Abstract
Approximately one-half of all species of amphibians occur in the New World tropics, which includes South America, Middle America, and the West Indies. Of those, 27% (801 species) belong to a large assemblage, the eleutherodactyline frogs, which breed out of water and lay eggs that undergo direct development on land. Their wide distribution and mode of reproduction offer potential for resolving questions in evolution, ecology, and conservation. However, progress in all of these fields has been hindered by a poor understanding of their evolutionary relationships. As a result, most of the species have been placed in a single genus, Eleutherodactylus, which is the largest among vertebrates. Our DNA sequence analysis of a major fraction of eleutherodactyline diversity revealed three large radiations of species with unexpected geographic isolation: a South American Clade (393 sp.), a Caribbean Clade (171 sp.), and a Middle American Clade (111 sp.). Molecular clock analyses reject the prevailing hypothesis that these frogs arose from land connections with North and South America and their subsequent fragmentation in the Late Cretaceous (80–70 Mya). Origin by dispersal, probably over water from South America in the early Cenozoic (47–29 million years ago, Mya), is more likely.
Keywords: amphibian, anura, biogeography, Eleutherodactylus
The evolutionary tree of amphibians is now being revealed at a rapid pace, largely from DNA sequence analyses (1–5). However, the evolutionary history of a major assemblage of frogs is not well understood. These are the eleutherodactylines and the related genus Brachycephalus, which comprise 13% (812 sp.) of all known species of amphibians and 27% of those occurring in the New World tropics (6). Unlike most temperate species, these frogs reproduce on land and undergo direct development, bypassing the tadpole stage (7). Most are relatively small, typically 20–50 mm in length. A majority of the species has been placed in Eleutherodactylus and, together with several other genera, assigned to the tribe Eleutherodactylini of the neobatrachian family Leptodactylidae (8), superfamily Hyloidea (9). However, molecular phylogenies of small sets of representative species over the last two decades have suggested that both the family-level and genus-level classification is in need of revision (2, 10–14).
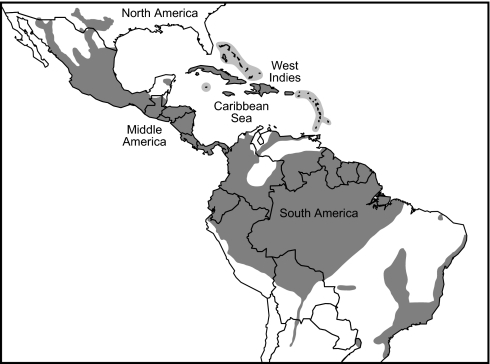
Terrestrial breeding and direct development have allowed eleutherodactyline frogs to occupy a diversity of ecological niches and have facilitated their wide distribution (Fig. 1). Eleutherodactylines occur on almost every island in the Caribbean and display near total endemicity to single-island banks. Their elevational range also is broad, with some species occurring up to 4,400 m in the Andes of South America. Thus, they are a model group for studying Neotropical biogeography and evolution. With this in mind, we assembled samples and available sequences of 276 species of eleutherodactylines and Brachycephalus for several mitochondrial and nuclear genes. Our goal was to identify the major groups of species and their times of divergence, to better understand the historical biogeography of eleutherodactyline frogs and the region in general. Our results revealed several major and, for the most part, geographically isolated, clades of eleutherodactyline frogs and showed that the Middle American and West Indian eleutherodactylines owe their origin to Cenozoic over-water dispersal, not from land connections in the Mesozoic.
Fig. 1.
Composite distribution of eleutherodactyline frogs and Brachycephalus (812 sp.). “Middle America” refers to Central America and Mexico. No evolutionary groupings are implied.
Results
Major Clades of Eleutherodactylines.
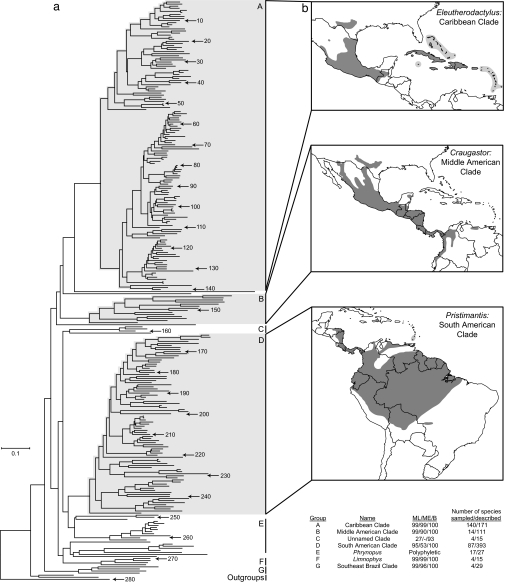
After alignment and removal of ambiguous regions, the 280-species data set encompassed 1,206 sites. The 146- and 65-species data sets included 2,578 and 3,709 sites, respectively. Maximum likelihood (ML), minimum evolution (ME), and Bayesian methods defined the same major clades for all data sets [Figs. 2 and 3; and see supporting information (SI) Figs. 5–13]. Support values for these three groups were variable in the 280-species data set, but were uniformly significant for all methods when the data sets encompassing more nucleotide sites were used.
Fig. 2.
Major clades of eleutherodactyline frogs. (a) ML phylogeny of 280 species of frogs including eleutherodactylines, Brachycephalus, and three out-group species. Species are numbered according to SI Table 4. Major groups with support values (ML bootstrap/ME bootstrap/Bayesian posterior probability), number of species sampled, and total number of described species per clade are indicated. ML, ME, and Bayesian trees including taxon names and all confidence values are available (SI Figs. 5–7). (b) Distribution of Caribbean, Middle American, and South American clades.
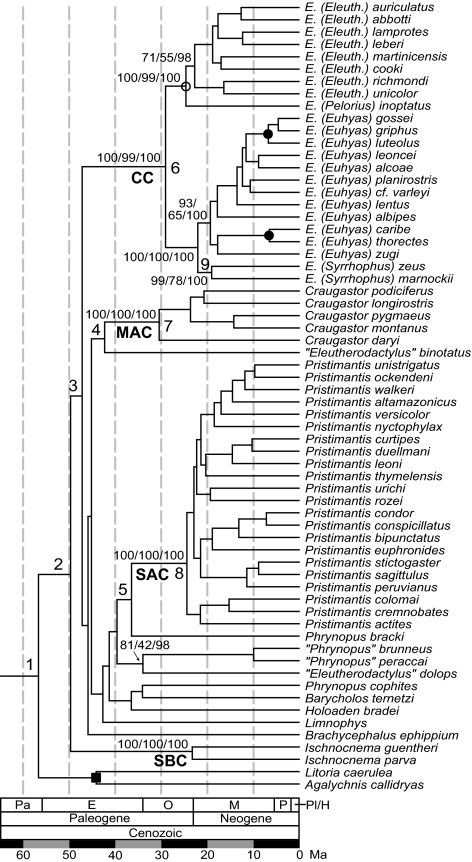
Fig. 3.
A time tree of eleutherodactyline frogs. The tree topology is derived from a ML analysis of 61 eleutherodactylines, Brachycephalus, and three out-group species. Support values for groups mentioned in the text are indicated at nodes (ML/ME/Bayesian posterior probability). Calibration nodes are indicated by open circle (minimum constraint), filled circles (maximum constraint), or filled square (minimum and maximum constraints). The two proposed oceanic dispersal events are on the branches leading to the Caribbean Clade (CC) and the Middle American Clade (MAC). [The South American Clade (SAC) and Southeast Brazil Clade (SBC) are indicated.] Times and credibility intervals for numbered nodes are shown in Table 1. Geologic epochs are abbreviated as follows: Paleocene (Pa), Eocene (E), Oligocene (O), Miocene (M), Pliocene (P), Pleistocene (Pl), Holocene (H).
The three largest and most diverse groups of species are largely defined by geography, with one dominant group each in the Caribbean region, Middle America, and northern South America. A smaller fourth group is found in southeast Brazil. By using past species–group affiliations, it was possible to assign species not included in this study to these major genetically defined clades (SI Table 2). The first major group, which we call the Caribbean Clade (Eleutherodactylus), consists of the West Indian members of the subgenus Eleutherodactylus (47 sp.), the subgenus Pelorius of Hispaniola (6 sp.), the West Indian subgenus Euhyas (91 sp.), and the subgenus Syrrhophus (26 sp.) of southern North America, Middle America, and Cuba.
A second large group (111 sp.) of eleutherodactyline frogs occurs in Middle America, and already has been recognized as the subgenus or genus Craugastor (2, 11, 12, 15). Our analyses indicate a slightly different composition of this Middle American Clade. Previous definitions included some primarily South American species (16), which we find to form a separate clade that is, instead, most closely related to other South American eleutherodactylines (see below). The single remaining South American endemic, the distinctive C. biporcatus, warrants further study with DNA sequences to verify its placement in Craugastor (17).
The third and largest group defined in our analyses includes nearly 400 species centered in the Andes but with species also occurring elsewhere in northern South America. A few species in this group extend into Central America, including nine endemic to southern portions of that region (see SI). Also, two species occur in the southernmost islands of the Lesser Antilles. This South American Clade includes species formerly placed in the Eleutherodactylus unistrigatus, conspicillatus, and 13 other species groups (7). We use the available name Pristimantis Jiménez de la Espada, 1870 for this previously undefined clade.
Besides these three major clades, our analyses suggest that most of the 31 species in southeastern Brazil formerly placed in Eleutherodactylus form a separate, smaller clade (Figs. 2–3). Our sparse taxonomic sampling from this region makes it difficult to determine the composition of this group, but the joining of four diverse species (E. guentheri, E. hoehnei, E. parvus, and E. juipoca) in a well supported group, suggests that other species from the region believed to be closely related to them also are part of that group, which takes the available name Ischnocnema Reinhardt and Lütken, 1862 (see SI Text). Four southeast Brazilian species in our analysis that are not part of that clade are E. binotatus, which has an unusual karyotype (18), Holoaden bradei, Barycholos ternetzi, and Brachycephalus ephippium. These species also branch basally among eleutherodactylines but are not closely related to other species or groups.
These major clades of species with definitive geographic patterns account for 87% of the 812 species of eleutherodactyline frogs and Brachycephalus. The remaining 106 species are all native to South America, mostly Andean, and are best characterized by their basal position in the phylogenetic trees (Figs. 2–3), suggesting that they represent an early stage of evolution of the group. Their relationships and those of the three major clades remain unresolved. Among these are the representatives of the (formerly Craugastor) anomalus and bufoniformis groups, which cluster strongly with a species in the E. sulcatus group. For this clade, we apply the available name Limnophys Jiménez de la Espada, 1871. The genus Phrynopus is polyphyletic, with species forming several independent groups, as was found elsewhere (19). Two species of Phrynopus cluster with species of the E. nigrovittatus and E. dolops groups. Other genera in this category of deeply branching lineages include Oreobates and Phyllonastes.
Times of Divergence.
Dates of divergence obtained by using nuclear data, mitochondrial data, or all data are similar for most nodes (Table 1 and SI Table 3). The eleutherodactyline lineage diverged from other hyloid frogs near the Mesozoic–Cenozoic boundary (57 Mya, C.I. = 78–44 Mya), as found elsewhere (20), with initial divergences occurring among eleutherodactylines ≈50 Mya (Fig. 3). The Caribbean Clade (Eleutherodactylus) diverged from its extant mainland relatives ≈47 Mya and began diversification ≈29 Mya, setting upper and lower bounds for the date that the West Indies was colonized. Assuming no extinction of the mainland source lineage, the dispersal most likely occurred early in that time interval rather than later. Similarly, the Middle American Clade (Craugastor) diverged 42 Mya and began diversification 31 Mya. Middle American and Cuban Syrrhophus split ≈19 Mya. The Southeast Brazil Clade diverged from other eleutherodactylines ≈50 Mya. The South American Clade (Pristimantis) diverged from other eleutherodactylines 37 Mya and began an explosive diversification 24 Mya.
Table 1.
Times of divergence (Mya) for major nodes in Fig. 3
| Node | Divergence | Time | 95% C.I.* |
|---|---|---|---|
| 1 | Eleutherodactylines plus Brachycephalus/ hylid frogs | 56.79 | (43.52, 78.13) |
| 2 | Southeast Brazil Clade (SBC)/other species | 49.79 | (37.18, 68.67) |
| 3 | Caribbean Clade (CC)/other eleutherodactylines | 47.28 | (35.09, 65.26) |
| 4 | Middle American Clade (MAC)/other eleutherodactylines | 42.39 | (30.99, 58.99) |
| 5 | South American Clade (SAC)/other eleutherodactylines | 36.52 | (26.56, 50.81) |
| 6 | Last common ancestor of Caribbean Clade | 29.09 | (20.95, 40.35) |
| 7 | Last common ancestor of Middle American Clade | 30.51 | (21.67, 43.17) |
| 8 | Last common ancestor of South American Clade | 24.45 | (17.30, 34.82) |
| 9 | Middle American Syrrhophus/Cuban Syrrhophus | 19.05 | (13.06, 26.92) |
Times are based on the combined nuclear and mitochondrial data set (3,709 bp) and measure the divergence of the two identified lineages separated by a slash. Divergence times for all nodes (combined, nuclear, and mitochondrial data sets) are presented in SI Table 3 with a guide tree available as SI Fig. 14. The dates of the two proposed dispersal events are constrained by nodes 3 and 6 (for the Caribbean Clade) and nodes 4 and 7 (for the Middle American Clade).
*Bayesian credibility interval.
Discussion
Major Clades of Tropical Frogs.
The discovery of three major and geographically defined groups of these tropical amphibians was unexpected. Previous studies on eleutherodactylines had been hampered by too few useful morphological characters and too few samples for molecular analysis. Although the Middle American Clade was known (12, 15), it had included species in South America shown here to be misclassified based on our sequence analyses. The Caribbean and South American clades, on the other hand, were unpredicted. Previous studies had assumed a close relationship between West Indian members of the subgenus Eleutherodactylus and the species-rich unistrigatus group (now in Pristimantis) in South America (7, 10, 11, 21). In part, this was based on shared morphological characters that may be associated with climbing habits (11). Our results show, however, that diverse morphologies and habits have evolved independently in the Caribbean and South American Clades. The geographical separation of these large clades highlights a general pattern, the greater importance of geography, revealed in many molecular phylogenetic studies (e.g., refs. 5 and 22).
Middle America and the Caribbean.
The origin of the Middle American and West Indian terrestrial vertebrates has focused on two competing models in the context of current geologic models for the region (23–25). The vicariance model suggests that they arose in the Late Cretaceous (80–70 Mya) by fragmentation of a continuous land mass (proto-Antilles) and its biota located between North and South America (26–28). This occurred as the Caribbean tectonic plate moved eastward, carrying the West Indian fauna and isolating the Middle American fauna from its South American counterparts. This is in contrast to an origin of these faunas by dispersal, on flotsam from continental source areas. One difficulty for the vicariance model has been the great age (Cretaceous) of the groups required for this model, which is largely unsupported by the fossil record (29). Also, the fauna of the West Indies is peculiar in missing many higher-level groups, indicative of dispersal (30). Geologic evidence does not rule out the possibility of a proto-Antillean island chain or corridor, but does not favor the substantial emergence of land in the Antilles before the mid-Eocene (37–49 Mya) (24).
Molecular clock analyses have yielded mixed results, although most groups have shown Cenozoic divergences with their closest relatives on the mainland (23, 31, 32). Estimates of Cretaceous divergence between West Indian and mainland representatives of insectivores (33), xantusiid lizards (33, 34), and (in past studies) eleutherodactyline frogs (10, 35), suggested that those groups may be vicariant relicts of the proto-Antilles even if most others are not. However, the relictual nature of the distribution of xantusiid lizards and West Indian insectivores raises the possibility of Cenozoic dispersal to the West Indies and subsequent extinction of those mainland source populations (34). Studies indicating Cretaceous ages for Middle American and West Indian eleutherodactylines either assumed proto-Antillean vicariance (12) or used geologic calibrations that have since been revised (10).
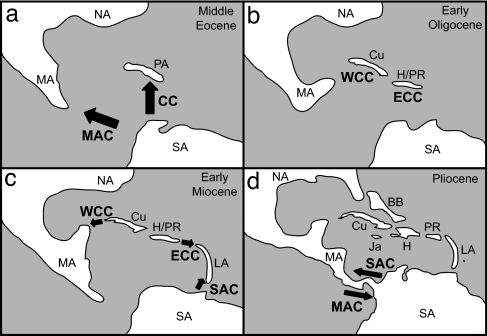
Based on our results, the Middle American and Caribbean clades of eleutherodactylines originated through dispersal from South America during the Cenozoic. For these clades to have originated through proto-Antillean vicariance, Mesozoic ages (e.g., 80–70 Mya) are required for divergences between these groups and their South American relatives. Instead, our data (Table 1) indicate that a single event 42–31 Mya established eleutherodactylines in Middle America, and another 47–29 Mya established eleutherodactylines in the West Indies (Fig. 4a). Early speciation in the Caribbean Clade was confined to Hispaniola and Cuba. The paleogeography of the West Indies in the mid-Cenozoic was substantially different from that today (24). Land connections between Cuba, northern Hispaniola, and Puerto Rico probably existed in the Late Eocene (≈35 Mya), facilitating dispersal among the islands. A proposed dry-land connection to South America at this time (24) lacks geologic support and remains controversial (23, 34). After subsidence in the Oligocene (23–34 Mya), land connections were broken, probably isolating the western Caribbean lineage (subgenera Euhyas plus Syrrhopus) in Cuba from the eastern Caribbean lineage (subgenera Eleutherodactylus plus Pelorius) in northern Hispaniola and Puerto Rico (Fig. 4b).
Fig. 4.
Biogeographic model showing the origin of the Middle American and Caribbean clades of eleutherodactyline frogs. Location of exposed land is conjectural and based on a synthesis of models (24, 25, 50). Landmasses are abbreviated as follows: North America (NA), Middle America (MA), South America (SA), Proto-Antilles (PA), Cuba (Cu), Hispaniola (H), Puerto Rico (PR), Lesser Antilles (LA), Jamaica (Ja), Bahama Bank (BB). (a) Middle Eocene (49–37 Mya), when dispersal over water from South America probably occurred, leading to the origin of the Middle American Clade (MAC) and Caribbean Clade (CC). (b) Early Oligocene (≈30 Mya), when land subsidence and higher sea levels led to isolation of a western Caribbean (WCC) lineage on Cuba and an eastern Caribbean (ECC) lineage on Hispaniola and Puerto Rico. (c) Early Miocene (≈20 Mya), when dispersal from Cuba to the mainland led to a radiation of the subgenus Syrrhopus in southern North America and northern Middle America and when the Lesser Antilles were colonized by members of the ECC and South American Clade (SAC). (d) Pliocene (≈3 Mya), when closing of the Isthmus of Panama allowed overland dispersal of species of the MAC to South America and species of the SAC to Middle America.
In the Early Miocene (19 Mya), an over-water dispersal occurred from western Cuba to southern North America within the subgenus Syrrhophus (Fig. 4c), as indicated by some earlier molecular studies (10, 11). It is possible that this lineage initially evolved in isolation to the north of the Middle American Clade, although the distributions of these two groups currently overlap. Dispersal from the Greater Antilles to the mainland has been found in other vertebrate groups, including turtles (36, 37) and anoline lizards (38). Other Miocene dispersals of eleutherodactylines, most probably over water, occurred among islands in the West Indies (Fig. 3). The direction of some of these dispersal events would have been against the present-day water currents, which flow primarily from southeast to northwest. However, current flow within the Caribbean may have been different in the past, before the emergence of the Isthmus of Panama (39).
A striking pattern in these results is the absence of subsequent successful colonizations of eleutherodactyline frogs in Middle America and the West Indies from South America after their origin in the early Cenozoic. Of the few exceptions, two species of the South American clade now occupy the southernmost Lesser Antilles (St. Vincent and Grenada) and 18 species of the South American Clade now occur in Middle America (see SI Text). In the latter case, the presence of some or all of those species may be explained by dispersal over land after the emergence of the Isthmus of Panama (≈3 Mya). Whether or not there were failed colonizations to Middle America and the West Indies as a result of competition (40) is unknown. Also, if the Middle American Clade and Caribbean Clade are later found to be closest relatives, the possibility that there was a stepwise dispersal (South America to one clade and from that clade to the other) should be considered.
South America.
Most of the basal branches of eleutherodactylines, with some dating to the early Cenozoic, occur in South America (Fig. 3). This indicates that South America was the place of origin for the group, as it was for hyloid frogs in general (13, 14). However, the great diversity of species, including the South American Clade of 393 species, is associated with Andes. The Andean uplift is relatively recent, occurring mostly in the last 10–20 million years (41, 42). Rapid diversification within the South American Clade, which began 24 Mya and has continued to the present, was probably linked with this uplift. Mountain-building and associated climatic changes resulted in repeating patterns of habitat isolation, which, in turn, probably resulted in genetic isolation and speciation in these amphibians (7).
Despite the large number of South American species included in this analysis (123 sp.), we are missing a majority of species including many from southeastern Brazil. Our results indicate that the eleutherodactyline fauna of southeastern Brazil is distinct and includes several basal clades. This region is an isolated area of montane rainforest and is a region of endemism for other amphibians (43).
Methods
Taxon Sampling.
Our data set encompasses ≈34% of known eleutherodactyline diversity with 276 species in 12 of 18 genera, including at least one representative of every genus with more than five described species. Included species were concentrated in the largest genera, with 140 species of Eleutherodactylus, 87 of Pristimantis, 14 of Craugastor, 17 of Phrynopus, and four of the Southeast Brazil Clade. Two hylid species, Agalychnis callidryas (South America) and Litoria caerulea (Australia), were included for calibration of divergence times. Seven additional hyloid species and a more distant ranoid species (Rana catesbeiana) were included as out groups.
Data Collection.
Our study included data from three mitochondrial genes: 12S ribosomal RNA (12S), 16S ribosomal RNA (16S), and intervening tRNA-Valine. In addition, fragments from two nuclear protein-coding genes were sequenced: recombination-activating gene 1 (Rag-1) and the tyrosinase gene (Tyr). Approximately 90% of the sequences used are previously uncharacterized. Data were collected as overlapping sets (SI Table 4) of 280 species (two genes), 146 species (three genes), and 65 species (five genes).
For the 280-species data set, partial 12S and 16S sequences were assembled for 277 in-group and three out-group species and used to define major clades (here recognized as genera and subgenera). This data set consists of an ≈350-bp fragment of 12S concatenated with a ≈800-bp fragment of 16S. For the 146-species data set, complete 12S and 16S sequences (≈2.5 kb), including the intervening tRNA and fragments of the flanking tRNA sequences, were assembled for 136 species representing all major groups as defined by the partial data set, the same three out-group species, and seven additional hyloid out-group species. This data set was used to test groups found with the 280-species data set, confirm rooting within eleutherodactylines by using additional out groups, and define subgroups within the largest clades. For the 65-species data set, we also included sequences from a 493-bp region of Tyr and a 639-bp region of Rag-1. This sample included representatives of most major clades and subclades, except where specimen availability or quality were limiting. Methods of sample collection, DNA extraction, amplification, and sequencing are presented in the SI Text, along with a list of primers (SI Table 5). When available, sequences for species of interest were obtained from GenBank (SI Table 4).
Phylogenetic Analyses.
Reconstructions of phylogenies for all data sets were performed by using ME, ML, and Bayesian methods. For ML and Bayesian analyses, the 65-species data set was divided into three partitions: 12S and 16S, Rag-1, and Tyr. ME analyses were implemented in MEGA 3.1 (44) by using the TN + Γ model of evolution. PAUP 4b10 (45) was used to estimate the γ-parameter, and branch support was assessed with 2,000 bootstrap replicates. ML analyses used RAxML-VI-HPC v.2.0 (46), accessed at the San Diego Supercomputing Center. For each data set, 100 alternative runs were performed under the GTRMIX model of evolution. Other parameters were maintained at default settings. Nonparametric bootstrap analysis (1,000 replicates) was used to provide branch support values for the most likely tree of 100 found in each data set. MrBayes 3.1 (47) was used to perform Bayesian analyses. Bayesian analyses used the GTR + I + Γ model of evolution, with all parameters unlinked in partitioned analyses. For the 65-species data set, all phylogenetic analyzes were performed by using only the two nuclear genes in addition to analyses employing both the mitochondrial nuclear data, to ensure that mitochondrial and nuclear data produced results that were not significantly divergent. Additional details of analyses are available in the SI.
Divergence Timing.
Times of divergence were estimated for the 65-species data set by using the T3 version of Multidivtime (48, 49). The assumed topology was from the five-gene ML analysis. The data were divided into three partitions, as in the phylogenetic analyses. In addition to estimating times by using all available data, timing analyses were also performed by using mitochondrial and nuclear data separately. A total of five calibrations, including both upper and lower bounds within and outside the eleutherodactylines, were used based on geologic and fossil evidence. These included the earliest divergence in the Jamaican radiation of the subgenus Euhyas (10 Mya, maximum constraint), the earliest divergence in the Hispaniolan South Island radiation of Euhyas (10 Mya, maximum), the divergence of the subgenera Eleutherodactylus and Pelorius (15 Mya, minimum), and the divergence of South American and Australian hylids (35 Mya, minimum; 70 Mya, maximum). Additional details for these and other priors are available in SI Text.
Supplementary Material
Acknowledgments
S.B.H. especially thanks Lori Fritz, Jennifer R. Grubb, Genna M. Lutz, Molly E. Means, Rebecca Montrose, Michael R. Tracy, and Cecelia Youngblood for their contributions to the initial generation of data for this project. We thank Jonathan A. Campbell and Eric N. Smith (University of Texas, Arlington, TX), David Cannatella (University of Texas, Austin, TX), Luis A. Coloma (Catholic University of Ecuador, Quito, Ecuador), Andrew J. Crawford (Smithsonian Tropical Research Institute, Balboa, Republic of Panama), Luis Diaz (Museum of Natural History, Havana, Cuba), Ronald Heyer and Addison Wynn (Smithsonian Institution, Washington, DC), Edgar Lehr (University of Kansas), John D. Lynch (National University of Colombia, Bogata, Colombia), Linda R. Mason (University of Iowa, Iowa City, IA), Lily Rodriguez (University of San Marcos, Lima, Peru), Michael Schmid (University of Würzburg, Würzburg, Germany), and Steven Werman (Mesa State College, Grand Junction, CO), for providing some samples and sequences; Paul Mann for discussion of geological history; and Bernard Moret for providing computer time on the NSF-funded CIPRES cluster at the San Diego Supercomputer Center. Richard Thomas (University of Puerto Rico, San Juan, Puerto Rico) contributed significantly to field work in the West Indies. This research was supported by National Aeronautics and Space Administration Grant NNA04CC06A (to S.B.H.) and by National Science Foundation (NSF) Grants 8906325, 9123556, 9525775, and 9615643 (to S.B.H.), 8307115 (to Richard Highton, University of Maryland, College Park, MD), 8805920 (to W.E.D.), and especially by NSF-Assembling the Tree of Life (AToL) Amphibia-Tree Project Grant 0334952 (to David Cannatella and David Hills, University of Texas, Austin, TX).
Abbreviations
- 12S
12S ribosomal RNA
- 16S
16S ribosomal RNA
- ME
minimum evolution
- ML
maximum likelihood.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EF493342–EF493828).
This article contains supporting information online at www.pnas.org/cgi/content/full/0611051104/DC1.
References
- 1.Bossuyt F, Brown RM, Hillis DM, Cannatella DC, Milinkovitch MC. Syst Biol. 2006;55:579–594. doi: 10.1080/10635150600812551. [DOI] [PubMed] [Google Scholar]
- 2.Frost DR, Grant T, Faivovich J, Bain RH, Haas A, Haddad CFB, De Sa RO, Channing A, Wilkinson M, Donnellan SC, et al. Bull Am Mus Nat Hist. 2006;297:1–371. [Google Scholar]
- 3.Wiens JJ, Fetzner JW, Parkinson CL, Reeder TW. Syst Biol. 2005;54:719–748. doi: 10.1080/10635150500234625. [DOI] [PubMed] [Google Scholar]
- 4.Parra-Olea G, Garcia-Paris M, Wake DB. Biol J Linn Soc. 2004;81:325–346. [Google Scholar]
- 5.Zhang P, Chen YQ, Zhou H, Liu YF, Wang XL, Papenfuss TJ, Wake DB, Qu LH. Proc Natl Acad Sci USA. 2006;103:7360–7365. doi: 10.1073/pnas.0602325103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.AmphibiaWeb. AmphibiaWeb: Information on Amphibian Biology and Conservation. Berkeley, CA: University of California; 2007. http://amphibiaweb.org. [Google Scholar]
- 7.Lynch JD, Duellman WE. U Kansas Spec Publ. 1997;23:1–236. [Google Scholar]
- 8.Lynch JD. U Kansas Mus Nat Hist Misc Publ. 1971:1–238. [Google Scholar]
- 9.Frost DR. Amphibian Species of the World: An Online Reference Version 5.0. New York: American Museum of Natural History; 2007. http://research.amnh.org/herpetology/amphibia/index.php. [Google Scholar]
- 10.Hass CA, Hedges SB. J Zool. 1991;225:413–426. [Google Scholar]
- 11.Hedges SB. In: Biogeography of the West Indies: Past, Present, and Future. Woods CA, editor. Gainesville, FL: Sandhill Crane; 1989. pp. 305–370. [Google Scholar]
- 12.Crawford AJ, Smith EN. Mol Phylogenet Evol. 2005;35:536–555. doi: 10.1016/j.ympev.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Darst CR, Cannatella DC. Mol Phylogenet Evol. 2004;31:462–475. doi: 10.1016/j.ympev.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 14.Ruvinsky I, Maxson LR. Mol Phylogenet Evol. 1996;5:533–547. doi: 10.1006/mpev.1996.0048. [DOI] [PubMed] [Google Scholar]
- 15.Lynch JD. Herpetologica. 1986;42:248–258. [Google Scholar]
- 16.Lynch JD. Rev Acad Colomb Cienc Exac Fisic Nat. 2000;24:129–156. [Google Scholar]
- 17.Savage JM, Myers CW. Am Mus Novit. 2002:1–48. [Google Scholar]
- 18.Siqueira S, Ananias F, Recco-Pimentel SM. Genet Mol Biol. 2004;27:363–372. [Google Scholar]
- 19.Lehr E, Fritzsch G, Muller A. Zool Scr. 2005;34:593–603. [Google Scholar]
- 20.Roelants K, Gower DJ, Wilkinson M, Loader SP, Biju SD, Guillaume K, Moriau L, Bossuyt F. Proc Natl Acad Sci USA. 2007;104:887–892. doi: 10.1073/pnas.0608378104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joglar RL. Biogeography of the West Indies: Past, Present, and Future. Gainesville, FL: Sandhill Crane; 1989. pp. 371–408. [Google Scholar]
- 22.Bossuyt F, Meegaskumbura M, Beenaerts N, Gower DJ, Pethiyagoda R, Roelants K, Mannaert A, Wilkinson M, Bahir MM, Manamendra-Arachchi K, et al. Science. 2004;306:479–481. doi: 10.1126/science.1100167. [DOI] [PubMed] [Google Scholar]
- 23.Hedges SB. In: Biogeography of the West Indies: Patterns and Perspectives. Woods CA, Sergile FE, editors. Boca Raton, FL: CRC; 2001. pp. 15–33. [Google Scholar]
- 24.Iturralde-Vinent MA, MacPhee RDE. Bull Am Mus Nat Hist. 1999;238:1–95. [Google Scholar]
- 25.Pindell JL. In: Caribbean Geology: An Introduction. Donovan SK, Jackson TA, editors. Kingston, Jamaica: The University of the West Indies Publishers' Association; 1994. pp. 13–39. [Google Scholar]
- 26.Rosen DE. Syst Zool. 1975;24:431–464. [Google Scholar]
- 27.Rosen DE. Ann MO Bot Gard. 1985;72:636–659. [Google Scholar]
- 28.Savage JM. Ann MO Bot Gard. 1982;69:464–547. [Google Scholar]
- 29.Pregill GK. Syst Zool. 1981;30:147–155. [Google Scholar]
- 30.Williams EE. In: Biogeography of the West Indies: Past, Present, and Future. Woods CA, editor. Gainesville, FL: Sandhill Crane; 1989. pp. 1–46. [Google Scholar]
- 31.Hedges SB, Hass CA, Maxson LR. Proc Natl Acad Sci USA. 1992;89:1909–1913. doi: 10.1073/pnas.89.5.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hedges SB. Annu Rev Ecol Syst. 1996;27:163–196. [Google Scholar]
- 33.Roca AL, Bar-Gal GK, Eizirik E, Helgen KM, Maria R, Springer MS, O'Brien SJ, Murphy WJ. Nature. 2004;429:649–651. doi: 10.1038/nature02597. [DOI] [PubMed] [Google Scholar]
- 34.Hedges SB. Ann MO Bot Gard. 2006;93:231–244. [Google Scholar]
- 35.Hedges SB. In: Contributions to West Indian Herpetology: A Tribute to Albert Schwartz. Powell R, Henderson RW, editors. Ithaca, NY: Society for the Study of Amphibians and Reptiles; 1996. pp. 95–127. [Google Scholar]
- 36.Seidel ME. Am Mus Novit. 1988;2918:1–41. [Google Scholar]
- 37.Seidel ME. In: Contributions to West Indian Herpetology: A Tribute to Albert Schwartz. Powell R, Henderson RW, editors. Ithaca, NY: Society for the Study of Amphibians and Reptiles; 1996. pp. 169–174. [Google Scholar]
- 38.Nicholson KE, Glor RE, Kolbe JJ, Larson A, Hedges SB, Losos JB. J Biogeogr. 2005;32:1–10. [Google Scholar]
- 39.Droxler AW, Burke KC, Cunningham AD, Hine AC, Rosencrantz E, Duncan DS, Hallock P, Robinson E. In: Tectonic Boundary Conditions for Climate Reconstructions. Crowley TJ, Burke KC, editors. New York: Oxford Univ Press; 1998. [Google Scholar]
- 40.Williams EE. Q Rev Biol. 1969;44:345–389. [Google Scholar]
- 41.Gregory-Wodzicki KM. Palaeogeogr Palaeoclimatol Palaeoecol. 2002;180:331–348. [Google Scholar]
- 42.MacFadden BJ. Trends Ecol Evol. 2006;21:157–165. doi: 10.1016/j.tree.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 43.Duellman WE. In: Patterns of Distribution of Amphibians. Duellman WE, editor. Baltimore: The Johns Hopkins Univ Press; 1999. pp. 255–328. [Google Scholar]
- 44.Kumar S, Tamura K, Nei M. Briefings Bioinf. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 45.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony and Other Methods Version 4. Sunderland, MA: Sinauer; 2003. [Google Scholar]
- 46.Stamatakis A. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 47.Ronquist F, Huelsenbeck JP. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 48.Thorne JL, Kishino H. Syst Biol. 2002;51:689–702. doi: 10.1080/10635150290102456. [DOI] [PubMed] [Google Scholar]
- 49.Yang Z, Yoder AD. Syst Biol. 2003;52:705–716. doi: 10.1080/10635150390235557. [DOI] [PubMed] [Google Scholar]
- 50.Pindell JL, Kennan L. Plate Model for the Caribbean. Austin, TX: University of Texas; 2002. www.ig.utexas.edu/CaribPlate/forum/pindell/pindell_kennan.htm. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.