Abstract
Studies of mouse models of tuberculosis (TB) infection have indicated a central role for MHC class I-restricted CD8+ T cells in protective immunity. To define antigens and epitopes of Mycobacterium tuberculosis (MTB) proteins that are presented by infected cells to CD8+ T cells, we screened 40 MTB proteins for HLA class I A*0201-binding motifs. Peptides that bound with high affinity to purified HLA molecules were subsequently analyzed for recognition by CD8+ cytotoxic T lymphocytes. We identified three epitopes recognized by CD8+ T cells from patients recovering from TB infection. Those three epitopes were derived from three different antigens: thymidylate synthase (ThyA30–38), RNA polymerase β-subunit (RpoB127–135), and a putative phosphate transport system permease protein A-1 (PstA175–83). In addition, CD8+ T cell lines specific for three peptides (ThyA30–38, PstA175–83, and 85B15–23) were generated from peripheral blood mononuclear cells of normal HLA-A*0201 donors. These CD8+ T cell lines specifically recognized MTB-infected macrophages, as demonstrated by production of IFN-γ and lysis of the infected target cells. Finally, CD8+ cytotoxic T lymphocytes reduced the viability of the intracellular MTB, providing evidence that CD8+ T cell recognition of MHC class I-restricted epitopes of these MTB antigens can contribute to effective immunity against the pathogen.
Because tuberculosis (TB) causes three million deaths annually, development of a vaccine to control and eradicate the infection is an important unmet medical need (1). Analysis of the mechanisms of protective immunity in mouse models has indicated that both MHC class I- and class II-restricted T cells contribute to immunity against TB. MHC class II-restricted CD4+ T cells release lymphokines such as IFN-γ and tumor necrosis factor-α that result in macrophage activation. More importantly, β2-microglobulin (β2m)-deficient mice are unable to develop MHC class I-restricted cytotoxic T lymphocytes (CTL) and rapidly succumb to Mycobacterium tuberculosis (MTB) infection (2). This pathway of protective immunity appears to involve CD8+ T cells (2–4) and transporters associated with antigen processing (TAP)-dependent presentation of peptide antigen (5). Furthermore, CD8+ T cells not only are able to lyse MTB infected cells but also can simultaneously kill intracellular bacteria by the release of the antimicrobial peptide granulysin (6, 7).
Several laboratories have begun to identify MTB peptides that are presented by human MHC class I molecules to T cells as well as the role these T cells play in containing infection. For instance, peptides derived from 19-kDa protein (membrane-bound lipoprotein) and ESAT 6 protein (early secretory protein) have been identified as immunogenic for class I-restricted CD8+ CTL (8, 9). MHC class I-restricted CTL specific for MTB proteins can be generated by an alternative class I antigen-processing pathway (10). Additional pathways of peptide presentation may be involved (11).
Here, we have defined several new T cell epitopes derived from distinct protein antigens that are presented by the prevalent MHC class I molecule, HLA-A*0201. Furthermore, epitope-specific CD8+ T cells recognize MTB-infected targets, can secrete IFN-γ, lyse the target cells, and kill the intracellular bacteria. Therefore, MHC class I-restricted CD8+ T cell responses to MTB antigens can contribute to effective immunity against the pathogen.
Materials and Methods
Study Subjects.
In this study, the reactivity of peripheral blood mononuclear cells (PBMC) from 11 HLA-A*0201 individuals was investigated. Six of these subjects had culture-proven TB and were seen at the Los Angeles County–University of Southern California Medical Center. Five subjects were healthy normal blood donors, one was bacillus Calmette–Guérin vaccinated, and the others were not vaccinated. PBMC from each individual were isolated by Ficoll/Hypaque density gradient separation (Pharmacia). To define HLA type, flow cytometry analysis were performed by using anti-HLA-A2 monoclonal antibody (MA2.1) followed by FITC-labeled goat anti-mouse F(ab′)2 fragments. The HLA subtype, A*0201, was confirmed by the PCR amplification technique specifically designed for the HLA-A2 subtyping (Dynal, Great Neck, NY).
Cell Lines.
.221A2 (provided by Robert DeMars, University of Wisconsin–Madison) is an Epstein–Barr virus-transfected B-cell line mutagenized and selected for loss of HLA antigens, then transfected with HLA-A*0201. K562 cells [American Type Culture Collection (ATCC)], a human erythroleukemia line, were added to all CTL assays as a cold target cell line to inhibit natural killer cell activity. THP-1 cells, premyelomonocytic cell line (ATCC), were used as targets for the MTB infection experiments.
Peptide Synthesis.
Peptides were either synthesized as previously described (12) or, for large epitope libraries, were purchased as crude material from Mimotopes (Chiron). Peptides were purified to >95% homogeneity by reverse-phase HPLC and the purity determined by using an analytical reverse-phase column. The sequence was confirmed by amino acid analysis and/or mass spectrometry analysis.
HLA-A*0201-Binding Assays.
Purified human class I molecules (5–500 nM) were incubated with 1–10 nM 125I-radiolabeled probe peptide and iodinated by the Chloramine T method (13) for 48 h at room temperature in the presence of 1 μM human β2m (Scripps Laboratories, San Diego, CA) and a mixture of protease inhibitors. The final concentrations of protease inhibitors were: 1 mM PMSF/1.3 nM 1.10 phenanthroline/73 μM pepstatin A/8 mM EDTA/200 μM N-α-tosyl-lysine chloromethyl ketone.
Class I peptide complexes were separated from free peptide by gel filtration on TSK200 columns (TosoHaas, Montgomeryville, PA) and the fraction of bound peptide calculated as previously described (14). In preliminary experiments, the HLA class I prep was titered in the presence of fixed amounts of radiolabeled peptides to determine the concentration of class I molecules necessary to bind 10–20% of the total radioactivity. All subsequent inhibition and direct binding assays were then performed by using these class I concentrations. In the inhibition assays, peptide inhibitors were typically tested at concentrations ranging from 120 μg/ml to 1.2 ng/ml. The data were then plotted, and the dose yielding 50% inhibition was measured. Peptides were tested in two to four completely independent experiments. Because under these conditions [label]<[MHC] and Kd<[MHC], the measured IC50s are reasonable approximations of the true Kd values. As a radiolabeled control probe peptide, A F6→Y analog of the HBV core 18–27 peptide (sequence FLPSDYFPSV) (12) was used. The average IC50s of F6→Y analog of the HBV core 18–27 peptide for the A*0201 assay were 5.0 nM.
Peptide Sequences.
Peptide sequences and their corresponding affinities are shown in supplementary tables (see www.pnas.org). Sequences of peptides that were used for the generation of CTL lines are as follows: ThyA30–38 is RLPLVLPAV (IC50 = 5.1 nM), derived from thymidylate synthase (GenBank accession no. X59273), RNA polymerase β-subunit protein (RpoB127–135) is MTYAAPLFV (IC50 = 13.8 nM), derived from RNA polymerase β-subunit (GenBank accession no. L27989), 85B15–23 is LMIGTAAAV (IC50 = 79.0 nM), derived from antigen 85B (GenBank accession no. P31952), and phosphate transport system permease protein A-1 (PstA1); PstA175–83 is SLYFGGICV (IC50 = 10.6 nM), derived from putative phosphate transport system permease protein A-1 (GenBank accession no. X75297). In this study, the translational start site for the PstA1 gene was tentatively defined as the 312-bp upstream region, because the translational start site in this upstream region contained high-affinity binders for the HLA-A*0201 molecules, and this upstream region cannot be excluded from the following reading frame (15).
In Vitro Induction of Recall CTL Responses from TB Patients.
PBMC were isolated from the blood of patients by density gradient centrifugation and resuspended in RPMI 1640 medium (Life Technologies, Grand Island, NY) supplemented with 2 mM l-glutamine/1 mM sodium pyruvate/100 units/ml penicillin/100 μg/ml streptomycin plus 10% pooled human serum and plated in a 24-well plate at 3 × 106 cells/well. Synthetic peptides were added to the PBMC cultures at a final concentration of 10 μg/ml. On days 3 and 6, recombinant IL-2 (rIL-2; Chiron) was added to each well at a concentration of 10 units/ml. On day 7, the cultures were restimulated with irradiated (3,500 rad) autologous monocytes that were pulsed with peptide in the presence of 3 μg/ml of β2m for 2 h. On days 10 and 12, 10 units/ml of rIL-2 was added into each well. The cytolytic activity of cultured PBMC was tested on day 14.
CTL Assay.
Cytolytic activity was measured by a standard 4-h 51Cr release assay. Approximately 2 × 106 target cells (.221A2 cells or macrophages) were prepulsed with peptide (10 μg/ml) and β2m (3 μg/ml) overnight. These cells were then labeled with 100 μCi Na51CrO4 for 1 h at 37°C. After washing, target cells were incubated with different ratios of effector cells in a 200-μl total reaction volume of RPMI supplemented with 10% FBS (Life Technologies) in a microtiter plate. To inhibit NK-cell activity, a 20-fold excess of K562 cells was added into each well. After 4-h incubation, 100 μl of supernatant was removed from each well, and its radioactivity was counted in a γ counter. Specific lysis (%) was calculated by using the formula 100 × (experimental release–spontaneous release)/(maximal release–spontaneous release). Maximal release was determined from supernatants of target cells lysed by the addition of 1% Triton X-100. Spontaneous release was determined from supernatants of target cells incubated with media only.
Generation of CTL Lines from Normal HLA-A*0201 Donors by in Vitro Immunization.
Primary in vitro CTL induction experiments were performed according to two different methods. The first method was described by Tsai et al. (16) and generated antigen-specific CTL in 50% of attempts. Briefly, dendritic cells (DC) were generated by culturing adherent monocytes with 800 units/ml recombinant granulocyte/macrophage colony-stimulating factor (Genetics Institute, Cambridge, MA) and 1,000 units/ml rIL-4 (Schering–Plough) for 1 week. On day 7, DC were pulsed for 4 h with 40 μg/ml peptide and 3 μg/ml β2m. CD8+ T cells were enriched with immunomagnetic beads (Dynal, Great Neck, NY). DC were irradiated (4,200 rads) and cultured with CD8+ T cells at a ratio of 1:20 DC to CD8+ T cells in the presence of 10 ng/ml of rIL-7 (Genzyme). On the next day, human rIL-10 (10 ng/ml, DNAX) was added. Each individual culture was stimulated weekly with peptide-pulsed autologous monocytes, and rIL-10 and rIL-2 were added at days 1 and 2, respectively, during each restimulation cycle. After three to four rounds of restimulation, a sample from each culture was tested for cytotoxic activity with the cultures that displayed specific cytotoxicity expanded for further experiments.
The second method used for in vitro CTL induction followed a protocol by Plebanski et al. (17) and yielded greater numbers of T cells for analysis, as well as providing antigen-specific CTL in approximately 70% of attempts. Briefly, PBMC from HLA-A*0201 donors were pulsed with 50 μg/ml peptide at 3 × 107 cells/ml in Iscove's modified Dulbecco's medium (Life Technologies) at 37°C for 90 min. These cells were washed and plated at 3 × 106 cells/well in 10% pooled human serum with rIL-7 (10 ng/ml) and keyhole limpet hemocyanin (5 μg/ml, Sigma). Cultures were restimulated weekly with peptide-pulsed adherent-irradiated autologous and supplemented with rIL-2 at 10 units/ml every 3–4 days. After four to five cycles of restimulation, CD4+ T cells were depleted, and the remaining CD8+-enriched T cells (80–90%) were restimulated weekly as described above.
Cytotoxicity Assay for MTB-Infected Macrophages.
Macrophages were generated by culturing adherent monocytes in RPMI with 10% human AB serum (Omega Scientific, Tarzana, CA) for 2–4 days. These macrophages were then infected with MTB (Erdman strain) at a multiplicity of infection in the range of 5–10. After 18–20 h after infection, extracellular MTB were removed by differential centrifugation to remove nonphagocytosed bacteria, and the infected cells were cultured for 4 days before use as targets in CTL assays. Approximately 40–60% of the cells were infected according to the Kinyoun method for acid-fast bacteria (Difco). For cytotoxicity experiments, 1–2 × 106 macrophages were labeled with 100 μCi of 51Cr for 1 h at 37°C, then added to wells of 96-well U-bottom plates at 4 × 103 cells/well. CD8+ T cell lines were added at various effector/target ratios. Specific lysis (%) was determined as described above.
MTB Viability Count and IFN-γ Release Assay in MTB-Infected THP-1 Cells.
THP-1 cells were pretreated with phorbol 12-myristate 13-acetate (PMA) (10 ng/ml) (Calbiochem) for 24 h to induce differentiation and adherence. Monolayers of PMA-treated THP-1 cells were pulse infected with MTB at a multiplicity of infection of five bacilli per cell for 6 h and washed to remove extracellular bacteria. MTB peptide specific CD8+ T cells were coincubated with MTB-infected THP-1 cells or in the presence of MTB-derived peptides at CD8+ T cells to THP-1 ratio as 20:1 for 24 h. A blocking anti-HLA class I monoclonal antibody (W6/32) and an isotype-matched control antibody (IgG2a) were used to demonstrate the MHC class I-restricted recognition of infected cells. IFN-γ release by CD8+ T cells was measured by ELISA (Endogen, Cambridge, MA) after stimulation with peptide-pulsed or unpulsed THP-1 cells for 20–24 h. For assay of MTB viability, cells were lysed with saponin (0.3% final concentration) and 10-fold serial dilutions of the cell lysates were plated on 7H11 Middlebrook agar plates and colony-forming units (CFU) after 21 days of incubation were enumerated.
Results
Synthesis of Potential Immunodominant MTB Peptides for HLA-A*0201 Restricted CD8+ CTL.
In this study, we report the results of the analysis of approximately 40 available MTB protein sequences from the GenBank database. In 1995, when this study was initiated, we analyzed these MTB proteins for potential T cell epitopes according to HLA-A*0201 class I molecule-binding motifs by using a previously described computer algorithm (18). From this analysis, a total of 222 sequences (either 9 or 10 mers) were identified to contain the binding motifs for the HLA-A*0201 molecules. Peptides were synthesized corresponding to the candidate sequences, and binding affinities of each peptide to soluble HLA-A*0201 were measured as described previously (14). From these experiments, we determined that 27 peptides bound with high affinity (3.3 < IC50 ≤ 50 nM), and 58 peptides bound with intermediate affinity (50 < IC50 ≤ 500 nM) (refer to supplementary Tables 1 and 2, www.pnas.org). Previous studies have indicated these affinity levels are associated with antigenicity and immunogenicity for human CTL (14, 19). Accordingly, these 85 peptides were selected for the further studies directed at the identification of immunodominant antigens and peptides derived from MTB proteins.
Screening for Immunogenicity of MTB-Derived Peptides by Recall CTL Responses from TB Patient PBMC.
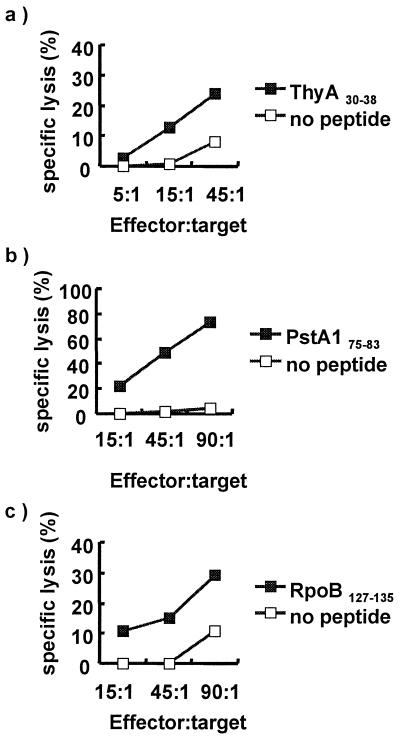
To test the immunodominance of these peptides during the natural course of infection, the existence of a recall CTL response specific for each peptide was evaluated by using PBMC from HLA-A*0201 TB patients. Memory CTL responses were recalled by stimulating whole PBMC from TB patients with each peptide for 1 week and by restimulating these cells with autologous feeder cells pulsed with the same peptide the following week. On day 14, the CTL activity was determined by 4-h chromium release assay. Previous studies have demonstrated that this assay strategy detects recall responses (20), whereas repeated stimulation by using DC induces primary CTL responses (16). Recall CTL responses were detected from PBMC cultures induced with three peptides derived from ThyA30–38, PstA175–83, and RpoB127–135 (Fig. 1). Recall CTL responses specific for ThyA30–38 and RpoB127–135 peptides were observed in one of two patients tested. PstA175–83-specific responses were observed in two of three patients tested. The demonstration of recall CTL responses suggests that these peptides are processed and presented to T cells during natural infection. Interestingly, these epitopes are derived from somatic proteins, demonstrating that somatic proteins derived from MTB replicating within intracellular compartments are processed and presented by the MHC class I presentation pathway.
Figure 1.
CTL responses from TB patients. PBMC from TB patients (HLA-A*0201) were stimulated with individual MTB peptides for 1 week and restimulated with peptide-pulsed autologous monocytes for an additional week. The cytolytic activity of these cultured PBMC was tested on day 14 by 51Cr release assay. (a) CTL activity specific for 9-mer peptide derived from thymidylate synthase (amino acids 30–38). (b) CTL activity specific for 9-mer peptide derived from RNA polymerase β-subunit (amino acids 127–135). (c) CTL activity specific for 9-mer peptide derived from PstA1 (amino acids 75–83).
Generation of CD8+ T Cell Lines Specific for MTB-Derived Peptides.
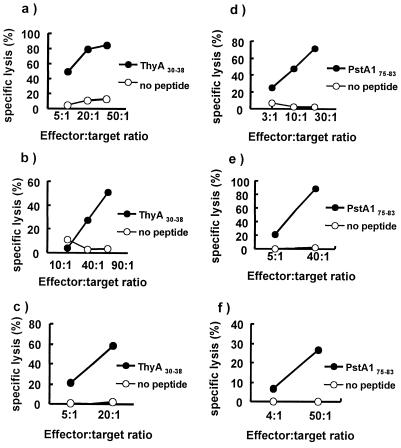
To verify that CD8+ T cell responses against these epitopes can also be induced in uninfected individuals, in vitro CD8+ CTL induction experiments were performed. PBMC from uninfected HLA-A*0201 individuals were stimulated weekly for 5–6 weeks with autologous PBMC pulsed with either ThyA30–38 or PstA175–83. After this protocol, CD8+ lines cytotoxic for either ThyA30–38 or PstA175–83 peptide were generated (Fig. 2). These CTL lines were specific, with greater cytotoxic activity against pulsed vs. unpulsed target cells. In addition, there was little cytotoxic activity against target cells pulsed with an irrelevant A*0201-binding epitope derived from influenza A virus matrix protein (NP58–66). Furthermore, this cytotoxic activity appeared to be class I restricted, because these peptide-specific responses were inhibited by the HLA class I-specific antibody, W6/32 (data not shown).
Figure 2.
Cytolytic activity of CTL lines generated from PBMC of uninfected subjects by in vitro immunization. PBMC from MTB-uninfected HLA-A*0201 donors were in vitro immunized to generate CTL lines specific for peptides, ThyA30–38 (a–c) and PstA175–83. (d–f). Cell lines (a) N1–091, (c) M7–091, and (f) M7–029 were generated by the method of Tsai et al. (16). Cell lines (b) M23–091, (d) M22–029, and (e) M23–029 were generated by the method of Plebanski et al. (17).
CD8+ T Cells Recognize MTB-Infected Macrophages: Release of IFN-γ.
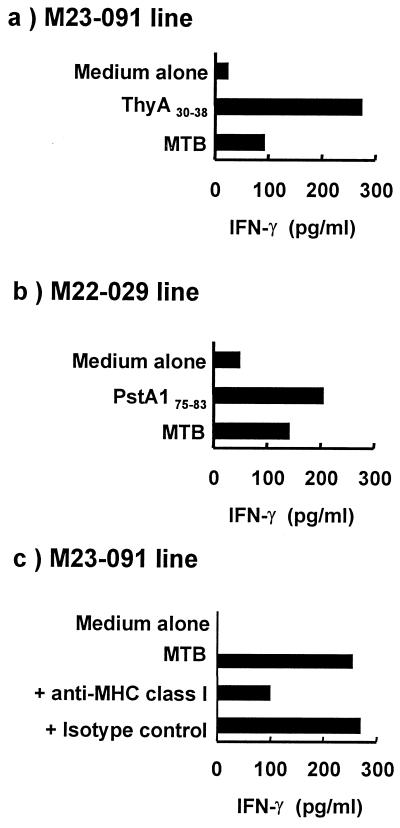
To demonstrate that these epitopes are processed and presented in infected cells, we infected THP-1 cells, a monocytoid cell line, with MTB, and measured recognition by specific CTL lines, as demonstrated by the release of IFN-γ. Two T cell lines, M23–091 and M22–029, specific for ThyA30–38 and PstA175–83, respectively, secreted IFN-γ specifically in response to MTB-infected cells (Fig. 3 a and b). The amount of IFN-γ induced by MTB-infected targets was similar to activation with the corresponding peptide for M22–029 but less in the case of M23–091. For one of the lines, we demonstrated that this response was inhibited by the HLA class I antibody, W6/32 (Fig. 3c). These data provide evidence that human MHC class I-restricted CD8+ T cells specific for ThyA30–38 and PstA175–83 antigens recognize MTB-infected target cells.
Figure 3.
IFN-γ release by MTB peptide-specific CTL after stimulation by MTB-infected and MTB peptide-pulsed cells. CD8+ T cell lines specific for (a and c) ThyA30–38 peptide (M23–091). (b) PstA175–83 peptide were coincubated with MTB-infected THP-1 cells at a ratio of 20:1. In some cultures, the peptide, either ThyA30–38 or PstA175–83, was added to the cultures. The supernatant was collected and assayed for IFN-γ. (c) The MHC restriction was evaluated by using the anti-HLA class I antibody (W6/32). The isotype-matched antibody (IgG2a) was used as a control.
Cytotoxicity of CTL Lines for MTB-Infected Macrophages.
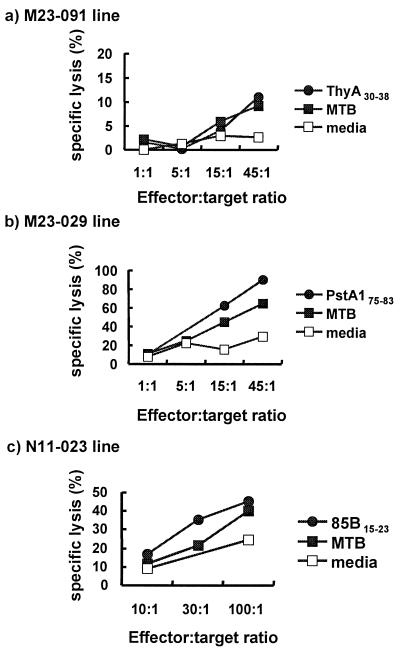
The ability of CTL to lyse infected targets is thought to facilitate control of the infection by releasing the bacilli so they can be taken up at low multiplicity by freshly activated macrophages and destroyed (21). To determine whether these CD8+ T cells can lyse infected target cells, heterologous HLA-A*0201 monocytes were infected with MTB and used as targets for CD8+ T cell lines (Fig. 4). In the case of the ThyA30–38-specific line, approximately 10% specific lysis was shown in MTB-infected target cells in the effector-to-target ratio of 45:1, and 12% specific lysis was observed in peptide-pulsed cells. In contrast, only weak lysis was observed in unpulsed cells. The T cell line (M23–029) specific for PstA175–83 exhibited greater specific lysis in both infected (65%) and peptide-pulsed (90%) macrophages. In addition, a CD8+ T cell line to peptide 15–23 of antigen 85B, one of the most abundant culture filtrate proteins, also recognized the MTB-infected target cells, with lysis of 40%, compared with the lysis of 24% in unpulsed target cells. It should be pointed out that approximately 40–60% of the targets were infected, such that the specific killing can reach only that level. In summary, these data indicate that CD8+ T cells can recognize and lyse MTB-infected target cells.
Figure 4.
Cytotoxicity of CD8+ T cell lines against MTB-infected macrophages. Macrophages from a heterologous HLA-A*0201 donor were infected with MTB at a multiplicity of infection of 5–10 bacilli per cell (a and b), or THP-1 were infected at a multiplicity of infection of 5–10 bacilli per cell (c). MTB-infected and corresponding peptide-pulsed THP-1 cells were tested for lysis by 51Cr release assay by incubation with (a) ThyA30–38 peptide-specific line (M23–091), (b) PstA175–83 peptide-specific line, and (c) 85B15–23 peptide-specific line (N11–023).
Activity of MTB Peptide-Specific CD8+ Lines on the Viability of Intracellular MTB.
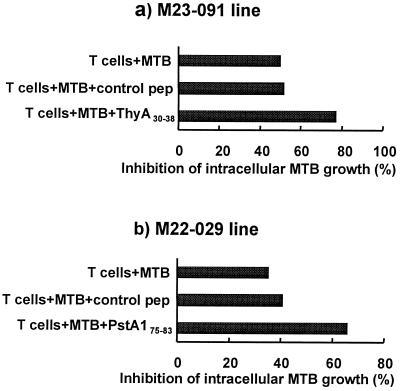
Our previous data indicated that CD8+, CD1-restricted CTL can lyse MTB-infected macrophages and reduce the viability of the intracellular MTB by the granule-dependent release of granulysin (7). We next determined whether the MTB-reactive MHC class I-restricted CD8+ CTL could also mediate an antimicrobial activity. Immunolabeling of the MHC class I-restricted CD8+ T cell lines (data not shown) illustrated the presence of cytolytic granules containing perforin and granulysin. After coculture of CD8+ CTL with the infected targets for 20 h, the viability of the intracellular MTB was measured by CFU. CD8+ CTL specific for ThyA30–38 (M23–091) directly reduced the viability of intracellular MTB by 52% as determined by CFU obtained from MTB-infected THP-1 cultured with or without CD8+ T cells. The addition of peptide ThyA30–38 to these cultures enhanced the inhibition of MTB growth (Fig. 5). Similarly, the CD8+ CTL specific for PstA175–83 peptide induced 34% growth inhibition of MTB, and the addition of peptide PstA175–83 to the culture enhanced growth inhibition. Taken together, these data demonstrate that MTB-reactive MHC class I-restricted CD8+ CTL mediate an antimicrobial activity against the tubercle bacillus.
Figure 5.
Inhibition of the growth of intracellular MTB by peptide-specific CD8+ T cells. MTB peptide-specific CD8+ T cell lines were coincubated with MTB-infected THP-1 cells in the presence or absence of MTB-derived peptides or a control peptide at an effector-to-target ratio of 20:1. Cells were lysed and CFU determined. The CFU of infected THP-1 without T cells at 24 h was 167,000 for the experiment in which M23–091 was tested and 66,400 for the experiment in which M22.029 was tested.
Discussion
TB presents a major public health problem and economic burden worldwide because of the morbidity and mortality associated with the disease (1). The incidence has been increasing because of the emergence of multidrug-resistant strains of MTB (22). This had led to intense efforts to develop a vaccine to prevent this deadly disease. Studies in mouse models have indicated that MHC class I-mediated CD8+ cytotoxic T cells have a critical role in protective immunity to TB. These CD8+ T cells can contribute to the immune response against MTB infection by at least three possible pathways: the release of IFN-γ, lysis of infected targets, and a direct antimicrobial activity. The present study provides evidence that human MHC class I-restricted CD8+ T cells recognize MTB-infected cells and can release IFN-γ, lyse infected target cells, and kill the intracellular bacteria.
There is compelling evidence to implicate CD8+ T cells as a critical mechanism in protective immunity against TB: (i) Adoptive transfer and cell depletion studies in vivo have demonstrated that CD8+ T cells are involved in controlling MTB infection (3, 23, 24); (ii) CD8+ T cell lines and clones can recognize MTB antigens in vitro, lyse MTB-infected macrophages in an antigen-specific manner, and restrict the growth of MTB in macrophages (25–29); and (iii) β2m-deficient mice as well as TAP-deficient mice that fail to express MHC class I molecules succumb to MTB infection (2, 5, 30). For these reasons, there has been an intense effort to define MTB epitopes that can be presented by MHC class I molecules to CD8+ T cells. In the present study, we screened 40 secreted and somatic MTB proteins to identify HLA-A*0201-binding peptides with the capability of recalling memory CD8+ T cell responses. Our studies revealed four new epitopes, three of which are derived from somatic proteins and one of which is derived from a secreted protein. CD8+ T cells that recognized these epitopes also recognized MTB-infected target cells, suggesting that the proteins containing these epitopes gain entry into the host cell cytoplasm, allowing processing and presentation via the class I pathway. Previous studies indicate a mechanism of transport, perhaps a pore, between the phagosome in which the bacteria reside and the host cell cytosol that provides access to the MHC class I presentation pathway (31, 32).
The MTB peptide epitope-reactive CD8+ T cell lines released IFN-γ on recognition of MTB-infected target cells, providing a mechanism by which this T cell subset might contribute to immunity against infection. Studies of mouse models have indicated an essential role for IFN-γ in a successful response to the pathogen. IFN-γ knock out (gko) mice on a genetically resistant C57BL/6 background rapidly succumbed to a fatal MTB infection with a median survival time of 17 days (33, 34), whereas 90% of littermate controls survived this infection more than 20 weeks. Increases in CFU of greater than 1 logarithm were seen in liver, spleen, and lung by 15 days in the gko mice. This established that IFN-γ is a necessary condition for protection against MTB challenge in the mouse. IFN-γ and tumor necrosis factor-α synergize for induction of inducible nitric oxide synthase and production of nitric oxide from murine macrophages, which we showed to be cytocidal to MTB (35).
A second mechanism by which MHC class I-restricted T cells can contribute to host defense against MTB infection is their ability to lyse infected target cells. In our study, the CD8+ T cells lysed MTB-infected targets of the monocyte/macrophage lineage. The continued lysis of infected cells could lead to the release of bacteria from this safe intracellular harbor so they can be taken up at a low multiplicity by freshly activated macrophages and destroyed (21).
Ultimately, the immune response must destroy and dispose of the pathogen to successfully defend the host against the microbial invader. CD8+ T cells can mediate an antimicrobial activity by the release of perforin and granulysin from their cytotoxic granules. The presence of both perforin and granulysin in these CD8+ T cells was demonstrated by histochemical staining. These CD8+ T cells simultaneously lysed the MTB-infected targets and reduced the number of viable bacteria by 30–50%. Although the magnitude of the reduction in CFU was not enormous, this level of reduction may have a biologically significant role as MTB infection in humans requires only 10–100 bacilli. These data provide new evidence that MTB-reactive MHC class I-restricted T cells recognize MTB-infected targets and kill the intracellular organisms.
In summary, we demonstrate that human MHC class I-restricted CD8+ T cells specific for MTB protein epitopes contribute to host defense by the release of IFN-γ, by lysing infected target cells, and by directly killing the intracellular bacteria. The detection of peptide-reactive CD8+ T cells in persons infected with MTB suggests that these epitopes are presented during the natural course of infection. These cells could also be induced in normal individuals with HLA-A*0201 subtype, suggesting they could be useful for immunoprophylaxis against TB infection. Interestingly, several of these epitopes were derived from somatic proteins of MTB, providing evidence that nonsecreted MTB antigens have access to the MHC class I processing and presentation pathway. Further analysis will determine whether incorporation of these epitopes into a vaccine provides protective immunity against this deadly pathogen.
Supplementary Material
Acknowledgments
We thank Lisa Butterfield and James Economou for their technical and scientific advice. We also thank Kevin Moore (DNAX) for IL-10. This work was supported in part by grants from the National Institutes of Health (AI 22553, AR 40312, AI 07118, AI 95362, and AI 27285) and the German Cancer Research Institute (D.K.F.Z.).
Abbreviations
- MTB
Mycobacterium tuberculosis
- TB
tuberculosis
- ThyA
thymidylate synthase
- PstA1
phosphate transport system permease protein A-1
- PBMC
peripheral blood mononuclear cells
- CTL
cytotoxic T lymphocytes
- CFU
colony-forming units
- β2m
β2-microglobulin
- rIL
recombinant IL
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.210391497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.210391497
References
- 1.Bloom B R, Murray C J L. Science. 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 2.Flynn J L, Goldstein M M, Triebold K J, Koller B, Bloom B R. Proc Natl Acad Sci USA. 1992;89:12013–12017. doi: 10.1073/pnas.89.24.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muller I, Cobbold S P, Waldmann H, Kaufmann S H E. Infect Immun. 1987;55:2037–2041. doi: 10.1128/iai.55.9.2037-2041.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serbina N V, Liu C C, Scanga C A, Flynn J L. J Immunol. 2000;165:353–363. doi: 10.4049/jimmunol.165.1.353. [DOI] [PubMed] [Google Scholar]
- 5.Behar S M, Dascher C C, Grusby M J, Wang C R, Brenner M B. J Exp Med. 1999;189:1973–1980. doi: 10.1084/jem.189.12.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stenger S, Mazzaccaro R J, Uyemura K, Cho S, Barnes P F, Rosat J P, Sette A, Brenner M B, Porcelli S A, Bloom B R, Modlin R L. Science. 1997;276:1684–1687. doi: 10.1126/science.276.5319.1684. [DOI] [PubMed] [Google Scholar]
- 7.Stenger S, Hanson D A, Teitlebaum R, Dewan P, Niazi K R, Froelich C J, Ganz T, Thoma-Uszynski S, Melian A, Bogdan C, et al. Science. 1998;282:121–125. doi: 10.1126/science.282.5386.121. [DOI] [PubMed] [Google Scholar]
- 8.Mohagheghpour N, Gammon D, Kawamura L M, van Vollenhoven A, Benike C J, Engleman E G. J Immunol. 1998;161:2400–2406. [PubMed] [Google Scholar]
- 9.Lalvani A, Brookes R, Wilkinson R J, Malin A S, Pathan A A, Andersen P, Dockrell H, Pasvol G, Hill A V. Proc Natl Acad Sci USA. 1998;95:270–275. doi: 10.1073/pnas.95.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canaday D H, Ziebold C, Noss E H, Chervenak K A, Harding C V, Boom W H. J Immunol. 1999;162:372–379. [PubMed] [Google Scholar]
- 11.Lewinsohn D M, Alderson M R, Briden A L, Riddell S R, Reed S G, Grabstein K H. J Exp Med. 1998;187:1633–1640. doi: 10.1084/jem.187.10.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruppert J, Sidney J, Celis E, Kubo R T, Grey H M, Sette A. Cell. 1993;74:929–937. doi: 10.1016/0092-8674(93)90472-3. [DOI] [PubMed] [Google Scholar]
- 13.Buus S, Sette A, Colon S M, Miles C, Grey H M. Science. 1987;235:1353–1358. doi: 10.1126/science.2435001. [DOI] [PubMed] [Google Scholar]
- 14.Sette A, Buus S, Colon S, Miles C, Grey H M. J Immunol. 1989;142:35–40. [PubMed] [Google Scholar]
- 15.Braibant M, Lefevre P, de Wit L, Ooms J, Peirs P, Huygen K, Wattiez R, Content J. FEBS Lett. 1996;394:206–212. doi: 10.1016/0014-5793(96)00953-2. [DOI] [PubMed] [Google Scholar]
- 16.Tsai V, Southwood S, Sidney J, Sakaguchi K, Kawakami Y, Appella E, Sette A, Celis E. J Immunol. 1997;158:1796–1802. [PubMed] [Google Scholar]
- 17.Plebanski M, Allsopp C E, Aidoo M, Reyburn H, Hill A V. Eur J Immunol. 1995;25:1783–1787. doi: 10.1002/eji.1830250645. [DOI] [PubMed] [Google Scholar]
- 18.Gulukota K, Sidney J, Sette A, DeLisi C. J Mol Biol. 1997;267:1258–1267. doi: 10.1006/jmbi.1997.0937. [DOI] [PubMed] [Google Scholar]
- 19.Sette A, Vitiello A, Reherman B, Fowler P, Nayersina R, Kast W M, Melief C J, Oseroff C, Yuan L, Ruppert J. J Immunol. 1994;153:5586–5592. [PubMed] [Google Scholar]
- 20.Bertoni R, Sidney J, Fowler P, Chesnut R W, Chisari F V, Sette A. J Clin Invest. 1997;100:503–513. doi: 10.1172/JCI119559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Libero G, Flesch I, Kaufmann S H. Eur J Immunol. 1988;18:59–66. doi: 10.1002/eji.1830180110. [DOI] [PubMed] [Google Scholar]
- 22.Bloom B R, McKinney J D. Nat Med. 1999;5:872–874. doi: 10.1038/11309. [DOI] [PubMed] [Google Scholar]
- 23.Orme I M, Collins F M. Cell Immunol. 1984;84:113–120. doi: 10.1016/0008-8749(84)90082-0. [DOI] [PubMed] [Google Scholar]
- 24.Orme I M. J Immunol. 1987;138:293–298. [PubMed] [Google Scholar]
- 25.DeLibero G, Flesch I, Kaufmann S H E. Eur J Immunol. 1988;18:59–66. doi: 10.1002/eji.1830180110. [DOI] [PubMed] [Google Scholar]
- 26.Zhu X, Venkataprasad N, Thangaraj H S, Hill M, Singh M, Ivanyi J, Vordermeier H M. J Immunol. 1997;158:5921–5926. [PubMed] [Google Scholar]
- 27.Turner J, Dockrell H M. Immunology. 1996;87:339–342. doi: 10.1046/j.1365-2567.1996.512590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan J S, Canaday D H, Boom W H, Balaji K N, Schwander S K, Rich E A. J Immunol. 1997;159:290–297. [PubMed] [Google Scholar]
- 29.Tascon R E, Colston M J, Ragno S, Stavropoulos E, Gregory D, Lowrie D B. Nat Med. 1996;2:888–892. doi: 10.1038/nm0896-888. [DOI] [PubMed] [Google Scholar]
- 30.Sousa A O, Mazzaccaro R J, Russell R G, Lee F K, Turner O C, Hong S, Van Kaer L, Bloom B R. Proc Natl Acad Sci USA. 2000;97:4204–4208. doi: 10.1073/pnas.97.8.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazzaccaro R J, Gedde M, Jensen E R, Van Santen H M, Ploegh H L, Rock K L, Bloom B R. Proc Natl Acad Sci USA. 1996;93:11786–11791. doi: 10.1073/pnas.93.21.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teitelbaum R, Cammer M, Maitland M L, Freitag N E, Condeelis J, Bloom B R. Proc Natl Acad Sci USA. 1999;96:15190–15195. doi: 10.1073/pnas.96.26.15190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooper A M, Dalton D K, Stewart T A, Griffin J P, Russell D G, Orme I M. J Exp Med. 1993;178:2243–2247. doi: 10.1084/jem.178.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flynn J L, Chan J, Triebold K J, Dalton D K, Stewart T A, Bloom B R. J Exp Med. 1993;178:2249–2254. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan J, Xing Y, Magliozzo R S, Bloom B R. J Exp Med. 1992;175:1111–1122. doi: 10.1084/jem.175.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.