Abstract
The kinetics of biomolecular isomerization processes, such as protein folding, is governed by a free-energy surface of high dimensionality and complexity. As an alternative to projections into one or two dimensions, the free-energy surface can be mapped into a weighted network where nodes and links are configurations and direct transitions among them, respectively. In this work, the free-energy basins and barriers of the alanine dipeptide are determined quantitatively using an algorithm to partition the network into clusters (i.e., states) according to the equilibrium transitions sampled by molecular dynamics. The network-based approach allows for the analysis of the thermodynamics and kinetics of biomolecule isomerization without reliance on arbitrarily chosen order parameters. Moreover, it is shown on low-dimensional models, which can be treated analytically, as well as for the alanine dipeptide, that the broad-tailed weight distribution observed in their networks originates from free-energy basins with mainly enthalpic character.
Energy landscape theory provides a framework for the description of the thermodynamics and kinetics of complex systems. Since the publication of the seminal ideas almost 40 years ago (1), the energy landscape paradigm has been successfully applied to the study of a broad range of systems (2, 3). The potential energy function of a multibody system such as a protein is a multidimensional and often very complex surface. At nonzero temperature, entropic contributions become relevant, and therefore the free-energy landscape governs the thermodynamics and kinetics. A common way to investigate and display the free energy involved in biomolecular isomerization and protein folding is to study it as a function of one or more order parameters, i.e., suitably chosen macroscopic quantities that distinguish the different states of the system (4). States are associated with local free-energy minima of the projected landscape. The depth of the minima is considered proportional to the stability of the states associated to them, and the barriers between different minima indicate activation energies between states. Due to the complexity of the process and the large number of degrees of freedom involved, order parameters are often arbitrarily chosen. Moreover, using free-energy projections for the study of the kinetics requires knowledge of a good reaction coordinate for the isomerization process, which is a difficult and unsolved problem (5–11). For this reason, new approaches based on graph theory have been explored for the analysis of free-energy landscapes. Recently, Krivov and Karplus introduced a method based on the disconnectivity graphs (DGs) for analyzing the unprojected free-energy surface of short peptides using an equilibrium molecular dynamics (MD) trajectory (12). They have developed the free-energy DG approach by exploiting an isomorphism between the total rate between two free-energy minima (considering all possible pathways) and the maximum flow through a network with capacitated edges, i.e., edges directly or indirectly connecting two nodes and having a certain flow capacity. The mincut and balanced mincut procedures have been used for the analysis of the configuration space of a tetrapeptide (12) and the folding of a simple hairpin of protein G (9, 13), respectively. At the same time, energy landscapes have been represented as complex networks (for a review about complex networks, see refs. 14–16). In ref. 17, the transitions of a short lattice polymer have been mapped onto a network. Doye has applied graph analysis for the study of the potential energy minima of a Lennard–Jones cluster of atoms (18). The free-energy landscape of a three-stranded β-sheet peptide sampled by MD simulations has been represented as a configuration space network (CSN) where configurations and direct transitions between them are the nodes and the links of the network, respectively (19). Recently, a similar approach has been applied to the investigation of the folding of a set of helical proteins (20). The network framework has been shown to be very effective for the visualization and representation of free-energy landscapes. However, the usefulness of the method for obtaining a quantitative description of the energy basins and barriers of the landscape has not been yet investigated.
The CSN shares a modular structure with most other networks representing systems as diverse as cell function (21), scientific collaborations (22), and the World Wide Web (23): some groups of nodes are more highly connected to each other than to the rest of the network. To unravel this intriguing property, several cluster-detection algorithms have been recently developed, each of them attempting to find a meaningful partition of the network (24–30). In CSNs characterizing high-dimensional energy landscapes with several basins, it is likely that nodes in the same energy basin are well connected among each other, whereas nodes in different basins are loosely connected. This argument suggests that finding the cluster structure of a CSN might reveal the topography of the underlying free-energy landscape. Detecting the cluster structure of a CSN thus opens a way to extract from the simulations the main features of the free-energy landscape at a more coarse-grained level, thus reducing the overall complexity of the problem (9, 10). Because of the complexity of CSNs, cluster detection is not straightforward (31). In other words, clusters defined using existing algorithms might not correspond to free-energy basins but simply to groups of nodes connected among each other more than average.
The present study was motivated by three main questions: Is network analysis useful for investigating pathways of molecular isomerization reactions? In particular, are cluster-detection algorithms able to obtain a quantitatively correct description of the free-energy basins and barriers? Can a simple analytical model of stochastic processes be formulated to explain the origin of broad-tailed weight distributions observed in CSNs? Our results indicate that the three questions can be answered affirmatively in the case of the alanine dipeptide. This system is of interest because it represents the minimal unit that still has the most relevant degrees of freedom of a polypeptide chain (6).
Results and Discussion
Free-Energy Basins.
Low-dimensional models.
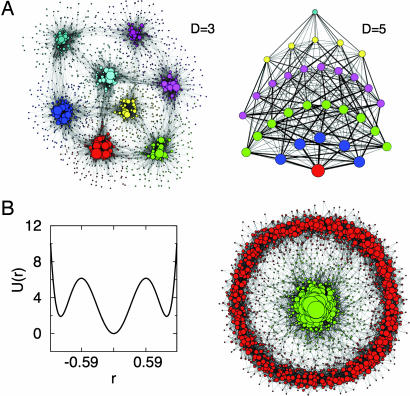
To illustrate the network approach, it is useful to start with an example where the surface is known a priori but its representation is not simple. The multidimensional double-well is defined by the energy function U(x) = 5 Σi=1D (xi4 − 2xi2 − εxi + 1), where ε = 0.05 gives an asymmetry between the minima. This landscape is characterized by 2D minima, where D is the dimensionality of the system. Given U(x), the system dynamics is simulated using a Monte Carlo protocol on a D-dimensional lattice. The CSN obtained with D = 3 and D = 5 is made of 1,752 and 2,815 nodes, respectively. The Markov clustering (MCL) algorithm (32, 33) with p = 1.2 finds the 8 (D = 3) and 32 (D = 5) expected clusters (Fig. 1A), where p is a parameter of MCL tuning the granularity of the clustering (see Methods and Models). Although these landscapes cannot be naturally embedded in a bidimensional space, the network representation illustrates the topography of the surface and its dynamic connectivity.
Fig. 1.
Energy landscape models. (A) Asymmetric double-well potential U(x) = 5 Σi=1D (xi4 − 2xi2 − 0.05xi + 1) CSN for D = 3 (Left) and network of clusters determined by MCL for D = 5 (Right). Line widths are proportional to the number of transitions. (B) One-dimensional plot of the Mexican-Hat landscape (Left) and its CSN for D = 2 dimensions (Right). Node size is proportional to the weight w. Colors are according to the cluster structure found by the MCL algorithm with granularity parameter p = 1.2 for the two CSNs, and they show clusters of the same size for the cluster network in D = 5. Node coordinates are automatically generated by the program visone, which minimizes the number of links intersections for the projection (http://visone.info).
In the previous example, energy basins are mainly enthalpic (because every basin is characterized by a pronounced bottom). Interestingly, a cluster analysis can detect the presence of entropic basins, i.e., regions in the free-energy surface without a single predominant attractor yet separated from the rest of the configurations of the system. An illustrative example is given by the Mexican-Hat landscape of Fig. 1B, which is defined in polar coordinates by the energy function U(r) = 40(r6 − 1.95r4 + r2). There is a pronounced minimum for r < 0.59 and a shallow entropic minimum along the solid angle Ω for r > 0.59. Two basins are correctly identified by the MCL algorithm [see supporting information (SI) Text for a quantitative analysis of the two basins].
Alanine dipeptide.
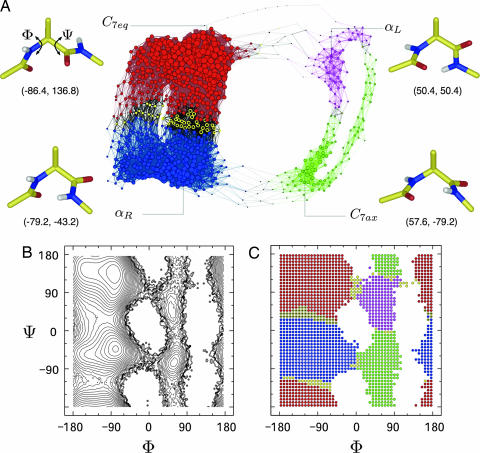
Analysis of the architecture of free-energy landscapes characterizing biomolecular isomerization sampled by equilibrium MD simulations is the object of multiple research efforts (13, 19, 34). The alanine dipeptide is a useful system for evaluating new methods for reaction coordinate identification (6, 7, 35). In the united atom representation the blocked alanine dipeptide is defined by 12 atoms (see Fig. 2A). The main degrees of freedom are the dihedral angles φ and ψ of its two rotatable bonds. In the continuum solvent approximation used here the projection of the free-energy landscape onto φ and ψ shows four minima (see Fig. 2B): C7eq, αR, C7ax, and αL.
Fig. 2.
Alanine dipeptide free-energy landscape. (A) Plot of the CSN. Node size and link width are proportional to node and link weight, respectively. Node coordinates are generated by the program visone (http://visone.info). Configurations lying at the bottom of the four basins are represented in cylinders with φ and ψ values in parentheses. (B) Free-energy projection on the dihedral angles φ and ψ. Isolines are drawn every 0.3 kcal/mol. (C) Ramachandran-map representation of the configurations (nodes) used for building the network. Colors are according to the cluster structure found by the MCL algorithm using p = 1.2. Yellow nodes represent unstable configurations identified by introducing noise in the MCL algorithm. For a better visualization of the main saddle between C7eq and αR basins, unstable nodes are shown in yellow with black in the middle of the circle.
Several discretization approaches can be used to define the nodes of a CSN from an MD simulation (see Methods and Models and SI Text). For the alanine dipeptide the most natural discretization consists of partitioning the (φ, ψ) space (Ramachandran-map) into cells of equal size (see Methods and Models) and labeling every snapshot visited during the simulation according to its (φ, ψ) value. Cells are the nodes of the network, and direct transitions between them observed during the simulation are the links. A 50 × 50 division of the Ramachandran-map gives a network of 1,832 visited nodes and 54,339 links. The CSN of the alanine dipeptide provides qualitative insight on the topography and dynamic connectivity of the landscape (Fig. 2A). The network shows four densely connected regions that correspond to the free-energy basins of the dipeptide. Moreover, multiple pathways between basins emerge from the picture. C7eq is connected to αR by two independent pathways characterized by different populations, where the statistically more (less) significant pathway corresponds to decreasing (increasing) values of ψ. There are also two independent pathways connecting C7ax and αL and two pathways between αL and C7eq, one of which (via increasing φ) was observed only once in the five 200-ns simulations. Notably, there is a striking similarity between the dynamic connectivity in the alanine-dipeptide CSN (Fig. 2A) and the optimal free-energy pathways reported in a previous work (see figure 3 of ref. 35). It is worth noting that the network contains the dynamic connectivity, whereas the projection of the free energy onto (φ, ψ) does not illustrate pathways (Fig. 2B).
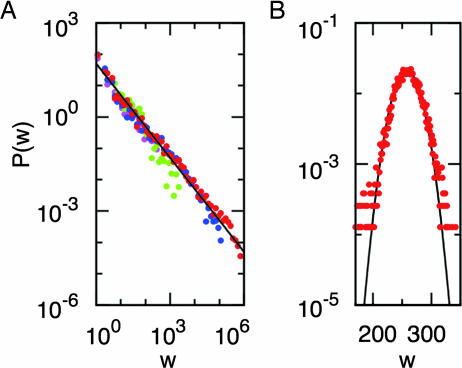
Fig. 3.
Distribution of node weights. (A) The four free-energy basins of the alanine dipeptide identified by MCL with p = 1.2. Each basin is represented by a different color, and the solid line is shown to visualize the 1/w behavior, which is typical of an enthalpic basin. Colors are the same as in Fig. 2. For clarity a logarithmic binning has been applied to the data. (B) Square well in D = 2 dimensions. Most of the nodes are characterized by the same weight, and P(w) follows a Gaussian (black line).
To obtain a quantitative description of the thermodynamics and kinetics of the system, the relation between the cluster structure of the network and the energy basins is investigated in more detail. The MCL algorithm, with a value of 1.2 for the granularity parameter p (32, 33), finds four clusters. Each of the C7eq, αR, C7ax, and αL minimum is grouped into separate clusters. In Fig. 2C, cells of the (φ, ψ) space are colored according to the clusters found by MCL. Interestingly, this cluster structure reflects very well the topography of the energy landscape, and cluster borders match the saddle points and isoline of the corresponding free-energy projection (see below for definition of yellow nodes). This result indicates that network clusters have the correct (φ, ψ) distributions of the free-energy basins of the dipeptide. Cluster structure is robust on the change of the number of cells used for the discretization of dihedral angles or when cells are directly defined as an array of raw interatomic distances (see SI Text). Provided that heterogeneous structures are not grouped to the same nodes, clusters defined by the MCL algorithm weakly depend on the discretization procedure. This result indicates that the network framework allows to identify the stable states of the dipeptide without a priori knowledge of the relevant coordinates of the system.
MCL results are encouraging and show that network clusterization can give a quantitative description of the free-energy basins of a complex system. However, correct partition of the network into free-energy basins is not obvious and might depend on the algorithm used for the clusterization. Neither the Potts Hamiltonian algorithm (26) nor a greedy optimization of the modularity cost-function Q (25) correctly detects the four free-energy basins (see SI Text). Interestingly, the latter yields a value of Q = 0.58, which is higher than the one obtained with MCL (Q = 0.15), indicating that an optimal value of modularity does not necessarily imply a good detection of free-energy basins (31).
The three clusterization approaches are very different in spirit with respect to each other and were not developed for any specific network target. A blind application of a clusterization algorithm may give unpredictable results, which suggests that a good knowledge of the characteristics of the system under study is fundamental for a correct interpretation of a network clusterization. MCL is based on the evolution of random walkers over the network. Hence, it mimics the natural kinetics thereby detecting “attractors.” In the MCL algorithm, varying the value of the parameter p changes the granularity of the clusterization. At p = 1, the network is considered as one single cluster by construction of the algorithm. Increasing the value of p results in the splitting of the C7eq and αR basins first (activation barrier ΔFαR→C7eq‡ ≈ 3.6 kcal/mol, Table 1) and then C7ax and αL(ΔF‡αL→C7ax ≈ 2.8 kcal/mol) as shown in SI Text. Note that ΔF‡ is estimated up to an additive constant (see below). Further increase of the value of p produces the detection of the marginally stable C5 basin [ΔF‡C5→C7eq ≈ 1.5 kcal/mol, C5 minimum has (φ, ψ) = (−140, 140)]. Hence, the value of p defines the minimum activation barrier height detected by the algorithm. This result indicates that MCL clusterization preserves activation energy barriers, which is a crucial feature for a correct assessment of the topography of the landscape. Note that p = 1.2 is the minimal value of the parameter allowing the detection of the four most relevant states. Although the Potts approach yields almost correct partitions into basins for γ = 0.001, its behavior for different values of the granularity parameter γ does not reflect the barrier heights of the landscape (see SI Text for details). Given the nature of the problem, MCL appears to be most suitable for the detection of free-energy basins in CSNs.
Table 1.
Relative free energies of the four basins and barriers as determined by the MCL (p = 1.2) clusterization of the alanine-dipeptide network
| Basin/barrier | |
|---|---|
| Basin* | ΔFi = Fi − FC7eq, kcal/mol |
| C7eq | 0.0 |
| αR | 1.5 |
| C7ax | 4.1 |
| αL | 5.0 |
| Direct transition† | ΔF‡, kcal/mol |
| C7eq → αR | 5.0 |
| C7eq → αL | 9.5 |
| αR → C7eq | 3.6 |
| αR → C7ax | 7.5 |
| C7ax → αR | 4.9 |
| C7ax → αL | 3.7 |
| αL → C7eq | 4.4 |
| αL → C7ax | 2.8 |
*The relative free energy of basin i is evaluated as ΔFi = −kBT log(Wi/WC7eq).
†Activation free energies are computed as the free energy of the barrier minus the free energy of the starting basin, e.g., ΔFαL→C7ax‡ = FαL→C7ax − FαL = −kBT log(A·wαL→C7ax/WαL). The free energy of the barrier is determined by counting the number of direct transitions from one basin to another, and A is set to unity (see text).
Hierarchical Organization of Free-Energy Basins.
A hierarchical organization is observed for nodes within free-energy basins. In fact, the minimum of the basin is defined as the most populated node of the corresponding cluster while nodes with small weights can be close to saddles especially in mainly enthalpic basins. The distribution P(w) of node weights inside the four basins of the alanine dipeptide detected by the MCL algorithm with p = 1.2 are shown in Fig. 3A. In Methods and Models, broad-tailed weight distributions were observed for enthalpic basins in low-dimensional energy surfaces. The alanine-dipeptide basins appear to follow a power-law w−δ with exponent δ ≈ 1, which is consistent with the behavior predicted for the bidimensional quadratic well. Hence, as a first approximation, the four free-energy basins of the alanine dipeptide might be reasonably considered as quadratic wells. On the other hand, in the case of a square well that resembles an entropic basin, all the nodes have similar weight, and P(w) follows a Gaussian curve peaked around the mean value of the node weight (see Fig. 3B and SI Text).
Low-weight nodes in mainly enthalpic basins are either saddles, as mentioned above, or “off-pathway” regions of unfavorable free energy. Unfortunately, it is not possible to directly identify saddles from the analysis of the P(w) because the weight of a node alone cannot define the presence of a saddle. For this reason, a stochastic algorithm is used in combination with MCL for the detection of unstable nodes, i.e., nodes that can be grouped to more than one basin (32). In Fig. 2 unstable nodes are colored in yellow. Especially for the well sampled transition C7eq ↔ αR unstable nodes characterize the saddle regions of the Ramachandran-map, showing that instabilities detection is able to determine interbasin transition regions (provided that free-energy basins are correctly identified) without the use of reaction coordinates.
Interbasin Transitions.
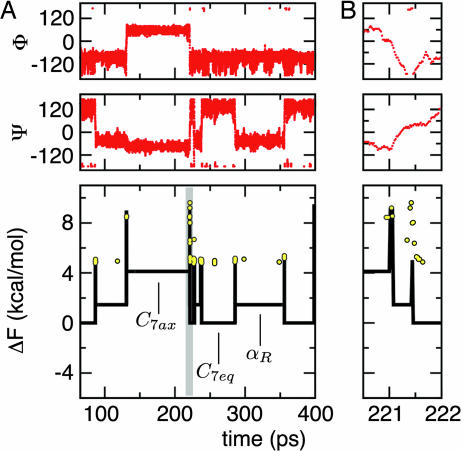
Once every snapshot sampled by the simulation has been assigned to a free-energy basin (e.g., using MCL), it is possible to quantitatively illustrate the thermodynamics and kinetics of the system. At equilibrium, the relative free energy of a basin is ΔFi = −kBT log(Wi/WC7eq), where Wi = Σa∈i wa is the total weight of basin i and C7eq is used as reference. In the same way, height of barriers relative to C7eq are estimated as ΔFi→j = −kBT log(A·wi→j/WC7eq), where wi→j is the number of direct transitions between basin i and basin j observed during the simulation and A is a constant. The exact value of A, which cannot be fixed unambiguously, depends on the snapshot saving frequency 1/ts [for example, in the harmonic approximation used in ref. 12, A = h/(tskBT), where h is the Planck constant]. Here, A is set to unity, which means that ΔFi→j is estimated up to an additive constant. ΔFi and ΔFi→j can be used as order parameters for the temporal analysis of the evolution of the system, as in Fig. 4Lower, where yellow circles represent configurations that have been identified as unstable nodes (see above). It is interesting to note that unstable nodes are typically located at the direct transition between two energy basins. Elsewhere, they indicate failed barrier crossings. The time series of interbasin transitions, embedding the system dynamics in one dimension, provides more useful and less noisy information than the time series of dihedral angles values (Fig. 4).
Fig. 4.
Alanine-dipeptide interbasin transitions. (A) Time series of a trajectory segment of the alanine dipeptide. The dihedral angles φ and ψ are shown in red. The black line shows the free energy of the basin or barrier visited by the system at every time step (see text and Table 1 for details) using as a reference the C7eq basin. Free energies are estimated from the MCL clusterization using p = 1.2. Black vertical spikes correspond to free-energy barriers whose height is evaluated using the number of interbasin transitions. Yellow circles show unstable configurations and their ΔF value is calculated using the weight of the most populated cell in C7eq (φ = −86.4, ψ = 136.8) as reference. (B) A zoom on the time series segment, which is gray in A Lower illustrating a very fast transition from C7ax to C7eq via αR.
Conclusions
A complex-network description and cluster-detection algorithms are used to obtain unprojected graphical representations of free-energy surfaces and quantitative analysis of free-energy basins, respectively. The results of the present study can be summarized in four main points. First, the network representation of configuration space sheds light on the topography of free-energy surfaces as well as the pathways between basins. Second, one of the three cluster-detection algorithms used in this work, which implicitly takes into account free-energy barriers, emerges as the most appropriate to quantitatively estimate the landscape topography of simple analytical models and the alanine dipeptide. These results indicate that preserving energy barrier heights sampled at equilibrium is crucial for defining basins. Moreover, the failure of the modularity cost-function commonly used in network cluster-detection algorithms (but that does not take into account barrier heights) suggests that the criteria to assess the quality of a clusterization are not a universal property of complex networks and might depend on the type of system under study. Third, provided a physically meaningful clusterization of the CSN, it is possible to compute free-energy differences for all the states of the system, activation energy barriers, as well as configurations participating in interbasin transitions. As a consequence, the free energy of basins and barriers is used as an order parameter, which, by naturally embedding the dynamics in one dimension, allows to follow the chronological evolution of the states of the system. Fourth, it is shown analytically, and illustrated by the alanine-dipeptide analysis, that the broad-tailed weight distribution observed in CSNs originates mainly from the enthalpic nature of the basins; whereas entropic basins, which lack a predominantly populated configuration, generate a Gaussian distribution. Finally, network clusterization has been shown to be an effective approach to the free-energy landscape dimensionality reduction problem. In the future, it will be interesting to generalize this analysis tool for applications to large biomolecules, such as structured peptides and proteins whose landscapes are characterized not only by enthalpic but also entropic basins.
Methods and Models
Free-Energy Landscapes from Low-Dimensional Models.
A stochastic process on an energy landscape is simulated by a Monte Carlo procedure on a D-dimensional lattice with a distance a between two neighbor sites. At each time step a neighbor site is chosen randomly and the system evolves according to the Monte Carlo rules. The trajectory consists of a chronological sequence of the sites visited during the dynamics (snapshots). This chronological sequence describes the dynamics at a microscopic level because only the nearest neighbor sites can be reached at each time step. Snapshots are taken every M steps. Sites are the nodes of the network, and two nodes are connected if the system evolved from one site to the next within M steps. For the systems investigated in this work snapshots are saved every M = 5 steps. For the multidimensional double-well the Monte Carlo is performed on a lattice of size length a = 0.2 and for D = 5 nodes are defined as boxes of size (3a)D. The total number of snapshots for D = 3 and D = 5 are N = 105 and N = 3·106, respectively (see Fig. 1A). In the case of the Mexican-Hat model in D = 2 dimensions, a = 0.05, and a total of N = 105 snapshots were saved (see Fig. 1B).
Free-Energy Landscapes from Atomistic MD Simulations.
Five Langevin dynamics simulations with a friction coefficient of 0.15 ps−1 of the alanine dipeptide were performed at 300 K for a total of 1 μs of simulation time. Snapshots were saved every ts = 0.02 ps (10 MD steps). Every trajectory was started from an extended configuration of the dipeptide. MD simulations were performed with the program CHARMM (PARAM19 force field) (36). A mean field approximation was used to describe the main effects of the aqueous solvent on the solute (ACE2) (37) with a nonbonding cutoff of 12 Å.
To define the nodes and links of the CSN of the alanine dipeptide a discretization of the space is needed. In this way, every snapshot sampled during the simulation is assigned to a cell of the discretized configuration space. Cells (i.e., similar configurations) are nodes of the network and the direct transitions between them are links. A weight w is assigned to each node to take into account the free energy of each conformation and is proportional to the number of snapshots within a given cell. The weight of a link from node i to node j corresponds to the number of transitions from site i to j visited during the simulation. The resulting network is directed, weighted, and can contain self-loops. A cell of the space is defined by the dihedral angles φ and ψ. In this discretization scheme, the (φ, ψ) space is discretized in n × n cells (see Fig. 2). Further discretization schemes and robustness analysis are presented in SI Text.
Weight Distribution: Analytical Derivation.
The node weight distribution is an important quantity in the understanding of complex networks, and it has been observed to differ from the one expected for random graphs (38, 39). In the following, it is shown that the energy landscape [U(x)] and the weight distribution [P(w)] of the CSN of U(x) are related by an analytical formula. The weight of a node is defined as the number of times the configuration is visited during the simulation. In the continuous approximation and spherical coordinates Pt(w) for w > 0 is written as
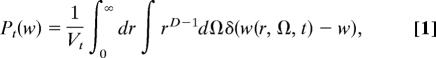 |
where Vt is the volume of the space visited in the simulation and D is the dimension. Ω is the solid angle in D-dimensional spherical coordinates and w(r, Ω, t) is the weight of the node at position (r, Ω) at time t. For simplicity, spherical symmetry of the energy landscape [U(x) = U(r)] will be assumed.
For large enough t, w(r, t) is proportional to the stationary solution w(r, t) ≈ exp[(−U(r)]. Taking U(r) in the units of kBT, U(0) = 0 and using the properties of the delta function, Eq. 1 becomes
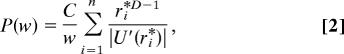 |
with C the appropriate normalizing factor and exp[(−U(ri✻)] = w/w(0) for all possible ri✻, i = 1, …, n. The first important remark is the w−1 factor in Eq. 2. This factor does not depend on the particular shape of the energy landscape or the dimension D. Thus, any weight distribution is expected to have a power-law P(w) ≈ w−1 multiplied by a modulating factor. In the particular case of the quadratic well with spherical symmetry, which is often the lowest order approximation of an energy basin, Eq. 2 reads
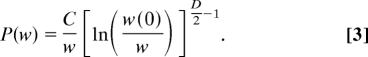 |
It is worth noting that for the case of an entropic basin like a square well, all the sites have the same weight and P(w) is peaked around a single value (see Fig. 3B and SI Text for the derivation and numerical comparisons).
Cluster-Detection Algorithms.
In this work, three different network clusterization approaches have been applied: the MCL algorithm (24, 33), Potts model clustering (26), and modularity optimization (25). The three algorithm are very different in spirit and implementation. In the following, the algorithms will be shortly introduced; the interested reader will find more detailed informations in the SI Text as well as in the papers referenced above.
MCL is based on the behavior of random walkers on the network. For this reason, it is very convenient to take into account directed weighted edges and even self-loops. The algorithm works as follows: (i) start with the transition matrix A of the network and normalize each column of the matrix to obtain a stochastic matrix S; (ii) compute S2; (iii) take the pth power (p > 1) of every element of S2 and normalize each column to one; and (iv) go back to step ii. After several iterations MCL converges to a matrix SMCL(p) invariant under transformations ii and iii. Only a few lines of SMCL(p) have some nonzero entries that give the clusters as separated basins (there is in general exactly one nonzero entry per column). Step iii reinforces the high-probability walks at short time scale at the expense of the low-probability ones. The parameter p tunes the granularity of the clustering. If p is large, the effect of step iii becomes stronger and the random walks are likely to end up in small “basins of attraction” of the network, resulting in several small clusters. On the other hand, a small p produces larger clusters. In the limit of p = 1, only one cluster is detected. Qualitatively, step iii plays a similar role as decreasing the temperature in simulated annealing, and a small value of p corresponds to a small rate of decrease. Yet, the similarity between MCL and simulated annealing has to be investigated in more detail. In all our examples, a small value of p (p = 1.2) was used to identify the largest and most significant energy basins.
Potts model clustering maps network communities onto the magnetic domains in the ground state or local minima of a modified Potts model Hamiltonian H = −Σ(i,j)∈E Jijδσi,σj + γ Σs=1q ns(ns − 1)/2. E is the set of edges, σi are the individual spins that can take q values, ns is the number of spins that have value s, and Jij is the weight of the link between node i and j. The parameter γ gives the strength of the coupling between the ferromagnetic and antiferromagnetic parts of the Hamiltonian.
Finally, the modularity optimization method is an agglomerative hierarchical clustering method and is based on the maximization of the modularity Q = Σi(eii − ai2), where eii is the fraction of edges between nodes of cluster i, ai = Σj eji is the fraction of edges attached to nodes of cluster i, and the sum runs over all the clusters. By definition, Q can be at maximum equal to 1.
Node Instabilities.
The detection of node instabilities probes the robustness of a cluster structure and reveals the presence of nodes that play a role in more than one cluster (26, 40). The idea is to add noise over the edges of the network and compare the clusters for different noisy realizations. Here, a modified version of the algorithm introduced in ref. 32, which takes into account link weights and degree heterogeneity, is applied. Noise was added on each edge with weight wij as +σij with probability 0.5 and −σij with probability 0.5, where ] and ki is the number of edges connected to node i. A node is considered as unstable if it is grouped <95% of the time to the same cluster.
Supplementary Material
Acknowledgments
We thank G. Caldarelli, S. Muff, E. Guarnera, and G. Settanni for interesting discussions; M. Karplus and S. Krivov for critical reading of the manuscript; and M. Seeber for the program Wordom used to analyze the MD trajectories. The simulations were performed on the Matterhorn cluster. This work was supported by a National Science Foundation grant (to A.C.) and grants COSIN, DELIS, and OFES-Bern (to D.G.).
Abbreviations
- CSN
configuration space network
- MD
molecular dynamics
- MCL
Markov clustering
- Q
modularity
- DG
disconnectivity graph.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0608099104/DC1.
References
- 1.Goldstein M. J Chem Phys. 1969;51:3728–3739. [Google Scholar]
- 2.Frauenfelder H, Sligar SG, Wolynes PG. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 3.Stillinger FH. Science. 1995;267:1935–1939. doi: 10.1126/science.267.5206.1935. [DOI] [PubMed] [Google Scholar]
- 4.Du R, Pande VS, Grosberg AY, Tanaka T, Shakhnovich ES. J Chem Phys. 1998;108:334–350. [Google Scholar]
- 5.Pande VS, Grosberg AY, Tanaka T, Rokhsar DS. Curr Opin Struct Biol. 1998;8:68–79. doi: 10.1016/s0959-440x(98)80012-2. [DOI] [PubMed] [Google Scholar]
- 6.Bolhuis PG, Dellago C, Chandler D. Proc Natl Acad Sci USA. 2000;97:5877–5882. doi: 10.1073/pnas.100127697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma A, Dinner AR. J Phys Chem B. 2005;109:6769–6779. doi: 10.1021/jp045546c. [DOI] [PubMed] [Google Scholar]
- 8.Best RB, Hummer G. Proc Natl Acad Sci USA. 2005;102:6732–6737. doi: 10.1073/pnas.0408098102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krivov SV, Karplus M. J Phys Chem B. 2006;110:12689–12698. doi: 10.1021/jp060039b. [DOI] [PubMed] [Google Scholar]
- 10.Das P, Moll M, Stamati H, Kavraki LE, Clementi C. Proc Natl Acad Sci USA. 2006;103:9885–9890. doi: 10.1073/pnas.0603553103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caflisch A. Curr Opin Struct Biol. 2006;16:71–78. doi: 10.1016/j.sbi.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Krivov SV, Karplus M. J Chem Phys. 2002;117:10894–10903. [Google Scholar]
- 13.Krivov SV, Karplus M. Proc Natl Acad Sci USA. 2004;101:14766–14770. doi: 10.1073/pnas.0406234101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albert R, Barabási A-L. Rev Modern Phys. 2002;74:47–97. [Google Scholar]
- 15.Newman MEJ. Siam Rev. 2003;45:167–256. [Google Scholar]
- 16.Boccaletti S, Latora V, Moreno Y, Chavez M, Hwang DU. Phys Rep. 2006;424:175–308. [Google Scholar]
- 17.Scala A, Amaral LAN, Barthélémy M. Europhys Lett. 2001;55:594–600. [Google Scholar]
- 18.Doye JPK. Phys Rev Lett. 2002;88:238701. doi: 10.1103/PhysRevLett.88.238701. [DOI] [PubMed] [Google Scholar]
- 19.Rao F, Caflisch A. J Mol Biol. 2004;342:299–306. doi: 10.1016/j.jmb.2004.06.063. [DOI] [PubMed] [Google Scholar]
- 20.Hubner IA, Deeds EJ, Shakhnovich EI. Proc Natl Acad Sci USA. 2005;102:18914–18919. doi: 10.1073/pnas.0502181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barabási AL, Oltvai ZN. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 22.Newman MEJ. Proc Natl Acad Sci USA. 2001;98:404–409. doi: 10.1073/pnas.021544898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibson D, Kleinberg J, Raghavan P. Inferring Web Communities from Link Topology. New York: ACM; 1998. [Google Scholar]
- 24.Enright AJ, Van Dongen S, Ouzounis CA. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clauset A, Newman MEJ, Moore C. Phys Rev E. 2004;70 066111. [Google Scholar]
- 26.Reichardt J, Bornholdt S. Phys Rev Lett. 2004;93:218701. doi: 10.1103/PhysRevLett.93.218701. [DOI] [PubMed] [Google Scholar]
- 27.Muff S, Rao F, Caflisch A. Phys Rev E. 2005;72 doi: 10.1103/PhysRevE.72.056107. 056107. [DOI] [PubMed] [Google Scholar]
- 28.Latapy M, Pons P. Proc Lect Notes Comp Sci. 2005;3733:285–293. [Google Scholar]
- 29.Palla G, Derényi I, Farkas I, Vicsek T. Nature. 2005;435:814–818. doi: 10.1038/nature03607. [DOI] [PubMed] [Google Scholar]
- 30.Danon L, Diaz-Guilera A, Duch J, Arenas A. J Stat Mech. 2005 P09008. [Google Scholar]
- 31.Massen CP, Doye JPK. Phys Rev E. 2005;71:046101. doi: 10.1103/PhysRevE.71.046101. [DOI] [PubMed] [Google Scholar]
- 32.Gfeller D, Chappelier JC, De Los Rios P. Phys Rev E. 2005;72:56135. doi: 10.1103/PhysRevE.72.056135. [DOI] [PubMed] [Google Scholar]
- 33.Van Dongen S. The Netherlands: Univ of Utrecht, Utrecht; 2000. Ph.D. thesis. [Google Scholar]
- 34.Cho SS, Levy Y, Wolynes PG. Proc Natl Acad Sci USA. 2006;103:586–591. doi: 10.1073/pnas.0509768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Apostolakis J, Ferrara P, Caflisch A. J Chem Phys. 1999;110:2099–2108. [Google Scholar]
- 36.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 37.Schaefer M, Bartels C, Leclerc F, Karplus M. J Comp Chem. 2001;22:1857–1879. doi: 10.1002/jcc.1137. [DOI] [PubMed] [Google Scholar]
- 38.Yook SH, Jeong H, Barabási A-L, Tu Y. Phys Rev Lett. 2001;86:5835–5838. doi: 10.1103/PhysRevLett.86.5835. [DOI] [PubMed] [Google Scholar]
- 39.Barrat A, Barthélémy M, Pastor-Satorras R, Vespignani A. Proc Natl Acad Sci USA. 2004;101:3747–3752. doi: 10.1073/pnas.0400087101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilkinson DM, Huberman BA. Proc Natl Acad Sci USA. 2004;101(Suppl 1):5241–5248. doi: 10.1073/pnas.0307740100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.