s, many moth pheromone components had been characterized, and a majority were acetates, alcohols, or aldehydes with long hydrocarbon chains (10–18C) containing 1–3 double bonds with variable positions and geometric configurations (1). Studies with labeled precursors were carried out to determine whether the pheromone components were produced via fatty acid precursors and what regulated the chain length and double bond positions. Data on the red-banded leafroller moth, Argyrotaenia velutinana, showed that the two major pheromone components, (Z)-11-tetradecenyl acetate (Z11–14:OAc) and (E)-11-tetradecenyl acetate (E11–14:OAc) were produced with Δ11 desaturation of myristic acid (14:Acid), followed by reduction and acetylation (2). Data on the cabbage looper, Trichoplusia ni, showed that the main pheromone component, (Z)-7-dodecenyl acetate (Z7–12:OAc), was produced by Δ11 desaturation of palmitic acid (16:Acid) followed by chain shortening to Z9–14:Acid and then to Z7–12:Acid, with subsequent reduction and acetylation (3).
As biosynthetic pathways were defined for more moth species, it became obvious that biochemical pathways involving two key enzymatic steps, desaturation and chain shortening, had evolved in the terminal segments (pheromone gland) of female moths. The limited chain-shortening steps added diversity by generating cascades of compounds of different chain lengths with double bond positions two carbons closer to the functional group with each round of chain shortening. It was soon found, however, that the integral membrane desaturases were able to add diversity by evolving unique substrate specificities, as well as regio- and stereospecificities. Thus, although many species were found to use rare Δ11 desaturases that produced Z11–16:Acid, mixtures of Z11/E11–14:Acid, or only E11–14:Acid, other species were found to use different desaturases. Primitive moth species in New Zealand are particularly interesting because the complex of morphologically similar species in the Planotortrix and Ctenopseutis genera produce pheromone components that involve at least Δ5, Δ7, Δ9, and Δ11 desaturases, either alone or in combination (4). How did these closely related species evolve such diverse desaturases, and how was it possible to mutate from one strongly stabilized pheromone system to another that used pheromone components with different double bond positions?
Desaturase Genes
The biosynthetic enzymes in the moth pheromone glands could have evolved from genes involved in normal fatty acid metabolism, but it was soon recognized (5) that the resolution powers of data on biosynthetic pathways for phylogenetic reconstruction are limited and do not necessarily impart evidence of evolutionary direction. A move to the molecular level was dictated in an attempt to determine homologies among the metabolic Δ9 desaturases and sex-pheromone desaturases.
A collaborative effort in the early 1990s was initiated on the gene that encodes a Δ11 desaturase in the sex-pheromone gland of female T. ni. Pheromone mRNA was isolated and reverse transcribed to make cDNA, which was used as template in PCR reactions with degenerate primers designed from conserved areas of rat and mouse acyl-CoA Δ9 desaturases (6, 7). Candidate desaturase clones were obtained but had to be assayed for functionality. This proved to be difficult with an in vitro reconstitutive biochemical assay because integral membrane desaturases were shown to use a complex consisting of the desaturase protein, NADH-cytochrome b5 reductase (a flavoprotein), and cytochrome b5 (a hemoprotein). However, a discovery that the yeast OLE1 desaturase can be functionally replaced in vivo by the rat desaturase (7) led the way to the development of yeast expression systems that could be used for the functional assay of pheromone desaturases (8).
The structures of Δ9 and Δ11 desaturase-encoding cDNAs from T. ni and the corn earworm moth, Helicoverpa zea, were first characterized and reported (8–10). Research on other selected species resulted in the characterization of structures of other pheromone desaturases with different regio- and stereospecificities, including Z/E11, E11, Z10, Z9, and Z/E14 desaturases (8–15). Cloning studies using mRNA isolated from fat bodies and pheromone glands revealed that there were at least two classes of Δ9 desaturases in the Lepidoptera. One produced a mixture containing palmitoleic acid (Z9–16:Acid) > oleic acid (Z9–18:Acid), and the other with a reverse ratio of Z9–18 > Z9–16. It also became obvious that there were desaturase genes present in the pheromone gland that were not functioning to produce unsaturated fatty acid product and some desaturase clones that showed no activity in any of the available functional assays. These results provided some insights into how this multigene family evolved and how these genes could play a role in the speciation process.
Corn Borer Speciation
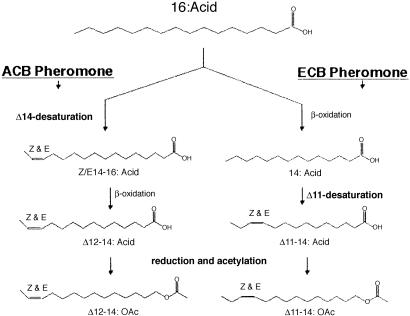
Research on the European corn borer (ECB), Ostrinia nubilalis, has shown that pheromone production (female) and response (male) are not genetically linked (16), which provides the basis for models on the evolution of new pheromones based on asymmetric tracking (17, 18) and includes examples wherein a large mutational effect in female pheromone production is subsequently tracked by male response. Desaturase cloning studies with ECB surprisingly revealed a way that the large change in female pheromone production could be effected. The studies (15) showed that the ECB pheromone gland contained mRNA for three different desaturases, but unsaturated products were only present in the gland from one of them, the Δ11 that makes precursors for the pheromone components, Z11-/E11–14:OAcs (19) (Fig. 1). Another one was a Δ9 desaturase, which is commonly present in pheromone glands, and the third was found to be a Δ14 desaturase. The presence of the Δ14 desaturase is significant because it is the desaturase used by the Asian corn borer (ACB), O. furnacalis (20), to produce its unusual mixture of Z/E12–14:OAcs pheromone components (21) (Fig. 1).
Fig. 1.
Pheromone biosynthetic pathways for ACB and ECB from hexadecanoic acid (16:Acid) and proceeding through different routes to the 14-carbon acetate pheromone components.
The ACB is the only Ostrinia species in the world known to use the Δ14 desaturase, with all others using a Δ11 desaturase (22). Research on mRNA from ACB pheromone glands showed that they had the same three desaturase clones as ECB, only in this case the only unsaturated product was that from the Δ14 desaturase. A sudden switch in pheromone components from those produced by Δ11 desaturation to the products of a different biosynthetic pathway involving Δ14 desaturation could help explain how the ACB population was derived about a million years ago. Studies of several other genes involved in the chemical communication system of ACB and ECB also seem to be unchanged, supporting the recent derivation of ACB. A single pheromone binding protein in male antennae has an identical structure in ACB and ECB males (23), and the reductase enzyme in ACB and ECB has similar specificities with a preference for Z11–14:Acid, even though ACB needs to reduce Z/E12–14:Acids (24).
The activation of a nonfunctional Δ14 desaturase in the pheromone gland of an ancestral Ostrinia species would provide the mutational shift in pheromone production needed to initiate evolution of this chemical communication system, but would there be any males to respond to this pheromone blend? A screening of ECB males in flight-tunnel assays (25) showed that there are rare males that exhibit a broad range of responses that include flight to the ACB pheromone blend. This was surprising because ECB males would not be expected to respond to Δ12 acetates and also because the ACB blend of Δ12 acetates contains 33% of the E isomer (33% of E11 acetate is antagonistic to this race of ECB males) (25). However, the rare males exhibited complete upwind flights to both the ECB (97:3 mix of Δ11 acetates) and ACB blend (2:1 mix of Δ12 acetates). The existence of these rare males supports the possibility for a major shift in pheromone blend being tracked by male response and fits well with simulation models (26), showing that sudden major switches in pheromone blend and male response seem more likely than accumulation of small changes.
Discovery of the Δ14 desaturase in ECB gives rise to many more questions on the origin of this gene and how long it has been carried in moths. Research in the postgenomic era is required to answer these questions.
Evolutionary Mechanisms
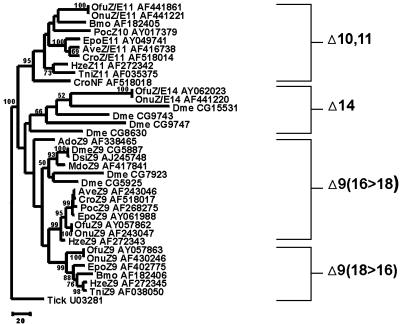
In a previous study (15), we identified several unique aspects of insect desaturase multigene family evolution. The first was the discovery that this family originated before the split among Lepidoptera, Diptera, and Orthoptera, which is estimated to have occurred ≈350 million years ago (27). Second, we found that the family is composed of at least four gene clusters, evolving at disparate rates that are correlated with the function of each group (15). For instance, the ΔZ9 (16 > 18) group has evolved the slowest and contains metabolic desaturases, which presumably represent the ancestral function of this gene family. The ΔZ9 (18 > 16) and Δ10,11 groups are composed of sex-pheromone desaturase genes and evolved at faster rates, which is coincident with a change in function from metabolism to reproduction. The Δ14 group is made up of the fastest-evolving sequences (15) that are nonfunctional (e.g., Δ14 of O. nubilalis), function in sex-pheromone biosynthesis (e.g., Δ14 of O. furnacalis), or are yet to be functionally characterized. Interestingly, we found (15) that several dipteran sequences cluster within this group (Fig. 2). However, until we know the function of the proteins these genes encode, we cannot be certain on the basis of phylogenetic data alone, whether or not these represent new groups distinct from the sequences classified in the Δ14 group.
Fig. 2.
Phylogeny of the desaturase genes of various insect species. Only species for which complete sequences were available were used. The tree was reconstructed from JTT amino acid distances (28) using the maximum likelihood method as implemented in the proml computer program in the phylip software package (http://evolution.genetics.washington.edu/phylip.html). Numbers along branches indicate bootstrap support from 1,500 replicates. The accession numbers for sequences are given after the species abbreviation, as per ref. 15. In the case of D. melanogaster, the gene names are given as they are listed in the Drosophila genome database (www.fruitfly.org). The tree is rooted with the desaturase gene from the tick (Amblyomma americanum), and the log likelihood was –14,878.90361.
Perhaps the most interesting finding of these studies is that reciprocal desaturase gene nonfunctionalization in O. furnacalis and O. nubilalis has resulted in a change in pheromone composition between these species. In this case, the Δ14 gene of O. furnacalis makes a functional desaturase gene product but the ΔZ/E11 does not, whereas the opposite is true in O. nubilalis. Because the ORFs of the ΔZ/E11 sequences are identical and the ORFs of the Δ14 differ by two amino acids in the Ostrinia species, the reciprocal nonfunctionalization event must have been relatively recent [i.e., at the time of their speciation, which is inferred to have occurred ≈1 million years ago (R. Harrison, personal communication), assuming a lepidopteran mtDNA substitution rate of 2% per million years (29)]. There are at least two possible mechanisms to account for the nonfunctionality of these genes. First, they may have recently become pseudogenes. The paradigm pseudogene is one that is no longer transcribed, possesses numerous nucleotide insertions or deletions resulting in frameshift mutations, and is highly divergent from its functional counterpart(s) (30). What is unusual about the Ostrinia nonfunctional genes is that they are well conserved between the two species (Fig. 2). However, recent pseudogenization could account for this. In addition, there are several other cases in which pseudogenes have been shown to be conserved and even possess intact ORFs. For example, the bacteria species Haemophilus aegyptius and Haemophilus influenzae are sister taxa that diverged very recently, and they each possess hap pseudogenes that are identical in sequence (31). In this case, not enough time has elapsed for substitutions to have occurred in either species' pseudogene. Highly conserved pseudogene sequences may also result among paralogues that duplicated recently, as has been shown to occur among the three aquaporin pseudogenes of humans (99% sequence similarity) (32) and the human desaturase functional/pseudogene pair (95% sequence similarity) (33). There are even numerous examples in which a pseudogene produces an mRNA transcript but not a translated protein (e.g., refs. 34–37).
Another possible explanation for the nonfunctionality of certain Ostrinia genes is that there is an epigenetic mechanism repressing their transcription and/or expression (38). For example, the genes nanos and pumilio interact to control the translational repression of genes involved in embryonic patterning and spermatogenesis–oogenesis switching (39). However, in cells in which the nanos protein is not required, it is translationally repressed through the action of at least two proteins known as Smaug and Bicaudal (reviewed in ref. 40). Such mechanisms for translational repression are found throughout eukaryotes, suggesting that a similar one might be involved in the control of moth sex-pheromone desaturase genes, although whether or not such a mechanism can account for the situation with Ostrinia remains to be shown.
Nevertheless, sequence conservation and/or preservation of an intact ORF do not necessarily ensure the functionality of a gene. The only reliable way to determine whether a protein-coding gene with an intact ORF is functional is to determine whether a functional protein product is made. Our work on the Ostrinia genes shows that in no instance has a protein product or predicted sex pheromone been detected in an experimental assay (15). The next step, which we are currently pursuing, is to determine how the genes became nonfunctional. In this regard, we draw attention to a study on the desat2 gene of Drosophila melanogaster, which showed that this gene has been nonfunctionalized in the Canton-S strain (41). In this case, a mutation in the promoter has inactivated the desat2 gene, rendering it a recently formed pseudogene. Follow-up studies showed that this nonfunctionalization of the desat2 gene has occurred in other Drosophila races and has led to an incipient speciation event among those with a functional desat2 versus those with a nonfunctional desat2 (42). This is because the functional desat2 races produce a different pheromone complement than the nonfunctional desat2 races, leading to a change in mating response. In addition to explaining how sex-pheromone desaturase gene nonfunctionalization has occurred in Drosophila, these studies reinforce our contention that desaturase gene nonfunctionalization can lead to speciation in insects (15). One question that remains is how gene duplication has played into the picture.
It is clear from the phylogenetic tree shown in Fig. 2 that there have been various points of desaturase gene duplication during the course of insect evolutionary history. The first duplication event gave rise to the Δ10,11 group followed by subsequent duplications that gave rise to the Δ14 and Δ9 (18 > 16) groups. As we stated previously (15), we believe that the ancestral gene that gave rise to these groups was a Z9 gene from the Δ9 (16 > 18) group based on the fact that (i) these genes function in metabolism in much the same way as in other animals and (ii) the involvement of desaturases in the production of sex pheromones is known to be a derived function found only in insects thus far. On further examination, it can be inferred that the ancestral moth Z9 gene duplicated in their common ancestor subsequent to the divergence from flies, roughly 330 million years ago (27), and gave rise to the lepidopteran Δ9 (18 > 16) group (Fig. 1). Similarly, the ancestral dipteran Z9 gene seems to have duplicated sometime after the dipteran–lepidopteran divergence followed by another duplication event, thus resulting in three dipteran paralogs that cluster within the Δ9 (16 > 18) group (Fig. 1). Likewise, a nonfunctional gene (CroNF) seems to have duplicated from the ancestral moth Z11 gene, long before Choristoneura rosaceana diverged from the other moth species shown in the Δ10,11 group.
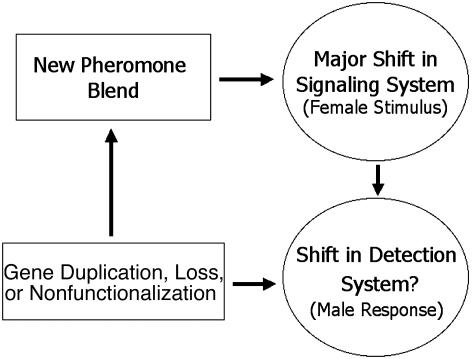
The above results, along with those of previous studies (15, 41, 42), indicate that gene duplication, gene loss, and pseudogene formation have influenced the evolution of desaturase genes in moths and flies. These processes are characteristic of the birth- and-death model of multigene family evolution (43–46). According to the birth-and-death model, multigene families are created through gene duplication, which gives rise to new member genes, but some genes become deleted from the genome or degenerate into pseudogenes, whereas others are maintained. As a result, the members of a multigene family subject to birth-and-death evolution will evolve more or less independently, and different paralogs will be shared by different species or lineages. Thus, in a phylogenetic analysis of a multigene family including member genes from relatively closely related species, we should see the following observations: (i) sequences will cluster by gene or duplication order and not by species; (ii) low levels of sequence homogeneity between genes will be observed (especially at noncoding sites), except in the case of recent duplicates; and (iii) evidence of gene loss/deletion or pseudogene formation will be apparent. As we have already shown, all of these occur in the insect desaturase multigene family. In addition, we believe that the male moth pheromone-receptor system, which is composed of olfactory receptor loci (47, 48), may also be subject to a birth-and-death model of evolution, perhaps as a result of coevolution with the desaturase multigene family. A large and diverse olfactory receptor multigene family would provide an adaptive advantage for male moths, allowing for the rapid evolution of male response to female pheromone blends (Fig. 3). Although such studies have not yet been conducted on moths, we predict that high levels of olfactory receptor gene duplication, gene loss, and pseudogenization will be found for these species on the basis of what has been found in other animals. For instance, a family of at least 60 olfactory receptor loci have been identified in D. melanogaster (49), and in vertebrates and Caenorhabditis elegans hundreds to thousands of olfactory receptor loci, including many pseudogenes, have been found (50, 51).
Fig. 3.
Proposed birth-and-death mechanism for speciation in O. furnacalis and nubilalis. Here, gene duplication, loss, or nonfunctionalization leads to a shift in pheromone blend (female stimulus), which, if it leads to speciation, is accompanied by a shift in the detection system (male response), which is also brought about through gene duplication, loss, or nonfunctionalization.
Future Directions: Genomics and Beyond
One particularly interesting avenue for future research involves studying how desaturase genes diversify once they duplicate. One possibility is that the genes originated under a subfunctionalization model (52–55) of gene family diversification (56). Under the subfunctionalization model, a generalized multifunctional gene/protein duplicate gives rise to one or more paralogs. The paralogous genes subsequently diverge and specialize in different functions that were previously all carried out by the ancestral, generalized multifunctional gene/protein. The problem is that direct evidence confirming multifunctionality of the ancestral/progenitor moth desaturase gene is lacking. An alternative to the subfunctionalization model is one in which sex-pheromone genes acquired their functions through rapid evolution subsequent to gene duplication via positive Darwinian selection (e.g., ref. 57). The ancient origin of insect desaturase genes and their increased substitution rates (15), which are highly pronounced in some cases (e.g., the Δ14 and Δ10,11 group genes in Fig. 2), are highly suggestive that nucleotide substitutions have become saturated. This obviously confounds attempts to detect positive Darwinian selection. For example, the estimate of synonymous substitution (57) between the O. furnacalis Z9 (16 > 18) and Z/E14 genes is 1.44 ± 0.44 and is 2.96 ± 1.20 between the Z/E11 and Z/E14 genes. Clearly, these values are well above the saturation level (58, 59). On the basis of our current state of knowledge, it is not possible to determine which of the above models best fits the insect desaturases. Further studies aimed at determining whether the desaturases of basal insects (e.g., Collembola) are multifunctional may help to answer this question.
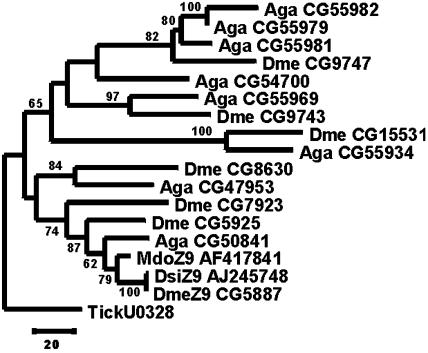
Another promising area of research is on the genomics of desaturase genes. Such studies are important for understanding the underlying basis of insect reproduction in terms of behavior as well as biochemistry. Consequently, until we obtain information on the number of desaturase genes and their genomic organization in various insect lineages, our understanding of the core elements of insect reproduction will be incomplete. Furthermore, such studies will provide insights into the evolution of insect desaturases. For example, now that their complete genome sequences are available, we know that the D. melanogaster and Anopheles gambiae genomes possess seven and eight desaturase genes, respectively (Figs. 4 and 5). By combining information from phylogeny (Fig. 4) and genomic organization (Fig. 5), we can make inferences regarding several aspects of desaturase evolution, genomics, and even gene function in dipterans. For instance, CG50841 of A. gambiae clusters significantly with the D. melanogaster genes CG5925 (desat2) and CG5887 (desat1). Because both of these genes are Z9 desaturases (Fig. 2), we predict that the A. gambiae CG50841 is also a Z9 desaturase.
Fig. 4.
Phylogeny of the desaturase genes of flies. The tree was reconstructed as in Fig. 1. Species and gene abbreviations are as per Fig. 2. The tree is rooted with the desaturase gene from the tick (A. americanum), and the log likelihood was –9,899.33096. Note that only partial sequences for the A. gambiae genes were available at the time this study was completed.
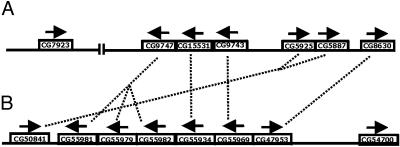
Fig. 5.
Genomic map of desaturase genes in D. melanogaster chromosome 3 (A) and A. gambiae chromosome 2 (B). The vertical lines bisecting the D. melanogaster chromosome represent the division between the left and right arms of chromosome 3. Arrows above genes represent the direction of transcription. Dotted lines connect orthologous counterparts between species, on the basis of clustering patterns depicted in Fig. 4.
The above approach can also be used to deduce the history of desaturase gene duplication and genomic organization. For example, the dotted lines connecting genes in Fig. 5 represent inferred orthologous relationships. Thus, D. melanogaster CG9747 and the ancestor of A. gambiae genes CG55981, CG55979, and CG55982 share an orthologous relationship. One piece of evidence for this comes from our phylogenetic analysis (Fig. 4), in which these genes cluster together with relatively high statistical support. Another piece of evidence comes from the conserved genomic organization of the fruit fly and mosquito desaturase genes (Fig. 5). In this case, the genomic location and direction of transcription of inferred orthologous gene pairs is conserved relative to other gene pairs. By using this line of reasoning in light of our results, we can also infer that there were four rounds of gene duplication that took place before the divergence of mosquitoes and flies. These events gave rise to (i) D. melanogaster CG8630 and A. gambiae CG47953, (ii)the ancestor of D. melanogaster CG5925/CG5887 and A. gambiae CG50841, (iii) D. melanogaster CG9743 and A. gambiae CG55969, (iv) D. melanogaster CG15531 and A. gambiae CG55934, and (v) D. melanogaster CG9747 and the ancestor of A. gambiae CG55981/CG55979/CG55982. Once the A. gambiae and D. melanogaster lineages diverged, subsequent gene duplications and, in at least one instance, gene loss can account for an inability to identify an orthologous counterpart of some genes. For instance, D. melanogaster CG7923 (fad2) seems to have duplicated from the ancestral gene that gave rise to CG5925 and CG5887 subsequent to the divergence from A. gambiae, thus accounting for the lack of an orthologous counterpart in that species. On the other hand, it seems that A. gambiae CG54700 has been lost from the D. melanogaster lineage, because the orthologous relationships of all other genes can be determined except for this one, and its phylogenetic position is uncertain as well, because of lack of statistical support (Fig. 4). Incidentally, these results show that dipteran desaturase genes undergo birth-and-death evolution, as discussed in the previous section.
The above discussion illustrates the power of combining phylogenetic and genomic analyses. This approach has enhanced our ability to reconstruct the evolutionary history of desaturase evolution in flies and allows us to make inferences that could not have been made by using either approach alone. In the same way, knowledge of the genomic organization of desaturase genes from moths would improve our understanding of this multigene family in moths and expand our opportunities to conduct comparative genomic studies across even deeper evolutionary divides. In that context, it will be interesting to see what sorts of evolutionary patterns characterize the desaturase genes of other insect species in which the reproductive biology of pheromones is different but no less complicated. Among these are the cockroaches, in which each species makes a single pheromone so chemically complex that it is not duplicated by other species (60). This stands in contrast to moths, which make blends of several different pheromones that are relatively simple chemical structures. Whether or not similar modes of evolution characterize the genes controlling these disparate, yet related, systems remains to be seen, but we anticipate that the evolutionary complexity of whatever model(s) are uncovered will reflect this functional and chemical disparity.
Acknowledgments
We thank C. Linn for contributions to the research and manuscript, W. Liu for conducting the molecular studies on desaturases, R. Harrison and T. J. Ward for providing helpful comments/discussion, and A. L. Hughes and J. Zhang for reviews of the manuscript.
Abbreviations: ACB, Asian corn borer; ECB, European corn borer; Z14–16:Acid, (Z)-14-hexadecenoic acid (fatty acids and acetates of other configurations, chain lengths, and position of unsaturation are named similarly).
References
- 1.Roelofs, W. L. & Bjostad, L. B. (1984) Bioorg. Chem. 12, 279–298. [Google Scholar]
- 2.Bjostad, L. B. & Roelofs, W. L. (1981) J. Biol. Chem. 256, 7936–7940. [PubMed] [Google Scholar]
- 3.Bjostad, L. B. & Roelofs, W. L. (1983) Science 220, 1387–1389. [DOI] [PubMed] [Google Scholar]
- 4.Foster, S. P. & Roelofs, W. L. (1996) Arch. Insect Biochem. Physiol. 32, 135–147. [Google Scholar]
- 5.Dugdale, J. S. (1997) in Insect Pheromone Research, eds. Cardé, R. T. & Minks, A. K. (Chapman & Hall, New York), pp. 463–472.
- 6.Thiede, M. A., Ozols., J. & Strittmatter, P. (1986) J. Biol. Chem. 261, 13230–13235. [PubMed] [Google Scholar]
- 7.Stukey, J. E., McDonough, V. M. & Martin, C. E. (1990) J. Biol. Chem. 265, 20144–20149. [PubMed] [Google Scholar]
- 8.Knipple, D. C., Miller, S. J., Rosenfield, C.-L., Liu, W., Tang, J., Ma, P. W. K. & Roelofs, W. L. (1998) Proc. Natl. Acad. Sci. USA 95, 15287–15292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu, W., Ma, P. W. K., Marsella-Herrick, P., Rosenfield, C.-L., Knipple, D. C. & Roelofs, W. L. (1999) Insect Biochem. Mol. Biol. 29, 435–443. [DOI] [PubMed] [Google Scholar]
- 10.Rosenfield, C.-L., You, K.-M., Marsella-Herrick, P., Roelofs, W. L. & Knipple, D. C. (2001) Insect Biochem. Mol. Biol. 31, 949–964. [DOI] [PubMed] [Google Scholar]
- 11.Liu, W., Jiao, H., Murray, N., O'Connor, M. & Roelofs, W. L. (2002) Proc. Natl. Acad. Sci. USA 99, 620–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hao, G., Liu, W., O'Connor, M. & Roelofs, W. L. (2002) Insect Biochem. Mol. Biol. 32, 961–966. [DOI] [PubMed] [Google Scholar]
- 13.Liu, W., Jiao, H., O'Connor, M. & Roelofs, W. L. (2002) Insect Biochem. Mol. Biol. 32, 1489–1495. [DOI] [PubMed] [Google Scholar]
- 14.Hao, G., O'Connor, M., Liu, W. & Roelofs, W. L. (2002) J. Insect Sci. 2, www.insectscience.org/2.26/. [DOI] [PMC free article] [PubMed]
- 15.Roelofs, W., Liu, W., Hao, G., Jiao, H., Rooney, A. P. & Linn, C. E., Jr. (2002) Proc. Natl. Acad. Sci. USA 99, 13621–13626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phelan, P. L. (1997) in Insect Pheromone Research, eds. Cardé, R. T. & Minks, A. K. (Chapman & Hall, New York), pp. 563–579.
- 17.Phelan, P. L. (1992) in Insect Chemical Ecology, eds. Roitberg, B. D. & Isman, M. B. (Chapman & Hall, New York), pp. 265–314.
- 18.Löfstedt, C. (1993) Philos. Trans. R. Soc. London B 240, 167–177. [Google Scholar]
- 19.Ma, P. W. K. & Roelofs, W. L. (2002) Zool. Sci. 19, 501–511. [DOI] [PubMed] [Google Scholar]
- 20.Zhao, C.-H., Löfstedt, C. & Wang, X. (1990) Arch. Insect Biochem. Physiol. 15, 57–65. [Google Scholar]
- 21.Klun, J. A., Bierl-Leonhardt, B. A., Schwarz, M., Litsinger, L. A., Barrion, A. T., Chiang, H. C. & Zhungxie, J. (1980) Life Sci. 27, 1603–1606. [DOI] [PubMed] [Google Scholar]
- 22.Ishikawa, Y., Takanashi, T., Kim, C., Hoshizaki, S., Tatsuki, S. & Huang, Y. (1999) Entomol. Exp. Appl. 91, 237–244. [Google Scholar]
- 23.Willett, C. S. & Harrison, R. G. (1999) Insect Biochem. Mol. Biol. 29, 277–284. [DOI] [PubMed] [Google Scholar]
- 24.Zhao, C.-H., Fang, L., Bengtsson, M. & Löfstedt, C. (1995) J. Chem. Ecol. 21, 1795–1810. [DOI] [PubMed] [Google Scholar]
- 25.Linn, C. E., Jr., Young, M. S., Gendle, M., Glover, T. J. & Roelofs, W. L. (1997) Physiol. Entomol. 22, 212–223. [Google Scholar]
- 26.Butlin, R. K. & Trickett, A. J. (1997) in Insect Pheromone Research, eds. Cardé, R. T. & Minks, A. K. (Chapman & Hall, New York), pp. 548–562.
- 27.Gaunt, M. W. & Miles, M. A. (2002) Mol. Biol. Evol. 19, 748–761. [DOI] [PubMed] [Google Scholar]
- 28.Jones, D. T., Taylor, W. R. & Thornton, J. M. (1992) Comput. Appl. Biosci. 8, 275–282. [DOI] [PubMed] [Google Scholar]
- 29.Brower, A. V. (1994) Proc. Natl. Acad. Sci. USA 95, 6491–6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nei, M. (1987) Molecular Evolutionary Genetics (Columbia Univ. Press, New York).
- 31.Kilian, M., Poulsen, K. & Lomholt, H. (2002) Mol. Microbiol. 46, 1367–1380. [DOI] [PubMed] [Google Scholar]
- 32.Kondo, H., Shimomura, I., Kishida, K., Kuriyama, H., Makino, Y., Nishizawa, H., Matsuda, M., Maeda, N., Nagaretani, H., Kihara, S., et al. (2002) Eur. J. Biochem. 269, 1814–1826. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, L., Lan, G. E., Parimoo, S., Stenn, K. & Prouty, S. M. (1999) Biochem. J. 340, 255–264. [PMC free article] [PubMed] [Google Scholar]
- 34.Kimura, M., Tokai, T., Matsumoto, G., Fujimura, M., Hamamoto, H., Yoneyama, K., Shibata, T. & Yamaguchi, I. (2003) Genetics, in press. [DOI] [PMC free article] [PubMed]
- 35.Lanteri, M., Giordanengo, V., Vidal, F., Gaudray, P. & Lefebvre, J. C. (2002) Glycobiology 12, 785–792. [DOI] [PubMed] [Google Scholar]
- 36.Moller, L. B., Petersen, C., Lund, C. & Horn, N. (2000) Gene 257, 13–22. [DOI] [PubMed] [Google Scholar]
- 37.Triglia, T., Thompson, J. K. & Cowman, A. F. (2001) Mol. Biochem. Parasitol. 116, 55–63. [DOI] [PubMed] [Google Scholar]
- 38.Wolffe, A. P. & Matzke, M. A. (1999) Science 286, 481–486. [DOI] [PubMed] [Google Scholar]
- 39.Parisi, M. & Lin, H. (2000) Curr. Biol. 10, R81–R83. [DOI] [PubMed] [Google Scholar]
- 40.Lipschitz, H. D. & Smibert, C. A. (2000) Genet. Dev. 10, 476–488. [DOI] [PubMed] [Google Scholar]
- 41.Dallerac, R., Labeur, C., Jallon, J. M., Knipple, D. C., Roelofs, W. L. & Wicker-Thomas, C. (2000) Proc. Natl. Acad. Sci. USA 97, 9449–9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fang, S., Takahashi, A. & Wu, C. I. (2002) Genetics 162, 781–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hughes, A. L. & Nei, M. (1989) Mol. Biol. Evol. 6, 559–579. [DOI] [PubMed] [Google Scholar]
- 44.Nei, M. & Hughes, A. L. (1992) in Proceedings of the 11th Histocompatibility Workshop and Conference, eds. Tsuji, K., Aizawa, M. & Sasazuki, T. (Oxford Univ. Press, Oxford), pp. 27–38.
- 45.Nei, M., Gu, X. & Sitnikova, T. (1997) Proc. Natl. Acad. Sci. USA 94, 7799–7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rooney, A. P., Piontkivska, H. & Nei, M. (2002) Mol. Biol. Evol. 19, 68–75. [DOI] [PubMed] [Google Scholar]
- 47.Laue, M., Maida, R. & Redkozubov, A. (1997) Cell Tissue Res. 288, 149–158. [DOI] [PubMed] [Google Scholar]
- 48.Krieger, J., Raming, K., Dewer, Y. M., Bette, S., Conzelmann, S. & Breer, H. (2002) Eur. J. Neurosci. 16, 619–628. [DOI] [PubMed] [Google Scholar]
- 49.Vosshall, L. B. (2001) Chem. Senses 26, 207–213. [DOI] [PubMed] [Google Scholar]
- 50.Glusman, G., Yanai, I., Rubin, I. & Lancet, D. (2001) Genome Res. 11, 685–702. [DOI] [PubMed] [Google Scholar]
- 51.Kratz, E., Dugas, J. C. & Ngai, J. (2002) Trends Genet. 18, 29–34. [DOI] [PubMed] [Google Scholar]
- 52.Hughes, A. L. (1994) Proc. R. Soc. London Ser. B 256, 119–124. [Google Scholar]
- 53.Hughes, A. L. (1999) Adaptive Evolution of Genes and Genomes (Oxford Univ. Press, Oxford).
- 54.Jensen, R. A. (1976) Annu. Rev. Microbiol. 30, 409–425. [DOI] [PubMed] [Google Scholar]
- 55.Lynch, M. & Force, A. (2000) Genetics 154, 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Knipple, D. C., Rosenfield, C. L., Nielsen, R., You, K. M. & Jeong, S. E. (2002) Genetics 162, 1737–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, J., Rosenberg, H. F. &. Nei, M. (1998) Proc. Natl. Acad. Sci. USA 95, 3708–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nei, M. & Kumar, S. (2000) Molecular Evolution and Phylogenetics (Oxford Univ. Press, Oxford).
- 59.Tanaka, T. & Nei, M. (1989) Mol. Biol. Evol. 6, 447–459. [DOI] [PubMed] [Google Scholar]
- 60.Roelofs, W. L. (1995) Proc. Natl. Acad. Sci. USA 92, 44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]