Abstract
A technique was developed for the detection of fluorescence signals from free single molecules for extended time periods and was applied to the characterization of the unfolded states of iso-1-cytochrome c (cyt c). Protein molecules labeled with fluorescent dye were slowly injected into a capillary at concentrations that allow for the observation of one molecule at a time. A laser was introduced into the capillary coaxially, and the fluorescence was imaged as traces by using a lens with a large focal depth and wide field of view. Thus, the traces reflect the time-dependent changes in the fluorescence signals from single proteins. Cyt c was labeled with Alexa Fluor 532 at the C-terminal cysteine (cyt c-Alexa). In bulk experiments, cyt c-Alexa was shown to possess different fluorescence intensity for the native state, the unfolded state (U), and the intermediate state. Single-molecule traces of cyt c-Alexa were recorded by using the device. Intensity histograms of the traces revealed two distributions with broad and narrow widths, which were interpreted to correspond to the U and intermediate state, respectively, observed in the bulk measurements. The broad width of the U suggested the existence of a relatively slow conformational dynamics, which might be consistent with the correlation time (≈15 ms) estimated from the traces assignable to the U. The technique was expected to reveal dynamics of proteins along the folding processes without artifacts caused by immobilization.
Keywords: fluorescence, heterogeneity, iso-1-cytochrome c, unfolded state
Proteins fold spontaneously and cooperatively from the unfolded state (U) to the native state (N) within biologically relevant time scales (1). The U is supposed to possess a myriad of extended conformations based on its apparent similarity to a random coil of homopolymer (2, 3). In contrast, the N possesses essentially a unique conformation that is composed of the secondary and tertiary structures stabilized by intraprotein interactions coded in the primary sequence of amino acid residues (4). Thus, the process of protein folding should involve complex dynamics that would lead the heterogeneous ensemble of unfolded conformations to form the single native conformation (5, 6). A recent consensus, however, supposes a surprising simplicity for the folding of small proteins (7, 8). The structures of the transition state ensemble for the folding kinetics of small proteins deduced by Φ-value analysis could be roughly explained by assuming Go's potential (9–11), which neglects the intraprotein interactions between residues not making contacts in the N (12). The statistical correlation between the rate of folding and the topological complexity of proteins also supported a simple folding mechanism (7, 13). However, the molecular mechanism that assures the approximation of protein folding based on Go's potential has yet to be clarified. Characterizations of the conformational heterogeneity of unfolded proteins and of the dynamics of the heterogeneous conformations are required to establish the rules that connect the U and the N.
Detection of the heterogeneity and dynamics of unfolded proteins has been limited, because the different conformations of the U are difficult to resolve in conventional experiments that rely on averaging the ensemble of molecules. In contrast, single-molecule observations have a great potential to resolve the conformational and dynamic properties of unfolded proteins by detecting signals from single molecules (14–16). Several pioneering experiments were performed for the observations of the conformational transitions at the single-molecule level by using fluorescence signals from labeled proteins (17–23). The selective detection of the unfolded conformations under physiological solution conditions was made possible (21). Furthermore, subpopulations in the unfolded conformations and the existence of heterogeneous transitions and pathways were revealed at the single-molecule level (18, 24).
Despite the significant success of the initial reports on the single molecule detection of protein folding, the experimental strategies used in these investigations have several drawbacks. One method utilizes the combination of total internal reflection microscopy and immobilization of proteins at the surface of optical plates (17, 25). The method can detect fluorescence from single proteins for extended time periods. Furthermore, low backgrounds due to the evanescent excitation enable the detection of signals with a good signal-to-noise ratio. The drawback of the method is an artifact on unfolded proteins caused by the interaction with the optical surface (19). The other method is based on confocal microscopy and the diffusion or flow of the labeled proteins at the focal volume of microscopic objectives (19–22). The method can eliminate background by using confocal alignment; however, it can monitor signals only for limited durations determined by the sample passage in the focal area. It is difficult to follow the entire folding processes of single proteins continuously by using this method. Several methods were proposed to extend the observation periods by tethering proteins with minimal artifacts (18, 24).
In this article, we propose a different approach for the detection of single proteins for extended time periods without artifacts originating from surface immobilization. We describe the principles of the method and demonstrate evidence for the detection of fluorescence signals from single molecules. As a demonstration of the method and the quality of the obtainable data, the properties of iso-1-cytochrome c (cyt c) in the unfolded conformations were investigated.
Results
Construction of the Single-Molecule Device.
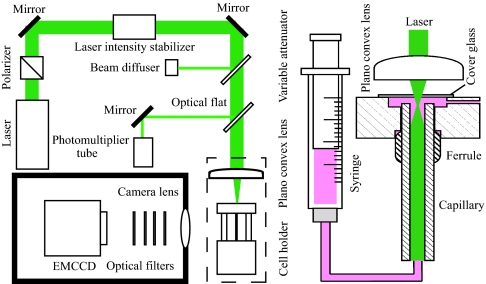
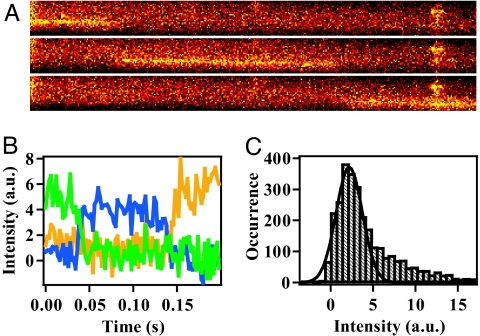
The system developed to observe free single molecules for extended time periods is shown in Fig. 1. A protein labeled with a fluorescent dye was introduced into the capillary cell at such a low concentration that only one molecule flows at a time on average in the observation area. The cell was made of fused silica with an internal diameter of 75 μm. The sample flow was adjusted at a low speed to keep small Reynolds numbers (≈0.1) and a laminar flow. The fluorescence from the labeled dye was excited by a laser introduced into the capillary coaxially and was imaged on an electron-multiplier CCD (EMCCD). The lens used for imaging has a moderate N.A. of 0.33, a large focal depth (≈11 μm), and a wide field of view (≈2 × 2 mm2). Examples of images recorded by the method are shown in Fig. 2A, which were detected by flowing Alexa Fluor 532 C5 maleimide (Alexa532) labeled to glutathione (G-Alexa) through the device. The time-dependent changes of fluorescence intensity shown in Fig. 2B were converted from the traces in Fig. 2A. A typical residence time of a molecule within a distance corresponding to one pixel of EMCCD was ≈500 μs. Considering the blurring of the traces, the method enables fluorescence detection of single molecules with a millisecond time resolution.
Fig. 1.
Schematic drawing of the apparatus developed for the flow detection of single molecules. (Right) A syringe pump was used to supply the solution of labeled protein into a capillary cell, which was made by fused silica with an inner diameter of 75 μm. (Left) A laser beam at 532 nm was passed through a polarizer and an intensity stabilizer and was focused to the sample exit of the capillary. The laser intensity was monitored by a photomultiplier tube. The florescence emitted from single molecules was collected by a camera lens, passed through four optical filters, and imaged on an EMCCD.
Fig. 2.
Examples of the fluorescence data from single molecules of G-Alexa dissolved in the buffer containing 4 M Gdm. (A) Typical images of the fluorescence traces from a single G-Alexa flowing inside the capillary. The excitation laser power was 600 mW. The images were recorded consecutively with an exposure time of 100 ms. The background intensity was not subtracted. (B) The time evolution of the fluorescent intensity from the single G-Alexa converted from the raw data presented in A. The residence time of one data point was adjusted to 2 ms per point. The green, blue, and yellow traces correspond to the time evolution converted from the upper, middle, and bottom images in A, respectively. (C) Intensity histogram of the fluorescence from single molecules of G-Alexa. The fluorescent intensities recorded from 73 images were plotted. The residence time of one data point of the traces was 2 ms per point. The black line shows the single Gaussian function fitted to the observed distribution with an average intensity of 2.17 and a standard deviation of 1.55.
A key component of the current system is the imaging lens with a moderate N.A. value. Conventionally, oil or liquid immersion objectives developed for microscopic observations were used for single-molecule detection, because the high N.A. values and magnifications of these objectives enable imaging at the diffraction limit with a high collection efficiency of photons (26). However, the field of view and the focal depth of the microscopic objectives are incompatible with these performances. In contrast, the method developed does not depend on the imaging at the diffraction limit but requires a wide field of view for the observation of flowing molecules for prolonged periods. Thus, although the light collection efficiency of the current lens is moderate, its low magnification and small spherical aberration ascertain a wide field of view. Furthermore, the large focal depth is advantageous for the reduction of the defocused fluorescence from molecules flowing outside the focal depth by imaging a larger cross-section of the flow.
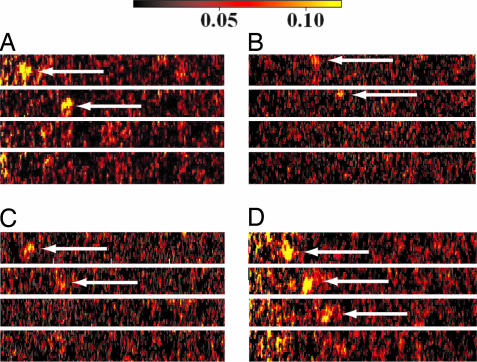
Several lines of evidence demonstrated that the data recorded by the device resulted from fluorescence originating from single molecules. First, the intensity histograms of G-Alexa consists of a single peak with a small shoulder, which is expected for a single fluorophore showing constant fluorescence intensity (Fig. 2C). The shoulder might be attributed to the spectral fluctuation of Alexa532, which was suggested for other fluorophores (27). Second, saturating behavior of the fluorescence intensity at the higher laser power was observed [supporting information (SI) Fig. 6]. The same saturation also was observed for the fluorescence intensity at the bulk concentration of G-Alexa and was caused by the involvement of the triplet state in the photocycle (SI Text and SI Fig. 7). Therefore, the saturation ascertains that the observed signals are not caused by light scattered by impurity particles, because the light scattering should demonstrate a linear power dependence. Third, photobleaching of dyes in a single step was detected by using modified systems that allowed the observation of fluorescence at a higher laser power and at an extremely slow flow speed (Fig. 3). The bleaching in a single step shows that the signals are not from clusters of molecules but from single molecules (28). These results demonstrate that the observed signals are fluorescence originating from single molecules. Thus, the device makes it possible to observe single proteins for the observation period of ≈100 ms and with the time resolution of ≈1 ms.
Fig. 3.
Photobleaching of G-Alexa observed at the single-molecule level. The exposure time for each panel was 10 ms, and the interval between adjacent panels was ≈30 ms. The single fluorescence spots of G-Alexa are indicated by the arrow. The fluorescence spot suddenly disappeared in a single step. Intensities are represented in color scaling from black to yellow. The fluorescence intensity was normalized by using the standard solution of G-Alexa.
Several features of the method are worth noting here. First, the sample–cell interface inside the capillary does not satisfy the condition of total reflection, because the refractive index of silica is higher than that of water. Therefore, the excitation laser was focused into the capillary with a narrow incident angle to minimize the dissipation of photons from the capillary. The photon flux is not uniform over the cross-section of the cell. Assuming that the flux is proportional to the intensity of background scattering in the absence of dye, the flux at the center was estimated to be approximately three times stronger than that of the uniform excitation. Although the nonuniformity of the flux might affect the distribution of fluorescence intensity, the effect is within the intensity distribution of G-Alexa shown in Fig. 2C. Second, the sample concentration is critically important for the observations of single molecules. The adsorption of samples to supply lines and the photobleaching of dyes before entering the observation area may decrease the concentration of the samples. On the other hand, excess concentration easily results in increased background due to the overlap of signals from many molecules. Thus, a careful adjustment of the sample concentration was necessary before recording single-molecule data. Third, the flow speed of individual proteins differs due to the laminar flow inside the cell. Therefore, the difference in the flow speeds was calibrated protein by protein based on the length of the fluorescence traces detected during the observation period of 100 ms. Proteins flowing slower than 4 ms per pixel were scarcely observed, which may be due to photobleaching before the proteins entered the observation area. Under the assumption of laminar flow, the proteins flowing faster than 4 ms per pixel should be located within 26 μm from the center of the capillary. Thus, the signals were not originating from proteins sticking to the inner surface of the capillary. Furthermore, the traces from proteins flowing slower than a certain speed (typically 2 ms per pixel) were not used for the analysis to improve the time resolution of the data.
Observation of Cytochrome c Folding at the Single-Molecule Level.
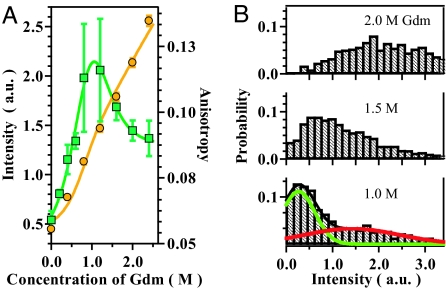
To observe protein folding dynamics at the single-molecule level, we chose cyt c from Saccharomyces cerevisiae (29, 30). The C-terminal cysteine of cyt c was labeled with Alexa532 (cyt c-Alexa). Cyt c possesses a heme group, which is covalently attached to cysteine residues 14 and 17 and acts as a quencher for the fluorescence from Alexa532 based on the fluorescence resonance energy transfer (31–33). Because heme is separated from Alexa532 along the polypeptide chain, the fluorescence is strong for the U. In contrast, the fluorescence is quenched in the N due to the proximity of the labeled fluorophore and heme. This result is confirmed in the fluorescence intensity of cyt c-Alexa upon the titration of guanidium chloride (Gdm) observed by bulk measurements (Fig. 4A). In addition, the bulk measurements of fluorescence anisotropy showed a peak at ≈1 M Gdm, suggesting the presence of a distinct state other than the N and U. As shown in Fig. 4A, the titration results could be analyzed by using a standard three-state model (1). We conclude that the folding of cyt c involves the U with strong fluorescence populated at a Gdm concentration of >2.0 M, an intermediate state (I) with moderate fluorescence maximally populated at 1.0 M Gdm, and the N with almost quenched fluorescence. The I likely corresponds to one of the two unfolded species described previously (30). A partially structured I was identified for horse cytochrome c under similar condition (34).
Fig. 4.
The fluorescent data of cyt c-Alexa recorded at the bulk and single-molecule levels in the presence of various concentrations of Gdm. (A) The fluorescence intensities (orange circles) and anisotropies (green rectangles) of cyt c-Alexa recorded by the bulk measurements. The orange and green lines are the fitting of the intensity and anisotropy data, respectively, based on the standard three-state model. (B) The fluorescence intensity histograms obtained for cyt c-Alexa at the single-molecule level. (Upper) Histogram obtained in the presence of 2.0 Gdm. (Middle) Histogram obtained in the presence of 1.5 Gdm. (Lower) Histogram obtained in the presence of 1.0 M Gdm. The excitation laser power was 545 mW for the data of 2.0 M and 480 mW for the data of 1.5 and 1.0 M, respectively. The numbers of the fluorescence images used for the calculation of the histograms for 2.0, 1.5, and 1.0 M of Gdm were 37, 57, and 32, respectively. The residence time of one data point of the original traces was 2 ms per point. The green and red curves represent the fitting of the data recorded in the presence of 1.0 M Gdm based on two Gaussian distributions. The fitted parameters are described in the text.
The results of single-molecule measurements on the equilibrium transitions of cyt c-Alexa confirmed the existence of distinct distributions corresponding to the different states. The intensity histograms of the single-molecule fluorescence from cyt c-Alexa, observed at 1.0, 1.5, and 2.0 M Gdm are demonstrated in Fig. 4B. Different from the distribution observed for G-Alexa (Fig. 2C), the distribution of cyt c-Alexa at 1.0 M Gdm consists of a narrow peak and a broad shoulder. The distribution changes at the higher Gdm concentration, where the broad distribution gains probability at the expense of the narrow distribution. Considering the bulk titration results, one can assign the narrow distribution to the I and the broad distributions to the U. The distributions for the N cannot be observed because of the low fluorescence intensity. Thus, the results of the single-molecule observation were consistent with the ensemble observations.
Discussion
Fluorescence from Free Single Molecules Can Be Detected for Extended Time Periods.
Previous methods for fluorescence detection of single molecules use either total internal reflection or confocal microscopies. These methods were developed to eliminate the background caused by the defocused fluorescence; however, the methods impose several limitations on single-molecule experiments. In the case of total internal reflection method, sample proteins need to be tethered to optical plates. The tethering sometimes causes undesirable interactions between the unfolded proteins and the surface (19). In the case of confocal methods, molecules can be observed without tethering but only for limited time spans determined by the residence time of a molecule in the focal volume (35). In contrast to these methods, we demonstrated here that the fluorescence signals from free single molecules can be detected for extended time periods by using an imaging lens possessing a large focal depth and wide field of view. The evidence for single-molecule detection is the presence of a single major peak in the intensity distribution, saturation behavior of the fluorescent intensity at higher excitation power, and photobleaching of fluorophore in a single step. The large focal depth and wide field of view of the optics used are advantageous for the reduction of the defocused background and for the detection of flowing molecules for extended time periods, respectively.
The drawback of the current method is the moderate collection efficiency of photons by the optics used, because commercial lenses with a large field of view do not possess large N.A. values. The light collection efficiency of the lens used in this study is ≈3%. Although the value is smaller than that of the microscopic objectives (on the order of several tens of percentage points), a high quantum efficiency of EMCCD enables the observation of fluorescence based on the current optics. At the incident laser power of 200 mW, the number of photons emitted as fluorescence from one Alexa532 can be estimated as ≈5.6 × 103 ms−1 based on the photon flux expected at the center of the capillary (≈3.6 × 1011 μm−2·ms−1), the absorption cross section (3.1 × 10−16 cm2) and the fluorescence quantum efficiency of Alexa532 (≈0.5). Because the light collection efficiency of the total optics including filters can be estimated as ≈1% by considering the collection efficiency of the lens (≈3%), the throughput of the optical filters (54% at 560 nm), and other losses, the number of photons expected to reach the EMCCD is ≈56 ms−1. In the current setting of the EMCCD, the conversion efficiency from photons to electrons, the amplification of the electrons, and the conversion ratio of the amplified electrons and the output counts were 0.9, 600, and 0.043, respectively. Thus, the photons arriving at the EMCCD will give ≈650 counts during the 500 μs after amplification and digitization by the EMCCD. For a single Alexa flowing at 500 μs per pixel, the expected fluorescence intensity observable at one pixel of the EMCCD is 100 counts after considering the broadening of the image. This value is close to the experimental fluorescence intensity (≈100–200 counts per pixel) of Alexa at the excitation of 200 mW. For comparison, the background intensity from the capillary cell observed at 200-mW excitation during the observation period of 100 ms is ≈210 counts per pixel. Thus, the typical signal-to-background ratio of the current experiments is ≈1:2 to 1:1. We note that the current experiment was preceded by several investigations based on optics with moderate N.A. values. For example, Lermer et al. (36) used a lens whose N.A. was 0.4 for the detection of single molecules in electrodynamically focused microdroplets. We conclude that the detection of fluorescence signals from single molecules without using microscopic objectives is feasible.
Conformational Dynamics of cyt c in the U.
The bulk titration experiment presented in Fig. 4A demonstrated that the folding transition of cyt c-Alexa can be analyzed by assuming an equilibrium involving the U, I, and N. The intensity histograms of the single-molecule fluorescence of cyt c-Alexa consist of both broad and narrow distributions, which were assigned to the U and I, respectively (Fig. 4B). In the distribution observed at 1.0 M Gdm, the peak intensities for the U and I estimated by the Gaussian fitting were 1.4 ± 0.5 and 0.27 ± 0.05, respectively. The peak intensities correspond to the extended and collapsed conformations for U and I, respectively, as deduced from the bulk experiment. In addition, the Gaussian fitting of the data showed that the distribution width for the U (1.0 ± 0.3) is significantly broader than that of the I (0.35 ± 0.06). A careful analysis of the width, as we will explain below, suggests that the conformational dynamics of cyt c in the U is relatively slow.
The distribution width of fluorescence intensities from single molecules is determined by several factors, including the noise present in backgrounds, the shot noise of fluorescence photons, and the dynamic heterogeneity of samples (21, 37). The background noise can be estimated to 0.13 from the regions of the images observed without the signal traces. The shot noise of fluorescence photons for the U and I can be estimated to 0.40 and 0.17, respectively, based on the peak intensities of the two distributions and the conversion ratio of the intensities and the number of photons (intensity:photon = 8.6:1). Thus, the expected widths without the effect of conformation dynamics are 0.53 and 0.30 for the U and I, respectively. Whereas the width expected for the I (0.30) is similar to that of the observation (0.35 ± 0.06), that for the U (0.53) is significantly narrower than that observed (1.0 ± 0.3). We note that the distribution widths might be further regulated by the nonuniformity of the excitation flux over the cross-section of the capillary; however, the effect should affect the widths for both the U and I. Hence, the broad width only for the U cannot be explained by the nonuniformity of the excitation laser. We suggest that the conformational dynamics of the unfolded cyt c is responsible for the broad width of the U. Because the distribution becomes broader if the dynamics is slower compared with the observation period of one data point (2.0 ms for the data in Fig. 4B), the conformational dynamics in the U is likely in the millisecond time domain.
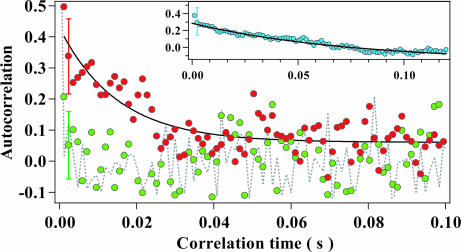
To further obtain information on the dynamic properties of cyt c-Alexa, we calculated autocorrelation functions from the single-molecule data. The single-molecule traces could be divided into two groups corresponding to the U and I. The red circles in Fig. 5 show the correlation for the U, which could be fitted by an exponential decay with a time constant of ≈15 ms (black line). In contrast, the correlation for the I denoted as green circles decays within one data point, which corresponds to 1.2 ms. For comparison, the gray dotted line shows the correlation calculated from the background noise, which decreases within 1.2 ms. The correlation calculated from the single-molecule traces of G-Alexa possesses a decay with a time constant of ≈75 ms (Fig. 5 Inset), which is partly caused by the involvement of the triplet state in the photocycle (SI Text and SI Fig. 8). The comparison of the autocorrelations for G-Alexa and for the U of cyt c-Alexa suggests that the presence of heme, which primarily acts as a quencher for the excited singlet state, causes the changes in the autocorrelation function. Further investigations are necessary for the analysis of the autocorrelation data; however, we propose that the conformational relaxation is partly responsible for the ≈15 ms correlation of the U.
Fig. 5.
The autocorrelation analysis of the single-molecule data for cyt c-Alexa recorded in the presence of 1 M Gdm. The autocorrelation functions shown were calculated from the fluorescence traces recorded in the presence of 1 M Gdm. The red and green circles represent the autocorrelation functions calculated from the traces assignable to the U and I, respectively. The dashed line represents the correlation function of the background noise, which was calculated by assuming a typical residence time of a molecule at one pixel (500 μs per pixel). The residence time of one data point was 1.2 ms per point. A typical error bar was presented at the point at 2.4 ms. The black line represents the single-exponential fitting of the function for the U, whose parameters are described in the text. (Inset) The autocorrelation function estimated for G-Alexa recorded in the presence of 4 M Gdm excited at 600 mW. The residence time of one data point was 1.2 ms per point.
It was shown that an extended form of cyt c was retained at 760 ms after the initiation of folding by observing the fluorescence lifetime of labeled cyt c (29), suggesting that the lifetime of the U is longer than the observation period of the current experiment (100 ms). Several time constants were proposed for the dynamic properties of polypeptides. The upper limit for the reconfiguration time of cold-shock protein was determined to be 25 μs (21). Conformational dynamics in the nanosecond time domain was reported recently (38, 39). In contrast, the rate of loop formation for 85-residue polypeptides corresponding to the segment between Cys-102 and heme in cyt c can be estimated to ≈4.0 ms based on the reported data (40). The presence of the slow dynamics in the millisecond time domain also was suggested for small proteins (41). Furthermore, the residual interactions in the U as reported for the immunity protein for colicin E7 (42), and a possible contribution of the different coordination states of heme in the unfolded cyt c might slow down the dynamics. The slow conformational dynamics is a possible explanation on the property of the unfolded cyt c.
Summary.
A method for the detection of fluorescence from free single molecules for extended time periods was proposed and applied to the single-molecule observation of unfolded cyt c. The observed traces for cyt c-Alexa resolved two distributions assignable to the U and I observed in the bulk measurements. The distribution and the correlation time observed for the U suggested a relatively slow conformational dynamics. The quality of the data obtainable by the method can be improved by adopting an elliptical mirror for a better collection of photons. Thus, the method is expected to be a powerful approach to investigate the dynamics of protein folding and other biophysical processes occurring in solutions.
Materials and Methods
Preparation of Labeled Samples.
cyt c from Saccharomyces cerevisiae was from Sigma (St. Louis, MO). Alexa532 was from Invitrogen (Carlsbad, CA). Gdm was from Nacalai (Kyoto, Japan). Other chemicals were of the highest grade available and used without further purification. Cyt c was labeled with Alexa at Cys-102 in 50 mM sodium phosphate at pH 7.0 containing 3 M urea. Labeling was conducted in the dark at room temperature. The labeled protein was purified by gel-filtration chromatography (Sephadex G-25; GE Healthcare, Buckinghamshire, U.K.) and by reverse-phase chromatography (Cosmosil 5C4; Nacalai, Kyoto, Japan). The concentration of cyt c-Alexa was determined by using the molar absorption coefficient at 408 nm (106,000 M−1·cm−1). Glutathione was labeled with Alexa and purified by the procedures similar to those used for cyt c.
Bulk Fluorescence Measurements.
The fluorescence intensity and anisotropy measurements in the bulk conditions were carried out by using a fluorescence spectrometer (F-4500; Hitachi, Tokyo, Japan). The fluorescence intensity was measured by excitation at 532 nm and by integrating emission from 535 nm to 600 nm. The anisotropy was measured by excitation at 532 nm and emission at 555 nm and calibrated by the grating factor of 1.625. The sample contained 335 nM cyt c-Alexa, 20 mM Hepes at pH 7.5, 40 mM sodium chloride, and various concentrations of Gdm.
Single-Molecule Measurements.
The sample solutions need to be prepared without any scattering particles or fluorescent impurities for the observation of single molecules. Deionized water freshly produced by milliQ (Millipore, Billerica, MA) was used. All glassware was cleaned by ultrasonication and confirmed to be free from the scattering particles upon inspection by filling dust-free water and irradiating laser. Cyt c-Alexa and G-Alexa were prepared at concentrations from several hundred attomolar to a few picomolar in 20 mM Hepes at pH 7.5, 0.001% polyoxyethylene sorbitan monolaurate (Surfact-Amp 20; Pierce, Rockford, IL), 40 mM sodium chloride, and various concentrations of Gdm. The buffer solutions were passed through a 0.22-μm-pore filter to remove particles mainly from Gdm and degassed by a vacuum pump with stirring at 60°C. The samples were prepared inside a clean bench.
The cell was constructed by assembling the fused-silica capillary (TSP075375, Polymicro Technologies, Phoenix, AZ) and the holder that possessed an optical window for the introduction of laser from the sample exit of the capillary. The lengths of the capillary and the observation region were 7.5 mm and ≈2 mm, respectively. The length between the entrance of the capillary and the observation area was ≈3 mm. The polyimide coating of the capillary was removed. The cell-holder assembly was ultrasonically cleaned for 15 min in a detergent solution containing Hellmanex 2 (Hellma, Müllheim, Germany) and washed with deionized water. This procedure was repeated twice. The assembly was then ultrasonically cleaned for 15 min in acetone and washed with deionized water. Finally, the assembly was ultrasonically cleaned for 15 min in deionized water twice and dried in a clean bench.
The output from a 532-nm diode-pumped frequency-doubled Nd:YAG laser (Green Ice; Kantum Electronics, Tokyo, Japan) was passed although a polarizer and a power stabilizer (LS-PRO-532–4W, Brockton Electro-Optics, Brockton, MA). The intensity fluctuation of the laser was continuously monitored and was within the rms of 1.3%. The laser was focused into the sample exit of the cell by a planoconvex lens (f = 40 mm). We estimated the changes in the laser flux in the capillary caused by the changes in the concentration of Gdm by monitoring the background intensity. The changes in the flux were within 12% in a Gdm concentration between 1.0 M and 2.0 M. The fluorescence was collected by a camera lens (F = 1.4 and f = 50 mm; Nikon, Tokyo, Japan), and imaged on a EMCCD (iXon; Andor Technology, Belfast, United Kingdom). The magnification of the image was ≈10-fold. A notch filter (Notch Plus; Kaiser, Ann Arbor, MI), two band-pass filters (S5868/50M; Chroma, Rockingham, VT, and BP580-60; Asahi-bunko, Tokyo, Japan), and a short-pass filter (SU0590; Asahi-bunko) were introduced between the camera lens and the EMCCD. The optical throughput of all of the filters was 54% at 560 nm. The images of the flow cell exposed on the EMCCD for 100 ms were digitized continuously with an EM gain setting of 240.
The following procedures were performed to adjust sample concentrations. The sample solution, whose concentration was initially adjusted at several hundred attomolar, was filled into a 250-μl syringe (Hamilton, Reno, NV) and introduced into the cell by a syringe pump (11Plus; Harvard Apparatus, Holliston, MA) at a rate of 1 μl/min. Tefzel tubes (Upchurch, Oak Harbor, WA) were used for the connection. If single-molecule traces could not be detected and the background intensity was the same as that of the buffer solution, the sample concentration was increased 10-fold. This procedure was repeated until single-molecule traces could be recorded. Single molecules were typically observed at the concentration of several tens of femtomolar and at the frequency of one molecule approximately every 50 s.
The fluorescence images from single molecules were converted into time-series data by the following procedure. From the series of images taken consecutively, those containing signals from single molecules were identified. The two images, recorded immediately before and after the image containing signals, were averaged and subtracted from the signal image as a background. The image recorded was divided by a flat-field image, which was taken at the bulk condition of G-Alexa (typically several picomolar), to correct the spatial differences in the detector response and in the light collection efficiency. Approximately four to six pixels along the perpendicular direction of the single-molecule trace in the flat-fielded image were binned to record the time-series intensity data. The flow speeds of individual molecules were estimated and used to adjust the time interval for one data point to either 1.2 or 2.0 ms per point.
The autocorrelation functions of the single-molecule data were calculated for each trace and were averaged. The error bars represented a standard deviation of the correlation at each time point. For the correlations of the U and I of cyt c-Alexa in the presence of 1.0 M Gdm, the regions of traces possessing the intensity larger and weaker than 0.83 were classified as the U and I, respectively. The regions of traces around 0.83 were discarded. The correlation times for the U and G-Alexa estimated by the single exponential fitting were 15 ± 2 ms for the U and 75 ± 9 ms for G-Alexa.
Observation of Photobleaching.
The observation of photobleaching was made possible based on the modified flow and optical systems that allow the detection of fluorescence from single molecules at an extremely slow flow speed. First, the excitation laser was irradiated perpendicularly to the flow direction to prevent the bleaching of dyes before entering the observation area. The excitation laser (Juno, Syowa Optronics, Tokyo, Japan) was illuminated to the observation area (≈20 μm × 2 mm) of the capillary by a cylindrical lens (f = 70 mm). The total power of the excitation laser was 4.7 W for Fig. 3D and 4.2 W for Fig. 3 A–C. Second, a sheath flow cell was used to confine the flow of samples within 5 μm from the center of the capillary and to maintain the flow speed of the sample constant. The cell was made of an outer capillary whose inner diameter was 76 μm and of an inner capillary whose outer and inner diameters were 72 and 20 μm, respectively. Both capillaries were from Polymicro Technologies. The basic design of the cell was adopted from ref. 43 with some modifications (K.K., M.K., and S.T., unpublished data). A two-channel syringe pump (μ-ton mini, Senshu Scientific, Tokyo, Japan) was used to inject buffer and sample solutions to the outer and inner capillaries, respectively. The speed of the inner flow was adjusted at 0.01 μl/min. The speed of the outer flow was adjusted at 0.3 μl/min for Fig. 3A, 0.5 μl/min for Fig 3B, and 0.1 μl/min for Fig. 3 C and D. The sample concentration filled in the syringe for the inner flow was 1 fM. The line speed of the flow was ≈0.40–1.9 mm/s. Third, the fluorescence was imaged on an image intensifier (VS4–1845; Videoscope, Sterling, Canada) by using a camera lens (F = 1.4 and f = 50 mm; Nikon). The multiplication of the intensifier was approximately ×340. A bandpass filter (PB0065, Asahi-bunko) was added to the same optical filters used for the flow system without the sheath. The output of the intensifier was optically coupled to a cooled CCD (CCD134; Princeton Instruments, Trenton, NJ). The exposure time and readout time of the charge coupled device were 10 ms and ≈30 ms, respectively. The spatial nonuniformity of the fluorescence intensity was normalized by using a standard solution of G-Alexa.
Supplementary Material
Acknowledgments
M.K. and S.T. thank Profs. Isao Morishima and Koichiro Ishimori for helpful discussions. This work was partially supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan (to S.T., Y.G., and T.K.).
Abbreviations
- Alexa532
Alexa Fluor 532 C5 maleimide
- cyt c
iso-1-cytochrome c
- cyt c-Alexa
cyt c labeled with Alexa532
- EMCCD
electron-multiplier CCD
- G-Alexa
glutathione labeled with Alexa532
- Gdm
guanidium chloride
- N
native state
- U
unfolded state
- I
intermediate state.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700267104/DC1.
References
- 1.Fersht AR. Structure and Mechanism in Protein Science. New York: Freeman; 1998. [Google Scholar]
- 2.Kohn JE, Millett IS, Jacob J, Zagrovic B, Dillon TM, Cingel N, Dothager RS, Seifert S, Thiyagarajan P, Sosnick TR, et al. Proc Natl Acad Sci USA. 2004;101:12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fitzkee NC, Rose GD. Proc Natl Acad Sci USA. 2004;101:12497–12502. doi: 10.1073/pnas.0404236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchner J, Kiefhaber T, editors. Protein Folding Handbook. Weinheim, Germany: Wiley–VCH; 2005. [Google Scholar]
- 5.Rao F, Caflisch A. J Mol Biol. 2004;342:299–306. doi: 10.1016/j.jmb.2004.06.063. [DOI] [PubMed] [Google Scholar]
- 6.Krivov SV, Karplus M. Proc Natl Acad Sci USA. 2004;101:14766–14770. doi: 10.1073/pnas.0406234101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker D. Nature. 2000;405:39–42. doi: 10.1038/35011000. [DOI] [PubMed] [Google Scholar]
- 8.Onuchic JN, Wolynes PG. Curr Opin Struct Biol. 2004;14:70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 9.Daggett V, Fersht AR. Trends Biochem Sci. 2003;28:18–25. doi: 10.1016/s0968-0004(02)00012-9. [DOI] [PubMed] [Google Scholar]
- 10.Munoz V, Eaton WA. Proc Natl Acad Sci USA. 1999;96:11311–11316. doi: 10.1073/pnas.96.20.11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galzitskaya OV, Finkelstein AV. Proc Natl Acad Sci USA. 1999;96:11299–11304. doi: 10.1073/pnas.96.20.11299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taketomi H, Ueda Y, Go N. Int J Pept Protein Res. 1975;7:445–459. [PubMed] [Google Scholar]
- 13.Plaxco KW, Simons KT, Baker D. J Mol Biol. 1998;277:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 14.Haran G. J Phys. 2003;15:R1291–R1317. [Google Scholar]
- 15.Schuler B. Chemphyschem. 2005;6:1206–1220. doi: 10.1002/cphc.200400609. [DOI] [PubMed] [Google Scholar]
- 16.Xie XS, Trautman JK. Annu Rev Phys Chem. 1998;49:441–480. doi: 10.1146/annurev.physchem.49.1.441. [DOI] [PubMed] [Google Scholar]
- 17.Arai Y, Iwane AH, Wazawa T, Yokota H, Ishii Y, Kataoka T, Yanagida T. Biochem Biophys Res Commun. 2006;343:809–815. doi: 10.1016/j.bbrc.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 18.Rhoades E, Gussakovsky E, Haran G. Proc Natl Acad Sci USA. 2003;100:3197–3202. doi: 10.1073/pnas.2628068100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talaga DS, Lau WL, Roder H, Tang J, Jia Y, DeGrado WF, Hochstrasser RM. Proc Natl Acad Sci USA. 2000;97:13021–13026. doi: 10.1073/pnas.97.24.13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deniz AA, Laurence TA, Beligere GS, Dahan M, Martin AB, Chemla DS, Dawson PE, Schultz PG, Weiss S. Proc Natl Acad Sci USA. 2000;97:5179–5184. doi: 10.1073/pnas.090104997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuler B, Lipman EA, Eaton WA. Nature. 2002;419:743–747. doi: 10.1038/nature01060. [DOI] [PubMed] [Google Scholar]
- 22.Lipman EA, Schuler B, Bakajin O, Eaton WA. Science. 2003;301:1233–1235. doi: 10.1126/science.1085399. [DOI] [PubMed] [Google Scholar]
- 23.Yang H, Luo G, Karnchanaphanurach P, Louie TM, Rech I, Cova S, Xun L, Xie XS. Science. 2003;302:262–266. doi: 10.1126/science.1086911. [DOI] [PubMed] [Google Scholar]
- 24.Kuzmenkina EV, Heyes CD, Nienhaus GU. Proc Natl Acad Sci USA. 2005;102:15471–15476. doi: 10.1073/pnas.0507728102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Funatsu T, Harada Y, Tokunaga M, Saito K, Yanagida T. Nature. 1995;374:555–559. doi: 10.1038/374555a0. [DOI] [PubMed] [Google Scholar]
- 26.Zander C, Enderlein J, Keller RA, editors. Single Molecule Detection in Solution. Berlin: Wiley–VCH; 2002. [Google Scholar]
- 27.Tinnefeld P, Herten DP, Sauer M. J Phys Chem A. 2001;105:7989–8003. [Google Scholar]
- 28.Tokunaga M, Kitamura K, Saito K, Iwane AH, Yanagida T. Biochem Biophys Res Commun. 1997;235:47–53. doi: 10.1006/bbrc.1997.6732. [DOI] [PubMed] [Google Scholar]
- 29.Lyubovitsky JG, Gray HB, Winkler JR. J Am Chem Soc. 2002;124:5481–5485. doi: 10.1021/ja017399r. [DOI] [PubMed] [Google Scholar]
- 30.Zuniga EH, Nall BT. Biochemistry. 1983;22:1430–1437. doi: 10.1021/bi00275a017. [DOI] [PubMed] [Google Scholar]
- 31.Schuler B, Lipman EA, Steinbach PJ, Kumke M, Eaton WA. Proc Natl Acad Sci USA. 2005;102:2754–2759. doi: 10.1073/pnas.0408164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stryer L, Haugland RP. Proc Natl Acad Sci USA. 1967;58:719–726. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S. Proc Natl Acad Sci USA. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Latypov RF, Cheng H, Roder NA, Zhang J, Roder H. J Mol Biol. 2006;357:1009–1025. doi: 10.1016/j.jmb.2006.01.055. [DOI] [PubMed] [Google Scholar]
- 35.Deniz AA, Laurence TA, Dahan M, Chemla DS, Schultz PG, Weiss S. Annu Rev Phys Chem. 2001;52:233–253. doi: 10.1146/annurev.physchem.52.1.233. [DOI] [PubMed] [Google Scholar]
- 36.Lermer M, Bames MD, Kung CY, Whitten WB, Ramsey JM. Anal Chem. 1997;69:2115–2121. doi: 10.1021/ac970093b. [DOI] [PubMed] [Google Scholar]
- 37.Gopich IV, Szabo A. J Phys Chem B. 2003;107:5058–5063. [Google Scholar]
- 38.Fierz B, Satzger H, Root C, Gilch P, Zinth W, Kiefhaber T. Proc Natl Acad Sci USA. 2007;104:2163–2168. doi: 10.1073/pnas.0611087104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nettels D, Gopich IV, Hoffmann A, Schuler B. Proc Natl Acad Sci USA. 2007;104:2655–2660. doi: 10.1073/pnas.0611093104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurchan E, Roder H, Bowler BE. J Mol Biol. 2005;353:730–743. doi: 10.1016/j.jmb.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 41.Merchant KA, Best RB, Louis JM, Gopich IV, Eaton WA. Proc Natl Acad Sci USA. 2007;104:1528–1533. doi: 10.1073/pnas.0607097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le Duff CS, Whittaker SB, Radford SE, Moore GR. J Mol Biol. 2006;364:824–835. doi: 10.1016/j.jmb.2006.09.037. [DOI] [PubMed] [Google Scholar]
- 43.Pabit SA, Hagen SJ. Biophys J. 2002;83:2872–2878. doi: 10.1016/S0006-3495(02)75296-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.