Abstract
Ab initio simulations of the folding pathways are currently limited to very small proteins. For larger proteins, some approximations or simplifications in protein models need to be introduced. Protein folding and unfolding are among the basic processes in the cell and are very difficult to characterize in detail by experiment or simulation. Chymotrypsin inhibitor 2 (CI2) and barnase are probably the best characterized experimentally in this respect. For these model systems, initial folding stages were simulated by using CA–CB–side chain (CABS), a reduced-space protein-modeling tool. CABS employs knowledge-based potentials that proved to be very successful in protein structure prediction. With the use of isothermal Monte Carlo (MC) dynamics, initiation sites with a residual structure and weak tertiary interactions were identified. Such structures are essential for the initiation of the folding process through a sequential reduction of the protein conformational space, overcoming the Levinthal paradox in this manner. Furthermore, nucleation sites that initiate a tertiary interactions network were located. The MC simulations correspond perfectly to the results of experimental and theoretical research and bring insights into CI2 folding mechanism: unambiguous sequence of folding events was reported as well as cooperative substructures compatible with those obtained in recent molecular dynamics unfolding studies. The correspondence between the simulation and experiment shows that knowledge-based potentials are not only useful in protein structure predictions but are also capable of reproducing the folding pathways. Thus, the results of this work significantly extend the applicability range of reduced models in the theoretical study of proteins.
Keywords: protein structure prediction, Monte Carlo simulations, protein denatured state, folding nucleus, residual structure
A large number of folded protein structures was determined by x-ray crystallography or NMR. For a few proteins, the folding intermediates were characterized by using protein engineering (1) and NMR techniques (2, 3). The major transition states (TS) of chymotrypsin inhibitor 2 (CI2) and barnase were mapped at the level of individual residues by protein engineering (4, 5). Much less is known, however, about early folding events. It is very important to understand how protein folding is initiated and how the native structure emerges subsequently. The denatured state, an ensemble of partially folded, highly mobile conformations, is very difficult to study, although there have been recent reports of NMR studies of residual structure in denatured proteins. Such structures, along with hydrophobic clusters, were discovered even under highly denaturing conditions (6, 7). Moreover, denatured proteins can exhibit a long-range ordering of native-like topology (8). Therefore, the folding process can be directed from the very beginning when starting from a specific structure (9, 10). It becomes evident that the denatured state plays a crucial role in all aspects of protein stability and folding mechanisms (11).
For small, single-domain proteins, two basic folding mechanisms can be traced. In the diffusion–collision mechanism (12), local secondary structure elements form independently, and their collisions eventually lead to the native structure. In the nucleation–condensation mechanism, a simultaneous consolidation of secondary and tertiary interactions follows a collapse around extended nuclei (13, 14). Frequently, a combination of these two mechanisms occurs (15). With increasing secondary propensities, the folding proceeds in a more stepwise manner and follows the diffusion–collision mechanism. In cases of an inherently unstable secondary structure, the nucleation–condensation mechanism is more likely to apply.
During the last 20 years, significant progress has been made in the field of protein folding (16). Molecular dynamics (MD) simulations became a common method for exploring the properties of intermediates (17), the TS (18), the unfolding reaction (19, 20), the energetics of folding (21), and, more recently, the denatured state (7, 22). Unfortunately, there is a gap between the time scales of MD simulation and the characteristic time of protein folding. Only small and ultrafast folding proteins (in the range of microseconds) are now fully tractable by classical MD simulations (23). An average protein folds slower by orders of magnitude.
So far, for larger proteins, simulations of the full folding process, from a random coil to the native state, have only been possible with Go models, in which merely the native interactions are taken into account. Neglecting the role of the nonnative interactions in the folding mechanisms (24, 25) is a serious shortcoming of this approach. Because of the limitations of the all-atom molecular mechanics, reduced models offer the most promising possibilities to study large scale protein rearrangements, as recently demonstrated by Liwo et al. (26).
This work describes the application of a high resolution reduced lattice model and MC dynamics for folding simulations (Fig. 1) at various folding stages, beginning from fully denatured state. The simplified representation significantly reduces the number of degrees of freedom treated explicitly (26, 27). With the implicit solvent and the time step of the MC dynamics being orders of magnitude larger than in MD the entire folding/unfolding process can be simulated.
Fig. 1.
CABS energy (E) and its standard deviation (Esd) as a function of T for barnase. Each point represents a single isothermal simulation. The transition temperature (Tt = 2,025) is identified by the steep drop of the energy and the peak of the heat capacity. Tt cannot be strictly identified with the TS. Sometimes, as for CI2, conformations observed at Tt may be relatively unstructured, with some features of a molten globule state. See also Figs. 2 and 3b.
The CA–CB–side chain (CABS) model (28) used for this experiment was tested successfully by Kolinski–Bujnicki group (29) in the sixth edition of Critical Assessment of Protein Structure Prediction (CASP6), a blind test for protein structure prediction. According to the CASP6 evaluation, the average score of models submitted by this group was second best among ≈200 groups participating. What seems important for application presented here is that our approach ranked second best for ab initio modeling after Rosetta (30). CABS employs knowledge-based statistical potentials. The only information specific to proteins studied here is the expected secondary structure (in a three-letter code), providing a weak bias for the local interactions. This input is standard for CABS ab initio modeling, which benefits from a high level of accuracy (80%) of the secondary structure prediction methods.
Results and Discussion
CI2 and barnase are two paradigms in experimental and theoretical studies of protein folding. They represent two classes of proteins, the former with a two-state kinetics, folding quickly, from a relatively unstructured denatured state, and the latter with multistate kinetics, folding more slowly, from a somewhat structured denatured state, passing through at least one intermediate state.
The simulations (Fig. 2) were compared with the three methods characterizing the folding process at the atomic level (31): NMR, protein engineering, and MD simulations. Site-directed mutagenesis in conjunction with kinetic measurements is the only method for analyzing the TS structures and rapidly formed intermediates, and the TS is the only state accessible to experimental study in a two-state folding (CI2). This method of analysis, introduced by Matouschek et al. (32), relies on the quantity of Φ: Φ = 0 suggests the absence of interaction in the TS, whereas Φ = 1 marks an interaction similar to that in the native state. However, it does not necessarily imply that the residues with high Φ values are kinetically more important then the residues with lower Φ values. Experimentally derived Φ values match the theoretical models of folding nicely; they can be correlated with the increase in the number of native-like contacts (18, 33).
Fig. 2.
Acquisition of structure elements in side-chain contact maps from simulations of barnase (a) and CI2 (b) at various temperatures: highly denaturing (hds), denaturing (ds), just before Tt, and at Tt. Native contact maps are provided for reference. For CI2, additional simulations below Tt (T = 1.6) are presented. The colors indicate the frequency of contacts. Short-range contacts (up to i, i+2) are omitted for clarity. (a) At T = 2.7, the most frequently appearing nonhelical turn (94, 97) is marked. Circled areas indicate α1 helix and interactions pattern of β3–β4 with the rest of the chain. (b) Cooperative substructures are circled in blue, interactions A16–L49 and A16–A58 for simulations at T = 1.6 are marked in red, and interactions of I20 with V47 and L49 are in gray.
Barnase.
Barnase is a 110-residue α+β protein (Fig. 3a) with three hydrophobic cores. It contains three helices in the first half of its sequence followed by the five-stranded antiparallel β sheet, and it is an example of a small multidomain protein. Its folding model includes at least one intermediate and has been described in detail previously (34).
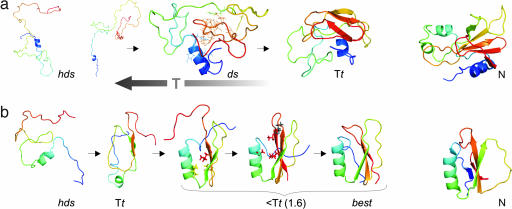
Fig. 3.
Folding pathways of barnase (a) and CI2 (b), as illustrated by snapshots from the simulation at different temperatures (see Fig. 2) and experimentally derived native (N) structures (Protein Data Bank ID codes: 1BNR and 2CI2, respectively). (a) Highly denaturing (hds) with residual α1 helix structure and β3–β4 turn (side chains W94 and Y97 marked with red sticks). A representative hydrophobic cluster is shown with the most frequently contacting side chains marked with lines (ds) and an example of a distorted structure at Tt, with a relatively loose, planar central part of the β sheet interacting with the helix. (b) Highly denaturing with residual α helix structure. The most nucleating area at Tt is β3–β4. Shown are the first stage of docking of the α helix to β3–β4 (I20 and V47 marked with yellow sticks, L49 in red), the conformation with properly ordered β3–β4 and β5 strands before the formation of the N-terminal strands (nucleus residues A16, L49, and A58 marked with red sticks, I57 with its side chain pointing opposite to the helix in dark gray), and the best-formed structure (at T = 1.6). Coordinate root mean square deviation, 3.8 Å.
Denatured state.
Barnase contains a considerable amount of residual structure in its denatured state, as investigated by MD, NMR, and other experimental techniques (35–37), especially with relation to the first and second helices and the central β strands: β3 and β4. In the later stages of folding, as evidenced by kinetic and engineering studies, structures consisting of the first helix and the β sheet center are frequently formed (34). Initiation sites for these residual structures develop into folding nucleation sites. The second helix forms very late (38), thus the initiation sites develop only when they enter into stabilizing long-range interactions (36).
NMR data for pH denatured barnase (39) are consistent with the native-like structure for the helix and a nonnative hydrophobic clustering around D93–Y97. There is weak evidence for a residual structure in the center of the first helix (D8–T16) and the tight turn between the central strands of the β sheet (W94–Y97) in the strongly denatured state (36).
The above data are in excellent agreement with our simulations (Fig. 2a). In strongly denaturing conditions (T = 2.7), two areas of structure were observed between residues 7–24 and 90–97 (with the most favorable helical contact F10–Y13 appearing in about ≈32% of snapshots; contact V10–L14, ≈25% of snapshots; and the most favorable turn contact W94–Y97, ≈27% of snapshots) (see Fig. 2a). Just above Tt (T = 2.2; Fig. 2a), a native-like residual structure of the first helix and the hydrophobic cluster (I88, L89, Y90, W94, L95, I96, and Y97) consolidated. Fragments of sequences 87–90 and 94–97 act like hydrophobic patches that tend to form tertiary interactions throughout the protein (especially with the hydrophobic residues W35, F56, W71, and I76) (Figs. 2a and 3a). Tertiary interactions begin to take place between central portions of the β sheet, which acts as the nucleation site.
Transition temperature.
The major hydrophobic core1 contains hydrophobic residues of α1 and a fragment of the β sheet. At Tt, the folding is nucleated by the native-like helix α1 and hydrophobic clusters located at β4–β3. This particular fragment of β sheet bears a stronger resemblance to the native state than the helix (Fig. 2a). The most frequently observed native contacts occur in the center of the hydrophobic core1, particularly I88 with I96 and Y97 (≈70%). According to experiments, the interactions of I88 are fully intact in the TS and weak (≈40%) in the intermediate (1). Core3 is formed by packing of loop3 (between β1 and β2: F56, L63, and P64) and loop5 (between β4 and β5: Y103) against β sheet (W71, L89, Y97, and F106). According to the results obtained by protein-engineering methods, core3 folds in the intermediate and is compact in the TS (1).
This picture is precisely what was observed in our simulations. It is interesting to note the large number of nonnative interactions of F56 and the neighboring residues with the β sheet F56 with L89 and Y90 ≈67%). F56 also interacts with W71 in the β2 strand. β2 exhibits strong native interactions: W71 with Y90 (≈64%) and L89 (≈56%). The structure of core2, the smallest of the three cores formed by residues within α2, α3, loop1, loop2, and β1, tends to be disrupted, and it definitely has the smallest number of native contacts. Once again, the results closely resemble those obtained in protein-engineering studies.
Nucleation site.
Our simulations show that the most frequent nonlocal contacts of the α1 helix (mainly F7 and Y13, L14, and Y17) form with the β3–β4 hairpin. The helix became significantly more ordered as the number of contacts with the central part of the β sheet increased.
The β-hairpin β3–β4 is conserved in the microbial nuclease family. Sequence analysis of this family shows that identical or homologous areas interact in the early stages of folding. Therefore, it can be concluded that these fragments may be evolutionary “programmed” to drive the folding process. It has been proven that the hairpin facilitates helix formation by a so-called “contact-assisted” secondary structure formation (22). There is vast experimental and theoretical evidence that the residual structure of the first helix and the central part of β sheet in the denatured state represent the initiation site of barnase folding (22, 35–37), which could be also predicted basing on the burial of the hydrophobic area in the native state (40).
As described above, it was discovered that these areas form a residual structure in strongly denaturing conditions. Remarkably, the map of standard deviations of the distances between amino acids, computed for the MC trajectory at Tt, show that the helix and β3–β4 hairpin form together a network of interactions that exhibits the lowest fluctuations (Fig. 4). This substructure is internally well defined and loosely coupled to edge strands. It acts as a nucleation site and serves as a scaffold on which the remaining β structure assembles. It was also reported that the α1 aids in the stabilizing of β3–β4. The resulting β structure is relatively planar and loosely packed (Fig. 3a) as in the postulated major intermediate (35). Looking at the number of native contacts within β3–β4 and long-distance tertiary contacts between β3–β4 and the rest of the chain, it may be concluded that β3–β4 is more important for barnase folding than the α1 helix. These findings are consistent with peptide fragment studies in which native-like structures persist on the removal of α1 helix (41). Thus, the average structures observed in our simulations at Tt exhibit characteristic features of the main TS and the main barnase folding intermediate.
Fig. 4.
Location of the main nucleation site of barnase at Tt. Shown is a map of distances between Cα displayed above the diagonal; color indicates average values in angstroms (color legend on the Left). The map shows close contacts of β3–β4 strands with the majority of the protein. Standard deviations of the Cα distances are in angstroms (color legend on the Right) are presented below the diagonal. The map shows that the α1 helix (7–17) and β3 (87–91)–β4 (96–99) form the most stable tertiary area. The main nucleation site is marked with the white circles.
CI2.
CI2 is a 64-residue protein that folds into a single α helix packed against parallel/antiparallel β sheet (Fig. 3b). The strands and helix form a single hydrophobic core; the side-group interactions are quite uniform over the structure. Unlike barnase, it is a single-module structure and, as such, a model for folding units in multidomain proteins. CI2 is structurally simpler than barnase, and, as a result, its folding mechanism is not as complex. It was the first protein for which a clear two-state folding without detectable intermediates (42) was experimentally demonstrated. CI2 was investigated thoroughly with the use of Φ value analysis (5, 13), which led to a hypothesis that the folding of CI2 protein proceeds by a chain collapse and condensation around an extended nucleus, containing portions of the helix and the β sheet followed by the simultaneous consolidation of secondary and tertiary structures. It is a classic example of the nucleation–condensation mechanism (14).
Denatured state.
Experimental NMR studies of protein fragments demonstrate that in denatured conditions CI2 is highly unstructured (43, 44), with a very slight tendency for the native helical structure and a minor hydrophobic clustering near the center of the chain. Likewise, a compilation of NMR experiments and MD simulations on a full-length CI2 indicate a highly unfolded denatured state with the residual structure mentioned above (7).
In our simulations, most of the native contacts detectable in the highly denatured state (T = 2.6) took place within the helix. Triplets of native contacts appearing most frequently (13–16, 16–19, 16–20, and 16–19, 16–20, 17–20) are present in ≈1% of snapshots. This fragment of helix is very active; it unfolds and refolds completely over time. A16 appears to be the most buried residue. The early occurrence of A16 interactions is in excellent agreement with protein-engineering studies. A16 is the only residue that has its full native interaction pattern in the TS (the highest Φ value = 1.1) (13). In strongly denaturing conditions, a minor hydrophobic clustering in the center of the chain could be observed in simulations (Fig. 2b) which is also in agreement with the NMR studies cited above (7). Closer to Tt, hydrophobic clustering between portions of β3 and β4 became more conspicuous, and just above Tt (T = 2.2), the most favorable pairs of native contacts appearing simultaneously (present in about ≈10% of snapshots) were I29, V47, and L49. These residues with fractional Φ values belong to the center of the hydrophobic core, partially formed in the TS (45). Marginal nonnative clustering occurred also between the edges of the active-site loop (G35–I44) with β4 (Y42–L49, Y42–V47) and β3 strand (L32–V38, V31–V38). However, these contacts exhibited high fluctuations compared with the hydrophobic clustering between β3–β4 strands. In the native state, the loop is very solvent-exposed, and mutations in this area did not destabilize the protein to any significant extent, except for V38 (13). Marginal tertiary interactions could also be observed between the hydrophobic cluster around β3–β4 hairpin and hydrophobic residues in the N-terminal part of the chain: the frequently populated turn W5–L8, V9 and α helix residues V19, I20, and L21.
Nucleation site.
Experiments show that residues forming the α helix have the highest average Φ values, followed by β strands 3 and 4 (13), suggesting a native-like structure of these protein fragments in the TS. Computationally predicted TS conformations (46) with a probability of ≈0.5 to reach the native state rapidly were verified by several independent simulations. Two types of alternative conformations were examined: one with a disrupted α helix and the other with disordered β3 and β4 strands. Surprisingly, α-disrupted states have a stronger tendency to fold (Pfold ≈0.3) than the β-disrupted states, which exhibit almost no tendency (Pfold ≈0) to fold, despite higher Φ values for the α helix in the TS. Thus, it was concluded that, relying exclusively on Φ values, it is not always possible to distinguish between the kinetically important and less important residues.
Our simulations confirm the importance of β3–β4 for CI2 folding, with this substructure being the main nucleation site. At Tt, hydrophobic clustering becomes significantly more intensive, mostly between the β3 and β4 strand (in ≈20% snapshots) (Fig. 2b). Persistent tertiary residual structure of the β3–β4 hairpin at Tt determines the main nucleation site.
From the most nucleating area to the near-native structure.
The best-formed CI2 structures observed at Tt were ≈7 Å from the native state, with unstructured and highly mobile terminal strands. To examine the last stages of folding more closely, several independent long-unfolding/folding simulations were performed at lower T (T = 1.6), starting from native-like structures. In all cases, temporary or permanent disruption of the edges of the β sheet (Fig. 2b, T = 1.6, above the diagonal) and sometimes even the disruption of the central part of the β sheet (β3–β4) were observed. Two simulations of 10, one of which is shown in Fig. 2b (T = 1.6, below the diagonal) resulted in correct native-like topology. The best-formed structure observed was 3.8 Å from the native (Fig. 3b). A native-like assembly of C-terminal and N-terminal strands was usually initiated by the contacts of A16, L49, and A58 (Fig. 2b). Strikingly, although protein-engineering studies for >100 mutants (13) demonstrated that mutations of A16, L49, and I57 dramatically decrease their stability and folding rate, the simulations suggested A58, and not I57, to be the interaction link between the helix and the N-terminal strand. Detailed analysis of the simulation trajectories shows that I57 is nearly always pointing away from the helix in the opposite direction (Fig. 3b) and maintains this orientation in native-like structures. Instead of I57, A16 frequently form contacts with a much smaller side chain of A58. Thus, both the experiments and our simulations show a high significance of the contacts between the same fragments of the polypeptide chain for the final stage of the folding. These findings support the idea that TS structures and folding mechanisms are determined by protein topology (47). Analyzing double-mutant cycles, Ladurner et al. (48) demonstrated a strain between A16 and I57 in the TS, whereas A16 and L49 interact favorably at the same stage. The strain between A16 and I57 is even more noticeable in the native state. Remarkably, these results are the exact opposite from those observed for barnase. An extensive, double-mutant cycle analysis of the major α helix (49), surface salt bridges (50), and hydrophobic cores of barnase did not reveal unfavorable interactions either in the major TS or in the native state. It was also noted that the residues forming the folding nucleus of CI2 are evolutionarily conserved in homologous proteins. Valine is more common than isoleucine in corresponding positions, which, according to Ladurner et al., suggests a preference for a smaller aliphatic side chain in the position 57. Thus, interactions between some of the closely packed residues in the folding nucleus of CI2 may have been evolutionarily optimized for the rate of folding rather than for protein stability.
In the observation of the sequential folding of CI2 it was noted that immediately after the β3–β4 and α helix formation, docking of α helix to β3–β4 begins with the helix C terminus, and a loosely defined hydrophobic cluster is formed. The final consolidation of the hydrophobic cluster takes place as the central part of the helix (A16) is bound to β4 (L49) and β5β6 (in the vicinity of A58) (Fig. 3b).
Description of hydrophobic core.
Studying 11 mutants in the hydrophobic core of CI2, Jackson et al. (45) found that residues with low Φ values (L8, V47, and I57) are all located at the edge of the core, whereas residues with fractional Φ values (0.3–0.65), e.g., I20, I29, L49, and V51, belong to the center of the core. The interactions are 50% weaker in the TS compared with the native state, suggesting that at TS the core has not attained the firm packing and could be partially exposed to the solvent (45). Our results complement these protein-engineering studies. These findings apply both to the central hydrophobic core interactions (I20 and L49), essential for docking the helix to β3–β4 and to the special role of the contacts at the edges of the hydrophobic cluster, created in the final stage of the folding simulations (V47 and A58, playing the role of I57), which is illustrated in the simulation snapshots (Fig. 3b). It was recorded that interactions of I20 with V47 and L49 are the first persistent and productive nucleation sites between the central part of the sheet (β3–β4 strands) and the helix. It is essential for the correct arrangement of the fold that strong interactions between β3–β4 and the α helix take place before any contacts occur between β5 and N-terminal strands (Fig. 2b).
Sequence of folding events: insights from MD and MC simulations.
MD unfolding simulations of the CI2 were usually conducted at very high temperatures (51–54). Consistent results were obtained for different force fields. At 500 K, the contacts between β1 and β5β6 disappeared very rapidly (51), followed by the contacts between β4 and β5β6 and contacts within the α helix. The cluster of contacts between β3 and β4 dissolved as the last. Comparable results were obtained in MD folding and unfolding simulations (52, 53). Moreover, the following cooperative substructures were identified: α, β1β2–β5β6, β3–β4, and β4–β5β6 (54). A characteristic sequence of events was observed: the unfolding of β1β2–β5β6 before the unfolding of clusters α, β3–β4, β4–β5β6. The unfolding of the last three clusters occurred essentially in parallel, especially at very high temperatures. Nevertheless, a preference of β3–β4 as the last to unfold could be noted.
An identical picture of the cooperativity of substructures as well as the overall sequence of folding events emerges from our studies with the reduced CABS model of CI2, with the exception of the fact that the events defined as parallel in time in the MD simulations are unambiguously sequential in ours.
Conclusions
In this work we used a simplified high-resolution lattice model and MC dynamics to study the dynamics of barnase and CI2 in the denatured state and at various stages of their folding processes. Barnase and CI2 fold with half-times of 50 and 10 ms, respectively (31). Classical MD simulations are currently limited to time scales of <1 μs, which is clearly far too short. Such a time scale can be introduced at the cost of a reduced representation of the protein conformational space and approximations in the force field. Despite these approximations, the folding mechanism of barnase was reproduced adequately, and there is a good agreement with the available experimental data for CI2. Thus, it can be stated that reduced models could be valuable tools for the theoretical study of protein dynamics and folding mechanisms. Moreover, knowledge-based potentials derived from the observation of structural regularities in folded proteins can be used not only in structure prediction but also in folding studies from the very beginning of the process.
General qualitative observations for the two proteins are consistent with experimental studies. The average number of native contacts at Tt for barnase (found to be folding by multistate kinetics from the denatured state containing a considerable amount of residual structure) was twice as large compared with CI2 (which folds from a relatively expanded denatured state). On the other hand, the fluctuation of the number of native-like contacts was two times larger for CI2 (two-state folding). Folding-initiation sites and nucleating areas were identified for both proteins. For instance, in the CI2 case, the hairpin β3–β4 was discovered to be the main nucleating area. This finding has not been observed before in any protein-independent force field study but is in agreement with MD unfolding simulations, which indicated that the α helix or β3β4 should be the last element to unfold. There is also a near-perfect correspondence between the long-range contacts observed most frequently in our simulations and the folding nuclei described in protein-engineering studies.
The folding pathway of barnase was adequately reproduced. The general picture of hydrophobic interactions is consistent with the experimental findings for the intermediate and TS structure and confirms their crucial role in the initiation and sustenance of protein folding (55). For CI2, even a quantitative relationship between the Φ values and CI2 structure formation can be observed. Our experiment showed the same structural cooperativity and folding mechanism as seen in the MD unfolding; however, it resulted in a better-defined sequence of events, especially with regard to the formation of α and β3–β4 as being clearly separated from the formation of the remaining substructures.
The presented approach goes far beyond simple analytical models or Go models, enabling the study of complete unfolding/folding pathways. Physically realistic folding mechanisms observed in the CABS simulations imply that the interactions in the denatured state have to be similar to those in the native structures. Consequently, the knowledge-based potentials from native structures can be considered a good approximation of the interactions in the denatured state. Therefore, the suggested model may be a useful tool for qualitative studies of entire folding pathways of large proteins and macromolecular assemblies.
Methods
The high-resolution reduced-protein model and simulation protocol have been described in detail recently (28). Each amino acid is represented in CABS by four interaction centers: Cα, Cβ, the center of a side-group mass, and the center of the peptide bond. Knowledge-based potentials of the force field include generic protein-like conformational biases, statistical potentials for the short-range conformational propensities, a model of the main-chain hydrogen bonds, and context-dependent statistical potentials describing the side-group interactions. The asymmetric Metropolis MC scheme controls the simulation process. MC moves have a local character. Therefore, their long random sequences simulate the long-time dynamics of a polypeptide chain. A single step of the MC algorithm consists of k*N attempts at various local conformational transitions, where k is an integer range of 20 and N is the number of residues.
Side-chain contact maps (Fig. 2) were derived from the distances between the gravity centers of the side chains, using the CABS force field cutoffs. Each contact map presented in this work is an average of five (except for the single simulations for CI2 at T = 1.6) independent isothermal simulations (different random seeds were enough to ensure complete lack of correlations between trajectories) consisting of 200,000 MC steps, where the second half of each trajectory (100,000 steps) was taken for the analysis. Longer simulations did not bring any significant changes to the results, and the differences of calculated observables between particular trajectories were negligible.
The BioShell package was very useful in managing and analyzing the large volume of simulation data (56). To prepare illustrations with some essential structural details, all-atom models were reconstructed from Cα backbone with the use of BBQ (57). The pictures, as well as the secondary structure assignment, were made by using the PyMOL (www.pymol.org).
Abbreviations
- CI2
chymotrypsin inhibitor 2
- MC
Monte Carlo
- MD
molecular dynamics
- T
reduced temperature
- TS
transition state
- Tt
transition temperature.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Matouschek A, Serrano L, Fersht AR. J Mol Biol. 1992;224:819–835. doi: 10.1016/0022-2836(92)90564-z. [DOI] [PubMed] [Google Scholar]
- 2.Bycroft M, Matouschek A, Kellis JT, Jr, Serrano L, Fersht AR. Nature. 1990;346:488–490. doi: 10.1038/346488a0. [DOI] [PubMed] [Google Scholar]
- 3.Udgaonkar JB, Baldwin RL. Nature. 1988;335:694–699. doi: 10.1038/335694a0. [DOI] [PubMed] [Google Scholar]
- 4.Serrano L, Matouschek A, Fersht AR. J Mol Biol. 1992;224:805–818. doi: 10.1016/0022-2836(92)90563-y. [DOI] [PubMed] [Google Scholar]
- 5.Otzen DE, Itzhaki LS, elMasry NF, Jackson SE, Fersht AR. Proc Natl Acad Sci USA. 1994;91:10422–10425. doi: 10.1073/pnas.91.22.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein-Seetharaman J, Oikawa M, Grimshaw SB, Wirmer J, Duchardt E, Ueda T, Imoto T, Smith LJ, Dobson CM, Schwalbe H. Science. 2002;295:1719–1722. doi: 10.1126/science.1067680. [DOI] [PubMed] [Google Scholar]
- 7.Kazmirski SL, Wong KB, Freund SM, Tan YJ, Fersht AR, Daggett V. Proc Natl Acad Sci USA. 2001;98:4349–4354. doi: 10.1073/pnas.071054398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shortle D, Ackerman MS. Science. 2001;293:487–489. doi: 10.1126/science.1060438. [DOI] [PubMed] [Google Scholar]
- 9.Dobson CM. Curr Biol. 1994;4:636–640. doi: 10.1016/s0960-9822(00)00141-x. [DOI] [PubMed] [Google Scholar]
- 10.Blanco FJ, Serrano L, Forman-Kay JD. J Mol Biol. 1998;284:1153–1164. doi: 10.1006/jmbi.1998.2229. [DOI] [PubMed] [Google Scholar]
- 11.Shortle D. FASEB J. 1996;10:27–34. doi: 10.1096/fasebj.10.1.8566543. [DOI] [PubMed] [Google Scholar]
- 12.Karplus M, Weaver DL. Protein Sci. 1994;3:650–668. doi: 10.1002/pro.5560030413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itzhaki LS, Otzen DE, Fersht AR. J Mol Biol. 1995;254:260–288. doi: 10.1006/jmbi.1995.0616. [DOI] [PubMed] [Google Scholar]
- 14.Fersht AR. Curr Opin Struct Biol. 1997;7:3–9. doi: 10.1016/s0959-440x(97)80002-4. [DOI] [PubMed] [Google Scholar]
- 15.Daggett V, Fersht AR. Trends Biochem Sci. 2003;28:18–25. doi: 10.1016/s0968-0004(02)00012-9. [DOI] [PubMed] [Google Scholar]
- 16.Ferguson N, Fersht AR. Curr Opin Struct Biol. 2003;13:75–81. doi: 10.1016/s0959-440x(02)00009-x. [DOI] [PubMed] [Google Scholar]
- 17.Daggett V, Levitt M. Proc Natl Acad Sci USA. 1992;89:5142–5146. doi: 10.1073/pnas.89.11.5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li A, Daggett V. Proc Natl Acad Sci USA. 1994;91:10430–10434. doi: 10.1073/pnas.91.22.10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daggett V, Levitt M. J Mol Biol. 1993;232:600–619. doi: 10.1006/jmbi.1993.1414. [DOI] [PubMed] [Google Scholar]
- 20.Li A, Daggett V. J Mol Biol. 1998;275:677–694. doi: 10.1006/jmbi.1997.1484. [DOI] [PubMed] [Google Scholar]
- 21.Boczko EM, Brooks CL., III Science. 1995;269:393–396. doi: 10.1126/science.7618103. [DOI] [PubMed] [Google Scholar]
- 22.Bond CJ, Wong KB, Clarke J, Fersht AR, Daggett V. Proc Natl Acad Sci USA. 1997;94:13409–13413. doi: 10.1073/pnas.94.25.13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duan Y, Kollman PA. Science. 1998;282:740–744. doi: 10.1126/science.282.5389.740. [DOI] [PubMed] [Google Scholar]
- 24.Rothwarf DM, Scheraga HA. Biochemistry. 1996;35:13797–13807. doi: 10.1021/bi9608119. [DOI] [PubMed] [Google Scholar]
- 25.Blanco FJ, Ortiz AR, Serrano L. Fold Des. 1997;2:123–133. doi: 10.1016/s1359-0278(97)00017-5. [DOI] [PubMed] [Google Scholar]
- 26.Liwo A, Khalili M, Scheraga HA. Proc Natl Acad Sci USA. 2005;102:2362–2367. doi: 10.1073/pnas.0408885102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levitt M, Warshel A. Nature. 1975;253:694–698. doi: 10.1038/253694a0. [DOI] [PubMed] [Google Scholar]
- 28.Kolinski A. Acta Biochim Pol. 2004;51:349–371. [PubMed] [Google Scholar]
- 29.Kolinski A, Bujnicki JM. Proteins. 2005;61:84–90. doi: 10.1002/prot.20723. [DOI] [PubMed] [Google Scholar]
- 30.Bradley P, Malmstrom L, Qian B, Schonbrun J, Chivian D, Kim DE, Meiler J, Misura KM, Baker D. Proteins. 2005;61:128–134. doi: 10.1002/prot.20729. [DOI] [PubMed] [Google Scholar]
- 31.Fersht AR, Daggett V. Cell. 2002;108:573–582. doi: 10.1016/s0092-8674(02)00620-7. [DOI] [PubMed] [Google Scholar]
- 32.Matouschek A, Kellis JT, Jr, Serrano L, Fersht AR. Nature. 1989;340:122–126. doi: 10.1038/340122a0. [DOI] [PubMed] [Google Scholar]
- 33.Salvatella X, Dobson CM, Fersht AR, Vendruscolo M. Proc Natl Acad Sci USA. 2005;102:12389–12394. doi: 10.1073/pnas.0408226102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fersht AR. FEBS Lett. 1993;325:5–16. doi: 10.1016/0014-5793(93)81405-o. [DOI] [PubMed] [Google Scholar]
- 35.Wong KB, Clarke J, Bond CJ, Neira JL, Freund SM, Fersht AR, Daggett V. J Mol Biol. 2000;296:1257–1282. doi: 10.1006/jmbi.2000.3523. [DOI] [PubMed] [Google Scholar]
- 36.Arcus VL, Vuilleumier S, Freund SM, Bycroft M, Fersht AR. J Mol Biol. 1995;254:305–321. doi: 10.1006/jmbi.1995.0618. [DOI] [PubMed] [Google Scholar]
- 37.Freund SM, Wong KB, Fersht AR. Proc Natl Acad Sci USA. 1996;93:10600–10603. doi: 10.1073/pnas.93.20.10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matthews JM, Fersht AR. Biochemistry. 1995;34:6805–6814. doi: 10.1021/bi00020a027. [DOI] [PubMed] [Google Scholar]
- 39.Arcus VL, Vuilleumier S, Freund SM, Bycroft M, Fersht AR. Proc Natl Acad Sci USA. 1994;91:9412–9416. doi: 10.1073/pnas.91.20.9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serrano L, Kellis JT, Jr, Cann P, Matouschek A, Fersht AR. J Mol Biol. 1992;224:783–804. doi: 10.1016/0022-2836(92)90562-x. [DOI] [PubMed] [Google Scholar]
- 41.Kippen AD, Sancho J, Fersht AR. Biochemistry. 1994;33:3778–3786. doi: 10.1021/bi00178a039. [DOI] [PubMed] [Google Scholar]
- 42.Jackson SE, Fersht AR. Biochemistry. 1991;30:10428–10435. doi: 10.1021/bi00107a010. [DOI] [PubMed] [Google Scholar]
- 43.De Prat Gay G, Ruiz-Sanz J, Neira JL, Itzhaki LS, Fersht AR. Proc Natl Acad Sci USA. 1995;92:3683–3686. doi: 10.1073/pnas.92.9.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neira JL, Itzhaki LS, Ladurner AG, Davis B, de Prat Gay G, Fersht AR. J Mol Biol. 1997;268:185–197. doi: 10.1006/jmbi.1997.0932. [DOI] [PubMed] [Google Scholar]
- 45.Jackson SE, elMasry N, Fersht AR. Biochemistry. 1993;32:11270–11278. doi: 10.1021/bi00093a002. [DOI] [PubMed] [Google Scholar]
- 46.Li L, Shakhnovich EI. Proc Natl Acad Sci USA. 2001;98:13014–13018. doi: 10.1073/pnas.241378398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alm E, Baker D. Curr Opin Struct Biol. 1999;9:189–196. doi: 10.1016/S0959-440X(99)80027-X. [DOI] [PubMed] [Google Scholar]
- 48.Ladurner AG, Itzhaki LS, Fersht AR. Fold Des. 1997;2:363–368. doi: 10.1016/S1359-0278(97)00050-3. [DOI] [PubMed] [Google Scholar]
- 49.Horovitz A, Serrano L, Fersht AR. J Mol Biol. 1991;219:5–9. doi: 10.1016/0022-2836(91)90852-w. [DOI] [PubMed] [Google Scholar]
- 50.Horovitz A, Serrano L, Avron B, Bycroft M, Fersht AR. J Mol Biol. 1990;216:1031–1044. doi: 10.1016/S0022-2836(99)80018-7. [DOI] [PubMed] [Google Scholar]
- 51.Lazaridis T, Karplus M. Science. 1997;278:1928–1931. doi: 10.1126/science.278.5345.1928. [DOI] [PubMed] [Google Scholar]
- 52.Ferrara P, Apostolakis J, Caflisch A. Proteins. 2000;39:252–260. doi: 10.1002/(sici)1097-0134(20000515)39:3<252::aid-prot80>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 53.Ferrara P, Apostolakis J, Caflisch A. J Phys Chem. 2000;104:4511–4518. [Google Scholar]
- 54.Reich L, Weikl TR. Proteins. 2006;63:1052–1058. doi: 10.1002/prot.20966. [DOI] [PubMed] [Google Scholar]
- 55.Dyson HJ, Wright PE, Scheraga HA. Proc Natl Acad Sci USA. 2006;103:13057–13061. doi: 10.1073/pnas.0605504103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gront D, Kolinski A. Bioinformatics. 2006;22:621–622. doi: 10.1093/bioinformatics/btk037. [DOI] [PubMed] [Google Scholar]
- 57.Gront D, Kmiecik S, Kolinski A. J Comput Chem. 2007;28:1593–1597. doi: 10.1002/jcc.20624. [DOI] [PubMed] [Google Scholar]