Abstract
Humans have two nearly identical copies of the survival motor neuron (SMN ) gene, SMN1 and SMN2. Homozygous loss of SMN1 causes spinal muscular atrophy (SMA). SMN2 is unable to prevent the disease due to skipping of exon 7. Using a systematic approach of in vivo selection, we have previously demonstrated that a weak 5′ splice site (ss) serves as the major cause of skipping of SMN2 exon 7. Here we show the inhibitory impact of RNA structure on the weak 5′ ss of exon 7. We call this structure terminal stem–loop 2 (TSL2). Confirming the inhibitory nature of TSL2, point mutations that destabilize TSL2 promote exon 7 inclusion in SMN2, whereas strengthening of TSL2 promotes exon 7 skipping even in SMN1. We also demonstrate that TSL2 negatively affects the recruitment of U1snRNP at the 5′ ss of exon 7. Using enzymatic structure probing, we confirm that the sequence at the junction of exon 7/intron 7 folds into TSL2 and show that mutations in TSL2 cause predicted structural changes in this region. Our findings reveal for the first time the critical role of RNA structure in regulation of alternative splicing of human SMN.
INTRODUCTION
Alternative splicing vastly increases the coding potential of the human genome by allowing individual genes to produce multiple proteins (1). However, very little is known about alternative splicing regulation that could be cell type-, developmental stage- or signaling pathway-specific (1–3). A fundamental problem in our understanding of alternative splicing is the lack of general rules by which exons are distinguished from introns and exon–intron boundaries are accurately defined (4). Pre-mRNA splicing involves two sequential transesterification reactions that are catalyzed by the spliceosome, the most complex macromolecular machine known: in addition to five snRNP particles (U1 snRNP, U2 snRNP, U4/U6 snRNP and U5 snRNP) the spliceosome contains several hundred non-snRNP splicing factors (5). Together they form a dynamic network of RNA–protein and RNA–RNA interactions responsible for accurate recognition of sequences at exon/intron junctions, called the 5′ and 3′ splice sites (ss). In addition to consensus splice site sequences, spliceosome assembly is guided by auxiliary cis-elements known as exonic and intronic splicing enhancers (ESE and ISE) and silencers (ESS and ISS), which promote or suppress splice-site selection, respectively. Enhancers and silencers are recognized by transacting factors, which include serine-arginine-rich proteins (SR proteins), SR-like proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs) (4,6–8). In addition, exon recognition can be influenced by exon length (9), intron length (10), the strength of the splicing signals (11) and the promoter structure (12–14). Furthermore, splicing is connected with other steps of gene expression, such as transcription (15,16), polyadenylation (17), transport (18) and translation (19).
In many cases, it has been reported that RNA secondary structure also influences the outcome of pre-mRNA splicing (20). There are several mechanisms through which RNA secondary structure can impact splicing: it can present binding sites for auxiliary splicing factors (21–23), it can bring widely separated cis-elements into juxtaposition (24,25), or it can directly affect accessibility of the splice sites (26–28). In the majority of cases the role of RNA structure in splicing has been studied using traditional approaches of individual model systems (21,24–37). Analysis of individual exons is now being complemented by more global computational approaches. For example, recent bioinformatics studies on the human CFTR gene have showed that pre-mRNAs with high versus low splicing efficiencies differ in their predicted secondary structures, providing the first statistical evidence that secondary structure elements may be required for correct splicing (38). In addition, statistical analysis of coding sequences in mRNAs from different species has revealed that coding regions exhibit biases in favor of local RNA structures (39). Analogous results have been reported for bacterial and Saccharomyces cerevisiae mRNA (40). Overall, the emerging picture implies that pre-mRNA sequences may harbor structural elements in vivo (20). However, the role of such structural elements in regulation of pre-mRNA splicing remains least understood.
Spinal muscular atrophy (SMA), the second most common autosomal recessive disorder, is caused by the absence of the survival motor neuron 1 (SMN1) gene (41), which encodes a ubiquitously expressed 38 kDa protein that plays an essential role in cellular metabolism (42–45). A nearly identical copy of the gene, SMN2, fails to compensate for the loss of SMN1 due to exon 7 skipping, producing a truncated protein SMNΔ7, which has been shown to be unstable (46). SMN1 and SMN2 differ by a critical C to T substitution (C6U transition in transcript of SMN2) at position 6 of exon 7 (47,48). C6U does not change the coding sequence, but it is sufficient to cause skipping of exon 7 in SMN1 (47,48). Two models have been proposed to explain the inhibitory effect of C6U. According to one model, C6U abrogates an ESE associated with SF2/ASF (49), whereas the other model implicates the creation of an ESS associated with hnRNP A1 (50). Recently, additional evidence has been forwarded to support the SF2/ASF model, while acknowledging the indirect role of hnRNP A1 in alternative splicing of exon 7 (51). Based on extensive mutations, we have shown that C6U creates an extended inhibitory context (Exinct) at the 3′ ss of exon 7 (52). In vivo selection of the entire exon 7 supported the existence of Exinct as well (53). It also revealed additional cis-elements, namely Conserved tract and 3′-Cluster that affect splicing of exon 7 [(53) and Figure 1]. The latter element (3′-Cluster) is located immediately upstream of the 5′ ss. Confirming the inhibitory role of 3′-Cluster, several deletions and point mutations within this element restored SMN2 exon 7 inclusion (53).
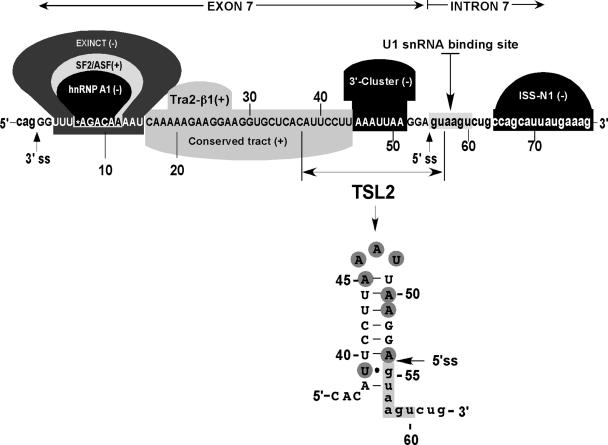
Figure 1.
Diagrammatic representation of exon 7 and adjacent intronic sequences. Capital letters represent exonic and small-case letters represent intronic nucleotides. Numbering starts from the first position of exon 7. The U1 snRNA binding site is indicated along with positive (+) and negative (−) cis-elements, which promote and inhibit exon 7 inclusion, respectively. Exinct, Conserved tract and 3′-Cluster were discovered by in vivo selection of the entire exon 7 (53). Binding sites for splicing factors SF/ASF2, hnRNP A1 and Tra2β have been described by others (49,50,54). ISS-N1 is a novel element we identified recently (60). Nucleotides involved in the formation of terminal stem–loop structure TSL2 are indicated. Based on the results of in vivo selection of the entire exon 7 (53), highly mutable nucleotides (shaded) are clustered within TSL2.
The accumulated experimental data indicate that splicing of SMN exon 7 involves complex interplay between positive and negative regulators. Most studied among these is SR-like protein Tra2-β1. It binds to a purine-rich ESE located in the middle of exon 7 (54). Overexpression of Tra2-β1 (54) or its associated proteins hnRNP G (55) and SRp30c (56) promoted exon 7 inclusion in SMN2. The increased expression of STAR (signal transduction and activation of RNA) family of proteins promote skipping of exon 7, suggesting tissue-specific regulation of exon 7 splicing (57). Proteins interacting with intronic sequences may also affect exon 7 splicing since cis-elements located in intron 6 and intron 7 have been shown to modulate exon 7 inclusion (58,59). Recently, we have discovered an intronic element (ISS-N1) located in the vicinity of the 5′ ss of exon 7 [(60) and Figure 1]. Confirming the silencer function of ISS-N1, an antisense oligonucleotide against ISS-N1 restored exon 7 inclusions in mRNAs derived from the SMN2 minigene and from endogenous SMN2, causing an increase in the levels of SMN protein in SMA-patient-derived cells (60).
In vivo selection of the entire exon 7 coupled with the discovery of ISS-N1 revealed that a weak 5′ ss is the limiting factor for exon 7 inclusion in SMN2 mRNA (53,60). Here we show that a local RNA structure that overlaps 3′-Cluster and partially sequesters the 5′ ss modulates SMN exon 7 splicing. We call this structure terminal stem–loop 2 (TSL2, Figure 1). A series of site-specific mutations confirmed the inhibitory nature of TSL2. Further, strengthening of TSL2 promoted exon 7 skipping even in SMN1. We demonstrate that the inhibitory effect of the strengthened TSL2 could be neutralized by increased base pairing between U1 snRNA and the 5′ ss of exon 7. To confirm the existence of TSL2, we performed partial structure probing of RNA that included the entire exon 7 and intronic sequences close to the splice sites. Our results for the first time point towards critical RNA structure as an additional level of control in a complex mechanism of SMN exon 7 splicing.
MATERIALS AND METHODS
Plasmid constructs and mutagenesis
Minigene splicing cassettes pSMN1ΔI6 and pSMN2ΔI6 were constructed by deleting ∼6 kb region within intron 6 from pSMN1 and pSMN2, respectively (52). In studies reported here we refer to pSMN1ΔI6 and pSMN2ΔI6 as SMN1 and SMN2, respectively. Mutations were generated by high-fidelity PCR as described earlier (53). Plasmids pUCBU1 and pUCBΔU1 were generous gifts from Dr H. Schaal (61). Site-specific mutations at the 5′ end of U1 RNA were generated by PCR, using a set of primers 5′pUCBU-mut (5′-CGAAGATCTCAGACTTACTCCGCAGGGGAGAT-3′) and 3′pUCBU1 (5′-ATCCTCGAGCCTCCACTGTAGGATTAAC-3′) and pUCBU1 plasmid as a template. The PCR product was cleaved with BglII and XhoI, gel purified and cloned into BglII–XhoI-digested plasmid pUCBΔU1. All mutations were confirmed by sequencing. All oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA).
Cell culture, transfection and in vivo splicing
Unless noted otherwise, all tissue culture media and supplements were purchased from Invitrogen. Human cervical carcinoma (C-33A) cell line was obtained from American Tissue Culture Collection (ATCC HTB-31). Cells were maintained in DMEM supplemented with 10% FBS. Transient transfections of C-33A cells were performed using Lipofectamine 2000 (Invitrogen) following manufacturer's instructions. Briefly, cells were plated one day before transfection, so that their density at the time of transfection was ∼90%. A total of 0.8 μg of plasmid DNA was used per each well of 24-well plate. In co-transfection experiments, 0.1 μg of an indicated minigene was used with 0.7 μg of one of the U1 snRNA expression vectors. For SMN1/36U37U39C construct U1 snRNA titration experiment was performed as follow: using 6-well plates 0.05 or 0.1 μg of SMN1/36U37U39C was co-transfected with the increasing amounts of either wild-type or mutant U1 snRNA expression vector. Total amount of transfected DNA was maintained constant (4.0 μg) by adding the appropriate amounts of the empty vector, pUCBΔU1. Lipofectamine 2000 was left in culture medium until cells were harvested for RNA isolation. Total RNA was isolated 20–24 h after transfection using Trizol reagent (Invitrogen) followed by treatment with RQ1 DNase (Promega). To generate cDNA, RT was performed using SuperScriptII reaction kit (Invitrogen) and oligo (dT) primer (Invitrogen). Generally, 0.5 μg of total RNA was used per 10 μl reaction. Minigene-specific spliced products were subsequently amplified using Taq polymerase (Invitrogen) and a set of primers, P1 and P2 (53). Analysis and quantitation of spliced products were performed using FPL-5000 Image Reader and ImageGauge software (Fuji Photo Film Inc.) (53). Results were confirmed by three independent experiments.
RNA structure probing
RNA secondary structure was probed using enzymes specific for single- and double-stranded RNA. RNA molecules were in vitro transcribed using T7 MEGAscript kit (Ambion) following manufacturer's suggestions. Templates for T7 transcription were generated as follow. First, a sequence that contained exon 7, 34 nt of intron 6 and 104 nt of intron 7, was amplified from the pSMN2ΔI6 minigene using primers 5′T7-EcoRI (5′-CACAGAATTCTAATACGACTCACTATAGGGTACCTATTTTTTTTAACTTC-3′) and 3′T7-HindIII (5′-CATTAACCAAGCTTCTTTTTAACATCTGAACTTTTTAAATG-3′). The resulting PCR fragment was digested with EcoRI and HindIII and cloned into pUC19 vector giving rise to the plasmid construct T7-SMN2-192. Subsequently, this plasmid was digested with KpnI and HindIII and used for cloning of PCR fragments, which contained the entire exon 7, 18 nt of intron 6 and 12 nt of intron 7. The PCR fragments were generated using SMN minigenes of interest as templates and a pair of primers 5′T7-84 (5′-CACTATAGGGTACCTTTATTTTCCTTACAGGG-3′) and 3′T7-84 (5′-CATTAACCAAGCTTGGCAGACTTACTCC-3′) or 5′T7-84 and 3′T7-84 + 54G (5′-CATTAACCAAGCTTGGCAGACTTACCCC-3′). Using plasmids linearized with HindIII, RNAs were T7-transcribed followed by dephosphorylation with calf intestinal phosphatase (NEB). Dephosphorylated RNA (∼2.7 μg in case of shorter transcript) was then 5′ end-labeled with [γ-32P]ATP (3000 Ci/mmol) and T4 polynucleotide kinase (NEB), ethanol precipitated and purified on denaturing 8% polyacrylamide gels by the ‘crush and soak’ method in 0.5 M NH4Ac, 1 mM EDTA plus 0.1% SDS (w/v). The elution was done for 1 h at 55°C. RNA was then ethanol precipitated, dissolved in 30 μl of water and centrifuged through a gel-filtration Bio-Spin column P-30 (Bio-Rad). A total of 1 μl of recovered RNA was used per each structure probing reaction. Prior to structure probing RNA was refolded in 50 mM Tris–HCl (pH 7.5), 10 mM MgCl2 either by heating at 65°C for 5 min and then slowly cooling to 37°C or by incubating at 37°C for 30 min. A total of 5 pg of RNase A (Ambion), 0.05 U of RNase T1 (Ambion), 0.0017 U of RNase V1 (Ambion), 0.354 U of nuclease S1 (Promega) and 0.67 ng of RNase U2 were used per 10 μl of structure probing reaction done at 22°C for 15 min. T1, A and V1 digestions were performed in 1× RNA Structure buffer (Ambion) in the presence of 0.1 μg/μl of yeast RNA (Ambion). S1 digestion was done in 50 mM sodium acetate (pH 4.5), 280 mM NaCl, 4.5 mM ZnSO4 and 0.1 μg/μl of yeast RNA. U2 cleavage was done in 50 mM sodium acetate (pH 4.5), 280 mM NaCl, 4.5 mM ZnSO4, 4 mM EDTA and 0.1 μg/μl of yeast RNA. The reactions were terminated by adding 20 μl of precipitation/inactivation buffer (Ambion) and the samples were ethanol precipitated, dissolved in formamide gel-loading buffer [90% formamide, 20 mM EDTA,0.05% (w/v) xylene cyanol with or without 0.05% (w/v) bromphenol blue], normalized for counts and analyzed in a denaturing 8% polyacrylamide gel, which was visualized by autoradiography using a FPL-5000 Image Reader (Fuji Photo Film Inc.). A control RNA sample was processed simultaneously and similarly as the digested RNAs, except the addition of enzymes was omitted. An alkaline hydrolysis ladder was prepared by incubating the 5′ end-labeled RNA in alkaline hydrolysis buffer (Ambion) at 95°C for 5 min. A G ladder was generated by incubating the 5′ end-labeled RNA in 1× Sequencing buffer (Ambion) for 5 min at 50°C, then 0.5 U of RNase T1 was added and the incubation continued for another 30 min. The reaction was terminated by adding precipitation/inactivation buffer (Ambion), and the samples were ethanol precipitated and dissolved in formamide gel-loading buffer. A C/U ladder was produced similarly except that 1 μg of yeast RNA (Ambion) was added to the reaction mixture and digestion with RNase A (5 pg) continued for 5 min. For an A ladder, U buffer [50 mM sodium acetate (pH 4.5), 200 mM NaCl, 4.5 mM ZnSO4, 20 mM EDTA, 7 M urea and 0.1 μg/μl of yeast RNA] was used and the digestion with RNase U2 (0.1 μg) continued for 30 min.
RESULTS
Site-specific mutagenesis supports the inhibitory role of RNA structure
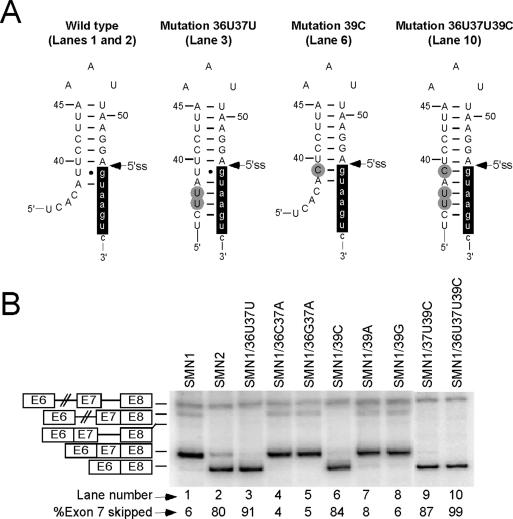
The RNA sequence at exon 7/intron 7 junction is predicted to fold into a terminal stem–loop structure, TSL2 [(62) and Figure 1]. TSL2 is comprised of an 8 bp-long stem and a triloop AAU. The first evidence of the inhibitory role of TSL2 in exon 7 splicing came from our previously reported observation that the double mutation 36U37U completely eliminated exon 7 inclusion in SMN1 [(53) and Figure 2]. At that time we maintained that the mutation abrogated a positive element, although simultaneous creation of a negative element, including an inhibitory RNA structure, was not ruled out. As shown in Figure 2A, non wild-type residues 36U and 37U together with the wild-type 35C can base pair with the third, fourth and fifth intronic positions, respectively, increasing the length of the stem from 8 to 11 bp and, as a consequence, making the proposed structural element more stable. Importantly, this structure sequesters all but one residue required for base pairing with U1 snRNA (Figure 2A).
Figure 2.
Effect of stem strengthening on splicing of SMN1 exon 7. (A) Mutations and their effect on the predicted TSL2 structure. Numbers and letters in each mutation's name represent the positions and types of substitutions within exon 7. Capital and small-case letters are used to indicate nucleotides of exon 7 and intron 7, respectively. Nucleotide numbering is the same as in Figure 1. Mutations are highlighted in gray. Nucleotides involved in base paring with U1 snRNA are highlighted in black. Canonical Watson–Crick base pairing and Wobble base pairing are indicated by continuous lines and filled circles, respectively. (B) In vivo splicing patterns of SMN1 mutants. Numbers and letters in each mutant's name represent the positions and the types of substitution within exon 7. Precursor mRNA, intron 7- and intron 6-retained splicing intermediates as well as exon 7-included and exon 7-skipped spliced products are indicated on the left. Abbreviations E6, E7 and E8 stand for exon 6, exon 7 and exon 8, respectively. Percentage of exon 7 skipping was calculated from the total value of exon 7-included and exon 7-skipped products.
To clarify whether the negative impact of 36U37U was due to an inhibitory structure, we made two additional mutants: SMN1/36C37A and SMN1/36G37A. Since 36C37Aand 36G37A mutations did not increase the size of TSL2 stem, they were able to maintain the splicing pattern of SMN1 (Figure 2B, lanes 4 and 5). The effects of mutations at positions 36 and 37 could not be explained based on the predicted ESEs. For example, ESEfinder [Release 2.0; (63)] predicts that all three double mutations (36U37U, 36C37A and 36G37A) abrogate an ESE associated with SF2/ASF. However, only 36U37U that strengthened the stem caused skipping of SMN1 exon 7 (Figure 2B, lane 3). To further investigate the effect of stem strengthening on exon 7 splicing, we generated mutants in which wild-type U at position 39 was changed to C, A or G. This U residue is predicted to form a Wobble base pair with the G residue at the first intronic position (Figure 2A). Therefore, a change from U to C at position 39 will strengthen the stem (Figure 2A), whereas a change from U to A or G will have a structure-destabilizing effect. Supporting the inhibitory effect of a strengthened stem, only 39C caused skipping of SMN1 exon 7, while 39A and 39G produced no effect on exon 7 splicing (Figure 2B, lanes 6–8). As shown in Figure 2B, different combinations of mutations that strengthened the stem also caused skipping of SMN1 exon 7 (lanes 9 and 10).
TSL2 competes with recruitment of U1 snRNP
The predicted TSL2 structure partially sequesters the 5′ ss of exon 7. Therefore, it is possible that TSL2 formation may interfere with U1 snRNP binding required at the early stages of spliceosome assembly. We hypothesized that improved binding of U1 snRNP at the 5′ ss of exon 7 would break the inhibitory structure, increasing inclusion of exon 7. To test this hypothesis we performed experiments in which the SMN2 minigene (wild-type TSL2) or SMN1 constructs with the strengthened stem were co-transfected with mutant U1 snRNA. This mutant has an increased capacity to hybridize with the 5′ ss of exon 7: it forms eleven consecutive base pairs with the 5′ ss as compared to only six formed by the wild-type U1 snRNA (Figure 3A). As shown in Figure 3B, mutant U1 snRNA was able to restore exon 7 inclusion in SMN2 (lane 15) and substantially improved inclusion of this exon in SMN1/39C, SMN1/36U37U and SMN1/37U39C (lanes 3, 6 and 9). However, it had no affect on exon 7 inclusion in SMN1/36U37U39C (Figure 3B, lane 12). It should be noted that mutant U1 snRNA failed to produce any changes in SMN1/36U37U39C exon 7 splicing even when co-transfected at 125-fold molar excess to the amount of SMN1/36U37U39C minigene (data not shown). SMN1/36U37U39C is predicted to have the most stable stem since it combines an increase in stem length and the substitution of a Wobble base pair with a more stable Watson–Crick base pair (Figure 2A). Note that neither empty vector, nor wild-type U1 snRNA was able to improve exon 7 inclusion in SMN2 or any of SMN1 mutants with strengthened TSL2 (Figure 3B).
Figure 3.
Effect of U1 snRNA base pairing on splicing of SMN1 mutants with strengthened TSL2. (A) Diagrammatic representation of base pairing formed between the 5′ ss of exon 7 and wild-type or mutant U1 snRNA. The last 6 nt of exon 7 are indicated by capital letters, while the first 8 nt of intron 7 are indicated by small-case letters. Numbering starts from the first position of exon 7. Mutations within U1 snRNA are highlighted in black. (B) In vivo splicing patterns of SMN1 mutants shown in Figure 2A in the presence of either wild-type or mutant U1 snRNA. Spliced intermediates and products are the same as indicated in Figure 2B.
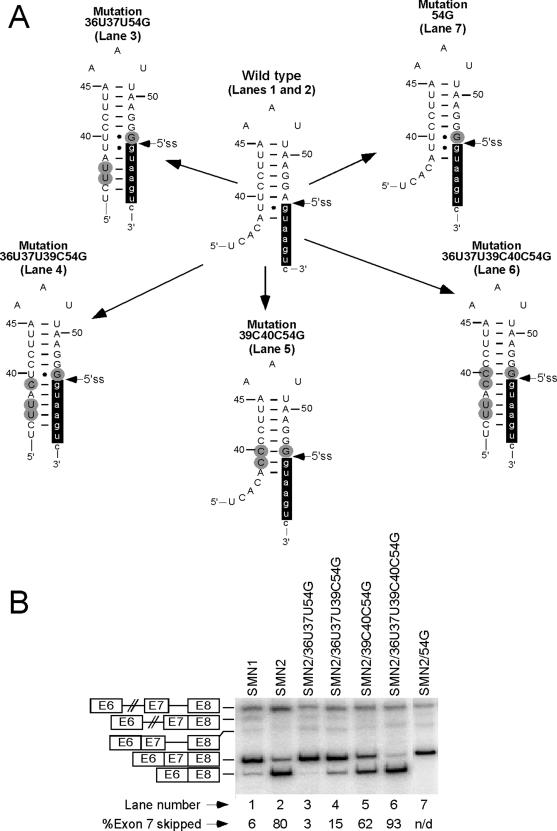
In the next set of experiments, we increased the number of base pairs formed between U1 snRNA and the 5′ ss of exon 7 by mutating the last position of exon 7 to G. We have earlier reported that 54G mutation strengthens the 5′ ss of exon 7 (53) causing complete restoration of exon 7 inclusion in SMN2 (Figure 4B, lane 7). As shown in Figure 4B, 54G was also able to overcome the negative effect of the stem strengthening even in combination with an inhibitory C6U mutation (note that the mutations were made in the SMN2 minigene). For example, in SMN2/36U37U39C54G that has an 11 bp-long stem (Figure 4A) exon 7 was predominantly included (Figure 4B, lane 4). Since 54G is predicted to forms a Wobble base pair with the wild-type 40U, substituting 40U with 40C will strengthen the stem (Figure 4A). Therefore, it was not surprising to see that the positive impact of 54G on exon 7 inclusion was reversed in the presence of 40C (Figure 4B, lanes 5 and 6). 40C mutation alone improved SMN2 exon 7 inclusion (Figure 6B, lane 6). The above results suggest that the inhibitory effect of TSL2 is directly proportional to its stability, which is affected by both, the number and the type of base pairs in the stem.
Figure 4.
Effect of 54G on splicing of exon 7 mutants with strengthened TSL2. (A) Effect of mutations on the predicted TSL2 structure. All mutations were made in SMN2. Numbers and letters in each mutation's name represent the positions and types of substitutions within exon 7. Numbering starts from the first position of exon 7. Capital and small-case letters indicate nucleotides of exon 7 and intron 7, respectively. Mutations, including 54G, are highlighted in gray. Nucleotides involved in base paring with U1 snRNA are highlighted in black. Canonical Watson–Crick base pairing and Wobble base pairing are indicated by continuous lines and filled circles, respectively. (B) In vivo splicing patterns of SMN2 mutants shown in (A). Spliced intermediates and products are the same as indicated in Figure 2B. N/D stands for not detectable.
Mutations that affect TSL2 stability modulate exon 7 splicing
To analyze the role of TSL2 in further detail, we generated a series of point mutations in both the stem and the loop regions of TSL2 using the SMN2 minigene: mutations in the stem rather than in the loop are predicted to destabilize the structure. We then tested the effect of these mutations on exon 7 splicing. A total of 23 out of 35 point mutations in the stem caused >10% improvement in inclusion of SMN2 exon 7 (Table 1). For some of these mutants the level of exon inclusion was comparable to the level observed for SMN1 exon 7. At the same time, 6 out of 12 mutations that did not improve exon 7 splicing caused even greater increase in exon skipping as compared to the wild-type SMN2 (Table 1), indicating that these mutations might have generated splicing silencers. For example, 50U substitution creates a UAGG motif associated with exon skipping (64–66). This substitution also creates a hexamer that is predicted to have a strong ESS activity (67). Accordingly SMN2/50U mutant showed no detectable amount of exon 7 inclusion (Table 1). A UAGG motif is also created by 51G producing a strong inhibitory effect on exon 7 splicing (Table 1). Further, 41U as well as 42U caused 10% increase in skipping of exon 7. Note that both mutations create the same motif, UCUU, which was found to be a core consensus sequence recognized by PBP/hnRNP I (68); however, in our case UCUU is not embedded in pyrimidine-rich context. PBP is known to act as a splicing repressor (69). Of note, when tested in the context of SMN1 exon 7, 41U as well as 42U produced almost 2-fold increase in exon 7 skipping (data not shown).
Table 1.
Effect of point mutations on SMN2 exon 7 skipping
Last 17 nt of exon 7 (capital letters) and first 2 nt of intron 7 (small-case letters) that form TSL2 are shown. Nucleotides that constitute the 5′ and 3′ side of the stem, as well as the loop, are marked. Mutations are highlighted in black. Percentage of exon 7 skipping is given for each mutant. Single mutations that improved exon 7 inclusion by >10% are highlighted in gray.
Substitutions in the loop region of TSL2 had the least effect on the splicing pattern of SMN2: only two out of nine mutations produced ∼10% improvement in inclusion of SMN2 exon 7 (Figure 5A, lanes 3 and 9). These mutations were 46C and 48C. Of note, for RNA loops of identical size, C residues produce a greater destabilizing effect than A and U residues (70). Another loop mutation, 48G, created a cryptic splice site A/gtaagg as confirmed by sequencing of a shorter band, which corresponded to the 47 nt-long exon 7-included product. In the SMN2 context, both the cryptic and the native splice sites produced exon 7-skipped transcript as a major product of splicing (Figure 5A, lane 10).
Figure 5.
Effect of loop mutations on exon 7 splicing. (A) In vivo splicing pattern of SMN2 mutants harboring point mutations within the loop portion of TSL2. Numbers and letters represent the positions and the types of substitution within exon 7. Splicing products are the same as in Figure 2B. ‘Cry’ refers to the cryptic splice site. (B) In vivo splicing pattern of SMN2 mutants in which the wild-type triloop was substituted with longer loops. Loop sequences are shown in the upper panel, where capital and small-case letters indicate nucleotides in the 5′-/3′-portion of the stem and the loop region, respectively. W.T. stands for wild-type. Splicing products are the same as in Figure 2B. (C) In vivo splicing pattern of SMN1 mutants harboring stable tetraloops. Splicing products are the same as in Figure 2B. Capital letters indicate loop sequnces, while small-case letters indicate nucleotides that form a closing base pair.
It has been shown that regardless of loop sequence, smaller loops are more stable than loops composed of 7 or 9 nt (70). To test how the increase in loop size might affect SMN2 exon 7 splicing, we generated a series of minigenes in which the AAU triloop was replaced by randomly selected 7 nt-long sequences. In vivo splicing patterns for these constructs are shown in Figure 5B (lanes 14–18, 20). In addition, we created YW-15 mutant in which the stem was shortened by 1 bp and the AAU triloop was substituted with the 10 nt-long sequence, AUACCAGCUA (Figure 5B). These changes would substantially weaken the structure. As shown in Figure 5B, in all cases destabilization of the structure was accompanied by marked improvement in SMN2 exon 7 inclusion. The varying degree of improvement can be explained by differences in loop sequences. For example, some of the sequences can display increased flexibility that would compensate for the loop size. In addition, it has been shown that for hairpin loops larger than three, stability also depends on the interaction of the first mismatch with the closing base pair and the type of the first mismatch within a loop (71). We also cannot exclude the possibility that some of the sequences used create binding sites for positive regulators of splicing.
It has been established that certain tetraloops can increase hairpin stability by providing an additional gain in free energy due to specific stacking and hydrogen bond interactions in the loop (72). Unusually stable tetraloops are generally represented by GNRA or UNCG sequence motifs, where N = A, C, G or U; and R = A or G (73). Interestingly, these tetraloops are abundant in natural RNAs and are well conserved in hairpins of rRNAs from different species. Another conserved tetraloop, CUUG, was also shown to be thermodynamically very stable (74). We decided to test whether TSL2 stabilization by any of the above-mentioned tetraloops will promote skipping of exon 7. For this purpose we generated SMN1 minigenes in which the AAU triloop was substituted with GAAA (GRNA family), UUCG (UNCG family) and CUUG tetraloops, thermodynamic stability and structure of which are well studied (74–77). The in vivo splicing results for these minigenes are shown in Figure 5C. As expected, these stable tetraloops promoted skipping of SMN1 exon 7 (Figure 5C, lanes 23, 24 and 26). Note that in the above constructs tetraloops have A-U as a closing base pair. Phylogenetic analysis revealed that UUCG loop is almost always closed by a C-G base pair, while CUUG has a bias for a G-C base pair (77). GRNA loops appear to have no preference for a closing base pair (77). Indeed, in their study Antao and Tinoco confirmed that the hairpins with the loop sequence GAAA closed by A-U were extra stable at least among other tetraloops closed by this base pair (76). To our knowledge there is no study addressing the stability of UUCG loop with A-U as a closing base pair. However, it has been reported that C-G makes a substantial contribution to the stability of the UUCG tetraloop since changing the UUCG loop-closing base pair from C-G to G-C resulted in significant decrease of the stability (76). In line with these results, combining the UUCG tetraloop and C-G as a closing base pair resulted in marked increase in SMN1 exon 7 skipping (Figure 5C, compare lanes 24 and 25). It should be noted that a C-G closing base pair alone (45C49G mutation) did not affect skipping of SMN1 exon 7 (data not shown). Similarly, combining the CUUG tetraloop with C-G or G-C as a closing base pair caused an additional increase in SMN1 exon 7 skipping (Figure 5C, compare lanes 27 and 28 with lane 26). Interestingly, a G-C closing base pair alone (45G49C mutation) caused restoration of exon 7 inclusion even in SMN2 (Figure 6B, lane 30).
Figure 6.
Effect of site-specific mutations on SMN2 exon 7 splicing. (A) Schematic representation of the predicted TSL2. Capital and small-case letters indicate nucleotides of exon 7 and intron 7, respectively. Numbering starts from the first position of exon 7. Nucleotides involved in base paring with U1 snRNA are highlighted in black. Mutations at indicated positions and their effect on structure are shown. Numbers and letters represent the positions and the types of substitution within exon 7. The lack of base pairing, canonical Watson–Crick base pairing and Wobble base pairing are indicated by dots, continuous lines and filled circles, respectively. (B) In vivo splicing pattern of SMN2 mutants harboring mutations shown in (A). Splicing products are the same as in Figure 2B. N/D stands for not detectable.
We also analyzed for the presence of ESE/ESS elements inadvertently created or disrupted by the described mutations using ESEfinder (63) and RESCUE-ESE (78) programs and aligning sequences of interest with 176 hexamers with ESS activity (67). We could not find a direct correlation between predicted ESE/ESS and our splicing results. For example, the 42G mutation was predicted to create an ESS [UUCGUU; (67)] but, in fact, it improved SMN2 exon 7 inclusion (Table 1 and Figure 6B, lane 15). Similarly, tetraloop sequences CUUG, c(CUUG)g, g(CUUG)c, UUCG and c(UUCG)g, where c and g represent nucleotides of a closing base pair, were predicted to create ESEs (63,78), but instead they promoted exon skipping. The fact that multiple point mutations that were predicted to destabilize TSL2 improved exon 7 inclusion, while TSL2 stabilization caused skipping of exon 7 even in SMN1 suggests that RNA structure is an important factor and an integral component of the regulatory network that controls SMN exon 7 splicing.
Compensatory mutations confirm the inhibitory role of TSL2
Since compensatory mutations provide the best supportive evidence for the role of structure, we decided to perform ‘rescue’ experiments. For this purpose we generated SMN2 double mutants in which substitutions on the complementary side of the stem were combined together to restore base pairing. We then tested whether this restoration of structure was able to ‘rescue’ skipping of exon 7. The results of in vivo splicing of such double mutants are shown in Figure 6B. Different point mutations at positions 40 and 54 caused improvements in SMN2 exon 7 inclusion (Table 1). Combining 40G and 54C in SMN2/40G54C mutant led to complete skipping of the exon (Figure 6B, lane 5). Note that in this mutant the structure becomes more stable since a U-A base pair is replaced by a G-C base pair (Figure 6A). Importantly, the restored base pair is located directly adjacent to the sequestered 5′ ss and would have a stabilizing effect on the neighboring 39U-55G Wobble base pair as well. On the other hand, SMN2 mutant generated by combining 40C and 54G showed no ‘rescue’ of exon 7 skipping (Figure 6B, lane 8). This was not totally unexpected, since as mentioned earlier, 54G strengthens the 5′ ss of exon 7, improving U1 snRNP recruitment. It appears that 40C–54G base pair becomes inhibitory only in the context of strengthened stem (see Figure 4, mutants SMN2/39C40C54G and SMN2/36U37U39C40C54G).
Rescue of exon 7 skipping was also observed for the mutant in which 51C was combined with 43G (Figure 6B, lane 20). Note that each mutation alone significantly restored inclusion of SMN2 exon 7 (Figure 6B, lanes 18 and 19). In contrast, combining 51C with 42G (both nucleotides belong to different base pairs in the stem) in SMN2/42G51C mutant produced no effect on skipping of exon 7 (Figure 6B, lane 21).
We obtained interesting results for the double mutation that restored the closing base pair for the predicted AAU triloop (positions 45 and 49). As shown in Figure 6B, single mutations at either side of this base pair promoted inclusion of exon 7 in SMN2. However, when these single mutations were combined to restore base pairing, splicing of exon 7 was affected differently. In SMN2/45G49C mutant with G-C as the closing base pair, the level of exon 7 inclusion remained high (Figure 6B, lane 30). At the same time, in SMN2/45C49G mutant that had C-G as the closing base pair the inclusion of exon 7 decreased noticeably (Figure 6B, lane 33). The observed differences could be attributed to several factors. For example, triloops with a C-G closing base pair are known to be the most stable (79). As far as the double mutation 45G49C is concerned, we hypothesize that a splicing enhancer that promoted inclusion of exon 7 might have been created.
Contrary to restoration of base pairing between positions 45 and 49, where the ‘rescue’ effect appeared to depend on whether it was C-G or G-C, restoration of base pairing between positions 42 and 52 failed to rescue exon 7 skipping in all mutants created (Figure 6B, lanes 14 and 17). Note that single mutations at either position improved SMN2 exon 7 inclusion (Figure 6B, lanes 12, 13, 15 and 16). In an attempt to explain these results we considered several possibilities. First, the 42A–52U base pair was not able to compensate for a more stable wild-type 42C–52G base pair. Second, point mutations at positions 42 and 52 may have created ESEs. Indeed, according to ESEfinder, both 52C and 52U are predicted to create ESE associated with SRp55. Either or both of these factors may have contributed towards the inability to produce the desired rescue effect in SMN2/42A52U and SMN2/42C52G. However, it should be mentioned that the 43G51C double mutation was able to regain skipping of exon 7 in SMN2 despite the fact that 51C just like 52C or 52U is predicted to create ESE associated with SRp55 with an even higher matrice score (data not shown).
As shown in Figure 6B, single mutations at position 44 (44G and 44C) but not at position 50 (50C and 50G) promoted inclusion of SMN2 exon 7 (Figure 6B, lanes 22, 23 and 25, 26). Since all but 50G mutations were expected to disrupt the stem, the reason for such a variable effect on splicing is unclear. Interestingly, restoration of base pairing between positions 44 and 50 in SMN2/44G50C and SMN2/44C50G was accompanied by a decrease in exon 7 inclusion (Figure 6B, lanes 24 and 27). The restoration of base pairing was particularly inhibitory in case of the latter mutant. Since positions 44 and 50 are separated by only 5 nt, we cannot rule out the possibility that these double mutations created ESSs. However, none of the generated decamer motifs matches ESSs reported in (67).
Finally, combining 41G and 53C substitutions together increased skipping of exon 7 as compared to the wild-type SMN2 minigene (Figure 6B, lanes 11 and 2, respectively), even though each substitution alone did not affect inclusion of SMN2 exon 7 (Figure 6B, lanes 9 and 10). Since 41G and 53C are separated by eleven nucleotides, the effect of double mutation cannot be explained by creation of ESSs, which are generally considered to be 6 nt-long motifs (67).
RNA structure probing confirms the presence of TSL2
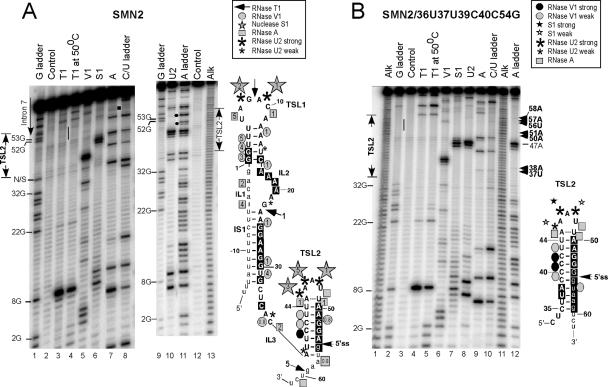
TSL2 is one of the possible structures predicted by mfold for SMN exon 7 (62). To investigate whether RNA sequence at the junction of exon 7 and intron 7 folds into TSL2, we performed RNA structure probing. In these experiments, 5′ end-labeled RNA molecules containing the entire exon 7 and adjacent intronic sequences were subjected to partial digestion with RNases specific for single and double-stranded regions. Single-stranded regions were probed by RNase T1, which cleaves after unpaired G residues; RNase U2, which cleaves after unpaired A and to some extent G residues; and RNase A, which cleaves after pyrimidines in non-base paired regions. We also employed nuclease S1 that cleaves indiscriminately within single-stranded regions. Double-stranded and stacked regions in RNA were identified by RNase V1 cleavage. It should be noted that nuclease S1 and RNase V1 generate 5′-fragments with 3′-hydroxyl groups. Therefore, S1 and V1 bands migrate slower than the corresponding sequencing bands. We began our probing experiments with RNA transcripts that contained the entire exon 7, 34 nt-long upstream sequence of intron 6 and 104 nt-long downstream sequence of intron 7. To decrease tertiary interactions that might complicate interpretation of structure probing results, we subsequently switched to shorter transcripts, in which the lengths of intron 6 and intron 7 sequences were decreased to 18 and 12 nt, respectively. A representative example of structure probing results for the entire SMN2 exon 7 (short transcript) is shown in Figure 7A. First, comparison of the digestion patterns generated by RNase T1 under denaturing (G ladder) and native conditions revealed that in folded RNA (native conditions), the majority of G residues were involved in secondary structure formation, as indicated by the lack of cleavage bands in comparison to a G ladder (Figure 7A, lanes 1 and 3). In particular, we observed the lack of bands corresponding to 52G, 53G and 55G residues that are involved in the formation of the stem in TSL2. The only strong T1 cleavage site corresponds to 8G, which falls within the predicted loop of TSL1 (Figure 7A, right panel). Since enzymatic digestion was performed under conditions when no more than one cut per molecule was produced, we interpreted weak T1 cleavages, such as 22G, 59G and 63G, as a consequence of occasional opening of base pairs, long enough to be trapped by the enzyme. The observation that these weak bands become stronger (relative to 8G) with the increase in T1 digestion temperature (50°C), when the least stable structural elements are expected to melt, confirms this ‘transient opening’ hypothesis. Note that the increase in temperature produced no changes in T1 digestion pattern (Figure 7A, lane 4).
Figure 7.
RNA secondary structure of exon 7 probed by partial enzymatic cleavage. (A) RNA structure probing of wild-type SMN2 exon 7. The 5′ end-labeled RNA was subjected to partial cleavage using the indicated enzymes followed by separation on denaturing 8% polyacrylamide gels. An alkaline hydrolysis ladder (alk) is included together with a partial RNase T1 (G ladder), RNase A (C/U ladder) and RNase U2 (A ladder) digestions under denaturing conditions. Positions corresponding to intron 7 and TSL2 are indicated. A bar indicates the lack of RNase T1 cuts at 52G, 53G and 55G. Filled circles indicate the lack of RNase U2 cuts at positions 50A, 51A and 54A. A square indicates 60U residue. Structure probing results were superimposed on the mfold predicted secondary structure of exon 7 shown on the right. Nucleotide numbers and the 5′ ss are marked. Numbering starts from the first position of exon 7. Structural elements are indicated as in (62). Employed symbols are shown in the insert. Relative susceptibilities of sites to nuclease S1, RNase T1, V1 and A are indicated by numbers. These numbers were generated by quantifying cleavage profiles and assigning an intensity of five to the highest peak; the remaining peaks were scaled accordingly. Since the intensities of 8G and 40U band (in T1 and V1 digestion, respectively) were very high compared to other bands, these sites were not included in quantifications. The bands located right next to 40U were obscured and, therefore, their intensity could not be quantified. Nucleotides protected from RNAses T1, A and U2 are shaded in black. A residue was considered to be protected if the corresponding band was present in denatured but absent or substantially weaker in folded RNA. All quantifications were corrected for the background represented by a control lane. N/S stands for non-specific. (B) RNA secondary structure probing of mutant SMN2/36U37U39C40C54G. RNA preparation and labeling details are the same as in (A). TSL2 region is indicated. Structure probing results were superimposed on the predicted TSL2 structure shown on the right. Employed symbols are shown in the insert. Nucleotides protected from RNAses T1, RNase A and RNase U2 are shaded in black. A residue was considered to be protected if the corresponding band was present in denatured but absent or substantially weaker in folded RNA.
As shown in Figure 7A, bands corresponding to RNase V1 cleavage sites were scattered along the RNA sequence and had different intensities (lane 5). V1 pattern of digestion is in a good accord with the results of cleavage produced by the single-strand specific nucleases with only a few, if any, overlapping bands. The strongest RNase V1 cleavage band corresponded to position 40 (Figure 7A, lane 5) confirming the presence of a stem in this region. 40U cleavage was followed by weaker cuts corresponding to positions 41C and 42C. This difference in cleavage intensity may suggest that the helix is not uniform in structure. The V1 cut on the opposite side (51A) of the stem was also weak. Such asymmetry in cleavage indicates that the enzyme might be sterically blocked by RNA folding or that the 5′ side of the stem is more exposed in solution.
Partial digestion with single-strand specific nuclease S1 gave rise to a series of strong cuts in the regions comprised of positions 7–9 and 45–47 (Figure 7A, lane 6). This pattern of S1 cleavage fits the predicted structure of exon 7 since both regions are located within the loops. Positions 46 and 47 were of particular interest because they are located within the loop portion of TSL2.
Next we compared the digestion patterns generated by RNase U2 under native (folded RNA) and denaturing (A ladder) conditions (Figure 7A, lanes 10 and 11). In folded RNA, adenosine residues in the central portion of the exon were protected from U2 cleavage. Also, note the absence of the bands corresponding to 50A, 51A and 54A (Figure 7A, lanes 10, marked by filled circles). This result is in line with the existence of a stem in this area. Three exonic regions that showed sensitivity to RNase U2 were comprised of positions 7 and 9, positions 36 and 38, and positions 45, 46 and 47 (Figure 7A, lanes 10). Interestingly, RNase U2 as well as nuclease S1 cleaved after 45A, which was predicted to form the closing base pair of the loop portion of TSL2. This cleavage would indicate little or no pairing between 45A and 49U. These results are expected for a closing base pair that is in transition between a stem and a loop.
The lack of base pairing between 45A and 49U or its instability was further confirmed by the results of structure probing with RNase A. Under native conditions the enzyme cleaved after 44U and 49U (Figure 7A, lane 7). At the same time, as per quantitative analysis, the intensity of these cuts was five times weaker than the 6U band that is located in the predicted loop. It should be mentioned that RNase A cleaved mostly after U and C residues that were followed by As, as judged by the results of RNase A digestion under denaturing conditions (C/U ladder, Figure 7A, lane 8). Indeed, it has been determined that the residue located 3′ to C and U affects the kcat of RNase A by a factor of >100 between CpA and CpU (80). Therefore, we could not determine structure status of pyrimidines followed by a residue other than A. Interestingly, RNase A cleaved after 60U under native but not denaturing conditions, indicating that this residue is highly exposed to the enzyme in the folded RNA (Figure 7A, lane 7, marked by a square).
Probing data were subsequently superimposed on the predicted exon 7 structure (Figure 7A, right panel), which is composed of two terminal stem–loops, including TSL2, an internal stem IS1 and three internal loops IL1, IL2 and IL3 (62). Here we attempted to distinguish between relative susceptibility of sites to probing enzymes, indicated by numbers. Quantitative results for the intron 6 portion were obtained from the gels, which were run for a shorter period of time. It appears that our experimental data in general support the validity of the predicted structure, including TSL2. A few exceptions from the predicted structure could be explained by bulge and loop sliding that are not uncommon. As shown in Figure 7A, the loops of TSL1 and TSL2 were highly sensitive to single-strand specific nucleases, while double-strand specific RNase V1 made cuts within the predicted stems. It is unclear whether the triloop closing base pair 45A–49U is formed. Also, RNase A cleaved at 44U, yet the 50A residue on the complementary side of the stem was protected from cleavage by RNase U2. Further, RNases U2 and A cut at 38A and 56U, respectively. These nucleotides are predicted to form a base pair at the end of the stem. Based on quantitative analysis, both cleavages were weak (Figure 7A). Therefore, it is possible that the base pair 38A–56U is transiently open/unstructured. Future chemical probing and NMR studies will add refinement to the structure of exon 7, in particular TSL2.
Enzymatic probing confirms structural changes in TSL2 mutants
Site-directed mutagenesis showed that substitutions that disrupted the stem of TSL2 improved inclusion of SMN2 exon 7, while substitutions that strengthened the structure caused an increase in exon 7 skipping. To confirm that these substitutions were indeed accompanied by structural changes, we performed enzymatic probing of secondary structure for a number of TSL2 mutants. We started with mutants that had the strengthened stem. A representative example of structure probing results for SMN2/36U37U39C40C54G is shown in Figure 7B. Even though we are interested in TSL2 area, probing results for the entire exon 7 are presented to confirm that the mutations within TSL2 did not affect the structure of other regions. In SMN2/36U37U39C40C54G, substitutions are predicted to increase the length of the stem from 8 to 11 bp. The existence of this extremely stable structure with four consecutive C-G base pairs in the middle was confirmed by the absence of T1 and U2 cleavages in the stem region even under denaturing conditions: 50°C and 7 M urea (Figure 7B, lanes 2 and 12). Note the lack of bands corresponding to 52G, 53G, 54G and 55G in the G ladder and the absence of bands corresponding to 38A, 50A, 51A, 57A and 58A in the A ladder (Figure 7B, lanes 2 and 12, indicated by a bar and arrowheads, respectively). We also observed that the band corresponding to 37U was extremely weak and the band corresponding to 56U was missing from the digestion patterns generated by RNase A under native and denaturing conditions (Figure 7B, lanes 9 and 10). At the same time, under native conditions the loop portion of TSL2 remained highly accessible to single-strand specific nucleases S1 and U2 (Figure 7B, lanes 7 and 8). The increase in stem size was indicated by changes in V1 cleavage pattern as compared to the wild-type RNA. Note two cuts at the 5′ side of the stem at positions 40C and 41C and a weaker cut at the 3′ side of the stem at position 56U in SMN2/36U37U39C40C54G mutant versus one strong cut at position 40U and a weak cut at position 51A in the wild-type RNA (Figure 7B, lane 6 and Figure 7A, lane 5, respectively). The results of structure probing were subsequently superimposed on the predicted TSL2 structure of SMN2/36U37U39C40C54G. As shown in Figure 7B, this structure was consistent with our enzymatic probing data.
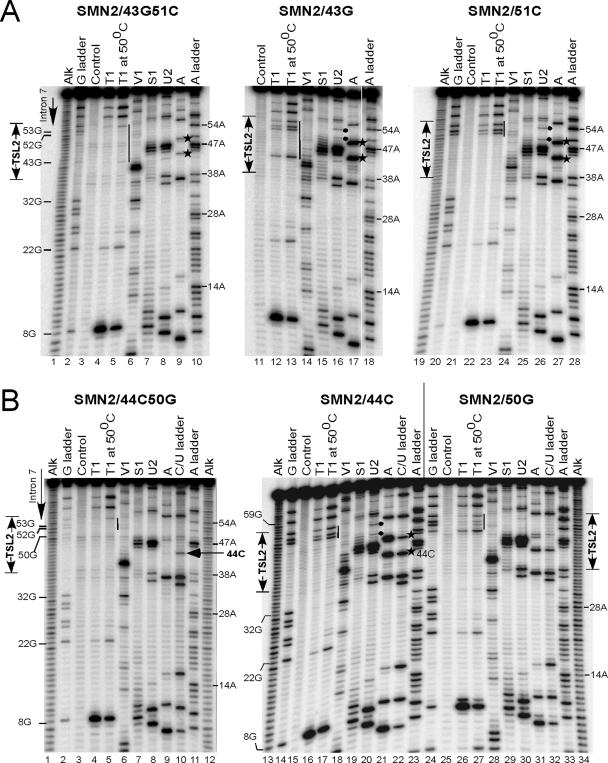
Next we probed the secondary structure of SMN2/43G, SMN2/51C and SMN2/43G51C (Figure 8A). Here, 43G and 51C substitutions are predicted to destabilize TSL2, while 43G51C is expected to reinstate the structure (Figure 6A). Accordingly, 43G and 51C caused improvement in SMN2 exon 7 inclusion, whereas 43G51C restored skipping of this exon (Figure 6B). As shown in Figure 8A, mutation at position 43 (43G) was accompanied by an increase in sensitivity to single-strand specific nucleases S1, U2 and A in the top portion of TSL2 (lanes 15–17). For example, there was a substantial increase in the intensity of bands corresponding to RNase A cleavage at positions 44 and 49 in SMN2/43G (Figure 8A, lane 17, indicated by stars) versus wild-type RNA (Figure 7A, lane 7). In SMN2/43G51C the intensity of these bands substantially decreased (Figure 8A, lane 9). Quantitative analysis confirmed this observation. The sensitivity to RNase A remained unchanged at positions located outside of TSL2. The appearance of bands, albeit weak, corresponding to RNase U2 cuts at 50A, 51A and 54A was consistent with opening/destabilization of TSL2 (Figure 8A, lane 16, indicated by filled circles). These bands were absent in the U2 cleavage pattern generated for wild-type RNA (Figure 7A, lane 10), while 50A and 54A bands were missing in the U2 cleavage pattern generated for SMN2/43G51C RNA (Figure 8A, lane 8). Destabilization of TSL2 structure in SMN2/43G RNA was also indicated by changes in T1 cleavage pattern as compared to the wild-type and SMN2/43G51C mutant. The difference became especially noticeable with the increase in cleavage temperature (50°C). Note the appearance of T1 cuts at positions 43, 52, 53 and 55G in SMN2/43G RNA (Figure 8A, lanes 13). In SMN2/43G51C RNA the intensity of cuts at 43G and 55G decreases dramatically, while bands corresponding to 52G and 53G disappear (Figure 8A, lane 5). Despite the destabilizing effect of 43G on TSL2 stem, C-G base pairing in the central part of the stem appears to be preserved, as judged by the strong V1 cleavage at 40U (Figure 8A, lane 14). There were multiple weaker cuts at positions located 5′ and 3′ to 40U, which overlapped with the cleavages produced by single-strand specific nuclease S1 (Figure 8A, lane 15). Two possibilities can account for susceptibility of this region to both double- and single-strand specific enzymes: (i) the region in question was only marginally stable and was easy to denature, (ii) multiple secondary structures coexisted in equilibrium. We also probed the structure of SMN2/51C, which showed complete restoration in exon 7 inclusion. As shown in Figure 8A, patterns of enzymatic cleavage for SMN2/51C were very similar to SMN2/43G construct (lanes 25–27 and 15–17, respectively). In both cases the top portion of TSL2 region showed increased sensitivity to single-strand specific nucleases. On the other hand, combining 43G and 51C substitutions in SMN2/43G51C reduced the accessibility of TSL2 to single-strand specific nucleases back to the levels observed for the wild-type SMN2 (Figure 8A, lanes 7–9). Also note that 43G residue was poorly accessible to T1 even under denaturing conditions, indicating the presence of a strong secondary structure (Figure 8A, lane 2). Also, multiple overlapping V1 and S1 cuts in TSL2 region disappeared (Figure 8A, lanes 6 and 7). As expected, 44C that restored SMN2 exon 7 inclusion, produced a destabilizing effect on TSL2 as well. For example, we observed increased cleavages with S1, U2 and A nucleases in the region surrounding 44C (Figure 8B, lanes 19, 20 and 21, respectively) and the appearance of T1 cuts at 52G, 53G and 55G, their intensity noticeably increased at 50°C (Figure 8B, lane 17, indicated by a bar). Interestingly, the effect of 50G, a single mutation on the complementary side of the stem, appeared to be less destabilizing than 44C, 43G and 51C. For example, 52G, 53G and 55G residues remained poorly accessible to T1 digestion even at 50°C (Figure 8B, lane 27). It is possible that 50G can form a Wobble base pair with 44U. Accordingly the 50G mutation produced <10% improvement in SMN2 exon 7 inclusion (Figure 6B). Combining 44C and 50G substitutions is predicted to restore base pairing formed within the stem portion of TSL2. Indeed, a 3-fold decrease in RNase A cleavage at position 44C in folded versus denatured RNA confirms this prediction (Figure 8B, lanes 9 and 10, marked by an arrow). Extremely poor RNase A cleavage at 49U in SMN2/44C50G RNA (Figure 8B, lane 10) is probably due to A50G mutation since, as mentioned earlier, the enzyme preferentially cuts after unpaired pyrimidines followed by As. Restoration of the structure was also indicated by the decrease in intensity of S1 and U2 cuts in the loop region at positions 46 and 47 (Figure 8B, lanes 7 and 8). Moreover, G residues at positions 50, 52, 53 and 55 and A residues at positions 51 and 54 were protected from RNase T1 and U2 cleavages (Figure 8B, lane 4 and 8). In summary, the above results confirm the expected structural changes in TSL2 region of SMN2 mutants.
Figure 8.
Analysis of TSL2 secondary structure in SMN2 mutants. (A) RNA secondary structure probing for mutants SMN2/43G51C, SMN2/43G and SMN2/51C. Note that single substitutions are predicted to destabilize TSL2 stem, while double substitutions are predicted to restore the stem. The 5′ end-labeled RNAs were subjected to partial cleavage using the indicated enzymes followed by separation on denaturing 8% polyacrylamide gels. An alkaline hydrolysis ladder (alk) as well as RNase T1 (G ladder), RNase A (C/U ladder) and RNase U2 (A ladder) digestion patterns under denaturing conditions are included. TSL2 region is indicated. Solid bars represent the absence or presence of T1 cleavage at G residues located in TSL2 area. Stars indicate RNase A cleavage at 44U and 49U residues. Filled circles mark RNase U2 cuts at 52A and 54A. (B) RNA secondary structure of mutants SMN2/44C50G, SMN2/44C and SMN2/50G. RNA preparation and other details are the same as in (A). The arrow marks RNase A cut that corresponds to 44C residue.
DISCUSSION
Regulation of SMN exon 7 splicing attracted attention due to the realization that SMA patients that lack SMN1 always carry at least one copy of SMN2; therefore, redirecting SMN2 exon 7 splicing represents a potential SMA therapy. Manipulation of splicing has been proposed for a number of diseases, including Duchenne muscular dystrophy, beta-thalassemia and certain cancers (81). Although many cis-elements and transacting factors are known to modulate SMN exon 7 splicing, the role of local RNA structure in regulation of exon 7 splicing has not been studied. Our interest in a potential role of RNA structure began with the results of in vivo selection that identified a number of highly mutable (inhibitory for exon 7 inclusion) residues that overlap the predicted stem–loop structure, TSL2 [(62) and Figure 1]. To demonstrate that the presence of TSL2 negatively affects exon 7 inclusion we used extended mutational analyses combined with enzymatic structure probing.
Recently, we have shown that a weak 5′ ss aids in SMN2 exon 7 skipping (53). Accordingly, improvement of the 5′ ss by 54G mutation or by deletion of the inhibitory element (ISS-N1) located in the vicinity of the 5′ ss promoted exon 7 inclusion in SMN2 (53,60). Interestingly, as mentioned in the recent review by Baralle and co-authors, several computational programs failed to detect a major improvement in the strength of the 5′ ss caused by 54G mutation indicating the importance of the ‘local context’ (82). Our results show that RNA structure (TSL2) provides this local context and is a new addition to the existing repertoire of cis-elements that contribute toward the weak 5′ ss of exon 7. Based on its location, size and base composition, TSL2 presents a unique structure locked within a short exon. Its triloop and 8 bp-long stem are formed almost exclusively by exonic sequences; only two intronic residues are involved in TSL2 folding (Figure 1). In contrast, a well studied stem–loop structure at the junction of exon 10 and intron 10 of tau contains a 6 nt loop entirely comprised of intronic sequences, and a bulged stem >70% of which are formed by intronic nucleotides (27). Compared to the tau stem with its high GC content, TSL2, in which the number of GC base pairs is only 25%, appears to be relatively weak.
Crucial evidence of the inhibitory function of TSL2 came from the observation that compensatory mutations ‘rescued’ skipping of exon 7 by restoring the predicted stem. The most convincing among these were 40G and 54C, separated from each other by thirteen nucleotides. When these two substitutions were combined, the effect of the compensatory mutation was fully realized (Figure 6). This result cannot be explained based on the concept of traditional cis-elements that are generally comprised of short sequences (63,67,78). An RNA structure, on the other hand, would provide the most plausible interpretation for the effect of the long-distance compensatory mutations, such as 40G54C, on exon 7 splicing. In fact, a single compensatory mutation involving long-distance interactions is sufficient to implicate the role of RNA structure (83).
To demonstrate the inhibitory impact of TSL2 on SMN exon 7 splicing we were confronted with the usual challenge of delineating the role of structure from the role of overlapping cis-elements. For example, the top portion of the stem and the entire loop of TSL2 coincide with 3′-Cluster, an inhibitory cis-element predicted by the results of in vivo selection of the entire exon (53). It is possible that several mutations in the region of TSL2 improved SMN2 exon 7 inclusion due to both, abrogations of 3′-Cluster and disruption of TSL2. This is illustrated by comparing the effect of 51C and 43G mutations located on the complementary sides of the TSL2 stem. The former mutation fully restored exon 7 inclusion in SMN2, whereas the latter one had only partial stimulatory effect (Figure 6). According to our structure probing results, both mutations have similar structure-destabilizing consequences (Figure 8). However, in contrast to position 43, position 51 is also the part of 3′-Cluster. Therefore, 51C mutation has an additional advantage of disrupting this negative cis-element. Our results imply that a potential transacting factor that recognizes and binds to 3′-Cluster may require both, nucleotide sequence and RNA secondary structure.
The inhibitory impact of the distal portion of TSL2 may be distinct from its proximal part that partially sequesters the 5′ ss of exon 7. Strengthening of the TSL2 stem by three additional base pairs, which results in increase of the sequestered portion of the 5′ ss from two to five nucleotides, causes complete skipping of exon 7 even in SMN1 (Figure 2). Of note, we believe the inhibitory effect of a stem–loop structure that sequesters the 5′ ss could not be over-generalized. For example, a 9 bp-long stem that sequestered the entire 5′ ss produced no in vivo effect on donor site selection in the reporter construct containing RP51A intron inserted in the lacZ gene (84). A 15 bp-long stem was necessary to achieve complete inhibition of splicing in the above system (84). In contrast, an 8 bp-long stem in TSL2 of SMN1/39C resulted in greater than 80% exon skipping (Figure 2). Interestingly, the inhibitory effect of TSL2 in SMN1/39C was gained due to a single substitution that changed a Wobble base pair to a stronger Watson–Crick base pair, causing stem strengthening without changing its size. This result highlights that the impact of RNA structure is not merely a matter of stem size but also stem composition. Further, presence of other negative cis-elements in the vicinity of TSL2 may contribute to the inhibitory role of this structure. For example, it is possible that transacting factors that interact with Exinct (52) and/or ISS-N1 (60) make secondary contacts with TSL2. In this case, the role of TSL2 goes beyond sequestering the 5′ ss of exon 7.
It has been shown that loop size rather than loop composition contributes to stability of a hairpin (70,77). Consistent with this observation, only two out of nine mutations in the loop caused some improvement in SMN2 exon 7 inclusions. However, when the TSL2 triloop was substituted with several known stable tetraloops, exon 7 skipping was promoted even in SMN1 (Figure 5). In this regard it should be noted that the above tetraloops would also disrupt 3′-Cluster. We believe that the created tetraloop hairpins form a rigid structure that is stable enough to cause skipping of exon 7.
The inhibitory effect of a relatively long stem in TSL2 correlated with the small size of an RNA duplex formed between the 5′ ss of exon 7 and U1 snRNA. Increasing the size of the 5′ ss:U1 duplex from 6 to 11 bp restored exon 7 inclusion even in mutants that had an 11 bp-long stem (Figure 3). Furthermore, a single intronic mutation at position 61 that extended the size of 5′ ss:U1 duplex from six to eight base pairs completely restored SMN2 exon 7 inclusion in the context of wild-type TSL2 (data not shown). In contrast, a 7 bp-long duplex formed between the 5′ ss of tau exon 10 and U1 snRNA did not overcome the inhibitory impact of a larger stem that sequesters this 5′ ss (27). These results further support the exon-specific relationship between the size of the 5′ ss:U1 duplex and the length of the stem that sequesters the 5′ ss of exon 7.
Our previous study indicated that 54G restores exon 7 inclusion in SMN2 (53). Here we show that 54G was able to restore inclusion of exon 7 in mutants that combined inhibitory C6U and strengthened TSL2. Several possibilities could account for such a strong positive impact of 54G on exon 7 inclusion. First, 54G improves complementarity between the 5′ ss of exon 7 and U1 snRNA. Second, it decreases the stability of TSL2 stem by changing a canonical (Watson–Crick) base pair to a Wobble base pair. In addition, 54G may help recruit U1C protein, which prefers a G residue at the last position of the exon (85). Importantly, 54G creates a 4 nt G run GGG/gu at the exon7/intron 7 junction. Experimental results accumulated by now indicate that G repeats play an important role in splice site recognition. Their effect on splicing appears to be position dependent. For example, it has been suggested that intronic G repeats increase the content of information used by the spliceosome to distinguish between authentic and cryptic splice sites (86). It has also been shown that intronic G triplets located immediately downstream of 5′ ss directly interact with U1 snRNA facilitating splice site recognition and functioning as ISE (87). On the other hand, G quadruplets adjacent to the 5′ ss have been shown to promote silencing of a brain-region-specific exon 19 of the GRIN1 gene (65) and inhibit the splicing of intron 3 in DQB1 reporter construct (88). Interestingly, the GGG/gu sequence similar to the one created by 54G was found to be inhibitory for NF1 exon 3 inclusion (89). This differential effect of GGG/gu on splicing stresses the importance of ‘local context’ and highlights the differences in the regulation of splicing at the level of individual exons. The stimulatory effect of 54G on splicing of SMN2 mutants with strengthened TSL2 was fully neutralized by an 11 bp stem with four consecutive C-G pairs located in the middle of this structure. Weakening of this strong stem by a single substitution that changed a Watson–Crick base pair to a Wobble base pair restored exon 7 inclusion in SMN2 (Figure 4).
Interestingly, 48G mutation created a cryptic splice site A/gtaagg (Figure 5). It is located only seven nucleotides upstream of the wild-type 5′ ss. Despite limited complementarity with U1 snRNA (only 5 bp), this cryptic 5′ ss was not discriminated in favor of the wild-type 5′ ss as judged by the amount of exon 7-included products generated from both splice sites. Due to the fact that both cryptic and wild-type donor sites were partially sequestered in TSL2, none was efficiently recognized. Once again, our results highlight the difficulty in evaluating the role of RNA structure using substitution mutations. In addition to creating and/or abrogating cis-elements, substitution mutations are also capable of creating cryptic splice sites. It is worth mentioning that a cryptic splice site is also created when the wild-type triloop is substituted with the tetraloop sequences UUCG or CUUG. However, as indicated by the results in Figure 5, this cryptic splice site is not activated in ‘tetraloop’ constructs. This is in line with our hypothesis that UUCG and CUUG stabilize the structure and as a consequence suppress the usage of the cryptic splice site.
As final demonstration of the role of RNA structure in SMN exon 7 splicing, we confirmed the existence of TSL2 by enzymatic structure probing. Our probing results were able to capture structural changes caused by mutations that either destabilized or reinstated TSL2. As expected, these mutations affected only the TSL2 region, while the remainder of exon 7 structure was unchanged. We also probed the structure of SMN2/36U37U39C40C54G, which was predicted to have a very strong stem. Consistently, this structure was not disrupted even at 50°C in 7 M urea. In agreement with the inhibitory effect of the strong stem, mutant U1 snRNA with increased base pairing to the 5′ ss failed to stimulate exon 7 inclusion in SMN2/36U37U39C40C54G (data not shown).
In comparison to other mammalian SMN exon 7, regulation of human SMN exon 7 splicing is distinct and unique (53,60). Some of these differences arise in part due to presence of an additional codon UUA (codes for 293Leu), which corresponds to positions 43, 44 and 45 of exon 7. Since 293Leu is the second last aminoacid in human SMN, it is unlikely that SMN protein containing or lacking 293Leu would have different functions. In fact, only the first eight (out of total 16) amino acids coded by exon 7, in particular QNQKE sequence, have been shown to be sufficient enough to restore SMN function (90). The presence of remaining coding sequences may have been necessary to increase the exon size to facilitate its recognition. Furthermore, 293Leu codon participates in constitution of both 3′-Cluster and TSL2, and may have contributed to human specific regulation of SMN exon 7 splicing. Of note, deletion of the codon that precedes 293Leu would also break TSL2 but showed no stimulatory effect on SMN2 exon 7 inclusion (53). It is likely that this deletion created alternative structure and/or abrogated the Conserved tract, a stimulatory cis-element (53). The conflicting effects of coding sequences on TSL2 formation reveal complexity of evolution in which only three positions (43rd, 44th and 45th) were available for UUA-codon insertion that produced the specific effect by constituting TSL2. The inhibitory effect of TSL2 must be viewed as complementary to other inhibitory cis-elements that participate in promoting SMN exon 7 skipping. Abrogation or deletion of these elements leads to SMN2 exon 7 inclusion despite the presence of TSL2 (53,60).
Pending further examination of additional factors that interact with 3′-Cluster and flanking inhibitory cis-elements, such as Exinct and ISS-N1, we can only infer that TSL2 takes advantage of a short 5′ ss:U1 duplex and facilitates SMN2 exon 7 skipping through a concerted effort of direct and indirect interactions that sequester the 5′ ss of exon 7. Since local RNA structures (or secondary structures) are formed instantaneously, TSL2 may provide the very first regulatory element aimed at weakening the 5′ ss of exon 7. To a larger significance, our results highlight TSL2 as a unique and essential component of the combinatorial control that modulates alternative splicing of SMN exon 7. Considering structure-specific compounds can be synthesized or selected from the available libraries, TSL2 provides an additional target for correcting the aberrant splicing of SMN2 exon 7 in SMA.
In summary, we have demonstrated that the 5′ ss of SMN exon 7 is sequestered in a stem–loop structure, TSL2. Disruption of TSL2 promoted exon 7 inclusion in SMN2 mRNA. Discovery of TSL2 as a regulatory element reveals for the first time the critical role of a RNA structure in alternative splicing of human SMN genes. It also provides a novel therapeutic target for correction of aberrant splicing of SMN2 in SMA.
Acknowledgments
The authors are grateful to Dr Heiner Schaal for providing plasmids and Dr Alvaro Martinez Del Pozo for a generous gift of RNase U2. The authors thank Drs Jonathan Cherry and Nirmal Singh for critical comments on the manuscript, and Yan Wang for technical support. This work was supported by a grant from Families of SMA (SINN05-06) to N.N.S. R.N.S. was supported by grants from FSMA (SINR05-06) and NIH (R01 NS055925). E.J.A. was supported by a grant from NIH (R01 NS40275). Funding to pay the Open Access publication charges for this article was provided by the University of Massachusetts Medical School, Worcester, MA, USA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 2.Rothrock C., Cannon B., Hahm B., Lynch K.W. A conserved signal-responsive sequence mediates activation-induced alternative splicing of CD45. Mol. Cell. 2003;12:1317–1324. doi: 10.1016/s1097-2765(03)00434-9. [DOI] [PubMed] [Google Scholar]
- 3.Matlin A.J., Clark F., Smith C.W.J. Understanding alternative splicing: towards a cellular code. Nature Rev. Mol. Cell. Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 4.Maniatis T., Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 5.Nilsen T.W. The spliceosome: the most complex macromolecular machine in the cell? Bioessays. 2003;25:1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- 6.Graveley B.R. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nature Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 8.Dreyfuss G., Kim V.N., Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nature Rev. Mol. Cell. Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 9.Berget S.M. Exon recognition in vertebrate splicing. J. Biol. Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 10.Fox-Walsh K.L., Dou Y., Lam B.J., Hung S.P., Baldi P.F., Hertel K.J. The architecture of pre-mRNA affects mechanisms of splice-site pairing. Proc. Natl Acad. Sci. USA. 2005;102:16176–16181. doi: 10.1073/pnas.0508489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green M.R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu. Rev. Cell. Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- 12.Cramer P., Pesce C.G., Baralle F.E., Kornblihtt A.R. Functional association between promoter structure and transcript alternative splicing. Proc. Natl Acad. Sci. USA. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pagani F., Stuani C., Zuccato E., Kornblihtt A.R., Baralle F.E. Promoter architecture modulates CFTR exon 9 skipping. J. Biol. Chem. 2003;278:1511–1517. doi: 10.1074/jbc.M209676200. [DOI] [PubMed] [Google Scholar]
- 14.Robson-Dixon N.D., Garcia-Blanco M.A. MAZ elements alter transcription elongation and silencing of the fibroblast growth factor receptor 2 exon IIIb. J. Biol. Chem. 2004;279:29075–29084. doi: 10.1074/jbc.M312747200. [DOI] [PubMed] [Google Scholar]
- 15.Proudfoot N.J., Furger A., Dye M.J. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 16.Kornblihtt A.R., de la Mata M., Federa J.P., Munoz M.J., Nogues G. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vagner S., Vagner C., Mattaj I.W. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end precessing and splicing. Genes Dev. 2000;15:403–413. [PMC free article] [PubMed] [Google Scholar]
- 18.Masuda S., Das R., Cheng H., Hurt E., Dorman N., Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J., Chang Y.F., Hamilton J.I., Wilkinson M.F. Nonsense-associated altered splicing: a frame-dependent response distinct from nonsense-mediated decay. Mol. Cell. 2002;10:951–957. doi: 10.1016/s1097-2765(02)00635-4. [DOI] [PubMed] [Google Scholar]
- 20.Buratti E., Baralle F.E. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol. Cell. Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Damgaard C.K., Tange T.O., Kjems J. hnRNP A1 controls HIV-1 mRNA splicing through cooperative binding to intron and exon splicing silencers in the context of a conserved secondary structure. RNA. 2002;8:1401–1415. doi: 10.1017/s1355838202023075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagel R.J., Lancaster A.M., Zahler A.M. Specific binding of an exonic splicing enhancer by the pre-mRNA splicing factor SRp55. RNA. 1998;4:11–23. [PMC free article] [PubMed] [Google Scholar]
- 23.Shi H., Hoffman B.E., Lis J.T. A specific RNA hairpin loop structure binds the RNA recognition motifs of the Drosophila SR protein B52. Mol. Cell. Biol. 1997;17:2649–2657. doi: 10.1128/mcb.17.5.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baraniak A.P., Lasda E.L., Wagner E.J., Garcia-Blanco M.A. A stem structure in fibroblast growth factor receptor 2 transcripts mediates cell-type-specific splicing by approximating intronic control elements. Mol. Cell. Biol. 2003;23:9327–9337. doi: 10.1128/MCB.23.24.9327-9337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graveley B.R. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell. 2005;123:65–73. doi: 10.1016/j.cell.2005.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blanchette M., Chabot B. A highly stable duplex structure sequesters the 5′ splice site region of hnRNP A1 alternative exon 7B. RNA. 1997;3:405–419. [PMC free article] [PubMed] [Google Scholar]
- 27.Grover A., Houlden H., Baker M., Adamson J., Lewis J., Prihar G., Pickering-Brown S., Duff K., Hutton M. 5′ Splice site mutations in tau associated with the inherited dementia FTDP-17 affect a stem–loop structure that regulates alternative splicing of exon 10. J. Biol. Chem. 1999;274:15134–15143. doi: 10.1074/jbc.274.21.15134. [DOI] [PubMed] [Google Scholar]
- 28.Jacquenet S., Ropers D., Bilodeau P.S., Damier L., Mougin A., Stoltzfus C.M., Branlant C. Conserved stem–loop structures in the HIV-1 RNA region containing the A3 3′ splice site and its cis-regulatory element: possible involvement in RNA splicing. Nucleic Acids Res. 2001;29:464–478. doi: 10.1093/nar/29.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chebli K., Gattoni R., Schmitt P., Hildwein G., Stevenin J. The 216-nucleotide intron of the E1A pre-mRNA contains a hairpin structure that permits utilization of unusually distant branch acceptors. Mol. Cell. Biol. 1989;9:4852–4861. doi: 10.1128/mcb.9.11.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Libri D., Piseri A., Fiszman M.Y. Tissue-specific splicing in vivo of the beta-tropomyosin gene: dependence on an RNA secondary structure. Science. 1991;252:1842–1845. doi: 10.1126/science.2063196. [DOI] [PubMed] [Google Scholar]
- 31.Domenjoud L., Gallinaro H., Kister L., Meyer S., Jacob M. Identification of a specific exon sequence that is a major determinant in the selection between a natural and a cryptic 5′ splice site. Mol. Cell. Biol. 1991;11:4581–4590. doi: 10.1128/mcb.11.9.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clouet d'Orval B., d'Aubenton Carafa Y., Sirand-Pugnet P., Gallego M., Brody E., Marie J. RNA secondary structure repression of a muscle-specific exon in HeLa cell nuclear extracts. Science. 1991;252:1823–1828. doi: 10.1126/science.2063195. [DOI] [PubMed] [Google Scholar]
- 33.Estes P.A., Cooke N.E., Liebhaber S.A. A native RNA secondary structure controls alternative splice-site selection and generates two human growth hormone isoforms. J. Biol. Chem. 1992;267:14902–14908. [PubMed] [Google Scholar]
- 34.Muro A.F., Caputi M., Pariyarath R., Pagani F., Buratti E., Baralle F.E. Regulation of fibronectin EDA exon alternative splicing: possible role of RNA secondary structure for enhancer display. Mol. Cell. Biol. 1999;19:2657–2671. doi: 10.1128/mcb.19.4.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muh S.J., Hovhannisyan R.H., Carstens R.P. A non-sequence-specific double-stranded RNA structural element regulates splicing of two mutually exclusive exons of fibroblast growth factor receptor 2 (FGFR2) J. Biol. Chem. 2002;277:50143–50154. doi: 10.1074/jbc.M207409200. [DOI] [PubMed] [Google Scholar]
- 36.Buratti E., Muro A.F., Giombi M., Gherbassi D., Iaconcig A., Baralle F.E. RNA folding affects the recruitment of SR proteins by mouse and human polypurinic enhancer elements in the fibronectin EDA exon. Mol. Cell. Biol. 2004;24:1387–1400. doi: 10.1128/MCB.24.3.1387-1400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McAlinden A., Havlioglu N., Liang L., Davies S.R., Sandell L.J. Alternative splicing of type II procollagen exon 2 is regulated by the combination of a weak 5′ splice site and an adjacent intronic stem–loop cis element. J. Biol. Chem. 2005;280:32700–32711. doi: 10.1074/jbc.M505940200. [DOI] [PubMed] [Google Scholar]
- 38.Meyer I.M., Miklos I. Statistical evidence for conserved, local secondary structure in the coding regions of eukaryotic mRNAs and pre-mRNAs. Nucleic Acids Res. 2005;33:6338–6348. doi: 10.1093/nar/gki923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seffens W., Digby D. mRNA have greater negative folding free energies than shuffled or codon choice randomized sequences. Nucleic Acids Res. 1999;27:1578–1584. doi: 10.1093/nar/27.7.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katz L., Burge C.B. Widespread selection for local RNA secondary structure in coding regions of bacterial genes. Genome Res. 2003;13:2042–2051. doi: 10.1101/gr.1257503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:1–5. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 42.Schrank B., Gotz R., Gunnersen J.M., Ure J.M., Toyka K.V., Smith A.G., Sendtner M. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl Acad. Sci. USA. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miguel-Aliaga I., Culetto E., Walker D.S., Baylis H.A., Sattelle D.B., Davies K.E. The Caenorhabditis elegans orthologue of the human gene responsible for spinal muscular atrophy is a maternal product critical for germline maturation and embryonic viability. Hum. Mol. Genet. 1999;8:2133–2143. doi: 10.1093/hmg/8.12.2133. [DOI] [PubMed] [Google Scholar]
- 44.Golembe T.J., Yong J., Dreyfuss G. Specific sequence features, recognized by the SMN complex, identify snRNAs and determine their fate as snRNPs. Mol. Cell. Biol. 2005;25:10989–11004. doi: 10.1128/MCB.25.24.10989-11004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eggert C., Chari A., Laggerbauer B., Fischer U. Spinal muscular atrophy: the RNP connection. Trends Mol. Med. 2006;12:113–121. doi: 10.1016/j.molmed.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 46.Lorson C.L., Strasswimmer J., Yao J.M., Baleja J.D., Hahnen E., Wirth B., Le T., Burghes A.H., Androphy E.J. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nature Genet. 1998;19:63–66. doi: 10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- 47.Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl Acad. Sci. USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Monani U.R., Lorson C.L., Parsons D.W., Prior T.W., Androphy E.J., Burghes A.H., McPherson J.D. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 49.Cartegni L., Krainer A.R. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nature Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 50.Kashima T., Manley J.L. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nature Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 51.Cartegni L., Hastings M.L., Calarco J.A., Stanchina E., Krainer A.R. Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am. J. Hum. Genet. 2006;78:63–77. doi: 10.1086/498853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh N.N., Androphy E.J., Singh R.N. An extended inhibitory context causes skipping of exon 7 of SMN2 in spinal muscular atrophy. Biochem. Biophys. Res. Commun. 2004;315:381–388. doi: 10.1016/j.bbrc.2004.01.067. [DOI] [PubMed] [Google Scholar]
- 53.Singh N.N., Androphy E.J., Singh R.N. In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA. 2004;10:1291–1305. doi: 10.1261/rna.7580704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hofmann Y., Lorson C.L., Stamm S., Androphy E.J., Wirth B. Htra2-β1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2) Proc. Natl Acad. Sci. USA. 2000;97:9618–9623. doi: 10.1073/pnas.160181697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hofmann Y., Wirth B. hnRNP-G promotes exon 7 inclusion of survival motor neuron (SMN) via direct interaction with Htra2β1. Hum. Mol. Genet. 2002;11:2037–2049. doi: 10.1093/hmg/11.17.2037. [DOI] [PubMed] [Google Scholar]
- 56.Young P.J., DiDonato C.J., Hu D., Kothary R., Androphy E.J., Lorson C.L. SRp30c-dependent stimulation of survival motor neuron (SMN) exon 7 inclusion is facilitated by a direct interaction with hTra2b1. Hum. Mol. Genet. 2002;11:577–587. doi: 10.1093/hmg/11.5.577. [DOI] [PubMed] [Google Scholar]
- 57.Stoss O., Novoyatleva T., Gencheva M., Olbrich M., Benderska N., Stamm S. p59(fyn)-mediated phosphorylation regulates the activity of the tissue-specific splicing factor rSLM-1. Mol. Cell. Neurosci. 2004;27:8–21. doi: 10.1016/j.mcn.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 58.Miyajima H., Miyaso H., Okumura M., Kurisu J., Imaizumi K. Identification of a cis-acting element for the regulation of SMN exon 7 splicing. J. Biol. Chem. 2002;277:23271–23277. doi: 10.1074/jbc.M200851200. [DOI] [PubMed] [Google Scholar]
- 59.Miyaso H., Okumura M., Kondo S., Higashide S., Miyajima H., Imaizumi K. An intronic splicing enhancer element in Survival Motor Neuron (SMN) pre-mRNA. J. Biol. Chem. 2003;27:15825–15831. doi: 10.1074/jbc.M209271200. [DOI] [PubMed] [Google Scholar]
- 60.Singh N.K., Singh N.N., Androphy E.J., Singh R.N. Splicing of a critical exon in human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol. Cell. Biol. 2006;26:1333–1346. doi: 10.1128/MCB.26.4.1333-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kammler S., Leurs C., Freund M., Krummheuer J., Seidel K., Tange T.O., Lund M.K., Kjems J., Scheid A., Schaal H. The sequence complementarity between HIV-1 5′ splice site SD4 and U1 snRNA determines the steady-state level of an unstable env pre-mRNA. RNA. 2001;7:421–434. doi: 10.1017/s1355838201001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singh N.N., Androphy E.J., Singh R.N. The regulation and regulatory activities of alternative splicing of the SMN gene. Crit. Rev. Eukaryot. Gene Expr. 2004;14:271–285. doi: 10.1615/critreveukaryotgeneexpr.v14.i4.30. [DOI] [PubMed] [Google Scholar]
- 63.Cartegni L., Wang J., Zhu Z., Zhang M.Q., Krainer A.R. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acid Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Del Gatto F., Gesnel M.C., Breathnach R. The exon sequence TAGG can inhibit splicing. Nucleic Acids Res. 1996;24:2017–2021. doi: 10.1093/nar/24.11.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han K., Yeo G., An P., Burge C.B., Grabowski P.J. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 2005;3:e158. doi: 10.1371/journal.pbio.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chabot B., LeBel C., Hutchison S., Nasim F.H., Simard M.J. Heterogeneous nuclear ribonucleoprotein particle A/B proteins and the control of alternative splicing of the mammalian heterogeneous nuclear ribonucleoprotein particle A1 pre-mRNA. Prog. Mol. Subcell. Biol. 2003;31:59–88. doi: 10.1007/978-3-662-09728-1_3. [DOI] [PubMed] [Google Scholar]
- 67.Wang Z., Rolish M.E., Yeo G., Tung V., Mawson M., Burge C.B. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 68.Perez I., Lin C.H., McAfee J.G., Patton J.G. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA. 1997;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- 69.Spellman R., Smith C.W. Novel modes of splicing repression by PTB. Trends Biochem. Sci. 2006;31:73–76. doi: 10.1016/j.tibs.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 70.Groebe D.R., Uhlenbeck O.C. Characterization of RNA hairpin loop stability. Nucleic Acids Res. 1988;16:11725–11735. doi: 10.1093/nar/16.24.11725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vecenie C.J., Serra M.J. Stability of RNA hairpin loops closed by AU base pairs. Biochemistry. 2004;43:11813–11817. doi: 10.1021/bi049954i. [DOI] [PubMed] [Google Scholar]
- 72.Jaeger J.A., Turner D.H., Zuker M. Improved predictions of secondary structures for RNA. Proc. Natl Acad. Sci. USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Antao V.P., Lai S.Y., Tinoco I., Jr A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 1991;19:5901–5905. doi: 10.1093/nar/19.21.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baumruk V., Gouyette C., Huynh-Dinh T., Sun J.S., Ghomi M. Comparison between CUUG and UUCG tetraloops: thermodynamic stability and structural features analyzed by UV absorption and vibrational spectroscopy. Nucleic Acids Res. 2001;29:4089–4096. doi: 10.1093/nar/29.19.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Heus H.A., Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991;253:191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- 76.Antao V.P., Tinoco I., Jr Thermodynamic parameters for loop formation in RNA and DNA hairpin tetraloops. Nucleic Acids Res. 1992;20:819–824. doi: 10.1093/nar/20.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Varani G. Exceptionally stable nucleic acid hairpins. Annu. Rev. Biophys. Biomol. Struct. 1995;24:379–404. doi: 10.1146/annurev.bb.24.060195.002115. [DOI] [PubMed] [Google Scholar]
- 78.Fairbrother W.G., Yeh R.F., Sharp P.A., Burge C.B. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 79.Shu Z., Bevilacqua P.C. Isolation and characterization of thermodynamically stable and unstable RNA hairpins from a triloop combinatorial library. Biochem. 1999;38:15369–15379. doi: 10.1021/bi991774z. [DOI] [PubMed] [Google Scholar]
- 80.Eun H.M. Enzymology Primer for Recombinant DNA Technology. San Diego, CA: Academic Press; 1996. pp. 170–180. [Google Scholar]
- 81.Wilton S.D., Fletcher S. RNA splicing manipulation: strategies to modify gene expression for a variety of therapeutic outcomes. Curr. Gene Ther. 2005;5:467–483. doi: 10.2174/156652305774329249. [DOI] [PubMed] [Google Scholar]
- 82.Buratti E., Baralle M., Baralle F.E. Defective splicing, disease and therapy: searching for master checkpoints in exon definition. Nucleic Acids Res. 2006;34:3494–3510. doi: 10.1093/nar/gkl498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bartel D.P., Zapp M.L., Green M.R., Szostak J.W. HIV-1 Rev regulation involves recognition of non-Watson–Crick base pairs in viral RNA. Cell. 1991;67:529–536. doi: 10.1016/0092-8674(91)90527-6. [DOI] [PubMed] [Google Scholar]
- 84.Goguel V., Wang Y., Rosbash M. Short artificial hairpins sequester splicing signals and inhibit yeast pre-mRNA splicing. Mol. Cell. Biol. 1993;13:6841–6848. doi: 10.1128/mcb.13.11.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Du H., Rosbash M. The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature. 2002;419:86–90. doi: 10.1038/nature00947. [DOI] [PubMed] [Google Scholar]
- 86.McCullough A.J., Berget S.M. G triplets located through a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol. Cell. Biol. 1997;17:4562–4571. doi: 10.1128/mcb.17.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McCullough A.J., Berget S.M. An intronic splicing enhancer binds U1 snRNPs to enhance splicing and select 5′ splice sites. Mol. Cell. Biol. 2000;20:9225–9235. doi: 10.1128/mcb.20.24.9225-9235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kralovicova J., Vorechovsky I. Position-dependent repression and promotion of DQB1 intron 3 splicing by GGGG motifs. J. Immunol. 2006;176:2381–2388. doi: 10.4049/jimmunol.176.4.2381. [DOI] [PubMed] [Google Scholar]
- 89.Buratti E., Baralle M., De Conti L., Baralle D., Romano M., Ayala Y.M., Baralle F.E. hnRNP H binding at the 5′ splice site correlates with the pathological effect of two intronic mutations in the NF-1 and TSHbeta genes. Nucleic Acids Res. 2004;32:4224–4236. doi: 10.1093/nar/gkh752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang H.L., Pan F., Hong D., Shenoy S.M., Singer R.H., Bassell G.J. Active transport of the survival motor neuron protein and the role of exon-7 in cytoplasmic localization. J. Neurosci. 2003;23:6627–6637. doi: 10.1523/JNEUROSCI.23-16-06627.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]