Abstract
To understand the mechanisms controlling vein patterning in Arabidopsis thaliana, we analyzed two phenotypically similar mutants, varicose (vcs) and trident (tdt). We had previously identified VCS, and recently, human VCS was shown to function in mRNA decapping. Here, we report that TDT encodes the mRNA-decapping enzyme. VCS and TDT function together in small cytoplasmic foci that appear to be processing bodies. To understand the developmental requirements for mRNA decapping, we characterized the vcs and tdt phenotypes. These mutants were small and chlorotic, with severe defects in shoot apical meristem formation and cotyledon vein patterning. Many capped mRNAs accumulated in tdt and vcs mutants, but surprisingly, some mRNAs were specifically depleted. In addition, loss of decapping arrested the decay of some mRNAs, while others showed either modest or no decay defects, suggesting that mRNAs may show specificity for particular decay pathways (3′ to 5′ and 5′ to 3′). Furthermore, the severe block to postembryonic development in vcs and tdt and the accompanying accumulation of embryonic mRNAs indicate that decapping is important for the embryo-to-seedling developmental transition.
INTRODUCTION
RNA decay is an essential process that allows rapid changes in a cell's gene expression profile, such as in response to environmental and developmental signals. Decay of an mRNA can be initiated by internal cleavage, or RNA decay can proceed from either its 5′ or its 3′ end. A large number of studies have elucidated the importance of microRNA (miRNA)–directed RNA cleavage for development in both animal and plant systems (Reinhart et al., 2002; Carrington and Ambros 2003). However, much less is known about the 5′ to 3′ and 3′ to 5′ decay pathways and their possible roles in the development of multicellular organisms.
In both mammalian and yeast cells, initiation of bulk mRNA decay typically requires the removal of the 3′ poly(A) tail (Couttet et al., 1997; reviewed in Parker and Song, 2004). The deadenylated mRNA can then enter one of two decay pathways: either the cytoplasmic exosome complex can extend decay from the 3′ end; or the 5′ cap can be cleaved (decapping), after which the XRN1 exoribonuclease hydrolyzes the RNA from its 5′ end. The decapping pathway is also used by specialized mRNA decay pathways. For example, deadenylation-independent decapping is used in yeast both for autoregulation of the RPS28B RNA and for nonsense-mediated decay, in which mRNAs with a premature stop codon are selectively degraded (Muhlrad and Parker, 1994; Badis et al., 2004). These RNA decay pathways have been characterized extensively in yeast, and studies using mammalian cells revealed that the mRNA-decapping pathway is largely conserved.
Decapping of mRNA in yeast is performed by DCP2, a NUDIX/mutT domain–containing protein whose activity is stimulated by interaction with a second protein, DCP1 (Beelman et al., 1996; Dunckley and Parker, 1999). In mammalian cells, decapping requires an additional protein, called either Ge-1 or HEDLS, which facilitates the binding of hDCP1 and hDCP2 (Fenger-Grøn et al., 2005; Yu et al., 2005). No HEDLS homolog was found in the yeast genome, indicating that there are differences between the decapping complexes in animals and yeast.
Yeast and mammalian decapping enzymes localize to dispersed cytoplasmic foci called processing bodies (P-bodies) (van Dijk et al., 2002; Sheth and Parker, 2003). In addition to roles in RNA decay, P-bodies are also important for sequestering translationally arrested mRNAs (Liu et al., 2005a, 2005b; Sen and Blau, 2005), and in yeast, sequestered RNAs can exit the P-body and return to polysomes (Brengues et al., 2005). Thus, the P-body is emerging as a dynamic structure with functions in multiple aspects of RNA metabolism and control.
In plants, bulk mRNA decay pathways and their possible roles in development are not well known. One indication that these pathways might have developmental roles comes from observations of Arabidopsis thaliana poly(A) ribonuclease mutants, which show defects in embryogenesis (Chiba et al., 2004; Reverdatto et al., 2004). However, the relative importance of 5′ versus 3′ decay and the specificity of mRNA targets for each pathway are largely unknown.
The 5′ to 3′ mRNA decay pathway in plants has been studied using mutants with defects in the Arabidopsis XRN4 exoribonuclease. In contrast with yeast and animals, Arabidopsis does not appear to contain an XRN1-like enzyme but instead has three XRN2-like enzymes (XRN2 to XRN4), and XRN4 appears to encode the functional 5′ exoribonuclease for the decay of cytoplasmic decapped RNAs (Kastenmayer and Green, 2000). Reverse genetics, ethylene insensitivity, and overexpression suppressor screens have led to the identification of xrn4 mutants (Gazzani et al., 2004; Souret et al., 2004; Olmedo et al., 2006; Potuschak et al., 2006). xrn4 mutants accumulate a small set of decapped RNAs, including fragments produced following the cleavage of some miRNA targets and RNAs encoding the ethylene-signaling components EBF1 and EBF2. These observations have led to the question of whether the 5′ to 3′ mRNA decay pathway in plants serves just for bulk decay or whether it might also be used for regulated destabilization of specific mRNA classes.
To identify genes that are required for normal leaf development, we are analyzing mutants with defects in leaf vein patterning. The vascular system is composed of two tissues, xylem and phloem, that are organized together into veins. Vascular tissues arise early in organogenesis, and leaves and cotyledons produce stereotypical patterns of dispersed veins. A large body of data supports a role for auxin as a positive signal (reviewed in Sieburth and Deyholos, 2006). In addition to auxin, molecular characterization of vascular mutants has revealed roles for polyamines, inositol (1,4,5) triphosphate, sterols, vesicle trafficking, and root-derived carotenoids in modulating vein patterning (Hanzawa et al., 2000; Carland et al., 2002; Geldner et al., 2003; Carland and Nelson, 2004; Van Norman et al., 2004; Clay and Nelson, 2005; Koizumi et al., 2005; Sieburth et al., 2006).
We previously described varicose (vcs) mutants and showed that VCS encodes a WD domain–containing protein of unknown function and that homologs are present in metazoan genomes (Deyholos et al., 2003). Five of our vcs alleles showed an identical temperature-dependent phenotype. When grown at 16°C, vcs mutants produce broad leaves with a pointed apex and a modestly diminished pattern of veins; at 22°C, they produce narrow leaves with a small number of abnormally thick veins; and when grown at 29°C, vcs leaves are highly reduced and contain only a single very thick vein. Molecular characterization of the lesions in these vcs mutants are consistent with their being null alleles. A highly similar gene, VARICOSE-RELATED (VCR), is located <1 kb from VCS. Although we could detect no phenotype for vcr mutants, we speculated that activity of this gene might be responsible for the suppressed phenotypes we observed at low growth temperatures. Recently, two studies characterized the VCS homolog from humans. In one study, hVCS (called Ge-1) was shown to colocalize with hDCP2 (Yu et al., 2005). DCP2 encodes the mRNA-decapping enzyme, which is located in P-bodies; this localization of hVCS suggested that VCS might also function in mRNA storage and decay. Another study found that interactions between hDCP2 and hDCP1 depended on the presence of hVCS (called HEDLS) (Fenger-Grøn et al., 2005). These studies raised the possibility that the developmental defects in vcs mutants might arise due to defects in the assembly of the mRNA-decapping complex.
Here, we report the characterization of the Arabidopsis trident (tdt) mutant and show that TDT encodes the Arabidopsis mRNA-decapping enzyme (DCP2 homolog). Furthermore, we show that VCS is also a component of the decapping complex. Both tdt and vcs mutants show defects in RNA decay, and our decay measurements revealed specific mRNA targets of this pathway.
RESULTS
The Phenotypic Severity of vcs Mutants Depends on Genetic Background
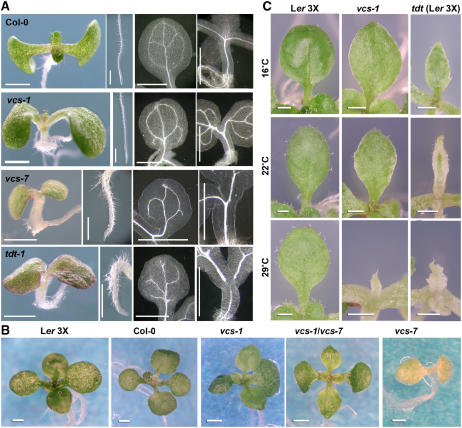
To gain insight into the molecular mechanisms controlling vein patterning, we are characterizing Arabidopsis mutants with vein patterning defects. We previously characterized five vcs alleles (vcs-1 to vcs-5) that were isolated in the Landsberg erecta (Ler) background and characterized vcs-1 in detail (Deyholos et al., 2003). All five produced identical temperature-dependent phenotypes, with increasingly severe phenotypes occurring when the mutants were grown at intermediate and high temperatures. We have now obtained additional vcs alleles from the SALK T-DNA insertion collection (Alonso et al., 2003), which were generated in the Columbia (Col-0) background. These insertion lines segregated for mutants with a more severe phenotype than vcs-1. We determined that one insertion allele, vcs-7, was an RNA null mutant (see Supplemental Figure 1 online), so we characterized this mutant in detail. When grown at 22°C, vcs-7 mutants had short roots, small chlorotic cotyledons, and typically failed to produce leaf primordia. By contrast, vcs-1 mutants were green, had slightly longer roots, and produced narrow pointed leaves (Figure 1A; see Supplemental Figure 2 online). In addition, vcs-7 mutants produced cotyledons that expanded slowly and accumulated anthocyanins around their margins, hypocotyls that were short and slightly swollen, and root hairs that were short and swollen. These traits were similar to those we reported previously for vcs-1 grown at 29°C (Deyholos et al., 2003). The vcs-7 mutant also showed cotyledon vein pattern defects. Cotyledons of wild-type and vcs-1 plants contain a primary vein and four secondary veins that form between two and four closed loops, whereas in vcs-7, the cotyledon secondary veins were mostly free-ending, resulting in a fork-like appearance (Figure 1A). This vcs-7 phenotype is similar to that of the recently described vcs-6 allele (Xu et al., 2006).
Figure 1.
Phenotypes of vcs and tdt Seedlings.
(A) Phenotypes of seedlings grown for 6 d at 22°C. Panels show seedling, root, cotyledon vein pattern, and hypocotyl vasculature (left to right).
(B) The vcs-1/vcs-7 transheterozygote shows a suppressed phenotype similar to that of vcs-1 homozygotes. These seedlings were grown for 15 d at 16°C.
(C) Leaf development of vcs-1, and tdt-1 in the Ler genetic background, depends on growth temperature.
Bars = 1 mm.
The vcs-7 mutant also differed from vcs-1 in its developmental response to growth temperature. The vcs-1 mutant, when grown at 16°C, produced broad pointed leaves with modestly disrupted vein patterns (Deyholos et al., 2003). By contrast, growth of vcs-7 at 16°C resulted in only modest suppression. For example, among 33 vcs-7 mutants grown at 16°C, 8 produced no visible leaf primordia, 15 produced numerous very small structures that resembled leaf primordia, and 10 produced small leaves (see Supplemental Figure 3 online). These data suggest that most, but not all, of the low-temperature suppression of the vcs phenotype was abolished in the vcs-7 mutant.
To determine why vcs-7 and vcs-1 mutants showed such different phenotypes, we constructed vcs-1/vcs-7 transheterozygotes. The transheterozygote was indistinguishable from vcs-1; it produced green organs, a longer root, and leaves that expanded to become broad and pointed (Figure 1B). This result indicated that vcs suppression arose due to a dominant activity in the Landsberg accession. A likely candidate for low-temperature suppression was VCR. Mutations in VCR result in no observable phenotype (Deyholos et al., 2003), and because VCR and VCS are separated by <1 kb, we have been unable to assess redundancy genetically. However, we were able to test whether the suppression of vcs-1 was linked to the VCS locus. To do this, we examined the mutant F2 progeny from crosses between vcs-7 heterozygotes and Ler and between vcs-1 heterozygotes and Col-0. The F2 progeny from both crosses segregated for both severe (vcs-7–like) and suppressed (vcs-1–like) mutants in ratios consistent with two or more unlinked suppressor loci. Because Ler suppressors were not linked to VCS, we conclude that VCR does not contribute substantially to low-temperature suppression. These observations, together with the transheterozygote phenotype, indicate that unlinked dominant loci within the Ler genome suppress the vcs phenotype.
tdt Mutants Appear Similar to vcs Mutants
Another mutant in our collection, tdt, had a severe phenotype similar to that of vcs-7, but it mapped to a different chromosomal location. Like vcs-7, tdt mutants produced no leaves, short roots with short and swollen root hairs, chlorotic cotyledons that accumulated anthocyanins around their margins, and a short and swollen hypocotyl (Figure 1A). In addition, the cotyledon–hypocotyl junction was extremely fragile, which caused cotyledons to frequently fall off, even with gentle handling. We attribute this fragility to a pinched region that we often observed at the proximal end of the cotyledon petiole (see Supplemental Figure 4 online).
Vascular defects in tdt mutants were also similar to those of vcs-7 in that cotyledon secondary veins mostly failed to form closed loops (Figure 1A; see Supplemental Figure 1 online). However, tdt mutants also showed a novel vascular phenotype within the vascular transition. The vascular transition is a region where the root-like vascular organization of the lower hypocotyl shifts to a shoot-like vascular organization (Esau, 1965). In tdt-1 mutants, this region ranged from being devoid of detectable xylem to containing xylem but with poorly aligned veins. This defect showed incomplete penetrance, but it was most prevalent when seedlings were germinated at higher temperatures (Table 1). By contrast, not only did we never observe a loss of vascular tissue within the vcs transition zone (n > 500 vcs-7 mutants), but vcs-1 (and other alleles in the Landsberg background) instead accumulate extra ectopic tracheary elements within their cotyledons and the upper hypocotyl (Deyholos et al., 2003).
Table 1.
Prevalence of the tdt-1 Transition Zone Defect Depends on Germination Temperature
| Growth Temperature | Vascular Transition Zone Defect | n |
|---|---|---|
| 16°C | 36% | 86 |
| 22°C | 69% | 42 |
| 29°C | 94% | 85 |
As with vcs-7, tdt was isolated in the Col-0 accession. To further explore possible similarities between vcs and tdt, we examined whether the phenotype of tdt mutants could also be suppressed by unlinked loci in Ler. Following a cross between tdt heterozygotes and Ler, the F2 mutants showed both suppressed and severe phenotypes (data not shown). We compared leaf development in vcs-1 (in the Ler accession) with leaf development of tdt after four sequential crosses to Ler (tdt MC4). As with vcs-1, tdt MC4 produced leaf primordia (Figure 1C). Furthermore, there was much greater leaf development in both vcs-1 and tdt MC4 when grown at 16°C, while growth at 22 and 29°C resulted in progressively more severe defects in leaf development. However, tdt MC4 showed less extensive leaf development than that shown by vcs-1. Although some details of the vcs and tdt phenotypes differed, the major phenotypic defects were similar, which suggested that VCS and TDT might function in related pathways.
VCS and TDT Are Required for the Formation of a Normal Shoot Apical Meristem
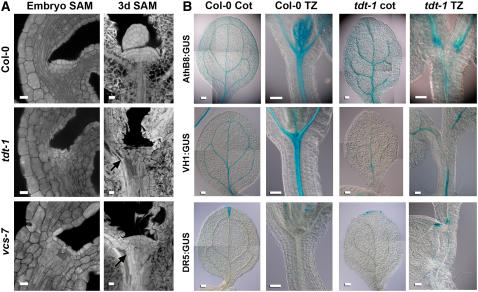
To gain insight into the leaf initiation defects in tdt and vcs-7 mutants, we used confocal microscopy to analyze shoot apical meristem (SAM) structure. In the wild type, the SAM arises during embryogenesis; in the mature embryo, it appears as a dome composed of three well-defined layers of small, dense cells (Barton and Poethig, 1993). Among the embryos derived from a tdt-1 heterozygote, 25% were clearly distinguishable by their misshapen cells (5 of 23). These embryos contained small dense cells in the SAM region; however, they were not organized into distinct layers and they did not appear as a dome (Figure 2A). The vcs-7 embryos were also clearly distinguished from their siblings based on their SAM morphology (seven vcs-7 embryos out of 25 embryos examined). These vcs-7 embryos contained small dense cells in the SAM region; however, this region was not dome-shaped, nor were the cells organized into distinct layers (Figure 2A).
Figure 2.
SAM and Vascular Defects in tdt and vcs-7.
(A) Confocal analysis of SAM organization in mature embryos and 3-d-old seedlings. Arrows indicate ectopic differentiating tracheary elements. Bars = 10 μm.
(B) Reporter gene expression in cotyledon and hypocotyl transition zones (TZ) of the wild type (Col-0) and tdt-1. AthB8:GUS confers expression in provascular, procambial, and vascular cells; VH1:GUS confers strong expression in provascular, procambial, and phloem cells; DR5:GUS confers expression in cells responding to auxin. Bars = 100 μm.
To determine how the SAM changed following germination, we also examined SAM of 3-d-old seedlings. The wild-type 3-d-old SAM appeared in its characteristic dome of layered cells and was accompanied by leaf primordia (Figure 2A). By contrast, the tdt 3-d-old seedlings still showed disorganized cell layers and no leaf primordia (n = 13). The SAM of the vcs-7 3-d-old seedlings varied: some were modestly rounded and layered (Figure 2A), while others were more similar to the tdt embryonic SAM; however, none had leaf primordia (n = 11). Thus, the severe defects in tdt and vcs SAMs might preclude the initiation of leaf primordia.
Vascular Defects in the tdt Transition Zone Arise Early
To understand the developmental basis for the tdt mutant's hypocotyl vascular defects, we characterized hypocotyl vascular development using well-characterized β-glucuronidase (GUS) reporters. We first used the AthB8:GUS reporter, which confers expression to provascular, procambial, and differentiated vascular tissues (Baima et al.,1995; Scarpella et al., 2004). To determine whether these tissues were present in the tdt transition zone, we compared GUS staining patterns of AthB8:GUS in the wild type with that of AthB8:GUS in tdt seedlings. In the wild type, we observed strong vascular-associated GUS staining (Figure 2B). In the tdt mutants, we observed strong AthB8:GUS expression in places where vascular tissues were present. However, in the tdt transition zone, the extent of GUS staining correlated with the extent of the vascular defect, and in individuals lacking transition zone vascular tissues, AthB8:GUS conferred no GUS staining. This observation suggested that TDT was required prior to provascular or procambial specification.
We also examined vascular development using the VH1:GUS reporter, which confers expression to provascular, procambial, and phloem cells (Clay and Nelson, 2002). We observed strong vascular expression in the wild type, including through the transition zone (Figure 2B). However, as with AthB8:GUS, the extent of GUS staining in the tdt-1 mutant transition zone correlated with the extent of transition zone vascular development. In tdt-1 mutants devoid of transition zone xylem, there was no VH1:GUS staining. These observations corroborated the AthB8:GUS expression patterns and suggested that the loss of xylem in the tdt-1 transition zone was accompanied by a loss of phloem.
Finally, we analyzed GUS staining patterns conferred by the auxin-responsive DR5:GUS reporter (Ulmasov et al., 1997), which is useful for detecting provascular and procambial cells (Mattsson et al., 2003). In tdt-1 mutants, we detected strong DR5:GUS expression at the proximal termini of cotyledon vascular strands but no expression within the transition zone (Figure 2B). This observation suggested that auxin was transported normally from the cotyledon but that transport pathways might terminate at the transition zone. Together, these reporter expression patterns suggested that vascular defects in the tdt transition zone arose early, prior to provascular cell specification.
TDT Encodes the Arabidopsis Homolog of DCP2
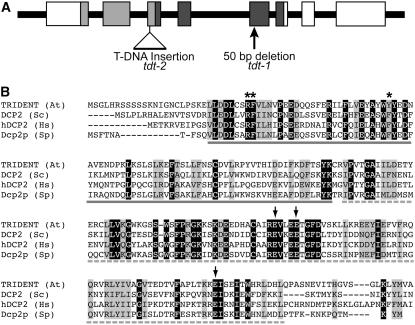
We mapped TDT to a 65-gene interval near the top of chromosome 5. We then screened T-DNA insertion lines (Alonso et al., 2003) mapping to genes within this interval, looking for mutants with a similar phenotype. Only one line, SALK_000519, segregated for a similar mutant, and this mutant was indistinguishable from tdt-1. Crosses between tdt-1 and the Salk line (heterozygotes) produced 25% mutant seeds (59 wild type and 18 tdt; χ2 = 0.108, df = 1, critical value = 3.841 at 95%), indicating that these were allelic mutants. SALK_000519 (tdt-2) contained a T-DNA insertion in exon 3 of gene At5g13570. We sequenced this gene from tdt-1 mutants and identified a 50-bp deletion within exon 5 (Figure 3A). The tdt-1 deletion occurred just after Val-176 and is predicted to result in a truncated protein with 17 novel C-terminal amino acids.
Figure 3.
TDT Encodes the mRNA-Decapping Enzyme.
(A) Diagram of the TDT gene. Boxes correspond to exons, and the 5′ end is to the left. Light gray boxes correspond to the DCP2 domain, and dark gray boxes correspond to the NUDIX domain. Locations of the T-DNA insertion and the lesion identified in tdt-1 are indicated.
(B) Alignment of DCP2 and NUDIX domains of TDT with those of Saccharomyces cerevisiae (Sc), Schizosaccharomyces pombe (Sp), and Homo sapiens (Hs). Arrows indicate conserved E residues that are required for catalytic activity (all are conserved in TDT), and asterisks indicate residues required for interaction with DCP1 (She et al., 2006). Solid gray underlining indicates the DCP2 domain, and the dashed underline indicates the NUDIX domain.
To determine whether we had identified the correct gene, we tested both a 35S:green fluorescent protein (GFP):TDT transgene and a genomic clone containing the TDT gene for their ability to rescue tdt-1 mutants. We observed full rescue of tdt-1 mutants by introduction of the TDT genomic clone (observed for 12 independent lines). For example, line 29 was identified by PCR as heterozygous at the TDT locus. Its self-pollinated progeny segregated 348 phenotypically wild-type seedlings and 19 tdt mutants (18.3:1), which is consistent with the 15:1 ratio expected for full rescue (χ2 = 0.742; critical value = 3.841 at 95%). The full rescue by the TDT genomic clone indicated that we had identified the correct gene. We observed partial rescue of the tdt mutant in lines carrying the 35S:GFP:TDT transgene. We followed the development of two homozygous tdt-1 transgenic seedlings. Both plants produced leaves; however, they were stunted, male-sterile, and showed limited female fertility. The partial rescue of tdt-1 by the GFP:TDT construct further confirmed that we had identified the correct gene.
TDT was predicted to encode a 374–amino acid protein with N-terminal DCP2 and NUDIX domains (Figure 3A). These two domains are highly conserved among mRNA-decapping enzymes (Figure 3B). The TDT N terminus, through the NUDIX domain, shared 49% identity (69% similarity) with the Schizosaccharomyces pombe sequence, 41% identity (65% similarity) with Homo sapiens, and 37% identity (62% similarity) with Saccharomyces cerevisiae. The C-terminal domains of these proteins are much less similar: both the S. cerevisiae and S. pombe proteins are much longer (741 and 970 amino acids, respectively, compared with 138 amino acids in TDT).
Studies of the yeast mRNA-decapping enzyme have identified three invariant Gln residues that are essential for catalytic activity (She et al., 2006); all three of these residues are present in the predicted TDT sequence (Figure 3B), and the lesions for tdt-1 and tdt-2 predict abolishing one and all three of these residues, respectively. Thus, the identical phenotypes of tdt-1 and tdt-2 are consistent with molecular predictions of a loss of enzyme activity. Characterization of the yeast DCP2 protein also led to the identification of three amino acids that are required for interaction with DCP1 (She et al., 2006). The Arabidopsis TDT sequence has mostly retained this motif; two of the amino acids are identical, and the third shows a conserved change (Figure 3B). The molecular identification of TDT and VCS as potential components of the same complex is consistent with their similar mutant phenotypes.
TDT and VCS Function in the Same Pathway and Can Physically Interact
In animal cells, Ge-1/HEDLS/hVCS provides a scaffold that stabilizes interactions between hDCP1 and hDCP2, and this stabilization is required for decapping activity (Fenger-Grøn et al., 2005). Sequence identity between TDT and DCP2 and between VCS and Ge-1/HEDLS suggested that these Arabidopsis proteins might also function together in mRNA decay. To test this possibility, we analyzed tdt-1 vcs-7 double mutants and tested whether we could detect interactions between the VCS and TDT proteins.
We reasoned that if VCS and TDT functioned in a single pathway, then the vcs-7 tdt-1 double mutant should appear similar to the single mutants. We generated several F2 lines that segregated nine phenotypically wild-type plants to seven mutants (n = 276; χ2 = 0.326), suggesting that they segregated for both vcs-7 and tdt-1. These F2 mutants all appeared similar, which was consistent with the double mutant resembling the single mutants. To be sure that the double mutant was represented among these similar mutants, we isolated DNA from individual mutant F2 seedlings and genotyped each at the VCS and TDT loci (Table 2). We found all of the expected genotypes, and each was present at its expected frequency, including the double mutant. Finding the double mutant among the phenotypically similar single mutants supports VCS and TDT functioning in the same pathway.
Table 2.
Genotype Analysis of tdt-1 vcs-7 Double Mutants
| Genotype | Number |
|---|---|
| VCS/VCS tdt/tdt | 21 |
| VCS/vcs tdt/tdt | 24 |
| vcs/vcs TDT/tdt | 30 |
| vcs/vcs TDT/TDT | 21 |
| vcs/vcs tdt/tdt | 15 |
| Total | 111 |
χ2 = 2.01, with a critical value = 15.507 (df = 4).
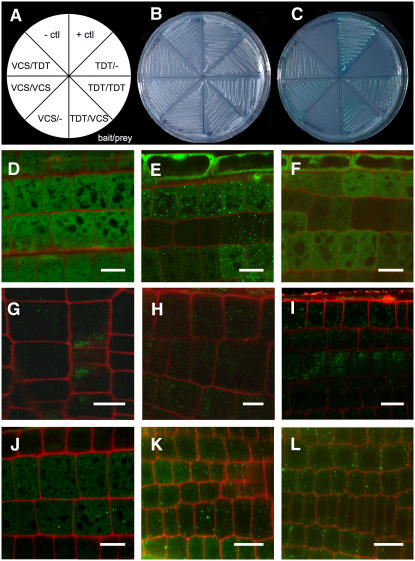
To explore possible physical interactions between TDT and VCS proteins, we performed a directed yeast two-hybrid analysis. While neither the VCS bait nor the TDT bait was capable of activating the selectable markers on its own, the selectable markers were activated in cells expressing both VCS bait and TDT prey and both VCS prey and TDT bait (Figures 4A to 4C). In addition, both VCS–VCS and TDT–TDT self-interactions were also detected. With the exception of the TDT–TDT interaction, these results are consistent with the in planta and in vitro interactions recently reported by Xu et al. (2006). The TDT–TDT self-interaction has not yet been verified in planta, but if confirmed, it might be difficult to integrate into the Xu et al. (2006) model for the decapping complex.
Figure 4.
VCS and TDT Interact, and TDT Localizes to P-Body–Like Cytoplasmic Foci.
(A) to (C) Yeast two-hybrid analysis of VCS and TDT interaction.
(A) Map of yeast strains shown in (B) and (C).
(B) trp-leu- medium shows growth of each transformed yeast line.
(C) trp-leu-his-ade-x-α-gal+ medium shows interaction in the positive control, combinations that include both VCS and TDT, and VCS–VCS and TDT–TDT self-interactions.
(D) to (L) Confocal micrographs of root elongation zones of 4-d-old seedlings. Bars = 10 μm.
(D) 35S:GFP in wild-type roots shows signal distributed throughout the cells.
(E) 35S:GFP:TDT in wild-type roots shows GFP signal that is largely localized to small spots dispersed through the cytoplasm.
(F) 35S:GFP:TDT in wild-type roots treated with cycloheximide no longer shows localized spots of GFP signal; instead, the GFP signal resembles that of 35S:GFP.
(G) to (I) 35S:GFP:TDT in roots of vcs-7 mutants.
(G) Untreated vcs-7 seedlings show weak and diffuse signal from 35S:GFP:TDT.
(H) DMSO-treated controls also show weak and diffuse staining.
(I) vcs-7 mutants treated for 1 h with the proteasome inhibitor MG132 show slightly brighter, but still diffuse, signal from the 35S:GFP:TDT transgene.
(J) to (L) 35S:GFP:TDT in wild-type roots shows similar localization and intensity in untreated (J), DMSO-treated (K), and MG132-treated (L) seedlings.
VCS-Dependent Localization of TDT to P-Bodies
In yeast and animal cells, mRNA decapping is localized to distinct cytoplasmic foci, called P-bodies (van Dijk et al., 2002; Sheth and Parker, 2003; Cougot et al., 2004). To determine whether TDT also localized to cytoplasmic foci, we analyzed the intracellular localization of the GFP:TDT fusion protein expressed from the 35S promoter. This construct conferred partial rescue to tdt mutants (see above), suggesting that its localization pattern is biologically relevant. In roots of 4-d-old seedlings, we found GFP expression that was largely localized to distinct cytoplasmic spots (Figures 4E and 4J to 4L) in contrast with the cytoplasmic localization of GFP lacking the TDT sequences (Figure 4D).
In yeast and animals, treatment with cycloheximide results in the loss of P-bodies (Sheth and Parker, 2003). Cycloheximide is a protein synthesis inhibitor that arrests ribosome movement, and its effect on P-bodies is thought to arise due to an interruption in the supply of mRNA destined for decay. To determine whether GFP:TDT localization was also sensitive to cycloheximide, we performed confocal analysis of cycloheximide-treated seedlings. Following a short cycloheximide treatment, no cytoplasmic foci were observed (Figure 4F); instead, we observed a cytoplasmic GFP signal similar to that of 35S:GFP. This loss of cytoplasmic foci following cycloheximide treatment supports the punctate pattern of GFP:TDT corresponding to P-bodies.
In mammalian cells, hVCS/Ge-1/HDLS is required for the assembly of the decapping complex. To test whether VCS was required for normal localization of TDT, we crossed GFP:TDT to vcs-7 and examined the intracellular pattern of GFP in mutant roots. We examined >25 GFP-positive vcs-7 mutants from three different crosses, and in every one, we found very weak and diffuse GFP expression (Figure 4G), while the phenotypically wild-type siblings still showed punctate spots within the cytoplasm. Incubation of the vcs-7 seedlings in the proteasome inhibitor MG132 resulted in slightly brighter, but still diffuse, GFP signal (Figure 4I), while a control incubation in DMSO (Figure 4H) had no discernible effect. The same construct in wild-type plants showed no altered pattern of GFP:TDT localization when incubated in DMSO or MG132 (Figures 4J to 4L). These data indicated that VCS is required for the normal localization of GFP:TDT and that in the absence of VCS, TDT is unstable. Together, these observations support the notion these two proteins functioning together in a complex.
RNA Decay Is Altered in vcs and tdt Mutants
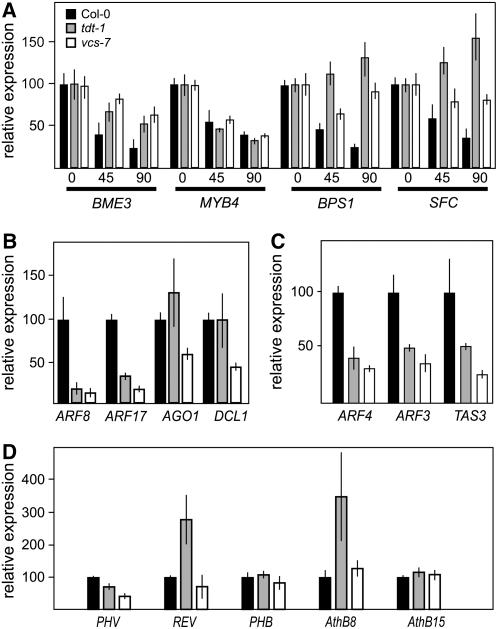
The molecular characterization of vcs and tdt led to the prediction that these mutants might have RNA decay defects. To test this prediction, we compared decay rates for four seedling-expressed mRNAs among the wild type, vcs-7, and tdt-1. We assayed RNA decay by comparing relative levels of specific seedling-expressed mRNAs following cordycepin-induced transcriptional arrest (Gutiérrez et al., 2002).
We analyzed the decay of BLUE MICROPYLAR END3 (BME3; At3g54810) RNA, which encodes a GATA-type zinc finger transcription factor that is required for germination (Gutiérrez et al., 2002; Liu et al., 2005c). This RNA was previously reported as unstable, and our decay assays confirmed its instability in the wild type (Figure 5A). In both tdt-1 and vcs-7, this mRNA's decay rate was approximately half that of the wild type. Reduction of decay in tdt and vcs suggested that the normal pathway for this mRNA's decay includes decapping. However, the fact that some decay still occurred suggests either that the loss of TDT and VCS did not provide a complete block to decapping or that BME3 mRNA's normal decay also uses another pathway. There is no evidence that this mRNA, or any included in this part of our investigation, is the target of a small RNA. Thus, the mostly likely candidate for an alternative decay pathway is exosome-mediated 3′ to 5′ decay.
Figure 5.
vcs and tdt Mutants Show Defects in RNA Decay and Aberrant Patterns of RNA Accumulation.
In these graphs, data for the wild type are represented by black bars, data for tdt-1 by gray bars, and data for vcs-7 by white bars. Four-day-old seedlings are represented in (A), and 3-d-old seedlings are represented in (B) to (D). Error bars indicate sd; n = 4.
(A) RNA decay monitored by analyzing the relative expression of four mRNAs following treatment with the transcription inhibitor cordycepin (0, 45, or 90 min after the addition of cordycepin).
(B) Relative expression levels of select miRNA target RNAs in 3-d-old seedlings grown at 22°C. In tdt-1 and vcs-7, these miRNA target RNAs show levels that are either similar to, or lower than, that of the wild type.
(C) Analysis of two trans-acting silencing RNA targets (ARF3 and ARF4) and the TAS3 RNA.
(D) Relative expression levels of the HD-ZIPIII RNAs in tdt-1 and vcs-7 reveal no consistent pattern of mRNA overaccumulation in tdt-1 and vcs-7 mutants.
We also analyzed the decay of MYB4 (At4g38620) mRNA. This mRNA showed similar decay rates in the wild type, tdt-1, and vcs-7 (Figure 5A). These similar mRNA decay rates suggested that decay of this mRNA might not require decapping; instead, it might specifically use the exosomal 3′ to 5′ pathway.
We also analyzed the decay of RNA encoding SCARFACE (SFC) and BYPASS1 (BPS1). SFC encodes an ARF-GAP protein that is required for normal vesicle trafficking between the plasma membrane and the endosome and is required for normal vein patterning and root growth (Sieburth et al., 2006). BPS1 encodes a plant-specific protein that is required to prevent the root from producing a novel graft-transmissible apocarotenoid that arrests shoot growth (Van Norman et al., 2004; Van Norman and Sieburth, 2007). Both of these RNAs showed decay in the cordycepin-treated wild type (Figure 5A). However, in tdt-1 mutants, the relative levels of these two RNAs actually increased. Because real-time RT-PCR detects relative mRNA levels, the apparent increase of SFC and BPS1 mRNAs in tdt-1 mutants indicates a slower rate of decay than that of the internal control (ACTIN2). These data indicate a stringent requirement for decapping for the decay of both of these mRNAs. The decay of SFC and BPS1 mRNA in vcs-7 mutants was also strongly reduced, but not to the same extent as in tdt-1 mutants. The simplest explanation for the decay of these RNAs in vcs-7 is that VCR might support the limited formation of a functional decapping complex in a few specialized cell types.
These data reveal that the requirement for the decapping complex for mRNA decay depends on the specific mRNA.
RNA Cleavage Targeted by miRNAs and trans-Acting Silencing RNAs Does Not Require VCS or TDT
In yeast and animal cells, P-bodies have been implicated in both RNA decay and the sequestration of translationally arrested mRNAs (reviewed in Bruno and Wilkinson, 2006). In animal cells, P-bodies also contain both ARGONAUTE2 (AGO2) and GW182 (Liu et al., 2005a; Behm-Ansmant et al., 2006), where they are likely to function in translational arrest of miRNA targets. Recently, though, GW182 has also been implicated in miRNA-targeted degradation of specific mRNAs (Behm-Ansmant et al., 2006), which suggested that miRNA-directed cleavage of mRNA targets might also occur in P-bodies.
To address whether VCS and/or TDT were required for the normal cleavage of miRNA targets, we used real-time RT-PCR to assess relative levels of several different uncleaved miRNA targets (Figure 5B). We reasoned that if miRNA-directed cleavage was disrupted, then we would observe elevated levels of targets in tdt-1 and vcs-7 mutants. By contrast, we found that vcs-7 and tdt-1 mutants accumulated either similar levels (AGO1 and DCL1) or reduced levels (ARF8 and ARF17) of miRNA targets. The lack of elevated miRNA targets in the mutants indicated that neither miRNA biogenesis nor target cleavage required TDT or VCS.
We also considered ARF3 and ARF4 as possible RNA substrates for VCS and TDT, because these RNAs are regulated by trans-acting silencing RNAs, and this regulation is important for normal leaf development (Adenot et al., 2006; Fahlgren et al., 2006; Garcia et al., 2006). We used real-time RT-PCR to compare the levels of uncleaved ARF3 and ARF4 RNAs and to detect unprocessed TAS3 RNA. We found that the relative levels of these RNAs were decreased in tdt and vcs mutants (Figure 5C). Although it is not readily apparent why these RNAs are reduced, the observation that their levels are not elevated indicates that neither the cleavage of ARF3 or ARF4 target RNAs nor the processing of TAS3 RNA required TDT or VCS.
Because of their roles in leaf and vascular development, we were particularly interested in whether the class III homeodomain leucine zipper (HD-ZIPIII) genes were misexpressed. The HD-ZIPIII RNAs are miRNA targets, and members of this family (PHB, PHV, REV, AthB15/CNA, and AthB8) play both positive and negative roles in vascular patterning (McConnell and Barton, 1998; Emery et al., 2003; Prigge et al., 2004). Real-time RT-PCR analysis revealed that uncleaved HD-ZIPIII RNAs were generally at normal levels, with the exception of REV and AthB8 in tdt-1 (Figure 5D), and we confirmed these results by real-time RT-PCR using alternative primers (also flanking the cleavage site), sequencing reaction products, and by analysis of microarray data (see below). The mostly unchanged levels of these miRNA targets further support TDT and VCS not being required for miRNA-directed cleavage of target RNAs. Our data are at odds, however, with a recent analysis by Xu et al. (2006), who found PHB, AthB8, PHV, and REV to be highly elevated in both tdt/dcp2 and vcs mutants. Possible explanations for the difference between our results include developmental timing (our study used 4-d-old seedlings to minimize developmental differences, while the Xu et al. [2006] study used 6-d-old seedlings) or the part of the RNA examined. We assayed uncleaved HD-ZIPIII products, while the portion of these RNAs analyzed by Xu et al. (2006) was not described. It is possible that the cleavage fragments accumulate in tdt and vcs mutants, as 3′ fragments of miRNA-cleaved target RNAs have also been shown to accumulate in xrn4 mutants (Souret et al., 2004).
Global Analysis of mRNA Profiles in tdt Mutants
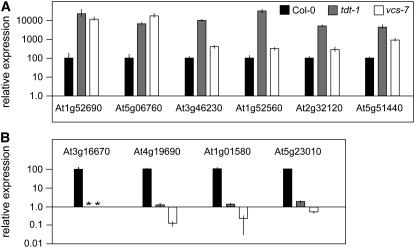
To gain broader insight into how the loss of mRNA decapping affected RNA levels, we performed microarray experiments to compare RNA profiles of tdt-1 and wild-type plants. To minimize the extent to which tdt's developmental defects would confound the results, we used RNA from 3-d-old seedlings. This analysis revealed a large number of mRNAs that were elevated in tdt mutants, and Supplemental Table 1 online lists the 142 mRNAs elevated by fivefold or higher. These elevated RNAs derived from many different functional classes, including signaling molecules, transcription factors, transporters, and metabolism-related. Two classes that were particularly well represented are embryo-expressed RNAs and those encoding heat-shock proteins.
To test whether these microarray results accurately reflected tdt-1 mRNA levels, and to explore whether RNA levels were affected similarly in vcs mutants, we performed real-time RT-PCR for six of these elevated mRNAs (Figure 6A). This analysis confirmed that each of these RNAs was elevated in tdt-1 and revealed even higher levels of accumulation than those reported by the microarray. Furthermore, each of these six RNAs was also elevated in vcs-7, although four of them showed more modest increases than that observed in tdt-1.
Figure 6.
RNA Accumulation and Depletion in tdt-1 and vcs-7 Mutants.
Real-time RT-PCR analysis of relative expression of genes with altered expression detected in the microarray. Data for the wild type are represented by black bars, data for tdt-1 by gray bars, and data for vcs-7 by white bars. Asterisks indicate not detectable levels; error bars indicate sd.
(A) The relative levels of six RNAs that microarray analysis indicated increased in tdt-1 show strong increases; the levels are also elevated in vcs-7.
(B) The relative levels of four RNAs that microarray analysis indicated as underaccumulated in tdt-1 show strong differences; the levels are also depleted in vcs-7.
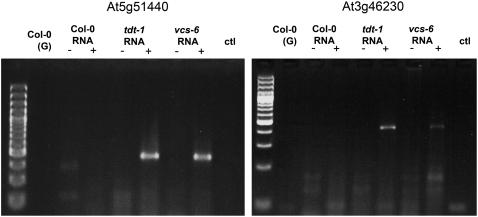
Molecular characterization of VCS and TDT led to the prediction that accumulated mRNAs would retain their 5′ cap. To determine whether this was the case, we assessed whether two of the RNAs that accumulated in tdt-1 and vcs-7 were capped. To do this, we used a modified rapid amplification of cDNA ends (RACE) assay and compared RNA ligation to an RNA primer before and after removal of the 5′ cap with Tobacco Acid Phosphatase (TAP) (Gazzani et al., 2004). Ligation products could only be detected in tdt and vcs RNAs that had been treated with TAP (Figure 7), indicating that in the untreated samples, the 5′ cap blocked ligation. These data provide strong evidence that VCS and TDT encode components of the mRNA-decapping machinery.
Figure 7.
tdt-1 and vcs-7 Accumulate Capped mRNAs.
Capped RNAs were assayed by ligation to a 5′ RNA adapter either before (−) or after (+) treatment with TAP to remove the cap. We detected an abundant product only in the mutants, and only after TAP treatment, confirming that these abundant mRNAs were capped. ctl indicates a reaction without template addition, and (G) indicates genomic DNA supplied as the template.
Our microarray analysis of tdt mutants also revealed a set of mRNAs that were present at lower levels in tdt-1, and Supplemental Table 2 online lists the 52 genes whose RNA levels were reduced by fivefold or more. These RNAs correspond to a wide range of functional groups, including signaling, transport (e.g., iron transport–related), and transcription factors. We had not predicted finding this group of severely reduced RNAs, and to characterize them further we examined four in detail using real-time RT-PCR. This analysis confirmed the very low expression of these RNAs in tdt-1 and revealed either similar or more severely depleted RNA levels in vcs-7 (Figure 6B).
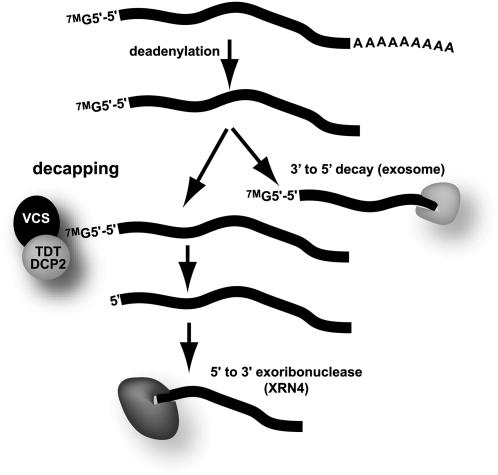
We compared the tdt overexpressed and underexpressed mRNAs with the 26 RNAs found to be elevated in xrn4 mutants (Souret et al., 2004; Olmedo et al., 2006). None of the RNAs elevated in xrn4 mutants was among our list of fivefold-overexpressed RNAs, and one, At4g00780, was among the tdt underexpressed mRNAs. The lack of congruence between gene expression changes in xrn4 and tdt RNAs was surprising, as models for mRNA decay depict these genes as acting in sequential steps of the same pathway (Figure 8).
Figure 8.
Model for mRNA Bulk Decay Pathways.
Two key features of an mRNA in the cytoplasm are the 5′-5′ 7-methylguanosine (7MG) cap and the 3′ poly(A) tail. Bulk mRNA decay is typically initiated by deadenylation. The deadenylated mRNA can then undergo further decay from its 3′ end by the activity of the exosome. Alternatively, the deadenylated mRNA can have its 5′ cap removed (decapped), followed by further decay from its 5′ end by XRN4 activity.
We noticed that many of the overexpressed mRNAs in tdt mutants encode embryogenesis-related proteins, such as Late Embryogenesis Early (LEA) and embryo-expressed heat shock proteins (Hughes and Galau, 1989; Wehmeyer and Vierling, 2000; Schmid et al., 2005). To further explore the connection with embryo-expressed mRNAs, we also analyzed the level of LEAFY COTYLEDON1 (LEC1) RNA. LEC1 encodes an embryo-expressed transcription factor that is sufficient to induce embryogenesis (Lotan et al., 1998). Using real-time RT-PCR, we found LEC1 RNA levels to be elevated by 30-fold in tdt-1 and by 12-fold in vcs-7. Elevated levels of translatable LEC1 in vcs and tdt mutant seedlings might contribute to the many overexpressed embryo-related RNAs, similar to the misregulation of LEC1 in pickle mutants that has also been linked to the expression of embryonic traits in mutant seedlings (Ogas et al., 1999).
DISCUSSION
Signal transduction pathways, whether communicating developmental or environmental information, typically result in altered gene expression. Although most commonly we think of these changes as occurring at the level of transcription, full responsiveness to a new signal is likely to also require removal of the earlier products of gene expression. One example of this comes from controlled proteolysis conferred by F-box proteins, for example the rapid proteolysis of AUX/IAA proteins following auxin binding to the TIR1 F-box protein (reviewed in Quint and Gray, 2006).
RNA decay can also be important for responses to both developmental and environmental signals. The best-known examples come from miRNA-directed RNA cleavage. For example, miR165/166 targets a family of five genes, known collectively as HD-ZIPIIIs, for cleavage. Loss of cleavage due either to mutations at target cleavage sites or to ectopic miRNA expression causes severe developmental defects (McConnell and Barton, 1998; Emery et al., 2003; Prigge et al., 2004; Kim et al., 2005; Williams et al., 2005). miRNA-directed cleavage also occurs in response to environmental signals, for example, in response to phosphate starvation (Fujii et al., 2005).
Here, we have described the early seedling defects that result from defects in TDT and VCS, and we present molecular, genetic, cell biological, and biochemical data implicating these genes as functioning in mRNA decapping.
mRNA Substrates Show Specificity for 3′ and 5′ Decay Pathways
The mRNA bulk decay pathways, worked out in yeast and animals, feature deadenylation followed by either 3′ to 5′ exoribonuclease activity (via the exosome) or decapping, followed by 5′ to 3′ exoribonuclease activity, which in yeast is performed by XRN1 (Figure 8). Our analyses of RNA decay in tdt and vcs mutants found that some mRNAs showed no decay, suggesting obligate decay via decapping. Other mRNAs showed a reduced decay rate, suggesting that these mRNAs normally decay using both pathways, and still others retained normal rates of decay in vcs and tdt, suggesting that their normal decay uses the 3′ to 5′ exosomal pathway. Thus, our data reveal that only some mRNAs show the promiscuous use of both decay pathways and suggest the presence of RNA populations that specifically decay via the 5′ to 3′ and the 3′ to 5′ decay pathways.
How specific mRNA substrates are targeted to a particular mRNA decay pathway is largely unknown. The most likely scenario is that proteins binding to specific mRNA sequences provide targeting. Some conserved sequence elements in 3′ untranslated regions have been observed, for example, in the auxin-induced SAUR genes (McClure et al., 1989), but no obviously conserved sequences were found among the unstable Arabidopsis RNAs (Gutiérrez et al., 2002).
Following decapping, a 5′ to 3′ exoribonuclease leads to RNA hydrolysis (Figure 8). In yeast, this activity is performed by XRN1 (Larimer and Stevens, 1990); however, in Arabidopsis, no XRN1-like gene has been identified, and instead cytoplasmic 5′ to 3′ exoribonuclease activity is thought to be performed by XRN4 (Kastenmayer and Green, 2000). Indeed, several different screens have led to the characterization of xrn4 mutants and have shown that xrn4 mutants accumulate some RNAs (Gazzani et al., 2004; Souret et al., 2004; Olmedo et al., 2006; Potuschak et al., 2006).
This model for bulk decay predicts that vcs, tdt, and xrn4 should accumulate the same set of mRNAs, albeit capped in vcs and tdt mutants and decapped in xrn4. However, we found that tdt/dcp2 mutants had many more RNAs with elevated levels than were found for xrn4 mutants (Souret et al., 2004; Olmedo et al., 2006), and we also found that some mRNAs were at severely reduced levels in tdt, which was not reported for xrn4. The more modest effect on RNA in xrn4 mutants agrees with that mutant's mild phenotype but is difficult to reconcile with the severe phenotype of decapping mutants and the prevailing model for bulk RNA decay (Figure 8). One possibility is that another, as yet unidentified, enzyme can also carry out 5′ to 3′ exoribonuclease activity. Alternatively, the phenotypic differences might arise due to the presence or absence of the cap; uncapped mRNAs accumulating in xrn4 might be silenced, while capped mRNAs accumulating in vcs and tdt might be translated, leading to secondary effects on gene expression. Finally, the difference might be related to P-bodies themselves. In addition to RNA decay, P-bodies also function in translational regulation (Brengues et al., 2005; Liu et al., 2005a, 2005b; Sen and Blau, 2005). In xrn1 mutants, P-bodies appear abnormal, yet they still form, whereas in vcs (and presumably tdt), P-bodies fail to form. If, like animal and yeast cells, plant P-bodies also function in translational regulation, then presumably vcs and tdt, but not xrn4, might regain the expression of translationally arrested mRNAs. This possibility might not only contribute to the severe tdt and vcs phenotypes but also might account for the unexplained reduction of uncleaved miRNA and trans-acting silencing RNA targets found in these mutants. If these RNAs are normally translationally regulated in P-bodies, then the loss of P-bodies in mutants might expose these RNAs to other decay pathways.
Uncovering Natural Variation in mRNA Decay Pathways
Natural variation has been shown for a variety of pathways, including flowering time and light responses (Johanson et al., 2000; Maloof et al., 2001). We found that vcs and tdt phenotypes were both strongly influenced by genetic background. Both vcs and tdt showed a suppressed phenotype when in the Ler accession, and this suppression is conferred by a dominant activity that is genetically unlinked to VCS or TDT.
Why both mutants are suppressed in Ler (but to different extents) remains an outstanding question. We first considered that Ler could have higher activity of genes related to VCS and TDT; however, TDT/DCP2 appears to have no close homologs, and VCR, the closest VCS homolog, is tightly linked to VCS and so inconsistent with an unlinked suppressor. Alternatively, Ler might express higher levels of an alternative scaffold, thus partially restoring decapping activity.
Suppression might also arise due to relief from the symptoms of decapping defects. We found that many capped mRNAs overaccumulate in vcs-7 and tdt-1. If the severe phenotype arises due to these abundant capped mRNAs, then any activity that reduces RNA levels (e.g., increased exosomal activity) might suppress the vcs and tdt phenotypes. It is also possible that only a few of the overaccumulated capped mRNAs cause the severe phenotype, and a suppressed phenotype might arise in a genotype in which these particular genes had lower transcription rates. We also found a set of mRNAs with lower than wild-type levels in vcs and tdt, and it is also possible that these reduced mRNAs are what leads to the severe vcs and tdt phenotypes. If so, perhaps in Ler, expression of these genes is not so severely depressed. We have begun analyzing the Ler suppressors and have found that the major Ler suppressors for vcs and tdt map to different loci.
Vascular Defects in Decapping Mutants
The striking vascular defects shown by both vcs and tdt mutants indicate that vascular patterning requires a normal mRNA decapping complex and presumably mRNA decay. Establishment of vein patterns has been linked to the movement of auxin through responsive cells, although genetic approaches have also revealed the importance of molecules such as sterols, polyamines, and inositol (1,4,5) triphosphate (reviewed in Fukuda, 2004; Sieburth and Deyholos, 2006). The flexibility of a set of cells to adopt a vascular fate can also be limited by differentiation (Scarpella et al., 2004), and vascular defects in vcs and tdt might also arise due to a lack of responsiveness to inductive signals.
In tdt mutants, vascular defects include the vascular transition. The vascular transition is a region within the upper hypocotyl connecting the root-like vascular system of the lower hypocotyl with the shoot's vascular system. This transition entails both a modification of vein position and a change in cell type organization—the lower hypocotyl shows exarch xylem with an alternating arrangement of xylem and phloem, whereas the shoot has endarch xylem and collaterally arranged vascular tissues (Esau, 1965). The developmental basis for the vascular transition is largely unknown, although proposals include it being simply one part of a single system or the vascular transition being the connection site for two separately established systems. Although careful anatomical analyses using Arabidopsis and other systems have revealed nearly simultaneous differentiation through this zone (Busse and Evert, 1999), the requirement for TDT/DCP2 for the establishment of a normal vascular transition might reflect an initially discontinuous origin of the embryo's vascular system.
Developmental Roles for mRNA Decapping?
The severe seedling defects in vcs and tdt mutants, together with the specificity shown by some mRNAs for decay using decapping, suggest that mRNA decapping might be used for developmental regulation of mRNA levels. Good candidates for developmentally regulated decapping are the heat-stress RNAs that are expressed during normal seed development (Wehmeyer and Vierling, 2000; Kotak et al., 2007). Recently, the levels of three heat-stress RNAs were shown to rise through seed development and then fall precipitously within hours of imbibition (Kotak et al., 2007), suggesting that imbibition might activate an mRNA decay pathway. We found these same three RNAs retained in 3-d-old tdt seedlings (two are listed in Table 1, and Hsp101 was reported as elevated fourfold in our microarray analysis). These results suggest that the imbibition-triggered decline of these RNAs requires mRNA decapping.
RNA decay directed by miRNA and trans-acting silencing RNA has been documented extensively in plants, and in zebrafish, decay via miRNA-directed deadenylation is required for normal progression through embryogenesis (Giraldez et al., 2006). Our data indicate that in plants, the developmental transition to early seedling development appears to be particularly sensitive to mRNA-decapping defects. Future characterization of decapping substrate specificity should help us to distinguish between developmental and housekeeping roles of this RNA decay process.
METHODS
Plant Growth and Microscopy
Growth of Arabidopsis thaliana plants, tissue preparation for light microscopy, and GUS staining were performed as described previously (Deyholos et al., 2003). The tdt mutant in the Landsberg background was derived from four sequential crosses to Ler, and vcs-7 corresponds to SALK_032031. Oligonucleotide sequences for genotyping are listed in Supplemental Table 3 online.
Confocal analysis for observation of embryo and seedling SAMs followed the aniline blue staining and tissue preparation methods described by Bougourd et al. (2000). The tdt and vcs mutants were easily identified by their misshapen cells. We attribute these misshapen cells to defects arising as a consequence of the tdt genotype, rather than fixation artifacts, because in replicate experiments phenotypically wild-type sibling embryos fixed alongside tdt (within the same tube) looked normal. Confocal analysis for GFP analysis used live roots of 4-d-old seedlings grown vertically on growth medium. To test the effect of cycloheximide, 4-d-old seedlings were incubated in liquid medium (0.5× Murashige and Skoog salts, 1% sucrose, and 0.5 g/L MES, pH 6.8) supplemented with 200 μg/mL cycloheximide (Sigma-Aldrich) and shaken for 80 min. Treatment with MG132 (Sigma-Aldrich) used a 50 μM solution and shaking for 60 min. We used a Zeiss 510 Meta laser scanning confocal microscope as described previously (Sieburth et al., 2006).
Analysis of transition zone defects in tdt-1 grown at different temperatures was performed on cleared tissue examined under a dark field using magnification up to ×90. The transition zone was considered defective if the primary vein of at least one cotyledon did not have differentiated xylem that connected with the hypocotyl vasculature.
Seedling images were obtained using an Olympus SZX12 microscope. Vascular patterns were observed using dark-field base, and tissue was fixed in Carnoys (3:1 ethanol:acetic acid) and cleared by incubation in saturated chloral hydrate (Sigma-Aldrich). Details of cellular anatomy and GUS staining patterns were examined using an Olympus BX-50 microscope.
Isolation of tdt and Molecular Analyses of Mutants
The tdt-1 mutant was isolated in a small pilot screen of T-DNA–mutagenized Col-0 seeds. This mutant was selected for analysis because it lacked leaves and its cotyledons showed a forked vein pattern. Subsequent analysis showed that the lesion was not linked to the T-DNA, so conventional mapping approaches were used to identify the TDT gene. The sequences of mapping primers are available upon request. The SALK lines corresponding to tdt-2 and vcs-7 were sequenced to verify their T-DNA insertion sites. We sequenced the entire gene from tdt-1, and we identified only one lesion, a 50-nucleotide deletion that was also readily detectable by PCR of genomic DNA.
Yeast Two-Hybrid Analysis
Directed yeast two-hybrid analyses followed the Clontech protocols. Full-length coding regions of VCS and TDT were amplified from seedling cDNA, sequenced, and cloned into both bait (pGBKT7) and prey (pGADT7) vectors.
Quantitative RT-PCR Analyses
We isolated total RNA using the Agilent RNA Isolation Mini kit or the Qiagen RNeasy Plant Mini kit (for the cordycepin experiments) and performed randomly primed cDNA synthesis with the Promega reverse transcription system. We performed real-time RT-PCR using the Roche Lightcycler and FastStart DNA Master SYBR Green 1 master mix. Each real-time RT-PCR experiment included internal controls (ACTIN2) for each template. Data shown are averages of four to six reactions and were confirmed by independent experiments. HD-ZIPIII expression was also confirmed by reactions using separately designed oligonucleotides. To determine the relative steady state level of RNAs, we calculated the relative expression 2[(average ΔCT (geneX, tissueX) – average ΔCT (geneX, WT)] and converted this number to a percentage. We tested each reaction's specificity using the machine's standard melt curve method and also analyzed product size by gel electrophoresis. Sequences of oligonucleotides used as primers are available in Supplemental Table 3 online. Some primer sequences were taken from Vazquez et al. (2004), Vaucheret et al. (2004), and Li et al. (2005); these primers are listed in Supplemental Table 3 online, along with sequences of primers designed by us.
RACE Analysis of Capped mRNAs
To determine whether accumulating mRNAs were capped, RNA ligase-mediated (RLM) RACE was performed using the First Choice RLM-RACE kit from Ambion. Total RNA was isolated from 4-d-old Arabidopsis seedlings (Qiagen RNeasy kit). Col-0, tdt-1, and vcs-7 RNAs (1 μg) were ligated to the 5′ RACE Adapter (5′-CUGAUGGCGAUGAAUGAACACUGCGUUUGCUGGCUUUGAUGAA-3′) either prior to or following treatment with TAP to remove the 5′ cap. Random decamers were used to prime the reverse transcription reaction, and PCR was performed using a primer specific to the RACE adapter (5′ RACE Outer Primer) and gene-specific primers.
GFP-TDT Fusion and TDT Rescue Constructs
The TDT rescue construct encompassed a 6019-bp PstI-EcoRI fragment from BAC T6I14, which was cloned into pCAMBIA1300. The plant DNA contained the entire At5g13570 coding region, the entire upstream (5′) and downstream (3′) regions, and small portions of the upstream and downstream genes. The GFP:TDT construct was made by placing the TDT cDNA coding region (clone U61209) between the EcoRI and BamHI cloning sites of pEGAD (Cutler et al., 2000). Constructs were sequenced prior to transformation into plants. Both constructs were introduced to wild type (Col-0) and tdt-1 heterozygotes using the floral dip method (Clough and Bent, 1998). Transgenic seeds were isolated based on their resistance (hygromycin for the pCAMBIA vector and BASTA for the pEGAD vector), and the GFP:TDT lines were introduced into the vcs-7 mutant background by standard genetic methods.
Analysis of tdt Rescue
We analyzed nine independently generated lines that segregated for tdt, and all showed similar ratios of phenotypically normal to mutant plants (∼15:1). Data presented in Results are from transgenic rescue line 29.
Microarray Analysis
Total RNA was isolated from 3-d-old wild-type and tdt-1 seedlings using the Agilent Plant RNA Isolation Mini kit. RNA samples (1 μg) were submitted to the University of Utah Microarray Core Facility and went through two steps of RNA quality control prior to the labeling protocol, which consisted of analysis of the sample on an Agilent bioanalyzer and validation of nucleic acid concentration on a NanoDrop spectrophotometer. Samples were labeled using the Agilent Low RNA Input Linear Amplification kit. RNA labeling involved first-strand reverse transcription primed using an oligo(dT)/T7 polymerase promoter sequence, second-strand cDNA synthesis, and generation of fluorescently labeled cRNA in an in vitro transcription reaction performed in the presence of Cy3-CTP or Cy5-CTP. Labeled samples were hybridized to the Arabidopsis 3 oligo expression array (44K) using the Agilent Gene Expression Hybridization kit and were incubated overnight (∼17 h) at 60°C. The microarray slide was washed (6× SSC [1× SSC is 0.15 M NaCl and 0.015 M sodium citrate] and 0.005% lauryl sarcosine at room temperature for 1 min, 0.6× SSC and 0.005% lauryl sarcosine at room temperature for 1 min, and a 5-s acetonitrile dip) and scanned using the Agilent G2565BA microarray scanner. Three biological replicates were performed, with a total of three arrays hybridized. One of these arrays was hybridized with reversed dyes. Data from the scanned microarrays were obtained using Agilent Feature Extraction software, version 8.5 (http://www.chem.agilent.com/Scripts/PDS.asp?lPage=2547). The quality of the data was assessed by visual inspection of M versus A plots. Some intensity-dependent bias was noted and was corrected in the Lowess-normalized data set generated by Agilent Feature Extraction. Control spots and spots for which there were data from two or fewer arrays were filtered from the data set. All pairwise differentially expressed genes were identified using SAM software (Tusher et al., 2001) using the data from the remaining 34,347 Arabidopsis probe sets. A false discovery rate parameter of 1% was used for the one-class SAM analysis. Average fold change for each gene was calculated from the log ratio data. Genes from the array were cross-referenced to The Arabidopsis Information Resource (http://www.Arabidopsis.org) using systematic names provided by Agilent. A single annotation was assigned to each gene using The Arabidopsis Information Resource annotations and the Gene Ontology biological process. All generated annotations were manually curated. All data from microarray experiments have been deposited in the Gene Expression Omnibus database under accession number GSE7359 (www.ncbi.nlm.nih.gov/geo).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: TDT (At5g13570), VCS (At3g13300), VCR (At3g13290), ACT2 (At3g18780), LEC1 (At1g21970), LEA (At1g52690, At5g06760), Hsp17.4 (At3g46230), At HSP90.1 (At5g52640), At HSP70 (At2g32120), HSP26.5 (At1g52560), HSP23.5-M (At5g51440), ARF4 (At5g60450), ARF3 (At2g33860), TAS3 (At3g17185), PHB (At2g34710), REV (At5g60690), AthB15/CNA (At1g52150), PHV (At1g30490), AthB8 (At4g32880), SFC (At5g13300), BPS1 (At1g01550), BME3 (At3g54810), MYB4 (At1g22640), DCL1 (At1g01040), ARF8 (At5g37020), ARF17 (At1g77850), and AGO1 (At1g48410). Microarray data have been deposited at the Gene Expression Omnibus site with accession number GSE7359.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. vcs-7 Is an RNA Null Allele.
Supplemental Figure 2. Phenotypes of vcs-1, vcs-7, and tdt-1.
Supplemental Figure 3. Low-Temperature Growth Confers Minimal Suppression to the vcs-7 Allele.
Supplemental Figure 4. Pinched Appearance of tdt Cotyledon Petioles.
Supplemental Table 1. Genes with at Least a Fivefold Increased RNA Level in tdt-1 Mutants.
Supplemental Table 2. Genes with at Least a Fivefold Decreased RNA Level in tdt-2 Mutants.
Supplemental Table 3. Oligonucleotide Sequences of Primers Used for PCR, Real-Time RT-PCR, and RACE in This Study.
Supplementary Material
Acknowledgments
This article is dedicated to John McN. Sieburth (1927–2006), whose scientific voyages on the R/V Trident inspired the naming of our tdt mutant. We thank Ed King and Ben Heaton for assistance in microscopy and mapping, respectively, and Brett Milash and Brian Dalley for assistance with microarray analysis and statistical analyses. We also thank the ABRC for supplying seed stocks and the National Science Foundation and the National Institutes of Health for support (National Science Foundation Grants IBN-0445723 and IBN-0642118 to L.E.S. and National Institutes of Health Training Grant 5 T32 GM-007464 to J.M.V.N.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Leslie E. Sieburth (sieburth@biology.utah.edu).
Online version contains Web-only data.
References
- Adenot, X., Elmayan, T., Lauressergues, D., Boutet, S., Bouché, N., Gasciolli, V., and Vaucheret, H. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16 927–932. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Badis, G., Saveanu, C., Fromont-Racine, M., and Jacquier, A. (2004). Targeted mRNA degradation by deadenylation-independent decapping. Mol. Cell 15 5–15. [DOI] [PubMed] [Google Scholar]
- Baima, S., Nobili, F., Sessa, G., Lucchetti, S., Ruberti, I., and Morelli, G. (1995). The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 121 4171–4182. [DOI] [PubMed] [Google Scholar]
- Barton, M.K., and Poethig, R.S. (1993). Formation of the shoot apical meristem in Arabidopsis thaliana: An analysis of development in the wild type and in the shoot meristemless mutant. Development 119 823–831. [Google Scholar]
- Beelman, C.A., Stevens, A., Caponigro, G., LaGrandeur, T.E., Hatfield, L., Fortner, D.M., and Parker, R. (1996). An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382 642–646. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant, I., Rehwinkel, J., Doerks, T., Stark, A., Bork, P., and Izaurralde, E. (2006). mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 20 1885–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougourd, S., Marrison, J., and Haseloff, J. (2000). An aniline blue staining procedure for confocal microscopy and 3D imaging of normal and perturbed cellular phenotypes in mature Arabidopsis embryos. Plant J. 24 543–550. [DOI] [PubMed] [Google Scholar]
- Brengues, M., Teixeira, D., and Parker, R. (2005). Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310 486–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno, I., and Wilkinson, M.F. (2006). P-bodies react to stress and nonsense. Cell 125 1036–1038. [DOI] [PubMed] [Google Scholar]
- Busse, J.S., and Evert, R.F. (1999). Vascular differentiation and transition in the seedling of Arabidopsis thaliana (Brassicaceae). Int. J. Plant Sci. 160 241–251. [Google Scholar]
- Carland, F.M., Fujioka, S., Takatsuto, S., Yoshida, S., and Nelson, T. (2002). The identification of CVP1 reveals a role for sterols in vascular patterning. Plant Cell 14 2045–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland, F.M., and Nelson, T. (2004). COTYLEDON VASCULAR PATTERN2-mediated inositol (1.4.5) triphosphate signal transduction is essential for closed venation patterns of Arabidopsis foliar organs. Plant Cell 16 1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C., and Ambros, V. (2003). Role of microRNAs in plant and animal development. Science 301 336–338. [DOI] [PubMed] [Google Scholar]
- Chiba, Y., Johnson, M.A., Lidder, P., Vogel, J.T., van Erp, H., and Green, P.J. (2004). AtPARN is an essential poly(A) ribonuclease in Arabidopsis. Gene 328 95–102. [DOI] [PubMed] [Google Scholar]
- Clay, N.K., and Nelson, T. (2002). VH1, a provascular cell-specific receptor kinase that influences leaf cell patterns in Arabidopsis. Plant Cell 14 2707–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay, N.K., and Nelson, T. (2005). Arabidopsis thickvein mutation affects vein thickness and organ vascularization, and resides in a provascular cell-specific spermine synthase involved in vein definition and in polar auxin transport. Plant Physiol. 138 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cougot, N., Babajko, S., and Séraphin, B. (2004). Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couttet, P., Fromont-Racine, M., Steel, D., Pictet, R., and Grange, T. (1997). Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc. Natl. Acad. Sci. USA 94 5628–5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler, S.R., Ehrhardt, D.W., Griffitts, J.S., and Somerville, C.R. (2000). Random GFP∷cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyholos, M.K., Cavaness, G.F., Hall, B., King, E., Punwani, J., Van Norman, J., and Sieburth, L.E. (2003). VARICOSE, a WD-domain protein, is required for leaf blade development. Development 130 6577–6588. [DOI] [PubMed] [Google Scholar]
- Dunckley, T., and Parker, R. (1999). The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 18 5411–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery, J.F., Floyd, S.K., Alvarez, J., Eshed, Y., Hawker, N.P., Izhaki, A., Baum, S.F., and Bowman, J.L. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13 1768–1774. [DOI] [PubMed] [Google Scholar]
- Esau, K. (1965). Plant Anatomy. (New York: John Wiley & Sons).
- Fahlgren, N., Montgomery, T.A., Howell, M.D., Allen, E., Dvorak, S.K., Alexander, A.L., and Carrington, J.C. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16 939–944. [DOI] [PubMed] [Google Scholar]
- Fenger-Grøn, M., Fillman, C., Norrild, B., and Lykke-Andersen, J. (2005). Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell 20 905–915. [DOI] [PubMed] [Google Scholar]
- Fujii, H., Chiou, T.-J., Lin, S.-I., Aung, K., and Zhu, J.-K. (2005). A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 15 2038–2043. [DOI] [PubMed] [Google Scholar]
- Fukuda, H. (2004). Signals that control plant vascular cell differentiation. Nat. Rev. Mol. Cell Biol. 5 379–391. [DOI] [PubMed] [Google Scholar]
- Garcia, D., Collier, S.A., Byrne, M.E., and Martienssen, R.A. (2006). Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 16 933–938. [DOI] [PubMed] [Google Scholar]
- Gazzani, S., Lawrenson, T., Woodward, C., Headon, D., and Sablowski, R. (2004). A link between mRNA turnover and RNA interference in Arabidopsis. Science 306 1046–1048. [DOI] [PubMed] [Google Scholar]
- Geldner, N., Richter, S., Vieten, A., Marquardt, S., Torres-Ruiz, R.A., Mayer, U., and Jürgens, G. (2003). Partial loss-of-function alleles reveal a role for GNOM in auxin transport-related, post-embryonic development of Arabidopsis. Development 131 389–400. [DOI] [PubMed] [Google Scholar]
- Giraldez, A.J., Mishima, Y., Rihel, J., Grocock, R.J., Van Dongen, S., Inoue, K., Enright, A.J., and Schier, A.F. (2006). Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312 75–79. [DOI] [PubMed] [Google Scholar]
- Gutiérrez, R.A., Ewing, R.M., Cherry, J.M., and Green, P.J. (2002). Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: Rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc. Natl. Acad. Sci. USA 99 11513–11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzawa, Y., Takahashi, T., Michael, A.J., Burtin, D., Long, D., Pineiro, M., Coupland, G., and Komeda, Y. (2000). ACAULIS5, an Arabidopsis gene required for stem elongation, encodes a spermine synthase. EMBO J. 19 4248–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, D.W., and Galau, G.A. (1989). Temporally modular gene expression during cotyledon development. Genes Dev. 3 358–369. [DOI] [PubMed] [Google Scholar]
- Johanson, U., West, J., Lister, C., Michaels, S., Amasino, R., and Dean, C. (2000). Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290 344–347. [DOI] [PubMed] [Google Scholar]
- Kastenmayer, J.P., and Green, P.J. (2000). Novel features of the XRN-family in Arabidopsis: Evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc. Natl. Acad. Sci. USA 97 13985–13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Jung, J.H., Reyes, J.L., Kim, Y.S., Kim, S.Y., Chung, K.S., Kim, J.A., Lee, M., Lee, Y., Kim, V.N., Chua, N.H., and Park, C.M. (2005). MicroRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J. 42 84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi, K., Naramoto, S., Sawa, S., Yahara, N., Ueda, T., Nakano, A., Sugiyama, M., and Fukuda, H. (2005). VAN3 ARF-GAP-mediated vesicle transport is involved in leaf vascular network formation. Development 132 1699–1711. [DOI] [PubMed] [Google Scholar]
- Kotak, S., Vierling, E., Bäumlein, H., and von Koskull-Döring, P. (2007). A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell 19 182–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimer, F.W., and Stevens, A. (1990). Disruption of the gene XRN1, coding for a 5′—3′ exoribonuclease, restricts yeast cell growth. Gene 95 85–90. [DOI] [PubMed] [Google Scholar]
- Li, H., Xu, L., Wang, H., Yuan, Z., Cao, X., Yang, Z., Zhang, D., Xu, Y., and Huang, H. (2005). The putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and microRNA 165/166 in Arabidopsis leaf development. Plant Cell 17 2157–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Rivas, F.V., Wohlschlegel, J., Yates, J.R., Parker, R., and Hannon, G.J. (2005. a). A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 7 1261–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Valencia-Sanchez, M.A., Hannon, G.J., and Parker, R. (2005. b). MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7 719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, P.-P., Koizuka, N., Martin, R.C., and Nonogaki, H. (2005. c). The BME3 (Blue Micropylar End 3) GATA zinc finger transcription factor is a positive regulator of Arabidopsis seed germination. Plant J. 44 960–971. [DOI] [PubMed] [Google Scholar]
- Lotan, T., Ohto, M., Yee, K.M., West, M.A., Lo, R., Kwong, R.W., Yarnagishi, K., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93 1195–1205. [DOI] [PubMed] [Google Scholar]
- Maloof, J.N., Borevitz, J.O., Dabi, T., Lutes, J., Nehring, R.B., Redfern, J.L., Trainer, G.T., Wilson, J.M., Asami, T., Berry, C.C., Weigel, D., and Chory, J. (2001). Natural variation in light sensitivity of Arabidopsis. Nat. Genet. 29 441–446. [DOI] [PubMed] [Google Scholar]
- Mattsson, J., Ckurshumova, W., and Berleth, T. (2003). Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol. 131 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure, B.A., Hagen, G., Brown, C.S., Geen, M.A., and Guilfoyle, T.J. (1989). Transcription, organization, and sequence of an auxin-regulated gene cluster in soybean. Plant Cell 1 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell, J.R., and Barton, M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125 2935–2942. [DOI] [PubMed] [Google Scholar]
- Muhlrad, D., and Parker, R. (1994). Premature translational termination triggers mRNA decapping. Nature 370 578–581. [DOI] [PubMed] [Google Scholar]
- Ogas, J., Kaufmann, S., Henderson, J., and Somerville, C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 96 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo, G., Guo, H., Gregory, B.D., Nourizadeh, S.D., Aguilar-Henonin, L., Li, H., An, F., Guzman, P., and Ecker, J.R. (2006). ETHYLENE-INSENSITIVE5 encodes a 5′ → 3′ exoribonuclease required for regulation of EIN3-targeting F-box proteins EBF1/2. Proc. Natl. Acad. Sci. USA 103 13286–13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, R., and Song, H. (2004). The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11 121–127. [DOI] [PubMed] [Google Scholar]
- Potuschak, T., Vansiri, A., Binder, B.M., Lechner, E., Vierstra, R.D., and Genschik, P. (2006). The exoribonuclease XRN4 is a component of the ethylene response pathway in Arabidopsis. Plant Cell 18 3047–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge, M.J., Otsuga, D., Alonso, J.M., Ecker, J.R., Drews, G.N., and Clark, S.E. (2004). Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17 61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint, M., and Gray, W.M. (2006). Auxin signaling. Curr. Opin. Plant Biol. 9 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. (2002). MicroRNAs in plants. Genes Dev. 16 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverdatto, S.V., Dutko, J.A., Chekanova, J.A., Hamilton, D.A., and Belostotsky, D.A. (2004). mRNA deadenylation by PARN is essential for embryogenesis in higher plants. RNA 10 1200–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella, E., Francis, P., and Berleth, T. (2004). Stage-specific markers define early steps of procambium development in Arabidopsis leaves and correlate termination of vein formation with mesophyll differentiation. Development 131 3445–3455. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Schölkopf, B., Weigel, D., and Lohmann, J. (2005). A gene expression map of Arabidopsis development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Sen, G.L., and Blau, H.M. (2005). Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 7 633–636. [DOI] [PubMed] [Google Scholar]
- She, M., Decker, C.J., Chen, N., Tumati, S., Parker, R., and Song, H. (2006). Crystal structure and functional analysis of Dcp2p from Schizosaccharomyces pombe. Nat. Struct. Mol. Biol. 13 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth, U., and Parker, R. (2003). Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth, L.E., and Deyholos, M.K. (2006). Vascular development: The long and winding road. Curr. Opin. Plant Biol. 9 48–54. [DOI] [PubMed] [Google Scholar]
- Sieburth, L.E., Muday, G.K., King, E.J., Benton, G., Kim, S., Metcalf, K.E., Meyers, L., Seamen, E., and Van Norman, J.M. (2006). SCARFACE encodes an ARF-GAP that is required for normal auxin efflux and vein patterning in Arabidopsis. Plant Cell 18 1396–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souret, F.F., Kastenmayer, J.P., and Green, P.J. (2004). AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol. Cell 15 173–183. [DOI] [PubMed] [Google Scholar]
- Tusher, V.G., Tibshirani, R., and Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk, E., Cougot, N., Meyer, S., Babajko, S., Wahle, E., and Séraphin, B. (2002). Human Dcp2: A catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 21 6915–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Norman, J.M., Frederick, R., and Sieburth, L.E. (2004). BYPASS1 negatively regulates a root-derived signal that controls plant architecture. Curr. Biol. 14 1739–1746. [DOI] [PubMed] [Google Scholar]
- Van Norman, J.M., and Sieburth, L.E. (2007). Dissecting the biosynthetic pathway for the bypass1 root-derived signal. Plant J. 49 619–628. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H., Vazquez, F., Crete, P., and Bartel, D.P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez, F., Gasciolli, V., Crete, P., and Vaucheret, H. (2004). The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14 346–351. [DOI] [PubMed] [Google Scholar]
- Wehmeyer, N., and Vierling, E. (2000). The expression of small heat shock proteins in seeds responds to discrete developmental signals and suggests a general protective role in desiccation tolerance. Plant Physiol. 122 1099–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, L., Grigg, S.P., Xie, M., Christensen, X., and Fletcher, J.C. (2005). Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 132 3657–3668. [DOI] [PubMed] [Google Scholar]
- Xu, J., Yang, J.-Y., Niu, Q.-W., and Chua, N.-H. (2006). Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell 18 3386–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J.H., Yang, W.H., Gulick, T., Bloch, K.D., and Bloch, D.B. (2005). Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA 11 1795–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.