Abstract
Low temperature inhibits the growth of maize (Zea mays) seedlings and limits yield under field conditions. To study the mechanism of cold-induced growth retardation, we exposed maize B73 seedlings to low night temperature (25°C /4°C, day/night) from germination until the completion of leaf 4 expansion. This treatment resulted in a 20% reduction in final leaf size compared to control conditions (25°C/18°C, day/night). A kinematic analysis of leaf growth rates in control and cold-treated leaves during daytime showed that cold nights affected both cell cycle time (+65%) and cell production (−22%). In contrast, the size of mature epidermal cells was unaffected. To analyze the effect on cell cycle progression at the molecular level, we identified through a bioinformatics approach a set of 43 cell cycle genes and analyzed their expression in proliferating, expanding, and mature cells of leaves exposed to either control or cold nights. This analysis showed that: (1) the majority of cell cycle genes had a consistent proliferation-specific expression pattern; and (2) the increased cell cycle time in the basal meristem of leaves exposed to cold nights was associated with differential expression of cell cycle inhibitors and with the concomitant down-regulation of positive regulators of cell division.
The growth of maize (Zea mays) seedlings is sensitive to low temperature, particularly during early spring in northern European and American environments. Delayed sowing to avoid this problem reduces the length of the growth season and the potential yield (Lyons, 1973; Graham and Patterson, 1982). Improved performance at low temperature would result in greater light interception early in the growth season and increase productivity through a timely onset of grain filling (Dugan, 1944; Iba, 2002). Breeding programs for cold-tolerant genotypes can benefit from improved knowledge of the plant's growth response to low temperature at the cellular and molecular level (Wang et al., 2003).
Cold in early spring has several distinct effects on the establishment of maize seedlings in the field. Severe cold (chilling stress) impairs chloroplast function, thereby reducing photosynthetic capacity (Allen and Ort, 2001), and may result in cell death (Gomez et al., 2004); prolonged chilling stress eventually may arrest growth. Interestingly, even before visible chilling effects occur, suboptimal temperature inhibits leaf growth (Ben-Haj-Salah and Tardieu, 1995). Monocotyledonous leaves have persistent developmental gradients along the leaf axis, with proliferating, expanding, and mature cells located at increasing distance from the leaf base (Fiorani and Beemster, 2006). In different organs and species, the responses to unfavorable environmental conditions are a consequence of inhibition of both cell proliferation and cell expansion (Pahlavanian and Silk, 1988; Beemster et al., 1996; Schuppler et al., 1998; Tardieu et al., 2000; Sharp et al., 2004). Although we know the basic molecular control mechanisms of the cellular growth processes, cell proliferation and expansion (for review, see Fry, 2004; Inzé, 2005; Sampedro and Cosgrove, 2005), our understanding of the impact of abiotic stress on regulatory molecular interactions is limited.
There is evidence that cell cycle regulation plays an important role in growth responses under unfavorable conditions. The activity of the A-type cyclin-dependent kinase (CDKA), one of the central regulators of cell cycle progression, is associated with the decrease in leaf growth rate of some species under stress conditions. In maize grown at contrasting temperature and water supply, there is a positive correlation between CDKA activity, cell division rates, and leaf growth (Granier et al., 2000). Similarly, in wheat (Triticum aestivum) leaves grown under mild drought stress, a reduction in the size of the leaf basal meristem is associated with a reduced CDKA activity, possibly due to an inhibitory phosphorylation on Tyr-15 residue (Schuppler et al., 1998). In Arabidopsis (Arabidopsis thaliana) roots, changes in the cell cycle machinery accompany a decreased cell production in response to salt stress. Severe salt stress transiently reduces the expression levels of the cyclins CYCA2;1 and CYCB1;1 (Burssens et al., 2000). Mild salt stress leads to loss of CDK activity and to reduced promoter activity of CYCB1;2 (West et al., 2004). Finally, the production of a CDK activity inhibitor, KIP-related protein (KRP), is stimulated by abscisic acid (ABA) in Arabidopsis (KRP1) and alfalfa (Medicago sativa; KRPMt), suggesting a role for this gene in the ABA-mediated stress response pathway (Wang et al., 1998; Pettko-Szandtner et al., 2006).
In this article, we studied a cold treatment that inhibits early leaf growth of maize but does not cause significant chilling symptoms. We compared the daytime growth (25°C) of seedlings exposed to control (18°C) and cold (4°C) nights and studied the cellular growth mechanisms that inhibit growth. We found a specific inhibition of cell cycle activity in the leaf basal meristem (hereafter referred to as leaf meristem). Therefore, we identified 43 putative maize homologs of cell cycle regulatory genes based on sequence database information. Finally, we analyzed the transcription of these genes by constructing expression profiles along the leaf growth zone using real-time PCR. Our results reveal a link between the observed leaf growth inhibition and the expression of specific cell cycle genes.
RESULTS
Effects of Low Night Temperature on Maize Leaf Growth
We investigated the effect of low night temperature on the growth of leaf 4 of maize seedlings by comparing plants grown under control conditions (25°C/18°C day/night [d/n]) and plants exposed to cold nights (25°C/4°C d/n). This treatment did not cause leaf discoloring or premature senescence. We confirmed these visual observations with measurements of leaf chlorophyll content, which was not significantly affected (Supplemental Fig. S1). Also, the transcript level of the large subunit of the Rubisco gene was similar in both control and treated leaves (data not shown). In contrast, severe cold stress, particularly occurring during the photoperiod, causes profound perturbation of chloroplast development and may lead to cell death (Allen and Ort, 2001; Gomez et al., 2004). Nevertheless, our treatment resulted in a 20% decrease of final length of leaf 4 (Table I; P < 0.001, n = 12).
Table I.
Effect of cold nights on cell division and cell expansion parameters during the photoperiod
| Growth Parametersa | Control | Cold Treated | Percent Changeb |
|---|---|---|---|
| Final leaf length (mm) | 658 ± 7 | 527 ± 5 | −20 |
| LER (mm/h) | 3.7 ± 0.2 | 2.9 ± 0.2 | −21 |
| Cell production (cells/h) | 30 ± 2 | 23 ± 1 | −22 |
| Division rate (cells/cell h) | 0.036 ± 0.004 | 0.022 ± 0.005 | −38 |
| Cell cycle duration (h) | 19 ± 2 | 32 ± 6 | 65 |
| Residence time in the meristem (h) | 191 ± 15 | 327 ± 43 | 71 |
| Dividing cells (no.) | 821 ± 94 | 1,060 ± 213 | 29 |
| Meristem size (mm) | 17 ± 6 | 16 ± 3 | NS |
| Mature cell size (μm) | 124 ± 10 | 127 ± 5 | NS |
| Residence time in the elongation zone (h) | 28 ± 1 | 37 ± 1 | 30 |
| Elongating cells (no.) | 705 ± 59 | 737 ± 22 | NS |
| Elongation zone size (mm) | 50 ± 2 | 45 ± 2 | NS |
Average ± se; n = 3, except for final leaf length and LER, n = 12. All parameters were determined at 48 h after leaf emergence.
Statistical significance based on one-way ANOVA tests (P < 0.05). NS, Not significant.
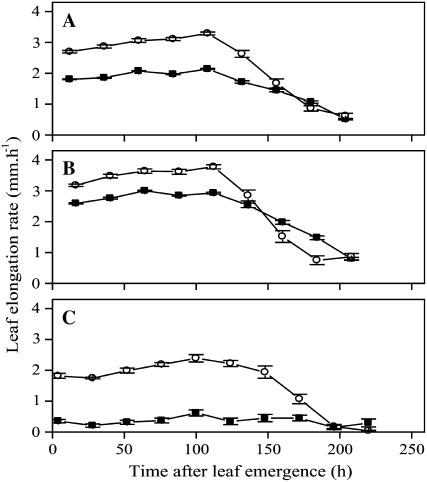
To analyze the developmental basis of reduced leaf size, we measured leaf elongation rate (LER) of leaf 4 from emergence to maturity. The average LER over a 24-h period was approximately constant during the first 120 h after emergence in both treatments. However, growth rates were about 35% lower in leaves exposed to cold nights (P < 0.001, n = 12; Fig. 1A). In contrast, we observed minimal effects on the growth during the periods before and after steady-state growth; leaves emerged from the sheath of the previous leaf at a similar length, and LER was negligible at 200 h after emergence irrespective of the treatment. Our measurements allowed us to estimate the magnitude of the two components contributing to the observed temperature response in our experimental setup: a general temperature effect, presumably due to temperature effects on enzyme kinetics and general physiological processes (Granier and Tardieu, 1999), and a cold stress effect. At night, cold-treated plants showed a growth reduction (LER × duration of measurement interval) of 90% compared to the control caused by both effects (1.5 mm versus 14.9 mm; P < 0.001, n = 12; Fig. 1C). During the photoperiod, this reduction was 20% due to the cold stress effect alone (45.9 mm versus 57.3 mm; P < 0.001, n = 12; Fig. 1B). Assuming the cold stress effect also accounts for 20% of growth inhibition at night (i.e. 3.0 mm), the total contribution of the cold stress effect to the temperature response ([57.3 − 45.9] + 3.0 = 14.4 mm) was larger than that of the general temperature effect ([15 − 1.5] − 3.0 = 10.5 mm).
Figure 1.
The effect of cold nights on leaf elongation. LER of leaf 4 was determined under control (25°C/18°C d/n; white circles) and low-temperature (25°C/4°C; black circles) conditions as a function of time from leaf appearance. A, LER measured over 24 h. B, LER during photoperiod. C, Ler during the night. Symbols are means ± se (n = 12).
Kinematic Analysis
Next, we set out to analyze the cold stress response at the cellular level by means of a kinematic analysis of leaf growth during daytime at steady-state growth (48 h after leaf emergence). For a detailed spatial and temporal characterization, we measured epidermal cell length as a function of position along the leaf growth zone. The cell length profiles of both control and cold-treated leaves were similar (Fig. 2, A and B). For both treatments, the cell length profile had a typical shape characterized by distinctive domains. At the leaf base, there was a zone of approximately constant cell size, in which cell growth and division occur simultaneously (leaf meristem). At more distal positions, cells sharply increased their length by expansion in the absence of mitotic activity (elongation zone). Finally, cell size did not vary throughout the mature part of the leaf.
Figure 2.
Effect of cold nights on epidermal cell length. A, Cell length profiles of files adjacent to stomatal rows in the abaxial epidermis measured for control (white circles) and cold-treated (black circles) plants. The data were obtained for leaf 4 from three plants for which the length of all cells in the basal 100 mm of two cell files adjacent to stomatal files were measured in function of position. To enable averaging across leaves, the data were smoothed and interpolated into 50-μm spaced data points as described in “Materials and Methods.” For each condition, the average and corresponding se is shown at each position (n = 3). Inset, detail of the cell size in the meristem. The arrows indicate the sampling positions for the transcript profiling experiments. B, Representative image of elongating epidermal cells on the leaf abaxial side. The arrow indicates the measured cell file. C, Representative image of DAPI-stained epidermal cells in the meristem used to identify mitotic figures and estimate the size of the leaf meristem.
Epidermal cells expanded to the same mature cell length (124 ± 10 μm and 127 ± 5 μm in control and cold-treated leaves, respectively), implying that overall postmeristematic cell expansion was unaffected and that reduced cell production was responsible for the observed growth reduction. Consistently, cell production decreased significantly by 22% in cold-treated leaves (Table I).
Because total cell production depends on the number of meristematic cells and on their division rate, we measured the size of the leaf meristem by staining nuclei and recording the position of the most distal mitotic figures (Fig. 2C). We found a leaf meristem size of 16 to 17 mm, irrespective of treatment (Table I). In contrast, the size of the proliferating epidermal cells was strongly reduced by the stress, particularly in the basal 5 mm (24.0 ± 2.7 μm in control versus 14.4 ± 0.8 μm in cold-treated leaves; Fig. 2A, inset; P < 0.05, n = 3). Combining these observations, we calculate that cold nights increased the number of cells in the leaf meristem (Table I). We concluded that the reduction in cell production due to low night temperature was due to a slower cell cycle progression. Indeed, the calculated average cell cycle duration was 65% longer in these leaves (P < 0.05).
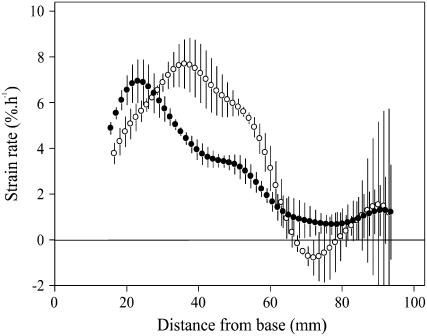
Finally, the reduced cell production in the leaf meristem of cold-treated leaves also affected the flux of cells passing through the expansion zone and resulted in altered cell expansion dynamics (Fig. 3). Except for the first 10 mm adjacent to the leaf meristem, cell elongation rates in cold-treated leaves were lower in most of the elongation zone. However, the longer residence time in the elongation zone in cold-treated leaves fully compensated for these lower expansion rates, resulting in identical mature epidermal cell size (Table I).
Figure 3.
Effect of cold nights on cell expansion. Profiles of cell expansion rates in leaf 4 of control (white circles) and cold-treated (black circles) plants were calculated based on average LER during the photoperiod and the derivatives of the smoothed cell length profiles of cell files adjacent to stomatal rows in the abaxial epidermis. These calculations are only valid outside the meristem, the size of which was determined from observations of mitotic figures. Symbols are averages ± se (n = 3).
Flowcytometry
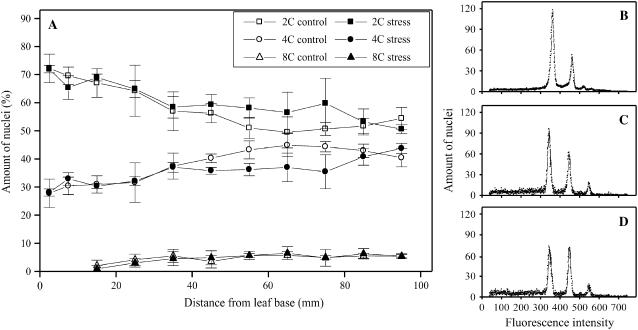
To investigate whether the cell cycle inhibition occurred during a specific cell cycle phase, we analyzed nuclear DNA content by flowcytometry. We sampled the growth zone in 10-mm segments and further subdivided the basal segment into two 5-mm segments. The distribution of nuclear DNA content was unaffected by the cold treatment (Fig. 4). In the leaf meristem, the 2C/4C ratio was similar (2.3 ± 0.2 versus 2.5 ± 0.4, average ± se, n = 3), where C is the haploid nuclear DNA content. This result indicated that the increased duration of the cell cycle in cold-treated leaves is associated with a comparable increase in duration of both G1 and G2 phases.
Figure 4.
Effect of cold nights on nuclear DNA content. Flowcytometry analysis of the ploidy level was performed at different positions of the growth zone. Fluorescence of nuclei was obtained from maize leaf segments (10 mm long, except the first 10 mm, which was divided in two 5-mm segments). A, Fractions of the amount of nuclei at different ploidy levels are shown for both control and low temperature. Representative fluorescence intensity distributions of meristem (B), elongation zone (C), and mature (D) samples under control conditions.
Our data also indicate that, irrespective of treatment, endoreduplication occurred in the elongation zone and stopped when cells matured (at distances >70 mm from the base), consistent with similar observations for Arabidopsis leaves (Beemster et al., 2005). However, endoreduplication was limited (5% of the cells reached 8C nuclear DNA content) and unaffected by the temperature treatment.
Annotation of Cell Cycle Genes
Because cold reduced growth through a prolonged cell cycle, we set out to investigate changes in the molecular cell cycle regulatory machinery (CDKs and cyclins and their interacting proteins). In maize, there are only a few cell cycle genes described to date (Table II). To define a more comprehensive gene set, we performed a sequence homology search on available maize expressed sequence tags (ESTs)/cDNA sequences. We used the cell cycle genes identified in the fully sequenced species Arabidopsis (Vandepoele et al., 2002) and rice (Oryza sativa; La et al., 2006; R.M. Barrôco, unpublished data) expanded with previously described maize homologs. After defining open reading frames on all sequences available in The Institute for Genomic Research (TIGR) maize Gene Indices (release 16.0), we used remote homology detection to identify new cell cycle genes based on hidden Markov model profiles (see “Materials and Methods”; Vandepoele et al., 2002; Eddy, 2004). Using this procedure, we identified 43 homologs of cell cycle genes representing all functional classes of cell cycle regulators (Table II).
Table II.
Overview of maize cell cycle genes
Based on one-way ANOVA analysis, genes were constitutively (V, P > 0.05, n = 3) and differentially expressed (P < 0.05, n = 3) and divided in four different classes based on hierarchical clustering (I, II, III, and IV; see Fig. 5).
Accession numbers for assembled ESTs were derived from TIGR tentative consensus (release 16.0).
Accession numbers for proteins were derived from UNIPROT.
Accession numbers for EST were derived from GenBank.
For all these classes, we performed a phylogenetic analysis comparing previously characterized cell cycle genes of Arabidopsis and rice with those newly identified in maize (Supplemental Fig. S2). Finally, we used the phylogenetic trees to build a nomenclature based on the one adopted for Arabidopsis (Vandepoele et al., 2002).
Effect of Low Temperature on Cell Cycle Gene Expression
To identify the molecular changes underlying the slower cell cycle progression in cold-treated leaves, we determined the expression profiles of the 43 identified cell cycle genes along the leaf growth zone. We collected samples from the leaf meristem (segment 0–5 mm), the elongation zone (segment 40–50 mm), and the mature part (segment 90–100 mm; arrows in Fig. 2A) during the steady-state growth of leaf 4. For each zone, we determined the relative transcript abundance by real-time PCR using three independent biological replicates.
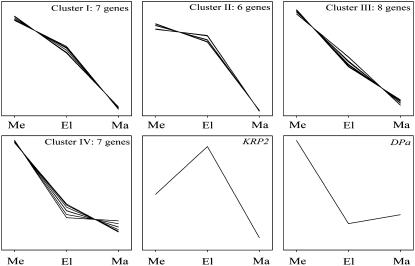
The expression levels of most of the cell cycle genes varied between the three analyzed cell developmental stages; 70% of the transcripts showed a significant change (P < 0.05, n = 3), the remaining 30% had a more stable expression pattern (Table II, class V). Hierarchical clustering of differentially expressed genes yielded four distinct classes and two outliers (Fig. 5; Table II). As expected, nearly all of these genes showed high expression in the leaf meristem and a pronounced decrease of transcript abundance toward the mature region. The inhibitor KRP2 represented an exception. The expression of KRP2 was highest in the elongation zone, suggesting a role in cell cycle exit. The four classes of genes with marked proliferation-specific expression displayed a different decline in transcript abundance in the elongation and mature zone. Cluster IV contained seven genes with the most abrupt decrease of transcript abundance in the elongation zone (Fig. 5; Table II). Although dimerization partner a (DPa) displayed the same pattern, it was not included in this cluster, probably because of the slight increase in expression after cell elongation. Clusters I and III contained seven and eight genes, respectively, showing a more gradual decrease of their expression (Fig. 5; Table II). In contrast, cluster II contained six genes characterized by a relatively high expression both in the leaf meristem and elongation zone. These latter genes most likely play a role in both cell division and processes involved in cell elongation and differentiation (Fig. 4). The comparatively high expression in the leaf meristem is in agreement with a regulatory role in cell proliferation for the identified cell cycle genes.
Figure 5.
Gene expression profiles of differentially expressed cell cycle genes in maize leaves. Hierarchical clusters of the expression profiles of cell cycle genes with a significantly different expression along the leaf growth zone are shown (Table II; P < 0.05; n = 3). Transcript abundance in the meristem (me), elongation zone (el), and mature zone (ma) were determined using real-time PCR on leaf segments of 5 mm in the meristem and of 10 mm in the elongation and mature zone (see Fig. 2A).
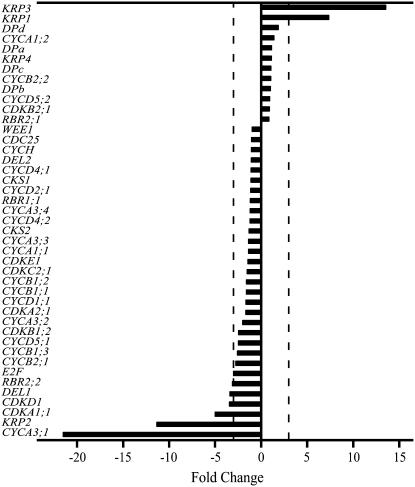
To understand the differences in cell division rates identified by kinematic analysis, we measured transcript levels in the leaf meristem following the cold treatment. The expression patterns were consistent between the three biological replicates despite the relatively high variability for some of the transcripts (Supplemental Table S1). Based on this observation, we chose a 3-fold cutoff threshold. By this criterion, 21% of the genes were differentially expressed (Fig. 6). Several members of the cyclin and CDK families, as well as members of the E2F/retinoblastoma-related (RBR) pathway, were down-regulated (Fig. 6). Specifically, an A-type cyclin (CYCA3;1) showed the largest down-regulation upon treatment, more than 20-fold on average, followed by CDKA1;1, CDKD1, DEL1, RBR2;2, and an E2F homolog. This group of transcripts includes members of the cell cycle-activating CDK/CYC complexes and of E2F/RBR pathway, which controls cell cycle entry. Additionally, two KRPs show a more constitutive expression profile in our data set (KRP1 and KRP3). These were strongly up-regulated in the leaf meristem, on average 8- and 14-fold, respectively. Combined with the observation that KRP2 was down-regulated by more than 10-fold in the leaf meristem, these results suggest functional divergence between the KRP genes. Taken together, these expression data indicate that a slower cell cycle progression in the leaf meristem of leaves grown at low night temperature was associated with concerted transcriptional changes of both positive and negative cell cycle regulators.
Figure 6.
Effect of cold nights on cell cycle gene expression in the meristem. RNA samples were extracted from 0.5-cm leaf segments at the base of leaf 4 from control and cold-treated plants. Expression levels were determined by real-time PCR and fold-changes between averages of three biological replicates are shown. Vertical dotted lines indicate 3-fold thresholds.
DISCUSSION
Low Night Temperature Inhibits Growth Rates during the Photoperiod
Temperature is one of the most important environmental factors influencing leaf growth (Lyons, 1973). Temperature directly affects enzyme kinetics of many biochemical reactions and thereby the growth rate of plant organs. Experimental data show that temperature also determines the activity of the cell cycle regulatory kinase CDKA in maize (Granier et al., 2000).
Here, we investigated whether cold has effects on growth and cell cycle regulation other than the general effects mediated by reaction kinetics. To interpret effects in a temperature-independent manner, growth rates are often modeled as a function of thermal time (Granier and Tardieu, 1998, and refs. therein). We used an alternative strategy to separate the general effects of temperature on enzyme kinetics from growth regulatory changes induced by cold stress. Only during nighttime, we lowered the temperature but maintained identical levels during daytime when we studied the growth effects. Using this method, we obtained results qualitatively similar to those obtained in previous studies in maize (Ben-Haj-Salah and Tardieu, 1995).
Low Night Temperature Inhibits Cell Cycle Progression
Using a kinematic analysis based on cell length profiles (indirect kinematic analysis; Fiorani and Beemster, 2006), we show that the leaf growth inhibition by low night temperature is tightly linked to the reduction of cell production. In turn, this reduction is a consequence of prolonged cell cycle duration and not of a reduced cell number in the leaf meristem (Table I). This result is significant, because cell cycle duration is constant across a wide range of experimental conditions (Baskin, 2000). Relatively few studies reported prolonged cell cycle duration, either transiently in cell cultures (Reichheld et al., 1999) and roots (West et al., 2004) or more persistently in leaves (Schuppler et al., 1998; Granier and Tardieu, 1999) and roots (Sacks et al., 1997). More frequently, a reduction of cell production was associated with a reduced meristematic cell number, e.g. in response to soil compaction (Beemster et al., 1996) and drought (Tardieu et al., 2000; Sharp et al., 2004). The flowcytometry data in our study indicate that low temperature apparently affects all cell cycle phases to a similar extent (Fig. 4). This finding is consistent with previous studies of temperature effects (Tardieu and Granier, 2000) and in contrast with other abiotic factors that act specifically on the G1-S transition (Sacks et al., 1997; Reichheld et al., 1999; West, et al., 2004). Moreover, growth reduction caused by abiotic factors is often due to a combination of both reduced cell production and mature cell size (Sacks et al., 1997; Granier et al., 2000; West et al., 2004), while our results show a specific effect on cell division only. A reduction of cell size at the base of the meristem accompanies this effect on cell division (Fig. 2A, inset). These observations suggest that our treatment primarily affects the growth of meristematic cells and that those smaller cells require a longer time to progress through the cell cycle. However, the size of cells at the transition between the meristem and elongation zone is similar in both control and cold-treated leaves, indicating that our treatment differentially affects cell division and expansion rates of actively dividing cells. To analyze the coordination between these two processes in more detail, local rates of cell division and expansion need to be measured throughout the meristem, which was not possible with the experimental approach used here.
In conclusion, reduced growth caused by low temperature treatment is associated with a prolonged cell cycle progression and not with a reduction of the meristematic cell number or a smaller mature cell size, as reported for other abiotic stresses. Therefore, we postulate that specific cold-inducible mechanisms regulating cell proliferation exist, as hypothesized previously based on ecophysiological data (Grime and Mowforth, 1982).
Transcriptional Analysis of Cell Cycle Genes under Environmental Stress
In this article, we present a set of 43 putative maize cell cycle genes, of which only 13 were previously annotated (Table II). This gene set shows overall the expected proliferation-specific transcription profile throughout leaf development, in agreement with a previous study in Arabidopsis (Beemster et al., 2005). Our choice of sampling by position, as opposed to the entire organ, is effective in capturing transcriptional gradients associated with cell developmental transitions encompassing proliferation, expansion, and mature phases in monocot leaves.
This survey of cell cycle gene expression in maize leaves provides an insight into potential mechanisms that regulate the cell cycle machinery in response to low temperature. Although previous studies showed a link between meristem activity and decreased growth rate during stress treatments (e.g. Ben-Haj-Salah and Tardieu, 1995; Beemster et al., 1996; Tardieu et al., 2000), few data are available on cell cycle gene regulation involved in tuning meristem activity to environmental signals (Burssens et al., 2000; Granier et al., 2000; West et al., 2004).
Our results show that transcriptional regulation of cell cycle gene expression plays an important role in the decreased cell production and growth during cold stress (Fig. 6). The most striking effect was a decrease in the transcript level of an A-type cyclin (CYCA3;1) that might be an important regulator in response to stress. This observation is consistent with previous studies of A- and B-type cyclins (Burssens et al., 2000; West et al., 2004). Taken together, these observations are particularly interesting considering that the large number of cyclin genes in plants compared with other eukaryotes may reflect the higher plasticity of plant growth (Inzé, 2005).
In addition to altered expression of positive cell cycle regulators, some inhibitory proteins (KRP1, KRP2, and KRP3) are also differentially expressed. KRP1 and KRP3 have a strongly increased expression, in contrast to KRP2. Only two of these KRPs (KRP1 and KRP2) were previously described and characterized in maize endosperm. Interestingly, in vitro assays suggested that KRP1 had a stronger inhibitory effect on CDK activity than KRP2 (Coelho et al., 2005), leading to the possibility that cold treatment preferentially leads to activation of more effective KRPs to tune the cell cycle to the adverse conditions. Previous work indicated a possible role for KRPs in stress response regulation. In Arabidopsis, KRP1 is known to respond to ABA (Wang et al., 1998), and an alfalfa KRP was induced both after mild salt treatment and ABA application (Pettko-Szandtner et al., 2006). Finally, we show a possible involvement of transcriptional regulation of the E2F/DP/RBR pathway in mediating responses to environmental conditions.
In conclusion, long-term exposure to low night temperature causes specific changes in the expression of cell cycle genes in maize leaves. Although the regulation of cell cycle progression involves several posttranslational mechanisms (Inzé, 2005), these results highlight that transcriptional changes also play a role, at least in long-term responses. A combination of opposite effects on positive and negative cell cycle regulators lead to prolonged cell cycle duration involving all cell cycle phases and result in a decreased cell production. In turn, this decreased cell production leads to slower growth rates and shorter final leaf size. Future experiments will provide further insight into the role of cell cycle machinery in cold stress responses and clarify which molecular pathways are essential for the modulation of the observed transcriptional changes.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Maize (Zea mays) cv B73 seeds (AgriObtention, Institut National de la Recherche Agronomique) were germinated in peat pellets (Jiffy International As) at 25°C, 70% humidity, a 16-h photoperiod, and a light intensity of 200 μmol m−2 s−1 photosynthetically active radiation. Thereafter, the seedlings were transferred to 2-L pots filled with soil (N°0, Structural) and placed in a growth chamber (type vb1014, Vötsch Industrietechnik) at 70% relative humidity, 400 μmol m−2 s−1 photosynthetically active radiation at plant level provided by a combination of fluorescent tubes (Osram-77 and Osram-31-830) in a 16-/8-h d/n cycle with a gradual decrease and increase of radiation intensity over 0.5 h. Temperature was kept at 25°C during the photoperiod and decreased to 18°C (control) or to 4°C (treatment) during the last 6 h of the night.
Growth Analysis
To calculate LER during the photoperiod and the night, we measured the length of leaf 4 (12 plants per treatment) at the beginning and end of the photoperiod from leaf emergence to maturity, using the soil level as a reference point.
To determine cell length profiles, we collected the basal 100 mm of leaf 4 during steady-state growth from three plants per treatment and prepared 10-mm segments for microscopy, as described previously (Fiorani et al., 2000). In these segments, we measured the cell length in two equivalent abaxial epidermal cell files adjacent to stomatal rows (Fig. 2B) by imaging with differential interference contrast microscopy (De Veylder et al., 2001). For samples of meristematic cells 40× (Achroplan, Zeiss; numerical aperture = 0.65) and for the elongating and mature cells 20× (Plan Apochromat, Zeiss; numerical aperture = 0.75) objectives were used, respectively.
The raw data obtained for individual leaves were smoothed and interpolated at an interval of 50 μm using the kernel smoothing function locpoly of the KernSmooth package (Wand and Jones, 1995) for the R statistical package (R Foundation for Statistical Computing), which allowed averaging between leaves and comparison between treatments.
To estimate meristem size, we isolated the basal 30 mm of leaf 4 from three plants per treatment during steady-state growth. Samples were fixed, as described previously (Fiorani et al., 2000), rinsed three times in a buffer containing 50 mm NaCl, 5 mm EDTA, and 10 mm Tris-HCl, pH 7, and stained by incubating with buffer containing 1 μg/mL 4′,6-diamidino-2-phenyindole (DAPI) for 5 min. Fluorescent nuclei were observed with a microscope (Axioskop, Zeiss) equipped with an epifluorescent condenser. ScionImage software (Scion) was used to measure the size of the leaf meristem defined as the distance between the base of the leaf and the most distal mitotic cell.
We calculated growth parameters through a kinematic analysis, as described previously (Fiorani et al., 2000).
Cell length profiles and LER were measured on separate batches of plants grown under identical conditions. To estimate variance for cell production, cell division, and cell cycle duration, these parameters were calculated for each sample separately based on their cell length profile (n = 3) and the average of the LER data (n = 12). Using the average LER rather than all possible combinations of the two sets of values avoids overestimation of the variance.
Chlorophyll Content Measurements
Four discs (113 mm2) were sampled from halfway along the blade of mature leaf 4 of different plants per treatment. They were placed in N,N-dimethylformamide and stored at 4°C in darkness. After 5 d of incubation, we determined spectrophotometrically chlorophyll a and b content (wavelengths 646.8 nm and 663.8 nm using 750 nm as baseline; Porra, 2002).
Flowcytometry
The basal 100 mm of steady-state growing fourth leaves was cut in segments of 10 mm. The most basal 10 mm was subdivided further into two 5-mm segments. To release the nuclei, the segments were chopped with a razor blade in 2 mL ice-cold buffer (200 mm Tris-HCl, pH 7.5, 4 mm MgCl2, and 0.1% Triton X-100), filtered over a 30-μm mesh, stained with DAPI (De Veylder et al., 2001), and analyzed with a PARTEC Cyflow analyzer. For each sample, we counted at least 10,000 nuclei and analyzed the relative abundance of nuclei with 2C and multiple DNA content using FloMax software (version 2.4 d, PARTEC).
Annotation of Putative Cell Cycle Genes
For all sequences available in the TIGR maize Gene Indices (release 16.0), the coding open reading frame was determined with FrameD software (Oryza Interpolated Markov model, parameters -E for eukaryotic EST analysis and -C for correcting frame shifts; Schiex et al., 2003). Subsequently, we constructed hidden Markov model profiles for all cell cycle gene families based on known dicotyledonous and monocotyledonous reference genes and used these profiles to screen the protein data set. Putative homologs were aligned to the reference sequences in the profile using ClustalW (Thompson et al., 1994) or T-coffee (Notredame et al., 2000). Family-specific signatures were analyzed to select valid maize homologs.
We used the neighbor-joining algorithm for phylogenetic analysis with the software package TREECON (Van de Peer and De Wachter, 1994). Distance matrices were calculated using the Poisson correction, and bootstrap analysis with 100 replicates was performed to estimate the significance of nodes. Based on the position in the tree, we classified genes according to the nomenclature rules for cell cycle genes (Renaudin et al., 1996; Joubès et al., 2000).
Transcriptional Analysis of Putative Cell Cycle Genes
To analyze cell cycle gene expression, we extracted RNA from the fourth leaf of three representative plants at 2 d after emergence. Each of these leaves served as a biological replicate for subsequent analyses and was separated into the leaf meristem (0–5 mm), elongation (30–40 mm), and mature zone (90–100 mm). Total RNA was isolated using TRI-reagent (Sigma-Aldrich). First-strand cDNA synthesis was performed on 3 μg of total RNA with the Superscript RT II kit (Invitrogen) and oligo(dT)18 according to the manufacturer's instructions. Based on the concentration of cDNA measured with a Spectrophotometer ND-1000 (NanoDrop Technologies), samples were normalized and 14 ng was used for each reaction.
We designed primers using the Beacon Designer 4.0 software (Premier Biosoft International; melting temperature = 59°C ± 1°C; amplicon length, 60–150 bp; Supplemental Table S2). Primer specificity was assessed by performing a BLAST search against all known maize sequences available at GenBank and TIGR. For 15 selected primer combinations, sequencing of the PCR products yielded the target sequence, further confirming their specificity. The transcripts were quantified with an iCycler (Bio-Rad) with the qPCR core kit for SYBR green I (Eurogentec). PCR reactions were performed in triplicate technical replicates, following manufacturer guidelines. For each PCR reaction, we observed product melting curves by heating from 60°C to 95°C at 0.2°C/s. For all transcripts, this procedure allowed identification of a single product, which we confirmed by analysis on 2% agarose gels (data not shown).
For relative quantification, we set a threshold cycle at the same level for each reaction within the exponential amplification phase. For normalization, we used the amount of total RNA, because all tested genes (including housekeeping genes) showed significant differences in expression level both for developmental zones and/or treatment. We calculated efficiency and corrected the crossing point values, as described previously (Ramakers et al., 2003).
We mean-centered, normalized, and clustered average crossing points of significantly different genes along the leaf developmental gradient using hierarchical clustering algorithms (Eisen et al., 1998) implemented in TMeV software (Saeed et al., 2003).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Leaf 4 chlorophyll content.
Supplemental Figure S2. Phylogenetic comparison of cell cycle genes.
Supplemental Table S1. Overview of transcript levels.
Supplemental Table S2. Real-time PCR primer sequences.
Supplementary Material
Acknowledgments
We thank Dr. R.M. Barrôco for sharing rice cell cycle sequence information prior to publication.
This work was supported by the Institute for the Promotion of Innovation by Science and Technology in Flanders (Contractueel Landbouwkundig Onderzoek no. 030816) and the Research Fund of Ghent University (Geconcerteerde Onderzoeksacties no. 12051403), by the Research Foundation-Flanders (postdoctoral fellowship to K.V.), and by the Marie Curie Intra-European Fellowship scheme (postdoctoral fellowship FP6–MEIF–CT–2004–009388 to F.F.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Dirk Inzé (dirk.inze@psb.ugent.be).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Ach R, Durfee T, Miller A, Taranto P, Hanley-Bowdoin L, Zambryski P, Gruissem W (1997) RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol Cell Biol 17 5077–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen DJ, Ort DR (2001) Impacts of chilling temperatures on photosynthesis in warm-climate plants. Trends Plant Sci 6 36–42 [DOI] [PubMed] [Google Scholar]
- Baskin TI (2000) On the constancy of cell division rate in the root meristem. Plant Mol Biol 43 545–554 [DOI] [PubMed] [Google Scholar]
- Beemster GTS, De Veylder L, Vercruysse S, West G, Rombaut D, Van Hummelen P, Galichet A, Gruissem W, Inzé D, Vuylsteke M (2005) Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol 138 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GTS, Masle J, Williamson RE, Farquhar GD (1996) Effects of soil resistance to root penetration on leaf expansion in wheat (Triticum aestivum L.): kinematic analysis of leaf elongation. J Exp Bot 47 1663–1678 [Google Scholar]
- Ben-Haj-Salah H, Tardieu F (1995) Temperature affects expansion rate of maize leaves without change in spatial distribution of cell length (analysis of the coordination between cell division and cell expansion). Plant Physiol 109 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burssens S, Himanen K, van de Cotte B, Beeckman T, Van Montagu M, Inzé D, Verbruggen N (2000) Expression of cell cycle regulatory genes and morphological alterations in response to salt stress in Arabidopsis thaliana. Planta 211 632–640 [DOI] [PubMed] [Google Scholar]
- Coelho CM, Dante RA, Sabelli PA, Sun Y, Dilkes BP, Gordon-Kamm WJ, Larkins BA (2005) Cyclin-dependent kinase inhibitors in maize endosperm and their potential role in endoreduplication. Plant Physiol 138 2323–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti J, Tyers M, Sundaresan V (1991) Isolation and characterization of cDNA clones encoding a functional p34cdc2 homologue from Zea mays. Proc Natl Acad Sci USA 88 3377–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GTS, Krols L, Terras F, Landrieu I, Van Der Schueren E, Maes S, Naudts M, Inzé D (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan DH (1944) Yield and bushel weight of corn as influenced by time of planting. J Am Soc Agron 36 166–170 [Google Scholar]
- Eddy SR (2004) What is a hidden Markov model? Nat Biotechnol 22 1315–1316 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorani F, Beemster GTS (2006) Quantitative analyses of cell division in plants. Plant Mol Biol 60 963–979 [DOI] [PubMed] [Google Scholar]
- Fiorani F, Beemster GTS, Bultynck L, Lambers H (2000) Can meristematic activity determine variation in leaf size and elongation rate among four Poa species? A kinematic study. Plant Physiol 124 845–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC (2004) Primary cell wall metabolism: tracking the careers of wall polymers in living plant cells. New Phytol 161 641–675 [DOI] [PubMed] [Google Scholar]
- Gomez LD, Vanacker H, Buchner P, Noctor G, Foyer CH (2004) Intercellular distribution of glutathione synthesis in maize leaves and its response to short-term chilling. Plant Physiol 134 1662–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafi G, Burnett RJ, Helentjaris T, Larkins BA, DeCaprio JA, Sellers WR, Kaelin WG Jr (1996) A maize cDNA encoding a member of the retinoblastoma protein family: involvement in endoreduplication. Proc Natl Acad Sci USA 93 8962–8967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham D, Patterson BD (1982) Responses of plants to low, nonfreezing temperatures: proteins, metabolism, and acclimation. Annu Rev Plant Physiol 33 347–372 [Google Scholar]
- Granier C, Inzé D, Tardieu F (2000) Spatial distribution of cell division rate can be deduced from that of p34(cdc2) kinase activity in maize leaves grown at contrasting temperatures and soil water conditions. Plant Physiol 124 1393–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granier C, Tardieu F (1998) Is thermal time adequate for expressing the effects of temperature on sunflower leaf development? Plant Cell Environ 21 695–703 [Google Scholar]
- Granier C, Tardieu F (1999) Water deficit and spatial pattern of leaf development: variability in responses can be simulated using a simple model of leaf development. Plant Physiol 119 609–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grime JP, Mowforth MA (1982) Variation in genome size-an ecological interpretation. Nature 299 151–153 [Google Scholar]
- Hsieh W-L, Wolniak SM (1998) Isolation and characterization of a functional A-type cyclin from maize. Plant Mol Biol 37 121–129 [DOI] [PubMed] [Google Scholar]
- Iba K (2002) Acclimative response to temperature stress in higher plants: approaches of gene engineering for temperature tolerance. Annu Rev Plant Biol 53 225–245 [DOI] [PubMed] [Google Scholar]
- Inzé D (2005) Green light for the cell cycle. EMBO J 24 657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubès J, Chevalier C, Dudits D, Heberle-Bors E, Inzé D, Umeda M, Renaudin J-P (2000) CDK-related protein kinases in plants. Plant Mol Biol 43 607–620 [DOI] [PubMed] [Google Scholar]
- La H, Li J, Ji Z, Cheng Y, Li X, Jiang S, Venkatesh P, Ramachandran S (2006) Genome-wide analysis of cyclin family in rice (Oryza Sativa). Mol Genet Genomics 275 374–386 [DOI] [PubMed] [Google Scholar]
- Lai J, Dey N, Kim C-S, Bharti AK, Rudd S, Mayer KFX, Larkins BA, Becraft P, Messing J (2004) Characterization of the maize endosperm transcriptome and its comparison to the rice genome. Genome Res 14 1932–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons JM (1973) Chilling injury in plants. Annu Rev Plant Physiol 24 445–466 [Google Scholar]
- Notredame C, Higgins DG, Heringa J (2000) T-coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302 205–217 [DOI] [PubMed] [Google Scholar]
- Pahlavanian AM, Silk WK (1988) Effect of temperature on spatial and temporal aspects of growth in the primary maize root. Plant Physiol 87 529–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettko-Szandtner A, Meszaros T, Horvath GV, Bako L, Csordas-Toth E, Blastyak A, Zhiponova M, Miskolczi P, Dudits D (2006) Activation of an alfalfa cyclin-dependent kinase inhibitor by calmodulin-like domain protein kinase. Plant J 46 111–123 [DOI] [PubMed] [Google Scholar]
- Porra R (2002) The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth Res 73 149–156 [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339 62–66 [DOI] [PubMed] [Google Scholar]
- Reichheld J-P, Vernoux T, Lardon F, Van Montagu M, Inzé D (1999) Specific checkpoints regulate plant cell cycle progression in response to oxidative stress. Plant J 17 647–656 [Google Scholar]
- Renaudin J, Colasanti J, Rime H, Yuan Z, Sundaresan V (1994) Cloning of four cyclins from maize indicates that higher plants have three structurally distinct groups of mitotic cyclins. Proc Natl Acad Sci USA 91 7375–7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaudin J-P, Doonan JH, Freeman D, Hashimoto J, Hirt H, Inzé D, Jacobs T, Kouchi H, Rouzé P, Sauter M, et al (1996) Plant cyclins: a unified nomenclature for plant A-, B- and D-type cyclins based on sequence organization. Plant Mol Biol 32 1003–1018 [DOI] [PubMed] [Google Scholar]
- Rossi V, Locatelli S, Lanzanova C, Boniotti MB, Varotto S, Pipal A, Goralik-Schramel M, Lusser A, Gatz C, Gutierrez C, et al (2003) A maize histone deacetylase and retinoblastoma-related protein physically interact and cooperate in repressing gene transcription. Plant Mol Biol 51 401–413 [DOI] [PubMed] [Google Scholar]
- Sabelli PA, Dante RA, Leiva-Neto JT, Jung R, Gordon-Kamm WJ, Larkins BA (2005) RBR3, a member of the retinoblastoma-related family from maize, is regulated by the RBR1/E2F pathway. Proc Natl Acad Sci USA 102 13005–13012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacks MM, Silk WK, Burman P (1997) Effect of water stress on cortical cell division rates within the apical meristem of primary roots of maize. Plant Physiol 114 519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed A, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34 374–378 [DOI] [PubMed] [Google Scholar]
- Sampedro J, Cosgrove D (2005) The expansin superfamily. Genome Biol 6 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiex T, Gouzy J, Moisan A, de Oliveira Y (2003) FrameD: a flexible program for quality check and gene prediction in prokaryotic genomes and noisy matured eukaryotic sequences. Nucleic Acids Res 31 3738–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuppler U, He P-H, John PCL, Munns R (1998) Effect of water stress on cell division and Cdc2-like cell cycle kinase activity in wheat leaves. Plant Physiol 117 667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp RE, Poroyko V, Hejlek LG, Spollen WG, Springer GK, Bohnert HJ, Nguyen HT (2004) Root growth maintenance during water deficits: physiology to functional genomics. J Exp Bot 55 2343–2351 [DOI] [PubMed] [Google Scholar]
- Sun Y, Dilkes BP, Zhang C, Dante RA, Carneiro NP, Lowe KS, Jung R, Gordon-Kamm WJ, Larkins BA (1999) Characterization of maize (Zea mays L.) Wee1 and its activity in developing endosperm. Proc Natl Acad Sci USA 96 4180–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardieu F, Granier C (2000) Quantitative analysis of cell division in leaves: methods, developmental patterns and effects of environmental conditions. Plant Mol Biol 43 555–567 [DOI] [PubMed] [Google Scholar]
- Tardieu F, Reymond M, Hamard P, Granier C, Muller B (2000) Spatial distributions of expansion rate, cell division rate and cell size in maize leaves: a synthesis of the effects of soil water status, evaporative demand and temperature. J Exp Bot 51 1505–1514 [DOI] [PubMed] [Google Scholar]
- Thompson J, Higgins D, Gibson T (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, De Wachter R (1994) TREECON: a software package for the construction and drawing of evolutionary trees. Comput Appl Biosci 9 177–182 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouze P, Rombauts S, Inzé D (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wand MP, Jones MC (1995) Kernel Smoothing. CRC Press, London
- Wang H, Qi Q, Schorr P, Cutler AJ, Crosby WL, Fowke LC (1998) ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J 15 501–510 [DOI] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Altman A (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218 1–14 [DOI] [PubMed] [Google Scholar]
- West G, Inzé D, Beemster GTS (2004) Cell cycle modulation in the response of the primary root of Arabidopsis to salt stress. Plant Physiol 135 1050–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Sanz-Burgos A, Hannon G, Gutierrez C (1996) Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. EMBO J 15 4900–4908 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.