Abstract
Diplomonads, parabasalids, as represented by trichomonads, and microsporidia are three protist lineages lacking mitochondria that branch earlier than all other eukaryotes in small subunit rRNA and elongation factor phylogenies. The absence of mitochondria and plastids in these organisms suggested that they diverged before the origin of these organelles. However, recent discoveries of mitochondrial-like heat shock protein 70 and/or chaperonin 60 (cpn60) genes in trichomonads and microsporidia imply that the ancestors of these two groups once harbored mitochondria or their endosymbiotic progenitors. In this report, we describe a mitochondrial-like cpn60 homolog from the diplomonad parasite Giardia lamblia. Northern and Western blots reveal that the expression of cpn60 is independent of cellular stress and, except during excystation, occurs throughout the G. lamblia life cycle. Phylogenetic analyses position the G. lamblia cpn60 in a clade that includes mitochondrial and hydrogenosomal cpn60 proteins. The most parsimonious interpretation of these data is that the cpn60 gene was transferred from the endosymbiotic ancestors of mitochondria to the nucleus early in eukaryotic evolution, before the divergence of the diplomonads and trichomonads from other extant eukaryotic lineages. A more complicated explanation requires that these genes originated from distinct α-proteobacterial endosymbioses that formed transiently within these protist lineages.
The diplomonad protist Giardia lamblia, a principal cause of diarrheal disease (1), is basal to all eukaryotes with mitochondria in phylogenies inferred from small subunit rRNAs (2, 3) and several protein-coding genes (4, 5). The early emergence of diplomonads, trichomonads, and microsporidia in molecular trees, coupled with their lack of mitochondria, supports the view that these organisms diverged before the endosymbiotic origin of mitochondria within eukaryotes (2, 5–8). However, discoveries of mitochondrial-like chaperonin 60 (cpn60), chaperonin 10 (cpn10), and heat shock protein 70 (hsp70) genes in a trichomonad (9–12) and hsp70 homologs in microsporidia (13, 14) challenge this interpretation. Mitochondrial cpn60, cpn10, and hsp70 proteins are encoded by nuclear genes that are specifically related to homologs in α-Proteobacteria, indicating that they are of endosymbiotic origin (9, 11, 15). The presence of mitochondrial-like homologs of these genes in trichomonads and microsporidia implies that the ancestors of these organisms once harbored mitochondria, or their endosymbiotic progenitors.
However, evidence for loss of mitochondrial functions from the diplomonad lineage is more tenuous (16). Phylogenies of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and triosephosphate isomerase (TPI) are poorly resolved but do show that eukaryotic homologs could have entered the eukaryotic nucleus by transfer from the ancestral mitochondrial endosymbiont (17, 18). If this interpretation is correct, the presence of typical eukaryotic GAPDH and TPI genes in G. lamblia suggests mitochondria or their ancestors were lost from the diplomonad lineage (17, 18). An ancestral mitochondrial endosymbiont in diplomonads was also supported by the report of a 60-kDa protein from G. lamblia that cross-reacts with mammalian mitochondrial cpn60 antibodies (19). Yet, none of these examples establishes a specific link with the mitochondrial lineage. Independent lateral transfer of genes from prokaryotes to eukaryotes (20) could explain the GAPDH and TPI phylogenies as well as the immunological cross-reactivity data.
Here we report the isolation and sequence analysis of a cpn60 gene from G. lamblia that is phylogenetically related to the mitochondrial cpn60 lineage. These data suggest that the α-proteobacterial endosymbiont that gave rise to mitochondria may have entered the eukaryotic lineage much earlier than previously thought, possibly before the divergence of all known eukaryotes.
MATERIALS AND METHODS
Cloning and Sequencing of G. lamblia cpn60.
Basic local alignment tool (BLASTX) searches of sequences from randomly selected G. lamblia cosmids identified the presence of a partial mitochondrial-like cpn60 gene (21). An ≈850-bp fragment from the end of cosmid CLM-8f8 was subcloned into the pBluescript plasmid vector (Stratagene) and used as a probe to screen a G. lamblia λZAPII genomic library (22) by using digoxigenin-labeling and detection methods (Boehringer Mannheim). After secondary screening, in vivo excision converted the positive clones into pBluescript plasmids (Stratagene). We determined the sequences of the G. lamblia cpn60 homolog as well as its immediate upstream and downstream regions on both DNA strands.
Cycle sequencing reactions were carried out on all plasmid and cosmid genomic clones by using the Sequitherm Long-read and Excel II kits (Epicentre Technologies, Madison, WI) with dye-labeled M13 forward, M13 reverse, T3, and T7 primers. Reactions were run on a LI-COR 4200 automated sequencer, and sequence data were collected and edited by using LI-COR software (LI-COR, Lincoln, NE).
We also determined the full-length sequence of Entamoeba histolytica cpn60, completing the partial sequence previously reported (15).
Growth of Cells.
G. lamblia strain WB (ATCC 30957), clone C6 trophozoites were grown, encysted, and excysted as previously described (23).
Preparation of RNA and DNA.
Total RNA was isolated from G. lamblia at the various stages of differentiation by extraction with RNazol B (Tel-Test, Friendswood, TX). Genomic DNA was isolated by using the Qiagen Blood and Culture DNA Kit (Qiagen, Chatsworth, CA).
Southern and Northern Analyses.
The probe used for Southern and Northern blots was a random primer-labeled PCR fragment amplified from G. lamblia genomic DNA with the G. lamblia-specific oligonucleotides GlcpnR1 (5′-AACCGGACAGGTTCATGAAG-3′) and GlcpnF1 (5′-ATAGGCAGACTATTGGGTAAG-3′). Southern hybridization was performed as described previously (22). For Northern hybridization, 20 μg of total RNA was fractionated in 1.5% formaldehyde-agarose gels, capillary blotted, and immobilized onto nylon membranes (Zeta-Probe, Bio-Rad). The membranes were prehybridized, hybridized and washed at high-stringency, and autoradiographed by using standard techniques.
Rapid Amplification of cDNA Ends (RACE) Analyses.
5′-RACE and 3′-RACE techniques (System 2; GIBCO/BRL) identified the starts of transcription and the polyadenylation site of cpn60. Oligonucleotide GlcpnR1 primed the first strand, and oligonucleotide Glcpn-2 (5′-GAGCAGCCCGGGGCTGCAGAGAGAG-3′) served as a nested primer. 3′-RACE was performed on cDNA generated from 10 μg total RNA by using Superscript II (GIBCO/BRL), using poly-T oligo SGS-10 (5′-CGAGCTGCGTCGACAGGC(T)17-3′) and gene-specific oligo GlcnpF1. PCR conditions were 94°C, 1 min; 55°C, 1 min; 72°C, 1 min for 30 cycles. For the production of DNA sequencing templates, the PCR products were cloned into the pGEM-T EASY vector (Promega).
Western Blot Analyses.
Western blots from encysting and excysting cells were prepared as described in ref. 24. Blots were reacted with rabbit anti-cpn60 (diluted 1:250) antibodies raised against Synechococcus sp. GroEL (StressGen Biotechnologies, Victoria, Canada) and probed with protein A-alkaline phosphatase conjugate. Controls for equal loading were reacted with monoclonal antibodies (diluted 1:250) to the G. lamblia lectin, taglin (25), and rabbit polyclonal antibodies to the endoplasmic reticulum protein BiP (26).
Stresses.
Attached trophozoites or 18-hr encysting cells were subjected to heat shock (40°C or 43°C) for 20 min, then allowed to recover at 37°C for 60 or 90 min. Encysting cells were also incubated in 3% ethanol for 20 min or DTT (7.5 mM) for 3 hr and allowed to recover for 0, 60, or 90 min.
Electron Microscopy.
Cells were harvested at the indicated times, and pellets were fixed and processed for cryosection immunoelectron microscopy, as described in ref. 27, then reacted with the Synechococcus sp. anti-cpn60 antibody followed by localization with 5 nm gold-labeled goat anti-rabbit antibodies.
Sequence Alignment.
A database containing 121 eubacterial GroEL, plastid and mitochondrial cpn60 homologs, archaebacterial thermophilic factor (tf) homologs, and eukaryotic t-complex polypeptide-1 (tcp) homologs was assembled from GenBank and Swiss-Prot databases. Sequences were aligned by using the clustalw program (28). Adjustments to the alignment were made by using the seqlab program (GCG), and regions of ambiguous alignment were removed to create datasets for phylogenetic analysis. The first dataset contained 179 aligned amino acid positions from 47 sequences representing all three domains of life. A second dataset contained 513 aligned amino acid positions and included only mitochondrial and eubacterial cpn60 homologs.
Protein Phylogeny.
Protein distance matrices were inferred by using the protdist program (Dayhoff PAM model), and trees were generated by using the fitch program with global rearrangements (29). Unweighted maximum parsimony analysis was carried out by 50 rounds of random stepwise addition heuristic searches with tree bisection reconnection (TBR) branch swapping, by using the paup* 4.0d56 program (30). Protein maximum likelihood (ML) trees were inferred by using the protein maximum likelihood program protml 2.2 (31) and the heuristic quick-add OTU searching method with the Jones, Taylor, and Thornton (JTT-f) amino acid replacement model. Deviations of amino acid frequencies in a given sequence from the overall frequencies in the dataset and estimates of the gamma shape parameter, α (describing the degree of rate variation among sites in the dataset), were evaluated by using the puzzle 3.1 program (32).
Nucleotide Phylogeny.
All nucleotide phylogenetic analyses were carried out by using the paup* 4.0d56 program (30). Nucleotide distance matrices were inferred from the first and second codon positions of corresponding nucleotide alignments by the maximum likelihood distance method employing the Hasegawa–Kishino–Yano substitution model with a four-category discrete approximation to the gamma distribution (HKY+ Γ). Maximum likelihood estimates of the transition/transversion (Ti/Tv) ratio and the gamma shape parameter (α) were based on the nucleotide distance/neighbor-joining topology. Trees were inferred by simple stepwise-addition heuristic searches with TBR branch swapping under the minimum evolution optimality criterion. Nucleotide maximum likelihood analysis was performed by using the HKY+Γ model and tree-searching methods as described above.
Bootstrap Analyses.
Bootstrap analyses for protein distance analysis utilized programs of the phylip package (29). The resampling estimated log-likelihood (RELL) method was used to estimate bootstrap values for protein maximum likelihood analyses (31). All other bootstrap analyses were carried out by using the paup* 4.0d56 program (30). Distance and parsimony analyses were based on 500 bootstrap resamplings whereas nucleotide maximum likelihood analysis was based on 250 resamplings.
RESULTS AND DISCUSSION
Characterization of the G. lamblia cpn60 Gene.
Of approximately 2,600 single-pass sequences from the ends of G. lamblia cosmids (21), two displayed significant similarity to the GroEL/cpn60 gene family. After secondary screening of a G. lamblia λZAPII genomic library (22), positive clones were subcloned and sequenced on both DNA strands, yielding a total of 2,777 bp containing the cpn60 gene and flanking regions.
The sequence of the ORF (547 codons) shows clear homology to both eubacterial GroEL and mitochondrial cpn60 over its entire length. No other ORFs were detected in the immediate upstream or downstream regions. Southern blots were consistent with a single-copy cpn60 gene in the G. lamblia genome.
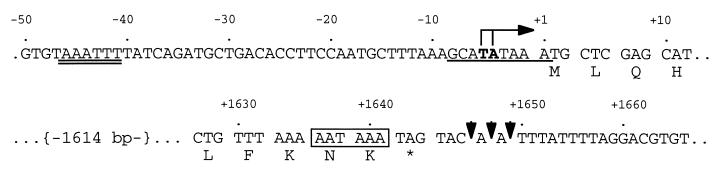
The sequence of 5′-RACE products indicated that the start site of the G. lamblia cpn60 gene lies at positions −5 and −4 upstream of the start codon (Fig. 1), well within the range of 1–11 nt observed for other G. lamblia 5′ untranslated regions (1, 22, 33). An AT-rich motif, corresponding to the 8- to 9-bp AT-rich initiator sites reported in other G. lamblia genes (33, 34), extends from positions −8 to +1 (Fig. 1). A second upstream motif, AAATTT, spans positions −46 to −41 (Fig. 1) and resembles the 6-base consensus motif CAATTT present upstream of other G. lamblia coding regions (33).
Figure 1.
Properties of the G. lamblia cpn60 gene and mRNA transcript. Upstream and downstream regions of the cpn60 gene are shown and numbered relative to the first base of the start codon (+1). The transcriptional start sites are shown in bold under vertical lines and rightward-pointing arrow (→). Two possible transcriptional signals are identified: an AT-rich transcription initiation signal (single underline), and an upstream promoter element (double underline) similar to the CAATTT signal reported for other G. lamblia genes (33). Possible sites of polyadenylation (↓), a putative polyadenylation signal (boxed), and the stop codon (∗) are also indicated.
Sequencing of 3′-RACE products revealed the presence of a poly(A) tail on the G. lamblia cpn60 transcript beginning 4 nt downstream of the stop codon (Fig. 1). A 6-nt motif, AATAAA, has only a single mismatch with the putative consensus polyadenylation signal AGTRAA for G. lamblia genes (1, 22) and it immediately precedes the stop codon (Fig. 1).
Expression of the cpn60 Gene Throughout G. lamblia Life Cycle.
Northern analysis of total G. lamblia RNA showed a single band of approximately 1.8 kb in length that strongly hybridized with the cpn60 probe (Fig. 2A). Because changes in expression of molecular chaperones are observed during differentiation of certain parasites (35), we followed the expression of cpn60 throughout the G. lamblia life cycle.
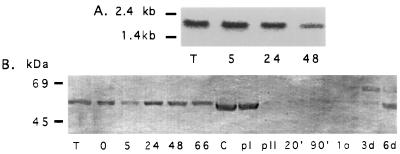
Figure 2.
Expression of cpn60 at different stages in the G. lamblia life cycle. (A) Northern blot analysis of total cellular RNA using a PCR fragment of the G lamblia cpn60 as a probe. Expression of the ≈1.8-kb cpn60 transcript was monitored in the vegetative trophozoite (T) stage, and after 5, 24, and 48 hr of encystation of G. lamblia. (B) Western blot analysis of total cellular protein isolated at different stages of encystation and excysation using a Synechococcus sp. anti-cpn60 antibody. Levels of the ≈60-kDa putative cpn60 protein were constant during the transition from the trophozoite stage (T) and throughout 0–66 hr of encystation. This protein was also present in the cyst phase (C) and after the first stage of induction of excystation (pI). A marked decrease in cpn60 protein was observed after the second stage of encystation (pII). Low levels of cpn60 persisted at 20 min, 90 min, 1 day, and 3 days after excystation, increasing after 6 days.
First, we determined whether the expression of the transcript was affected by differentiation from the vegetative trophozoite to cyst life-cycle stages in response to elevated pH and bile concentrations, which induce encystation. Levels of cpn60 transcript are nearly constant during the in vitro encystation process, with a slight decrease in expression late in encystation (48 hr) (Fig. 2A), mirroring the decrease in levels of many transcripts observed at this stage in the G. lamblia life cycle (22).
Western blots were prepared from total G. lamblia protein and probed with an anti-cpn60 antibody raised against Synechococcus sp. GroEL. The antibody reacted with a band in the 60-kDa range, corresponding to the expected size of the cpn60 gene product (Fig. 2B). The level of this protein did not change appreciably during the encystation process (Fig. 2B).
Excystation is necessary to initiate infection. When cysts are ingested they are exposed to an increase in ambient temperature of greater than 30°C and a large decrease in pH as they pass from cold water into the host stomach (1). Cysts exposed to these stimuli during the induction stage of excystation (pI) in vitro displayed a constant level of cpn60 protein (Fig. 2B). A dramatic drop in the level of cpn60 protein was observed after the second stage of excystation (pII) in vitro, which is induced by exposure to trypsin at pH 8 (modeling the passage of cysts into the host small intestine). Reduced cpn60 expression persisted for at least 3 days after excystation but returned to previous levels after 6 days (Fig. 2B). Expression of the G. lamblia taglin control was constant throughout both encystation and excystation (not shown), indicating that the trypsin treatment did not directly cause the decrease in cpn60 protein.
Expression of G. lamblia cpn60 After Stress.
Since cpn60 proteins often are regulated in response to stresses (36), we examined cpn60 protein abundance (via Western blot analysis) in G. lamblia cells exposed to heat shock and ethanol treatment for varying durations and intensity known to induce stress response in this organism (37). Transfer of cells to 40°C and 43°C did not alter the level of cpn60 protein, and neither did exposure of cells to 3% ethanol. Exposure of cultured cells to DTT can induce a stress response because of accumulation of misfolded proteins in the endoplasmic reticulum (38). However, G. lamblia cells exposed to DTT also showed no change in the level of cpn60 protein. Collectively, these results suggest that the synthesis of G. lamblia cpn60 is not responsive to generalized stresses. This is consistent with the absence of a heat shock protein in the 60-kDa range shown by metabolic labeling (37).
Localization of the G. lamblia cpn60 Protein.
In most eukaryotes, a cpn60 protein is localized in mitochondria and facilitates refolding of proteins after transport across mitochondrial membranes (36). In trichomonads, cpn60, cpn10, and hsp70 localize to the hydrogenosome, an unusual double-membraned organelle of anaerobic energy metabolism (9). However, G. lamblia lacks recognizable mitochondria or hydrogenosomes. Preliminary evidence suggests that a subcellular compartment (possibly a mitochondrial relic) may exist in the amitochondriate parasite Entamoeba histolytica (reviewed in ref. 15). It is possible that the cpn60 protein in G. lamblia may be targeted to a similar structure.
Organelle-targeted proteins often have N-terminal extensions that are cleaved during transport into the organelle. An alignment of the G. lamblia and E. histolytica cpn60 N-terminal regions with other homologs is shown in Fig. 3. The processed targeting peptides of the mitochondrial and hydrogenosomal cpn60s of Leishmania tarentolae and Trichomonas vaginalis are 8 and 15 aa in length, respectively (9, 39). The mature N termini of these proteins correspond roughly with the N termini of GroEL homologs in eubacteria. E. histolytica cpn60 possesses a 8- to 9-residue, serine-rich extension relative to eubacterial homologs that resembles an N-terminal extension encoded by the pyridine nucleotide transhydrogenase gene of this organism (15). In contrast, the G. lamblia cpn60 extends only 2–3 aa past the N termini of eubacterial homologs and is probably too short to represent a full targeting signal. However, because similarity to other cpn60 and GroEL homologs does not start until amino acid position 8 of the G. lamblia cpn60 (Fig. 3), we cannot rule out the possibility that the first 7 aa, or a subset of them, constitute a targeting peptide to an unknown organelle in G. lamblia.
Figure 3.
An alignment of the N termini of G. lamblia, E. histolytica, mitochondrial, hydrogenosomal, and eubacterial cpn60 homologs. The deduced N-terminal sequence of G. lamblia and E. histolytica cpn60s are aligned with homologs from L. tarentolae, T. vaginalis, and Caulobacter crescentus. The L. tarentolae and T. vaginalis targeting peptides (underlined) are removed during import into mitochondria and hydrogenosomes, respectively (9, 39). An N-terminal extension of the E. histolytica cpn60 homolog is suggestive of a targeting peptide for a cryptic organelle in this organism. G. lamblia cpn60 has a small, 2-aa N-terminal extension relative to C. crescentus. Amino acid identities of the G. lamblia cpn60 to other homologs are indicated by asterisks (∗) under the alignment.
We employed immunoelectron microscopy to localize the cpn60 protein in G. lamblia cells. The Synechococcus sp. anti-cpn60 antibody yielded a punctate labeling pattern dispersed throughout the G. lamblia cytosol (not shown), in agreement with a previous report (19). Neither study detected an association of anti-cpn60 antibodies with any specific membranous compartment. Further studies are needed to better determine the localization and function of the cpn60 protein within G. lamblia cells.
The Phylogenetic Position of the G. lamblia cpn60 Homolog.
An initial phylogenetic analysis was conducted on an alignment of 47 sequences of eubacterial, archaebacterial, eukaryotic cytosolic, and organellar cpn60 homologs. In the tree of highest log likelihood, the G. lamblia homolog shares a most recent common ancestor with mitochondrial-like cpn60 homologs from other eukaryotes (not shown). The relative branching order of the Gram-positive eubacterial, cyanobacterial, bacteroides, spirochete, chlamydial, proteobacterial, and mitochondrial lineages is similar to published small subunit rRNA phylogenies (40).
A second dataset was assembled containing cpn60 homologs that represent the diversity of mitochondrial and proteobacterial sequences, as well as their nearest outgroups. In the optimal trees recovered by protein distance, maximum parsimony, and protein maximum likelihood methods, the G. lamblia cpn60 homolog formed a clade with eukaryotic, mitochondrial-like cpn60s to the exclusion of the bacterial lineages (Fig. 4). In agreement with previous analyses (10, 12), all methods showed a specific relationship between the mitochondrial lineage and the subdivision of the α-Proteobacteria that contains the rickettsias.
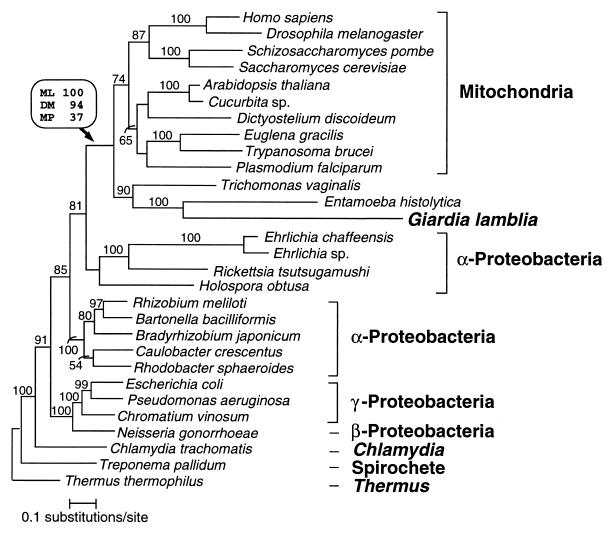
Figure 4.
Phylogenetic relationships of cpn60 homologs. Protein maximum likelihood analysis of 513 aligned amino acid positions yielded the tree shown (log likelihood = −18676.09). Optimal trees obtained by using protein distance and maximum parsimony methods differed from this topology in the branching order of the major eukaryotic groups and, to a lesser extent, within the α-, β-, and γ-proteobacterial clades. The branching order between these clades was identical for all methods. Bootstrap values obtained for the G. lamblia/mitochondrial-like cpn60 clade when using protein maximum likelihood (ML), protein distance matrix (DM), and maximum parsimony (MP) methods are shown in a box above the relevant node (indicated by an arrow). For all other nodes in the tree, protein ML bootstrap values (where >50%) are shown above each branch. The scale bar indicates estimated sequence divergence per unit branch length.
Bootstrap support for the monophyly of the clade containing G. lamblia and mitochondrial-like sequences was strong when using protein maximum likelihood (100%) and protein distance (94%) methods, and weak (37%) when using the maximum parsimony method. The branching order within the mitochondrial subtree varied according to the phylogenetic methods used and in many cases was not strongly supported by bootstrap analysis. However, several consistent features were apparent in protein maximum likelihood and distance trees, including the grouping of Metazoa with Fungi and the early divergence of the three amitochondriate groups (Fig. 4), in agreement with phylogenies of other molecules (2, 5, 41). Curiously, the cpn60s of the three amitochondriate protists, G. lamblia, E. histolytica, and T. vaginalis, form a clade in optimal trees obtained with each method. The G. lamblia/E. histolytica affinity seemed particularly strong with 100%, 92%, and 94% bootstrap support from protein maximum likelihood, distance, and maximum parsimony methods, respectively. This strong association was not expected because an affinity between G. lamblia and E. histolytica has not been observed in phylogenies of other genes such as small subunit rRNA, and elongation factors (2, 5). Because of this and the extremely divergent nature of both sequences, we suspect that this clade is an artifact. Although maximum likelihood methods are generally robust to the long branch attraction artifact, they may succumb to this problem when the evolutionary model is violated (42).
On further investigation, two violations of the maximum likelihood model became apparent. First, the amino acid frequencies in each sequence were compared with the overall frequencies in the dataset (incorporated in the model) by using a χ2 test. Of the 29 sequences in the dataset, only the G. lamblia, E. histolytica, and Thermus thermophilus sequences deviated significantly from the overall amino acid frequencies in the dataset (P < 0.0001 for G. lamblia, P < 0.02 for E. histolytica, and P < 0.02 for T. thermophilus). For 11 of the 20 amino acid types, the G. lamblia and E. histolytica sequences deviated from the overall frequencies in the same direction.
A second violation of the protein maximum likelihood model involved rate variation among sites in the cpn60 dataset. The maximum likelihood estimate of the gamma shape parameter (α) for the cpn60 amino acid dataset was 0.69, suggesting that rate variation among sites in this dataset is extreme (43).
Under the hypothesis that these two model violations coupled with high rates of substitution in the G. lamblia and E. histolytica lineages were responsible for the artifactual clustering of these sequences, we explored the effect of corrections for these problems. To combat the effects of biased amino acid composition, we analyzed the nucleotide sequences coding for the cpn60 proteins. Second, we used distance and likelihood methods with a model (the HKY+Γ model) that accounts for rate variation among sites. Nucleotide distance analysis with trees selected under the minimum evolution criterion by using the HKY+Γ model (with Ti/Tv = 0.85 and α = 0.75) showed that the G. lamblia/E. histolytica relationship was still recovered, but with lower bootstrap support (61%). By contrast, maximum-likelihood analysis by using the same model generated an optimal tree that did not display the G. lamblia/E. histolytica relationship. Bootstrap analysis by using this method on a taxonomically reduced dataset indicated that a G. lamblia/E. histolytica relationship is poorly supported (14% support). These results show that the G. lamblia/E. histolytica relationship observed in the amino acid phylogenies is probably artifactual. By contrast, optimal trees obtained with both nucleotide distance and maximum likelihood methods did recover a G. lamblia/T. vaginalis relationship, although it was poorly supported by bootstrap analysis (50% and 46% bootstrap support from distance and likelihood analyses, respectively). Support for the G. lamblia/mitochondrial-like cpn60 clade was somewhat stronger, gaining 66% bootstrap support from nucleotide distance analysis and 62% from nucleotide maximum likelihood analysis.
In addition to the nucleotide-level analyses, we reduced the effects of model violation and long-branch attraction artifacts by performing amino acid analyses on datasets with the divergent E. histolytica and T. vaginalis sequences removed. We observed strong bootstrap support for a G. lamblia/mitochondrial cpn60 clade in protein maximum likelihood (88%) and protein distance (82%) analyses, with lower support recovered by maximum parsimony methods (35%). Thus, the specific relationship of G. lamblia cpn60 to mitochondrial cpn60s is not an artifact because of the presence of the divergent E. histolytica and T. vaginalis cpn60 sequences.
The Evolutionary Origins of the G. lamblia cpn60 Gene.
The presence of a cpn60 gene related to the mitochondrial cpn60 lineage in G. lamblia is most parsimoniously explained if it was transferred to the nucleus from mitochondria or their endosymbiotic ancestors. However, several other scenarios consistent with the phylogenetic data warrant consideration. First, it is possible that G. lamblia has acquired a cpn60 gene by lateral transfer from another eukaryotic lineage. This scenario could be distinguished from the previous one by establishing the presence or absence of cpn60 homologs in other diplomonads, such as the distantly related, free-living flagellate Hexamita inflata (2). If lateral transfer to the Giardia lineage occurred recently, then Hexamita inflata and its close relatives will lack this gene.
It is also possible that the ancestors of diplomonads acquired the cpn60 gene from an α-proteobacterial endosymbiont that was related to, but distinct from, the ancestors of mitochondria. This scenario could be tested if a free-living or endosymbiotic α-proteobacterium were found that contained a GroEL homolog that robustly grouped with the G. lamblia cpn60 in GroEL/cpn60 trees to the exclusion of the mitochondrial homologs.
In the absence of concrete evidence for either of these scenarios it is probable that the ancestors of G. lamblia acquired the cpn60 gene directly from the genome of the mitochondrial endosymbiont. The lack of mitochondrial functions in G. lamblia (44) is probably a result of secondary loss in early diplomonad evolution. The concomitant loss of many mitochondrion-targeted proteins may have caused a relaxation of functional constraints on diplomonad cpn60 proteins leading to their rapid divergence from other homologs. The long branches leading to both the G. lamblia and E. histolytica sequences in the cpn60 tree (Fig. 4) are therefore excellent examples of accelerated molecular evolution as a result of changed or reduced constraints on the function of a protein.
We cannot rule out the presence of a membrane-bounded mitochondrial relic organelle to which cpn60 and other proteins could be targeted. An organelle with a protein-import mechanism would rationalize the existence of a cpn60 homolog in G. lamblia, because cpn60 typically functions in refolding proteins during import into organelles in other eukaryotes (36). We do not know what the function of such an organelle would be in G. lamblia, but it might be related to energy metabolism. For instance, pyruvate:ferredoxin oxidoreductase (PFOR), an enzyme of anaerobic energy metabolism, functions within hydrogenosomes of trichomonads. In G. lamblia, PFOR is reported to be associated with membranes (45, 46), perhaps indicating the existence of a related organelle in this organism. A second possible function could be the detoxification of peroxide, because a membrane-associated NADH peroxidase exists in G. lamblia (47).
It is also possible that cpn60 may not localize within a membranous organelle but instead functions in the cytosol of G. lamblia. If other mitochondrion-derived proteins have been coopted for use in the G. lamblia cytosol, they may still require cpn60 to fold properly. Whatever its function, the expression of the cpn60 protein throughout much of the G. lamblia life cycle suggests that its presence may be essential to this organism.
CONCLUSIONS
The presence of a cpn60 gene of mitochondrial origin in G. lamblia suggests that diplomonads might not be representatives of a premitochondrial phase of eukaryotic evolution (8, 48). Instead, they could have lost mitochondrial functions secondarily. This conclusion is also supported by a recent finding that G. lamblia, along with other eukaryotes, possesses a proteobacterial-like valyl-tRNA synthetase that might derive from the mitochondrial endosymbiosis (T. Hashimoto, L. B. Sánchez, T. Shirakura, M. Müller, and M. Hasegawa, personal communication). Since diplomonads and trichomonads are among the earliest branching lineages in phylogenies of small subunit rRNA (2), elongation factors (5, 49), and the largest subunit of RNA polymerase II (4), these data suggest that the mitochondrial endosymbiosis may have occurred very early in eukaryote evolution. Two other flagellated protist groups remain as possible candidates for primitively amitochondrial eukaryotes: retortamonads and oxymonads (8, 48). If these groups are shown to branch with or later than diplomonads and trichomonads in molecular phylogenies, or they are shown to contain genes of mitochondrial origin, then the mitochondrial endosymbiosis may have taken place before the divergence of all known surviving eukaryotic lineages.
Acknowledgments
We thank H. Ward for antibodies to taglin, R. Gupta for antibodies to BiP, M. Hetsko for technical assistance, and J. M. McCaffery for ultrastructural analysis. We thank D. L. Swofford for allowing us to perform analyses with the paup* 4.0d56 program and publish the results. For critical reading of the manuscript, we thank A. G. B. Simpson. This work was supported by Grants AI24285, DK35108, and GM53835 awarded to F.D.G. from the National Institutes of Health, a grant from the Wellcome Trust awarded to C.G.C., Grant GM32964 awarded to M.L.S. from the National Institutes of Health, and the G. Unger Vettlesen Foundation. A.J.R. is supported by a fellowship from the Natural Sciences and Engineering Research Council of Canada.
ABBREVIATIONS
- cpn
chaperonin
- hsp
heat shock protein
- HKY
Hasegawa–Kishino–Yano
Footnotes
References
- 1.Adam R D. Microbiol Rev. 1991;55:706–732. doi: 10.1128/mr.55.4.706-732.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leipe D D, Gunderson J H, Nerad T A, Sogin M L. Mol Biochem Parasitol. 1993;59:41–48. doi: 10.1016/0166-6851(93)90005-i. [DOI] [PubMed] [Google Scholar]
- 3.Sogin M L, Gunderson J H, Elwood H J, Alonso R A, Peattie D A. Science. 1989;243:75–77. doi: 10.1126/science.2911720. [DOI] [PubMed] [Google Scholar]
- 4.Stiller J W, Hall B D. Proc Natl Acad Sci USA. 1997;94:4520–4525. doi: 10.1073/pnas.94.9.4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hashimoto T, Hasegawa M. Adv Biophys. 1996;32:73–120. doi: 10.1016/0065-227x(96)84742-3. [DOI] [PubMed] [Google Scholar]
- 6.Margulis L. Proc Natl Acad Sci USA. 1996;93:1071–1076. doi: 10.1073/pnas.93.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavalier-Smith T. In: Endocytobiology II. Schwemmler W, Schenk H E A, editors. Berlin: De Gruyter; 1983. pp. 1027–1034. [Google Scholar]
- 8.Patterson D J. In: Progress in Protozoology. Hausmann K, Hülsmann N, editors. Stuttgart: Gustav Fischer Verlag; 1994. pp. 1–14. [Google Scholar]
- 9.Bui E T, Bradley P J, Johnson P J. Proc Natl Acad Sci USA. 1996;93:9651–9656. doi: 10.1073/pnas.93.18.9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horner D S, Hirt R P, Kilvington S, Lloyd D, Embley T M. Proc R Soc Lond B Biol Sci. 1996;263:1053–1059. doi: 10.1098/rspb.1996.0155. [DOI] [PubMed] [Google Scholar]
- 11.Germot A, Philippe H, Le Guyader H. Proc Natl Acad Sci USA. 1996;93:14614–14617. doi: 10.1073/pnas.93.25.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roger A J, Clark C G, Doolittle W F. Proc Natl Acad Sci USA. 1996;93:14618–14622. doi: 10.1073/pnas.93.25.14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirt R P, Healy B, Vossbrinck C R, Canning E U, Embley T M. Curr Biol. 1997;7:1–4. doi: 10.1016/s0960-9822(06)00420-9. [DOI] [PubMed] [Google Scholar]
- 14.Germot A, Philippe H, Le Guyader H. Mol Biochem Parasitol. 1997;87:159–168. doi: 10.1016/s0166-6851(97)00064-9. [DOI] [PubMed] [Google Scholar]
- 15.Clark C G, Roger A J. Proc Natl Acad Sci USA. 1995;92:6518–6521. doi: 10.1073/pnas.92.14.6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmer J D. Science. 1997;275:790–791. doi: 10.1126/science.275.5301.790. [DOI] [PubMed] [Google Scholar]
- 17.Henze K, Badr A, Wettern M, Cerff R, Martin W. Proc Natl Acad Sci USA. 1995;92:9122–9126. doi: 10.1073/pnas.92.20.9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keeling P J, Doolittle W F. Proc Natl Acad Sci USA. 1997;94:1270–1275. doi: 10.1073/pnas.94.4.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soltys B J, Gupta R S. J Parasitol. 1994;80:580–590. [PubMed] [Google Scholar]
- 20.Rosenthal B, Mai Z, Caplivski D, Ghosh S, de la Vega H, Graf T, Samuelson J. J Bacteriol. 1997;179:3736–3745. doi: 10.1128/jb.179.11.3736-3745.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith M W, Holmsen A L, Wei Y H, Peterson M, Evans G A. Nat Genet. 1994;7:40–47. doi: 10.1038/ng0594-40. [DOI] [PubMed] [Google Scholar]
- 22.Que X, Svärd S G, Meng T C, Hetsko M L, Aley S B, Gillin F D. Mol Biochem Parasitol. 1996;81:101–110. doi: 10.1016/0166-6851(96)02698-9. [DOI] [PubMed] [Google Scholar]
- 23.Meng T C, Hetsko M L, Gillin F D. Infect Immun. 1996;64:2151–2157. doi: 10.1128/iai.64.6.2151-2157.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hetsko, M. L., McCaffery, J. M., Svärd, S. G., Meng, T.-C., Que, X. & Gillin, F. D. (1998) Exp. Parasitol., in press. [DOI] [PubMed]
- 25.Ward H D, Lev B I, Kane A V, Keusch G T, Pereira M E. Biochemistry. 1987;26:8669–8675. doi: 10.1021/bi00400a027. [DOI] [PubMed] [Google Scholar]
- 26.Gupta R S, Aitken K, Falah M, Singh B. Proc Natl Acad Sci USA. 1994;91:2895–2899. doi: 10.1073/pnas.91.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCaffery J M, Faubert G M, Gillin F D. Exp Parasitol. 1994;79:236–249. doi: 10.1006/expr.1994.1087. [DOI] [PubMed] [Google Scholar]
- 28.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Felsenstein J. phylip, Phylogeny Inference Package, Seattle. Seattle: Univ. of Washington; 1993. , Version 3.57c. [Google Scholar]
- 30.Swofford D L. paup*, Phylogenetic Analysis Using Parsimony (*and other methods) Sunderland, MA: Sinauer; 1997. , Version 4.0d56. [Google Scholar]
- 31.Adachi J, Hasegawa M. Computer Science Monographs. Tokyo: Institute of Statist. Math.; 1996. , No. 28. [Google Scholar]
- 32.Strimmer K, von Haeseler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 33.Holberton D V, Marshall J. Nucleic Acids Res. 1995;23:2945–2953. doi: 10.1093/nar/23.15.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hilario E, Gogarten J P. Biochim Biophys Acta. 1995;128:94–98. doi: 10.1016/0005-2736(95)00130-u. [DOI] [PubMed] [Google Scholar]
- 35.Das A, Chiang S, Fujioka H, Zheng H, Goldman N, Aikawa M, Kumar N. Mol Biochem Parasitol. 1997;88:95–104. doi: 10.1016/s0166-6851(97)00081-9. [DOI] [PubMed] [Google Scholar]
- 36.Stuart R A, Cyr D M, Craig E A, Neupert W. Trends Biochem Sci. 1994;19:87–92. doi: 10.1016/0968-0004(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 37.Lindley T A, Chakraborty P R, Edlind T D. Mol Biochem Parasitol. 1988;28:135–144. doi: 10.1016/0166-6851(88)90061-8. [DOI] [PubMed] [Google Scholar]
- 38.Pahl H K, Baeuerle P A. Trends Biochem Sci. 1997;22:63–67. doi: 10.1016/s0968-0004(96)10073-6. [DOI] [PubMed] [Google Scholar]
- 39.Bringaud F, Peyruchaud S, Baltz D, Giroud D, Simpson L, Baltz T. Mol Biochem Parasitol. 1995;74:119–123. doi: 10.1016/0166-6851(95)02486-7. [DOI] [PubMed] [Google Scholar]
- 40.Olsen G J, Woese C R, Overbeek R. J Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wainright P O, Hinkle G, Sogin M L, Stickel S K. Science. 1993;260:340–342. doi: 10.1126/science.8469985. [DOI] [PubMed] [Google Scholar]
- 42.Lockhart P J, Larkum A W, Steel M, Waddell P J, Penny D. Proc Natl Acad Sci USA. 1996;93:1930–1934. doi: 10.1073/pnas.93.5.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z. Trends Ecol Evol. 1996;11:367–372. doi: 10.1016/0169-5347(96)10041-0. [DOI] [PubMed] [Google Scholar]
- 44.Müller M. Annu Rev Microbiol. 1988;42:465–488. doi: 10.1146/annurev.mi.42.100188.002341. [DOI] [PubMed] [Google Scholar]
- 45.Ellis J E, Williams R, Cole D, Cammack R, Lloyd D. FEBS Lett. 1993;325:196–200. doi: 10.1016/0014-5793(93)81072-8. [DOI] [PubMed] [Google Scholar]
- 46.Townson S M, Upcroft J A, Upcroft P. Mol Biochem Parasitol. 1996;79:183–193. doi: 10.1016/0166-6851(96)02661-8. [DOI] [PubMed] [Google Scholar]
- 47.Brown D M, Upcroft J A, Upcroft P. Mol Biochem Parasitol. 1995;72:47–56. doi: 10.1016/0166-6851(95)00065-9. [DOI] [PubMed] [Google Scholar]
- 48.Cavalier-Smith T. Microbiol Rev. 1993;57:953–994. doi: 10.1128/mr.57.4.953-994.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamamoto A, Hashimoto T, Asaga E, Hasegawa M, Goto N. J Mol Evol. 1997;44:98–105. doi: 10.1007/pl00006127. [DOI] [PubMed] [Google Scholar]