Short abstract
Analysis of Arabidopsis and rice polygalacturonases suggests that polygalacturonases duplicates underwent rapid expression divergence and that the mechanisms of duplication affect the divergence rate.
Abstract
Background
Polygalacturonases (PGs) belong to a large gene family in plants and are believed to be responsible for various cell separation processes. PG activities have been shown to be associated with a wide range of plant developmental programs such as seed germination, organ abscission, pod and anther dehiscence, pollen grain maturation, fruit softening and decay, xylem cell formation, and pollen tube growth, thus illustrating divergent roles for members of this gene family. A close look at phylogenetic relationships among Arabidopsis and rice PGs accompanied by analysis of expression data provides an opportunity to address key questions on the evolution and functions of duplicate genes.
Results
We found that both tandem and whole-genome duplications contribute significantly to the expansion of this gene family but are associated with substantial gene losses. In addition, there are at least 21 PGs in the common ancestor of Arabidopsis and rice. We have also determined the relationships between Arabidopsis and rice PGs and their expression patterns in Arabidopsis to provide insights into the functional divergence between members of this gene family. By evaluating expression in five Arabidopsis tissues and during five stages of abscission, we found overlapping but distinct expression patterns for most of the different PGs.
Conclusion
Expression data suggest specialized roles or subfunctionalization for each PG gene member. PGs derived from whole genome duplication tend to have more similar expression patterns than those derived from tandem duplications. Our findings suggest that PG duplicates underwent rapid expression divergence and that the mechanisms of duplication affect the divergence rate.
Background
The functions and regulation of cell wall hydrolytic enzymes have intrigued plant scientists for decades. These enzymes cleave the bonds between the polymers that make up the cell wall, and include polygalacturonases (PGs), beta-1, 4-endoglucanases, pectate lyases, pectin methylesterases, and xyloglucan endo-transglycosylases [1]. As a consequence of their action, cell wall extensibility and cell-cell adhesion can be altered leading to cell wall loosening that results in cell elongation, sloughing of cells at the root tip, fruit softening, and fruit decay [2-4]. Cell separation processes also contribute to important agricultural traits such as pollen dehiscence and abscission of organs including leaves, floral parts, and fruits [5-7]. In addition, these enzymes are hypothesized to be involved in general housekeeping functions in plants [8].
Among these hydrolytic enzymes, the PGs belong to one of the largest hydrolase families [9,10]. PG activities have been shown to be associated with a wide range of plant developmental programs such as seed germination, organ abscission, pod and anther dehiscence, pollen grain maturation, xylem cell formation, and pollen tube growth [5,11-13]. Over-expression of a PG in apple (Malus domestica) has resulted in alterations in leaf morphology and premature leaf shedding [14]. Interestingly, the functions of PGs are not restricted to the control of cell growth and development as they are also reported to be associated with wound responses [15] and host-parasite interactions [16]. These findings illustrate the divergent and important roles of PGs in plants.
PGs have been identified in various plants including Arabidopsis, pea and tomato [5,17]. In both tomato and Arabidopsis it has been determined that many PGs are located within tandem clusters [9,18]. In addition to tandem duplication, the Arabidopsis genome contains large blocks of related regions derived from whole genome duplication events [17,19,20]. In this study, we conducted a comparative analysis of PGs from Arabidopsis and rice to address several key questions on the evolution and function of this gene family. We compared the PGs from Arabidopsis and rice to determine the pattern of expansion and the extent of PG losses prior and subsequent to the divergence between these two species. To uncover the mechanisms that contributed to the expansion of this gene family, we examined the distribution of PGs on Arabidopsis chromosomes in conjunction with the large-scale duplicated blocks. Torki et al. [9] have suggested that a group of related PGs tend to be expressed in the flowers and flower buds, while PGs expressed in vegetative tissues belong to other groups. The implication is that the diverse functions of PGs may be a consequence of differential expression. This expression divergence and/or subfunctionalization most likely contribute to the retention of PG duplicates [21,22]. To evaluate the degree of spatial expression divergence between PGs, we conducted RT-PCR analysis on all 66 Arabidopsis PG genes in five non-overlapping tissue types. To supplement the RT-PCR expression data, we also examined expression tags generated from other large-scale sequencing projects. Finally, we analyzed expression at five stages of floral organ abscission to assess the degree of temporal expression divergence among members of this gene family.
Results and discussion
Expansion of the PG family in Arabidopsis and rice
To investigate the relationships among PGs and the extent of lineage-specific expansion in rice and Arabidopsis, we identified PGs from the GenBank polypeptide records and the genomes of Arabidopsis and rice (Oryza sativa subsp. indica). All PGs identified contain GH28 domains that are approximately 340 amino acids long and encompass approximately 75% of the average PG coding sequence (for lists of genes used in this analysis, see Figure 1 and Additional data files 1,2 and 8). According to the phylogenetic relationships of bacterial, fungal, metazoan, and plant PGs (Additional data file 3), we found that the 66 Arabidopsis and 59 rice PGs fall into three distinct groups (Figure 1, groups A, B, and C). Sixteen of the rice PGs contain more than one glycosyl hydrolase 28 (GH28) domain and were regarded as mis-annotated tandem repeats. It should be noted that the rice PGs were derived from the shotgun sequencing of the O. indica genome that was estimated to be 95% complete [23]. We identified the nodes that lead to Arabidopsis-specific and rice-specific clades and predict that these represent the divergence point between these two species. We have designated the clades defined by such nodes as AO (Arabidopsis-Oryza) orthologous groups. For example, in the A3 clade there exists one Arabidopsis subclade and one rice subclade, and we predict that only one ancestral A3 sequence was present before the divergence between Arabidopsis and rice. However, gene losses could have occurred and therefore some PGs may be present in the Arabidopsis-rice common ancestor but later lost in either Arabidopsis or rice (Figure 1, arrowheads). Therefore, Arabidopsis (A, indicating loss(es) in rice) and rice (O, indicating loss(es) in Arabidopsis) clades were also identified based on their sister group relationships to the AO clades. Since the clades that we defined are most likely orthologous groups (Figure 1, red circles), the number of clades reflects that there were at least 21 ancestral PGs before the Arabidopsis-rice split. Further expansion of this gene family occurred after the split as suggested by the duplication events in the lineage-specific branches that reside within each clade. It should be noted that some clades such as the A1 clade were not defined based on the AO clade-based criteria because the nodes within had relatively low bootstrap supports (<50%). If we assumed these less well-supported nodes are correct, there are 27 ancestral PGs.
Figure 1.
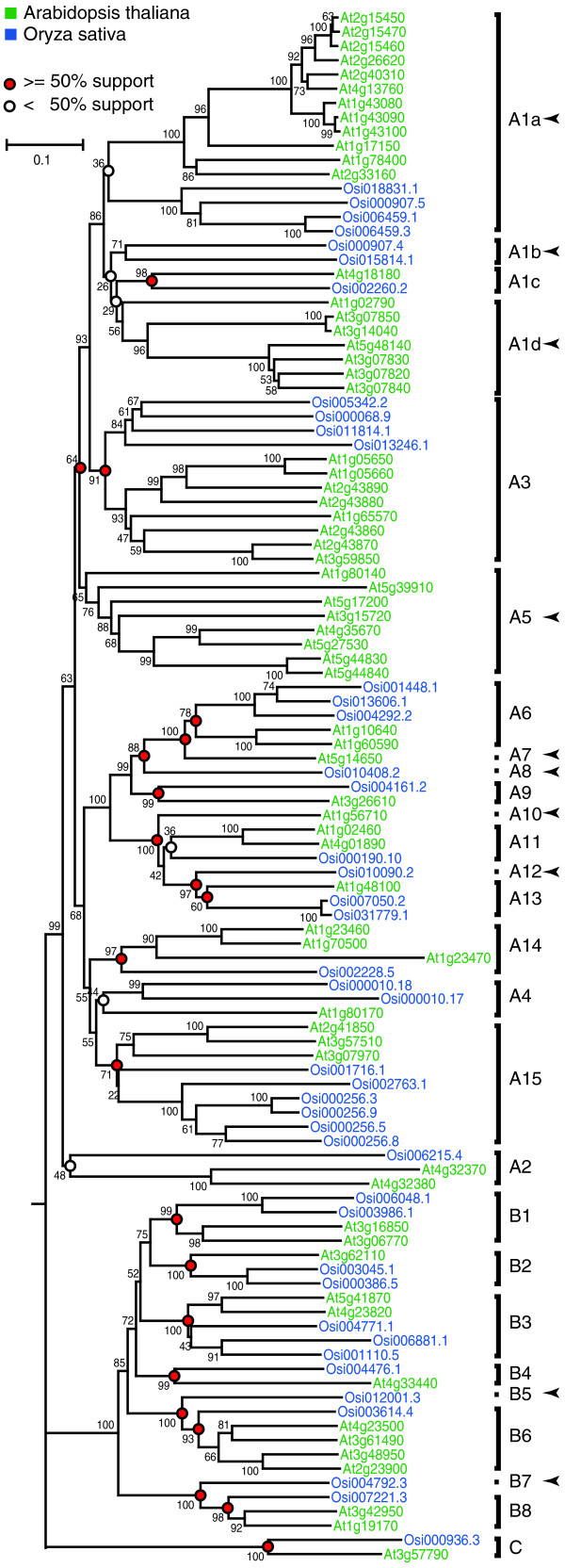
The phylogeny of Arabidopsis and rice PGs. The amino acid sequences for the glycosyl hydrolase 28 family motif were aligned. The phylogeny was generated using neighbor-joining algorithm with 1,000 bootstrap replicates. Sequences are color-coded according to the key. The plant PGs are classified into three major groups and multiple clades. The clades were defined by identifying nodes representing speciation events (circles, see Results section for criteria). For these nodes, red circles indicate that the bootstrap support for the subtending branches is higher than 50% and indicate the criteria for least number of common ancestral PGs between rice and Arabidopsis. The nodes are labeled with white circles if the bootstrap support is less than 50%. Arrowheads indicate clades that contain only sequences for one of the two plants.
Duplication mechanisms accounting for the PG family expansion
Examination of the distribution of the Arabidopsis PGs on all five chromosomes indicates a non-random distribution of many PGs (Figure 2). More than one third of the Arabidopsis PGs (24 of 66) have at least one related sequence within ten predicted genes, and these 24 genes fall into nine clusters that range from two to four genes per cluster (Figure 2, column cluster). In most cases, these physically associated PGs are from the same clades; however, there are five exceptions including genes in clusters 1d, 2b and 3a (Figure 2). In these cases, some members within the cluster are not closest relatives. Besides these 24 tandem repeated sequences, all remaining PGs are at least 100 genes apart. This bimodal distribution of PG physical distances and relationships between closely linked genes suggests that the 24 closely linked PGs are derived from tandem duplications.
Figure 2.
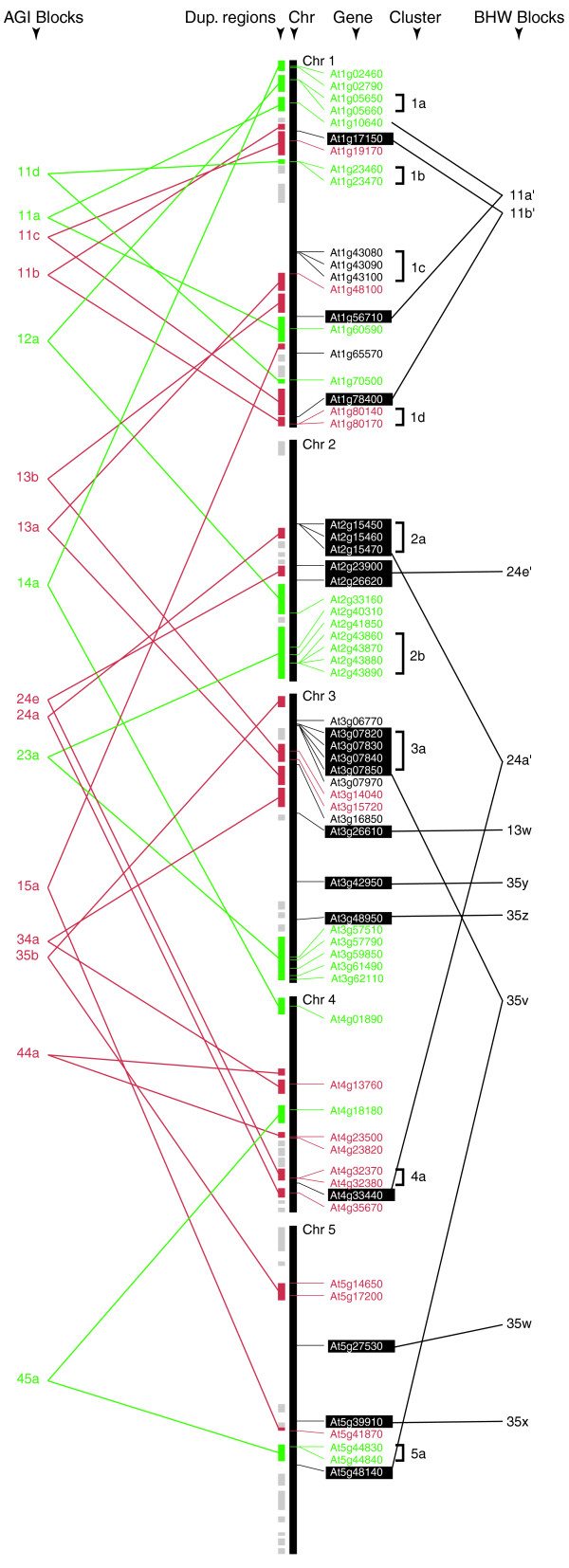
Mechanisms of Arabidopsis PG family expansion. The locations of Arabidopsis PGs are indicated on the Arabidopsis chromosomes. The tandem clusters are also indicated. They are color-coded based on the following scheme: PGs found in both duplicated regions of a block pair (green); PGs found in only one duplicated region of a block pair (red); and no PG is located in these blocks (gray). PGs covered by AGI blocks are either red or green, while PGs covered by BHW but not AGI blocks are with white text and black-boxed background. If PGs are found in both duplicated regions of a block, the gene names are linked. In addition, these gene names are italicized if they belong to the same clade. PGs that are not found in either AGI or BHW blocks are shown in black text. Tandem duplications are indicated by cluster designation. BHW block names were modified from the original designations of Blanc et al. [20]. BHW block names with a prime indicate that they overlap with AGI blocks of the same names. The reference for the block names can be found in Additional data file 2.
In addition to tandem duplications, it has been shown that the Arabidopsis genome is the product of several rounds of polyploidization or whole-genome duplications [17,19,20]. To determine the contribution of these large-scale duplications, we mapped Arabidopsis PGs to the duplicated blocks established in two independent studies. The first dataset from the Arabidopsis Genome Initiative [17] contains 31 blocks (AGI blocks), and forty Arabidopsis PGs fall in 16 of the AGI blocks (Figure 2, indicated in red and green). Blocks from the second dataset from Blanc et al. [20] are designated as BHW (after Blanc, Hokamp, Wolfe) blocks, and 19 PGs were found in 10 BHW blocks (Figure 2, shaded). The AGI and BHW blocks were identified using different approaches and their combined use increases the coverage of duplicated regions. As a result, nearly 90% (59 out of 66) of Arabidopsis PGs are covered in the 26 AGI and BHW blocks.
Within these 26 duplicated blocks, 29 PGs are found in both duplicated regions of ten block pairs. To investigate the origin of PGs in these ten block pairs, we conducted similarity searches between regions of each pair to determine if PGs mapped to the corresponding duplicated regions, and if their neighboring genes were arranged collinearly (Figure 3; see also (Additional data file 4) for all comparisons). Sixteen PGs in five of these block pairs are clearly located in such collinear regions, indicating that they were derived from large-scale duplication of their associated blocks. For example, AGI block 23a contains nine PGs in six corresponding duplicated regions that show extensive collinearity (Figure 3). In Figure 3b, At2g41850 and At3g57510 are flanked by paralogous genes that are arranged collinearly, indicating that they were products of a block duplication. This is also true for a tandem cluster of four PGs and a PG singleton shown in Figure 3d. Interestingly, At3g57790 corresponds to At2g43210, a potential pseudogene lacking the signal peptide and the bulk of the PG catalytic domain (Figure 3c). We also observed that there are 23 duplicated block pairs with asymmetrical distribution (Additional data file 4). Among them, 16 block pairs have PGs on only one of the blocks (Figure 2 and (Additional data file 4)): ten for AGI and six for BHW blocks. For the remaining seven block pairs, the PGs are found on both blocks but are not arranged in a collinear fashion. Taken together, these findings clearly indicate that many members of the PG family are derived from large-scale duplication events. However, quite a few of them were not retained.
Figure 3.
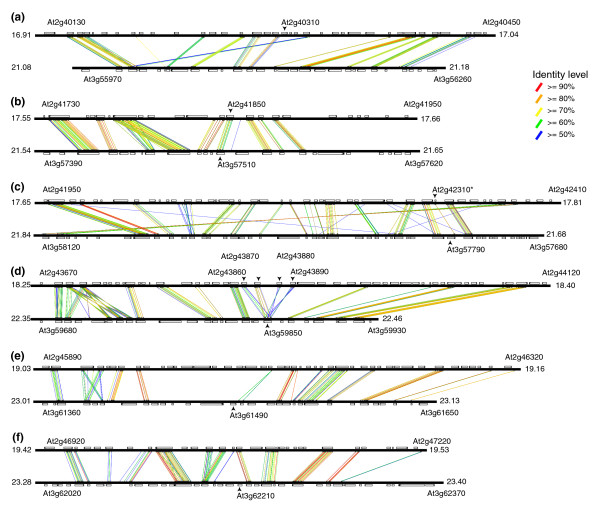
Collinearity of PGs in AGI block 23a. After locating areas with similarities in the block 23a (see also Additional data file 4), six distinct PG-containing regions were defined. (a) At2g40310 does not have PG in the collinear region. (b) At2g41850 and At3g57510 are located in collinear regions. (c) The 3' end of At3g57790 is highly similar to At2g42310*, a truncated PG that is likely a pseudogene. (d) A tandem of four PGs (At2g43860, At2g43870, At2g43880, At2g43890) is located in the collinear region with At3g59850. (e) At3g61490 does not have any PG in the corresponding collinear region. (f) At3g62210 does not have any PG in the collinear region. For each region pair, the solid black bars are the chromosomes (top: chromosome 2, bottom: chromosome 3) flanked by the starting and ending positions in Mb. The annotated genes are drawn to scale in a rectangular box on the chromosome and in each box the thicker black line indicates the 3' position of the gene. The names are only shown for PGs and the starting and ending genes in each block pair. The areas that are at least 30 amino acids long with at least 50% identity are linked by colored lines based on their identity levels (see key).
PG expression in Arabidopsis tissues
The size of the plant PG family and the patterns of PG duplication in Arabidopsis indicate that the PG family expanded in both Arabidopsis and rice after their divergence. The continuous expansion of this gene family raises an intriguing question on the mechanisms of duplicate retention and their functions in plants. Since retention may be due to functional divergence between duplicate copies, it is possible that PG functional divergence can be, in part, attributed to expression divergence. To evaluate the degree of expression divergence between PG duplicates, we analyzed the expression of all 66 Arabidopsis PGs in five tissue types (flowers, siliques, inflorescence stems, rosette and cauline leaves, and roots) with RT-PCR (Figure 4 and Additional data file 5). PCR reactions were repeated at least three times for each gene in each tissue type, and all primers were tested using genomic DNA as a positive control (see Figure 5). In addition, PCR products of 40 of the 43 PGs were sequenced to verify their identity. We found that 23 PGs did not have detectable RT-PCR products in any of the five tissue types tested. We further tested the expression of these 23 PGs in a T87 suspension culture cell line that had been previously shown to have >60% genes expressed [24]. Only one PG (At2g43860) was detected. To rule out the possibility of faulty primer designs, a second primer set was designed for each of these 23 PGs, but none led to detectable products.
Figure 4.
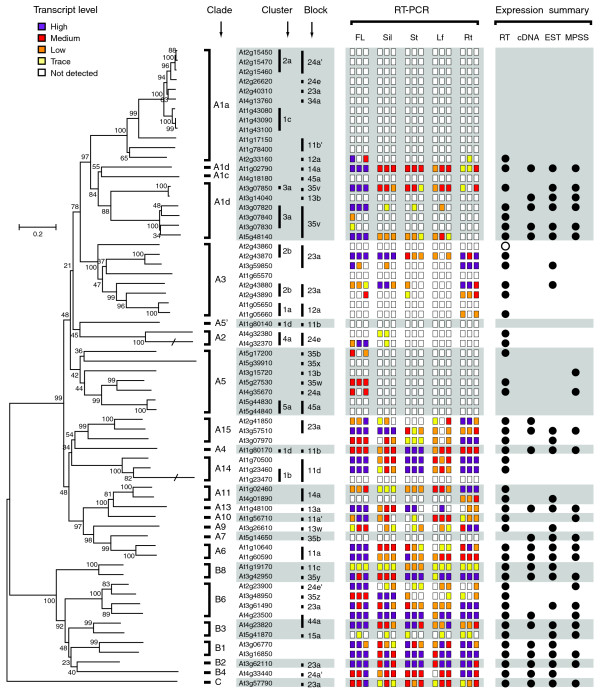
The phylogeny and expression patterns of Arabidopsis PGs. The phylogeny was generated using all Arabidopsis PGs with Erwinia peh1 as the outgroup. The clade classification, cluster and block designation are also shown. The levels of transcripts are classified into five categories as shown in the key. The tissue source abbreviations are as follows: Fl, flower; Si, silique; St, stem; Lf, rosette and cauline leaf; Rt, root; gDNA, genomic DNA. For each gene, three colored rectangles represent the level of RT-PCR products from three independent biological replications for each tissue type. On the right, the solid black circles indicate the presence of the four different expression tags. RT-PCR data are from this study and a solid circle represents repeatable expression from one or more of the six tissue types analyzed including expression in At2g43860 from suspension cultures. Open circles represent expression that was only detected in one of the RT-PCR reactions yet verified by sequencing. cDNAs, ESTs and MPSS tags were obtained from SIGnAL, GenBank, and the Arabidopsis MPSS project websites, respectively. Branches that were shortened are intersected with a solidus (/).
Figure 5.
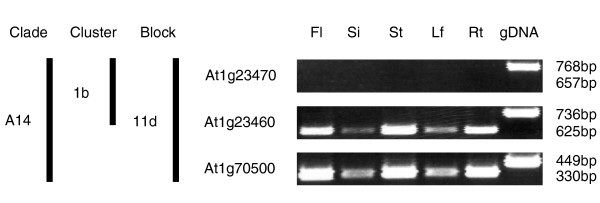
RT-PCR of PGs in five tissue types. The competitive RT-PCR, using both cDNA and gDNA templates, is demonstrated. The expression pattern of PGs in the clade A14 is variable except At1g23470, which has no detectable expression in all five tissue types. RT-PCR product sizes are indicated to the right of the figure. Tissue source abbreviations are as in Figure 4.
To complement the RT-PCR approach, we also examined the expression tags that were publicly available including full-length cDNAs, expressed sequence tags (ESTs), and massive parallel signature sequencing (MPSS) tags (Additional data file 6). The presence of RT-PCR products or other expression tags is shown in Figure 4 (far right-hand panel). Among these four different expression measures, the RT-PCR approach detects the highest number of PGs. In the 43 PGs with RT-PCR products, other expression tags support only 30 of them. In addition, only three PGs have cDNA, ESTs, and/or MPSS but not RT-PCR products. These findings indicate that RT-PCR is the most sensitive approach with a relatively low false-negative rate. For further analyses, we consider a PG expressed if two out of three of the RT-PCR reactions had detectable products (42) or if its expression is supported by the presence of either cDNA or EST (three). Based on these criteria, 45 PGs had detectable expression (Figure 4). Approximately 50% of these expressed PGs are found in all five tissues and 20% have relatively higher level of expression in more than one tissue. In addition, more than 50% of expressed PGs have high level of expression in floral tissues, 40% in root tissue, 16% in stem and 12% in silique. Only nine PGs (approximately 20%) are found in only one tissue type (Figure 4). These findings indicate that most PGs have rather wide expression patterns and the expression level seems to be generally higher in floral tissues. The complexity of expression patterns represented in Figure 4 emphasizes the need for additional interpretation, and is the basis for the statistical analyses described below for the expression data.
Effects of duplication mechanisms on gene expression
While it was anticipated that more closely related genes would tend to have similar expression patterns, we did not find significant correlation between the synonymous substitution rate (Ks) and the expression profile (Figure 6). In addition, to evaluate the relationships between Ks and expression correlation using all PG pairs, we also reached the same conclusion after partitioning the data as within clade (r = -0.119, p = 0.39), between clade (r = 0.002, p = 0.58), or reciprocal best matches (r = -0.4389, p = 0.12). This finding indicates that expression patterns have diverged quickly after PG duplications. In particular, significantly fewer PGs in tandem clusters were expressed when compared with those not in clusters (Table 1; Fisher's exact test; p = 0.0326). In several cases, the tandem duplicated regions have one relatively highly expressed gene while the rest have either low expression levels or no RT-PCR products. For example, in the 1b tandem cluster of clade A14, At1g23460 is highly expressed while At1g23470 does not have any detectable expression. Curiously, we found that related PGs found in duplicated blocks tend to have similar expression patterns at the tissue level. For example, in block 11d clade A14, At1g23460 and At1g70500 have nearly identical expression profiles (Figure 4). We selected 18 PG pairs that were derived from tandem or large-scale block duplication to compare their expression divergence. Among nine pairs in large-scale duplicated blocks, the expression pattern is significantly different in only one pair (Table 2). Among the nine pairs derived from tandem duplications, the t-test could only be conducted for four pairs because several of the tandem duplicates had no detectable expression. In addition to two pairs with significant differences (p < 0.05), three pairs with only one of the tandem duplicates expressed are also classified as pairs showing expression divergence. Therefore, excluding two pairs with no expression for both duplicates, five out of seven tandem pairs have divergent expression. Significantly fewer PG pairs derived from tandem duplications have similar expression patterns compared with those derived from large-scale duplications (Fisher's exact test; p < 0.01). Therefore, tandemly duplicated PGs have higher levels of expression divergence compared with PGs derived from large-scale duplications. These findings suggest that duplication mechanisms contribute to divergence of expression patterns differently.
Figure 6.
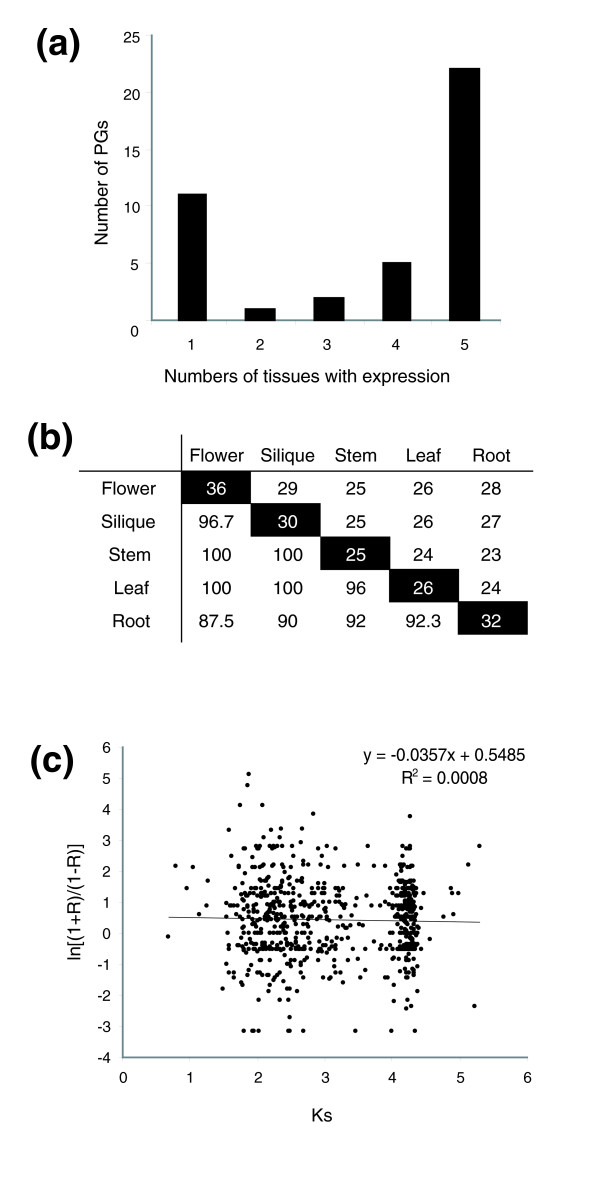
Expression of PGs shared among tissues and the correlation between expression patterns and the Ks. (a) Overlapping expression of PGs - the majority of expressed PGs are found in all five tissues tested. (b) Pairwise comparisons of tissues with PGs - the numbers in black boxes represent the number of PGs expressed in indicated tissues. The numbers in the upper-right half are the number of PGs expressed in both tissues specified in the top row and in the leftmost column. The numbers in the lower-left half are the percent overlap between two tissues. (c) The relationships between the Ks and transformed correlations in expression patterns - the Ks values were determined for all PG pairs. The correlations between expression patterns were calculated for all PG pairs and transformed as described in the Materials and methods. The formulae for the best fit and the correlation coefficient determined by linear regression are shown on the top right corner.
Table 1.
Distribution and expression of Arabidopsis PG genes in duplicated regions
| Out of duplicated regions* | Within duplicated regions* | |||||
| With match† | Without match† | |||||
| Number of genes | Expression‡ | Number of genes | Expression‡ | Number of genes | Expression‡ | |
| Singular | 4 | 3 | 11 | 9 | 27 | 21 |
| Tandem | 3 | 0 | 10 | 8 | 11 | 4 |
| Total | 7 | 3 | 21 | 17 | 38 | 25 |
*Duplicated regions are the regions that are covered by the AGI and BHW blocks. †The presence (with match) or absence (without match) of PGs in collinear regions of each duplicated block pair as shown in Figure 4 and Additional data file 4. ‡Expression detected in at least two out of three RT-PCR reactions or supported by the presence of cDNA or EST tags.
Table 2.
Expression (RT-PCR) of Arabidopsis PG genes in different clades
| Set* | Gene1 | Gene2 | Ks† | t‡ | p < 0.05‡ |
| B1 | At1g02460 | At4g01890 | 1.0564 | 3.09 | n |
| B2 | At1g10640 | At1g60590 | 1.252 | -0.32 | n |
| B3 | At1g23460 | At1g70500 | 0.8011 | -0.73 | n |
| B3 | At1g23470 | At1g70500 | 1.877 | -14.70 | y |
| B4 | At2g41850 | At3g57510 | 0.6805 | -1.43 | n |
| B5 | At2g43860 | At3g59850 | 2.1371 | -3.00 | n |
| B5 | At2g43870 | At3g59850 | 0.9534 | 2.13 | n |
| B5 | At2g43880 | At3g59850 | 1.8279 | 1.00 | n |
| B5 | At2g43890 | At3g59850 | 1.8308 | -1.41 | n |
| T1 | At1g05650 | At1g05660 | 0.2385 | ND§ | y |
| T2 | At1g23460 | At1g23470 | 0.878 | 6.53 | y |
| T3 | At2g43860 | At2g43870 | 1.4013 | -6.53 | y |
| T4 | At2g43880 | At2g43890 | 4.2072 | 2.83 | n |
| T5 | At3g07820 | At3g07830 | 0.5342 | ND§ | y |
| T5 | At3g07820 | At3g07840 | 0.4923 | ND§ | y |
| T5 | At3g07830 | At3g07840 | 0.457 | ND | ND |
| T6 | At4g32370 | At4g32380 | 2.6336 | 0.73 | n |
| T7 | At5g44830 | At5g44840 | 0.1626 | ND | ND |
*Each set contains genes that were duplicated through either local-scale block duplication (B) or tandem duplication (T). In duplicated blocks where a PG is collinear with a cluster, the one-to-many relationships are shown. For tandem clusters, all pairwise combinations are shown. †Ks, synonymous substitution rate. ‡Differences in expression patterns significant (y) or not (n) for t-test with df = 2, p < 0.05 [52]. ND, not determined since both genes do not have detectable RT-PCR product or §expression was documented for only one gene in the pair.
Developmentally regulated expression divergence among PGs expressed in abscission zone
So far, our expression analyses were performed in five widely different tissues. To further expand our understanding of PG expression, we took a close look at 43 of the expressed PGs in the abscission zones of flowers and developing siliques at five developmental stages during floral organ abscission (Figure 7a). During the abscission process there are discrete stages when cell wall loosening and cell wall dissolution occurs, thus providing an excellent biological system to look at more subtle changes in the regulation of cell separation. And indeed, this analysis allowed us to discern differences in expression between PGs that had been initially regarded as similar due to limitations in resolution (Figure 7). For example, at the tissue level, At1g23460 and At1g70500, from block 11d clade A14 were regarded as having nearly identical expression profiles. However, when we examined five stages of abscission, these genes have distinct profiles (Figure 7c and 7e, Additional data file 7).
Figure 7.
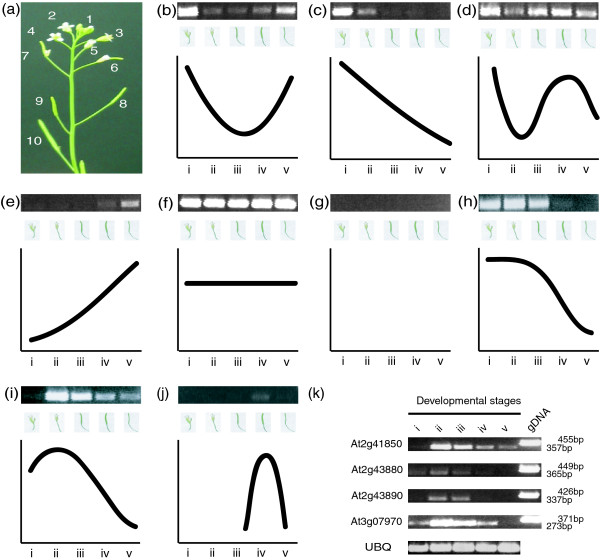
RT-PCR on floral organ abscission zones representing five unique stages of development. Expression of 43 PGs is examined in the abscission zones at five different stages of floral organ abscission as determined by position on the inflorescence, where position one represents anthesis and larger numbers are progressively older flowers (a). Five developmental stages were examined with the RT-PCR; i (position1/2) and ii (position 4/5), pre-abscission, iii (position 7/8), during abscission, iv (position 0/11), and v (position 13/14) post-abscission. Expression during the abscission process is classified into nine different unique patterns shown in (b) to (j); the gene names are provided in Additional data file 7. PGs specifically up-regulated during the abscission process are shown with RT-PCR products (k).
We determined that there are nine unique patterns of expression for the PGs during the five stages of abscission that are shown in Figure 7 and Additional data file 7. Eight PGs display high levels of expression at anthesis, low levels during the events of cell separation, and high levels post abscission as depicted in Figure 7b. These genes are all from independent clades except two sets: At1g19170 and At3g42950 (B8), and At2g23900 and At3g48950 (B6). In Figure 7c, 7 PGs show initial high expression at anthesis that decreases steadily during abscission, while in Figure 7d, PG expression (At1g02460, At1g56710, and At3g61490) initially decreases right before abscission and then increases after the loss of floral organs or during what is described as post abscission repair. In Figure 7e, two PGs (At1g23460 and At1g10640) have very low or undetectable expression during anthesis that goes up continually during abscission. Other patterns include ten PGs with constitutive expression (Figure 7f), and six PGs with no expression (Figure 7g). Last, we observed three patterns of expression that correlated with unique changes during the process of abscission (Figure 7h,i,j). In Figure 7h, high levels of gene expression correlate with cell wall loosening or the earliest steps of abscission, while in Figure 7i highest levels of gene expression correlate with cell separation or loss of floral organs. In Figure 7j, it is only at around positions 10 and 11 that we observe detectable gene expression, and this correlates with predicted stages of cell repair [25].
Taken together, expression divergence between PGs that show no difference at the tissue level were revealed when we examined PG expression at different developmental stages of abscission, thus indicating duplication mechanisms contribute to divergence of expression differently. Our findings also provide candidate PGs important for different abscission stages. More importantly, the expression divergence between duplicate genes in general appears to be under-estimated in expression studies due to the limitations in resolution.
Conclusion
PG family expansion history
PGs fall into several taxon-specific clades where eubacterial, fungal, and plant PGs organize into different clusters [10]. We have hypothesized that there were approximately 21 PGs present in the immediate common ancestor of Arabidopsis and rice, and when additional monocots and dicots are sequenced, we will be able to have a more accurate estimate of the ancestral family size. Since Arabidopsis and rice diverged more than 150 million years ago (MYA), gene conversion events that occurred soon after divergence of these two lineages will be much rarer than those that occurred in a lineage-specific fashion.
By examining the physical locations of Arabidopsis PGs and their relationships to the proposed large-scale duplication patterns, we found that tandem duplications and large-scale duplications were two of the major factors responsible for the expansion of the PG family in Arabidopsis. This is similar to other gene families such as the NBS-LRR [26] and the RLK/Pelle gene family [27]. Among duplicates in the same tandem cluster, nearly all belong to the same PG clades or are close relatives of each other. The only exception is At1g80140 and At1g80170 in cluster 1d, suggesting that they are tandem duplicates that formed before the Arabidopsis-rice split. Most of the PGs (59) are located within 26 duplicated block pairs (Table 1). However, the comparison of gene contents between duplicated blocks in each pair indicates that 22 PGs are distributed asymmetrically in ten of these duplicated block pairs, thus suggesting gene losses. The rest of the duplicated block pairs contain PGs in both duplicated regions. Since only 13 of these PGs are collinear, our findings suggest that large-scale duplications did contribute to some expansion of the PG family but gene losses occurred frequently. Members of each PG pair (either one-to-one or one-to-many) located in collinear regions are from the same clade. Since a clade is defined as the PG ancestral unit right before the divergence between Arabidopsis and rice, the blocks harboring these PGs would be duplicated after the split between these two plants. Blanc et al. [20] assigned duplicated gene pairs to blocks and used synonymous substitution rates to establish the block age. We found that 17 PGs were in 'recent' blocks that duplicated after the split between the Arabidopsis and rice lineages (Additional data file 4). This correlation is consistent with our interpretation based on a phylogenetic approach.
In the cases where PGs were present in only one of the collinear regions, it is likely that the absence of PGs was due to gene losses, and almost 80% of the PGs generated by large-scale duplications could have been lost in Arabidopsis. These findings are consistent with the high duplicate loss rate in the Arabidopsis genome [28,29]. In addition, the collinear regions flanking PGs are generally larger than the corresponding regions without PGs (considering the numbers of genes or physical distances between the two genes flanking the PGs that were collinear), thus suggesting that the deletion of chromosome regions contributes to PG loss. Another explanation for the asymmetrical distribution of PGs in blocks is that they were inserted de novo through an alternative mechanism such as retro-transposition; however, this is unlikely, as all of the plant PGs have multiple introns.
Divergence of expression pattern after duplications
Although a large number of PG duplicates were lost, there is a net gain in the PG family size after the split between Arabidopsis and rice, and thus, the immediate question is how were these duplicates retained? The fate of duplicated genes varies and depends on the selection constraints [21,22]. Since one third of the Arabidopsis PGs do not have any evidence of expression, these genes could be pseudogenes. However, some of them have diverged substantially from their closest relatives with large synonymous substitution rates and have most likely persisted beyond the time frame of pseudogenization in Arabidopsis proposed to be a million years [30]. Meanwhile, PGs without evidence of expression may be present in tissues not sampled or induced under untested conditions. A closer look at other developmental events involving cell wall degradation, cell separation or cell wall loosening may provide additional insights.
There is mounting evidence that retention of duplicated genes may be due to acquisition of novel functions, partitioning of original functions, or both. The contribution of differential expression in retaining duplicated genes has been hypothesized more than 25 years ago [31,32]. More recently, Force et al. [33] proposed the DDC (Duplication/Degeneration/Complementation) model predicting that genes sharing overlapping but distinct expression patterns will be retained due to the partitioning of ancestral expression profiles. In our study, we found that two thirds of the Arabidopsis PGs are expressed and almost three quarters of these expressed PGs are detected in at least three tissues. If the AtGenExpress microarray data for Arabidopsis is considered [34], five additional PGs are likely expressed using a stringent intensity cutoff (data not shown). Among the PGs that are expressed rather ubiquitously, related PGs in general have overlapping but distinct expression profiles, consistent with the prediction of the DDC model, although it is possible that some expression differences are due to gain of expression rather than loss. In any case, divergent expression among closely related PGs is evident in the different developmental stages of abscission. It has also been reported more recently that duplicated genes tend to have more similar expression patterns when the Ks is relatively small [35,36]. However, in the PG family, the more recent duplicates do not necessarily have more similar expression patterns. The expression correlation breaks down even more when we examine the expression profiles of PGs in different developmental stages of the abscission process. This lack of correlation may be attributed to relatively long divergence time (large Ks value) between PG duplicates and the lack of statistical power, because a much smaller number of genes are examined compared with an analysis of the whole genome. In addition, we suggest that the mechanism of gene duplication appears to contribute differently to expression divergence. The number of expressed PGs is significantly lower if they are located in tandem repeats. On the other hand, PGs with similar tissue expression patterns tend to be localized to corresponding large-scale duplicated blocks. One possible mechanism for this difference in expression pattern conservation may be the fact that tandem duplication may or may not allow the duplication of whole promoter regions and coding sequences. On the other hand, large-scale duplication involves the duplication of multiple genes together with their promoter and/or enhancer elements. Thus, tandem duplications will result in faster expression divergence than large-scale duplications, and that large-scale duplications ultimately lead to "fine tuning" of gene expression. Another potential explanation for the differences in expression may be due to differences in gene silencing. Homology-dependent gene silencing is a common phenomenon in plants [37]. Since the average sequence divergence between tandem repeats is smaller than that of large-scale duplications (data not shown), one might also argue that tandemly duplicated genes tend to be silenced at a higher frequency.
Functional studies have established that plant PGs are involved in diverse roles including plant growth and development, wounding responses, and plant-microbe interactions [4]. Although the PG family members have substantial overlap in tissue-level expression even between distantly related members, when we analyzed distinct developmental stages of abscission we were able to discern unique patterns of expression. These findings suggest that although even if there may be functional overlap between PGs, substantial expression divergence contributed to their retention and probably their functions. Given the number of PGs and the complexity of plant tissues and cell types, it is likely that PGs expressed in the same tissues have subtle differences in their temporal or spatial profiles. This is consistent with the PG expression patterns in different developmental stages of abscission. Alternatively, these seemingly co-expressed PGs may have also diverged at the biochemical levels, such as their catalytic properties. In this study, we used genome sequence information combined with gene expression to provide a framework to unravel the complexity of gene family function. By careful analysis we have been able to take a family of 66 genes and identify four members (Figure 7i) that have unique changes just as cell wall loosening and cell wall dissolution is predicted to occur; thus presenting a small subset of genes for further studies on abscission. Additional analyses in the temporal and spatial patterns of expression in other tissues, their biochemical properties, and in the biological functions of these genes will lead to novel insights regarding functional divergence and conservation in this gene family.
Materials and methods
Sequence selection, alignment, and phylogenetic analysis
Representative PGs were the sequences in the seed alignment of glycosyl hydrolase family 28 (GH28) from Pfam database [38]. The representative set was used as query sequences to conduct BLAST searches [39] against polypeptide sequences of A. thaliana for candidate PGs from Munich Information Center for Protein Sequences (MIPS) [40]. All sequences with E values less than one were regarded as candidate PGs and further analyzed with the Pfam HMM models from GenBank polypeptide sequences; The PGs of O. sativa subsp. indica were identified from predicted coding sequences obtained from Dr. W. Karlowski in MIPS Oryza sativa Database (MosDB) [41] with a similar procedure outlined above. The rice PG sequences appeared highly redundant, and thus almost 30% of the entries that were more than 99% identical at the nucleotide level were eliminated from further analysis. For a list of PGs, including redundant entries, see Additional data files 1 and 8. The protein sequences of PGs identified were aligned against the Pfam GH28 seed alignments using the profile alignment function of ClustalW [42]. The GH28 domain sequence alignments of rice and Arabidopsis PGs analyzed can be found in Additional data file 8. The phylogeny of all PGs identified was generated with MEGA2 [43] using the neighbor-joining algorithm [44] with 1,000 bootstrap replicates. Poisson correction for multiple substitutions was used. Sequence gaps were treated as missing characters. Both the Arabidopsis-rice and Arabidopsis-only trees were rooted with Erwinia peh1.
Mapping chromosome location and duplicated blocks
Two large-scale duplication datasets were used. The first is based on the analysis of the Arabidopsis Genome Initiative [17] that was provided by Heiko Schoof and MIPS/Institute of Bioinformatics, Germany. The correspondence between block names given in this study and those in the original analysis, and the starting and ending gene names for these blocks are given in Additional data file 2. The second is based on Blanc et al. [20] and is available from [45]. The collinearity of blocks that contain PGs in corresponding duplicated regions was determined using tBLASTn. For these blocks, the nucleotide sequences of one of the duplicated regions were used as query to search against a translated database built from the nucleotide sequence of the other region. To increase the number of High Scoring Pairs recovered, the query sequences were split into 5 kb windows. The matching areas (at least 50 amino acids long and 60% identical) of blocks that contain PGs in the corresponding duplicated regions are shown in Additional data file 4. After identifying the collinear regions surrounding PGs, we took at least 100 kb regions surrounding PGs and their corresponding duplication regions, regardless of the presence of PGs, and repeated the BLAST analysis splitting query sequences into 1 kb windows. Matching areas were defined as similar regions at least 30 amino acids long.
Plant materials and growth
Arabidopsis ecotype Columbia (COL) was used for this study and plants grown as described by Patterson and Bleecker [25]. T87 suspension-cultured cell lines were derived from COL ecotype [46,47] and provided by Sebastian Bednarek (University of Wisconsin, Madison, WI, USA). The abscission zones of developing flowers and siliques were collected by removing the primary inflorescence from the plant, and then trimming each individual sample within 0.75 mm +/- 0.25 of the floral abscission zone on both sides. Trimmed samples were immediately frozen in liquid nitrogen and stored at -80°C until further analysis.
Nucleic acid isolation and quantification
Plant tissue was frozen in liquid nitrogen, ground and added to TES-Lysis (50 mM Tris pH 8, 5 mM EDTA, 50 mM NaCl, 1% (w/v) SDS, 1% w/v sarkosyl) followed by extraction with a phenol:chloroform:isoamyl alcohol mix (25:24:1). Samples were centrifuged for 5 minutes at (12,000 g) and the resulting aqueous phase was extracted twice with chloroform:isoamyl alcohol (24:1). Nucleic acids were precipitated at 4°C with isopropanol and 10 M NH4OAc (one-third volume) and resuspended in TE. One-half volume of 6.0 M LiCl was added to the sample, incubated at 4°C for 4 hours, and then centrifuged 15 minutes at 12,000 g. DNA (supernatant fraction) was precipitated by adding 10 M NH4OAc (1/3 volume) and ethanol, and RNA (pellet) was washed with ethanol and resuspended in DEPC-treated H20 (1 μg/μl). DNA and RNA yields were quantified using a Smart Spec 3000 Biorad (Hercules, CA, USA). Nucleic acid quality was assessed by gel electrophoresis.
RT-PCR analysis
A quantity of 1 μg of each RNA sample was used to prepare cDNA by modifying standard procedures [48]. First strand synthesis was carried out using 500 μg/ml of an 18 mer oligo dT primer (IDT, Coralville, IA, USA). Resulting cDNAs were diluted 1:2 and 1 μl was added as template for a standard 20 μl PCR reaction. For each gene, primers were designed that flanked an intron in the genomic DNA similar to that described by Wang et al. [48]. Since the mRNA and genomic copy of a gene share identical primer sites, they had comparable amplification efficiencies in the PCR reaction and were distinguishable by size. Reactions were incubated at 95°C for 5 minutes, and cycled 28 or 36 times as follows: 94°C for 3 minutes, annealing temperature for 30 seconds, 72°C for 2 minutes. After the last cycle, reactions were incubated at 72°C for 7 minutes. Annealing temperatures and cycle numbers were optimized and are shown in Additional data file 5. A quantity of 10 μl of each PCR reaction was analyzed by gel electrophoresis, and the relative levels of PCR products were recorded.
DNA sequencing
PCR products were excised from the gel, cleaned using a Qiagen Gel extraction kit (Qiagen, Valencia, CA, USA) and sequenced directly as described below. Cycle sequencing reactions with a thermostable DNA polymerase and fluorescently labeled dideoxy terminators (BIG DYE Applied Biosystems, Foster, CA, USA) were carried out on each purified product or subcloned fragment. At2g43870 was subcloned into the PCR 4-TOPO vector (Invitrogen, Carlsbad, CA, USA) before sequencing. All reactions were outsourced to the UW-Madison Biotechnology Center and run on an ABI automated DNA sequencer.
Expression tags of PGs and analysis
The cDNA sequences released by the SIGnAL database [49] were retrieved from GenBank. The predicted protein sequences of PGs were used to search against the cDNA sequences. The cDNAs for PGs are listed in part I of Additional data file6. The Arabidopsis ESTs were retrieved from GenBank (part II of Additional data file 6), and a BLAST search was conducted using the predicted coding sequences of PGs. All matches with more than 80% identity were inspected. After eliminating gaps longer than three from the alignments, cognate ESTs were defined as those that were top matches to the gene in question with at least 97% identity. The accessions, source tissue information for the matching ESTs, can be found in part II of Additional data file 6. The MPSS tags matching the PG genes were retrieved using a batch query script from the Arabidopsis MPSS database [50]. Only tags matching exons in the crick strand with levels significantly different from 0 were regarded as evidence of expression.
The PG expression levels as determined by RT-PCR were converted into 5 categories: high (4), medium (3), low (2), trace (1), and none (0). For each gene, the median (M) of the converted expression levels was used for all subsequent analyses. For each gene pair, the synonymous and non-synonymous substitution rates were determined using the yn00 phylogenetic analysis by maximum likelihood program PAML [51]. The Pearson correlation coefficient (r) was determined for each gene pair and transformed into ln [(1+R)/(1-R)] for linear repression analyses [35,36]. For determining the differences in expression patterns between tandemly duplicated and block-duplicated genes, we conducted t-tests for 18 PG pairs. For each tissue, the expression levels were considered if both or either one of the genes in a pair were expressed.
Additional data files
The following additional data are available with the online version of this paper. Additional data file 1 lists PGs identified from Genbank protein records. Additional data file 2 is the BHW and AGI assignment of PGs to duplicated blocks in Arabidopsis. Additional data file 3 shows a phylogeny generated with all the PGs from fungi, bacteria, metazoa, and plants. Additional data file 4 shows a figure with the matching areas for duplicated blocks containing PGs in both regions. Additional data file 5 lists the primers used for the RT-PCR analysis. Additional data file 6 is summary of expression tags including a list of the PG cDNAs from Arabidopsis and a list of the PG cognate ESTs from Arabidopsis. Additional data file 7 lists PGs that are expressed in the floral organ abscission zones of Arabidopsis with their patterns of expression. Additional data file 8 shows GH28 domain sequence alignments of rice and Arabidopsis PGs analyzed.
Supplementary Material
Table containing PGs (except Arabidopsis proteins) sorted according to their taxa.
Table of assignment of PGs to duplicated blocks in Arabidopsis.
Figure showing a phylogeny generated with all the PGs from fungi, bacteria, metazoa, and plants.
Figure with the matching areas for duplicated blocks containing PGs in both regions.
Primer pairs and conditions used to amplify PG genes.
Table showing summary of expression tags and list of cDNAs for Arabidopsis PGs.
A list of PGs that are expressed in the floral organ abscission zones of Arabidopsis with their patterns of expression.
GH28 domain sequence alignments of rice and Arabidopsis PGs analyzed.
Acknowledgments
Acknowledgements
We thank Wojciech M Karlowski and MIPS for providing predicted indica gene sequences, Ronan O'Malley for helpful discussions on the statistical tests and technical advice for the expression work, Runsun Pan for providing software for evolutionary rate calculation, and Yun-Huei Tzeng for helpful discussions on the statistical tests. This work was supported by USDA (00-35301-9085) and NSF (DBI-0217552) to S.E.P., NIH National Research Service Award (5F32GM066554-01) to S-H.S., and NIH grants to W-H.L.
Contributor Information
Joonyup Kim, Email: joonyupkim@wisc.edu.
Shin-Han Shiu, Email: shius@msu.edu.
Sharon Thoma, Email: slthoma@wisc.edu.
Wen-Hsiung Li, Email: whli@chicago.edu.
Sara E Patterson, Email: spatters@wisc.edu.
References
- Carpita NC, McCann MC. The cell wall. In: Buchanan BB, Gruissem W, Jones R, editor. Biochemistry and Molecular Biology of Plants. Rockville: American Society Plant Physiologists; 2000. pp. 52–109. [Google Scholar]
- Rose JKC, Bennett AB. Cooperative disassembly of the cellulose-xyloglucan network of plant cell walls: parallels between cell expansion and fruit ripening. Trends Plant Sci. 1999;4:176–183. doi: 10.1016/S1360-1385(99)01405-3. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Expansive growth of plant cell walls. Plant Physiol Biochem. 2000;38:109–124. doi: 10.1016/S0981-9428(00)00164-9. [DOI] [PubMed] [Google Scholar]
- Roberts JA, Elliott KA, Gonzalez-Carranza ZH. Abscission, dehiscence, and other cell separation processes. Annu Rev Plant Biol. 2002;53:131–158. doi: 10.1146/annurev.arplant.53.092701.180236. [DOI] [PubMed] [Google Scholar]
- Hadfield KA, Bennett AB. Polygalacturonases: many genes in search of a function. Plant Physiol. 1998;117:337–343. doi: 10.1104/pp.117.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JA, Whitelaw CA, Gonzalez-Carranza ZH, McManus MT. Cell separation processes in plants: models, mechanisms, and manipulation. Ann Bot. 2000;86:223–235. doi: 10.1006/anbo.2000.1203. [DOI] [Google Scholar]
- Patterson SE. Cutting loose. Abscission and dehiscence in Arabidopsis. Plant Physiol. 2001;126:494–500. doi: 10.1104/pp.126.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Campillo E. Multiple endo-1,4-beta-D-glucanase (cellulase) genes in Arabidopsis. Curr Top Dev Biol. 1999;46:39–61. doi: 10.1016/s0070-2153(08)60325-7. [DOI] [PubMed] [Google Scholar]
- Torki M, Mandaron P, Mache R, Falconet D. Characterization of a ubiquitous expressed gene family encoding polygalacturonase in Arabidopsis thaliana. Gene. 2000;242:427–436. doi: 10.1016/S0378-1119(99)00497-7. [DOI] [PubMed] [Google Scholar]
- Markovic O, Janecek S. Pectin degrading glycoside hydrolases of family 28: sequence-structural features, specificities and evolution. Protein Eng. 2001;14:615–631. doi: 10.1093/protein/14.9.615. [DOI] [PubMed] [Google Scholar]
- Sitrit Y, Hadfield KA, Bennett AB, Bradford KJ, Downie AB. Expression of a polygalacturonase associated with tomato seed germination. Plant Physiol. 1999;121:419–428. doi: 10.1104/pp.121.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander L, Child R, Ulvskov P, Albrechtsen M, Borkhardt B. Analysis of a dehiscence zone endo-polygalacturonase in oilseed rape (Brassica napus) and Arabidopsis thaliana: evidence for roles in cell separation in dehiscence and abscission zones, and in stylar tissues during pollen tube growth. Plant Mol Biol. 2001;46:469–479. doi: 10.1023/A:1010619002833. [DOI] [PubMed] [Google Scholar]
- Demura T, Tashiro G, Horiguchi G, Kishimoto N, Kubo M, Matsuoka N, Minami A, Nagata-Hiwatashi M, Nakamura K, Okamura Y. Visualization by comprehensive microarray analysis of gene expression programs during transdifferentiation of mesophyll cells into xylem cells. Proc Natl Acad Sci USA. 2002;99:15794–15799. doi: 10.1073/pnas.232590499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson RG, Schroder R, Hallett IC, Cohen D, MacRae EA. Overexpression of polygalacturonase in transgenic apple trees leads to a range of novel phenotypes involving changes in cell adhesion. Plant Physiol. 2002;129:122–133. doi: 10.1104/pp.010986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco-Cardenas ML, Ryan CA. Polygalacturonase beta-subunit antisense gene expression in tomato plants leads to a progressive enhanced wound response and necrosis in leaves and abscission of developing flowers. Plant Physiol. 2003;133:693–701. doi: 10.1104/pp.103.023226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvana R, Kannaiyan S. Influence of cell wall degrading enzymes on colonization of N2 fixing bacterium, Azorhizobium caulinodans in rice. Indian J Exp Biol. 2002;40:369–372. [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Hong SB, Tucker ML. Genomic organization of six tomato polygalacturonases and 5' upstream sequence identity with tap1 and win2 genes. Mol Gen Genet. 1998;258:479–487. doi: 10.1007/s004380050758. [DOI] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–2117. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- Blanc G, Hokamp K, Wolfe KH. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003;13:137–144. doi: 10.1101/gr.751803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH. Evolutionary change of duplicate genes. Isozymes Curr Top Biol Med Res. 1982;6:55–92. [PubMed] [Google Scholar]
- Prince VE, Pickett FB. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet. 2002;3:827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- Stolc V, Samanta MP, Tongprasit W, Sethi H, Liang S, Nelson DC, Hegeman A, Nelson C, Rancour D, Bednarek S, et al. Identification of transcribed sequences in Arabidopsis thaliana by using high-resolution genome tilling arrays. Proc Natl Acad Sci USA. 2005;102:4453–4458. doi: 10.1073/pnas.0408203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson SE, Bleecker AB. Ethylene-dependent and -independent processes associated with floral organ abscission in Arabidopsis. Plant Physiol. 2004;134:194–203. doi: 10.1104/pp.103.028027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S-H, Bleecker AB. Expansion of the receptor-like kinase/pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003;132:530–543. doi: 10.1104/pp.103.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y. The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA. 2002;99:13627–13632. doi: 10.1073/pnas.212522399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16:1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E. Multilocus enzymes, gene regulation, and genetic sufficiency. J Mol Evol. 1978;12:57–89. doi: 10.1007/BF01732545. [DOI] [PubMed] [Google Scholar]
- Ferris SD, Whitt GS. Evolution of the differential regulation of duplicate genes after polyploidization. J Mol Evol. 1979;12:267–317. doi: 10.1007/BF01732026. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Gu Z, Nicolae D, Lu HH, Li WH. Rapid divergence in expression between duplicate genes inferred from microarray data. Trends Genet. 2002;18:609–613. doi: 10.1016/S0168-9525(02)02837-8. [DOI] [PubMed] [Google Scholar]
- Makova KD, Li WH. Divergence in the spatial pattern of gene expression between human duplicate genes. Genome Res. 2003;13:1638–1645. doi: 10.1101/gr.1133803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, Saedler H. Homology-dependent gene silencing in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:23–48. doi: 10.1146/annurev.arplant.47.1.23. [DOI] [PubMed] [Google Scholar]
- Pfam entry Glyco_hydro_28 http://www.sanger.ac.uk/cgi-bin/Pfam/getacc?PF00295
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIPS - Plant genome bioinformatics group http://mips.gsf.de/proj/thal/
- The MIPS Oryza sativa Database (MOsDB) http://mips.gsf.de/proj/plant/jsf/rice/index.jsp
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Large-scale duplication database http://wolfe.gen.tcd.ie/athal/all_results
- Axelos M, Curie C, Mazzolini L, Bardet C, Lescure B. A protocol for transient gene expression in Arabidopsis thaliana protoplasts isolated from cell suspension cultures. Plant Physiol Biochem. 1992;30:123–128. [Google Scholar]
- Kang BH, Busse JS, Dickey C, Rancour DM, Bednarek SY. The Arabidopsis cell plate-associated dynamin-like protein, ADL1Ap, is required for multiple stages of plant growth and development. Plant Physiol. 2001;126:47–68. doi: 10.1104/pp.126.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AM, Doyle MV, Mark DF. Quantitation of mRNA by polymerase chain reaction. Proc Natl Acad Sci USA. 1989;86:9717–9721. doi: 10.1073/pnas.86.24.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIGnAL Salk Institute Genomic Analysis Laboratory http://signal.salk.edu/
- Arabidopsis MPSS plus - about our database http://mpss.udel.edu/at/
- Yang Z, Nielsen R, Goldman N, Pedersen AM. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snedecor GW, Cochran WG. Statistical Methods. Ames, Iowa: Iowa State University Press; 1980. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table containing PGs (except Arabidopsis proteins) sorted according to their taxa.
Table of assignment of PGs to duplicated blocks in Arabidopsis.
Figure showing a phylogeny generated with all the PGs from fungi, bacteria, metazoa, and plants.
Figure with the matching areas for duplicated blocks containing PGs in both regions.
Primer pairs and conditions used to amplify PG genes.
Table showing summary of expression tags and list of cDNAs for Arabidopsis PGs.
A list of PGs that are expressed in the floral organ abscission zones of Arabidopsis with their patterns of expression.
GH28 domain sequence alignments of rice and Arabidopsis PGs analyzed.


