Abstract
Xanthomonas oryzae pv. oryzae strain PXO99A induces the expression of the host gene Os8N3, which results in increased host susceptibility to bacterial blight of rice. Here, we show that PXO99A affects the expression of two additional genes in a type III secretion system-dependent manner, one encoding a bZIP transcription factor (OsTFX1) and the other the small subunit of the transcription factor IIA located on chromosome 1 (OsTFIIAγ1). Induction of OsTFX1 and OsTFIIAγ1 depended on the type III effector genes pthXo6 and pthXo7, respectively, both encoding two previously undescribed members of the transcription activator-like (TAL) effector family. pthXo7 is strain-specific and may reflect adaptation to the resistance mediated by xa5, an allele of OsTFIIAγ5 encoding a second form of the TFIIA small subunit on chromosome 5 of rice. The loss of pthXo6 resulted in reduced pathogen virulence, and ectopic expression of OsTFX1 abrogated the requirement for pthXo6 for full virulence. X. oryzae pv. oryzae therefore modulates the expression of multiple host genes using multiple TAL effectors from a single strain, and evidence supports the hypothesis that expression of the associated host genes contributes to host susceptibility to disease.
Keywords: avrBs3, pthXo6, pthXo7, transcription activator-like, pthXo1
Gram-negative pathogenic bacteria often secrete multiple effector proteins through a type III secretion system (T3S) to modulate eukaryotic host cell functions for the benefit of the invasion process (1). X. oryzae pv. oryzae, the causal agent of bacterial blight of rice, harbors multiple genes for T3S effectors of the transcription activator-like (TAL) effector family (2, 3). TAL effectors each possess central repetitive regions that vary in repeat number and specific repeat sequence as well as three highly conserved C-terminal nuclear localization signals (NLS), and C-terminal acidic transcription activator-like domains (AD). TAL effectors are associated with phenotypes that contribute to strain virulence, disease symptoms, and host recognition and resistance responses, the latter also known as avirulence activity. With a few exceptions, the phenotypes conferred by the various TAL effector family members are associated with the differences in type and number of the repeats within the respective repetitive regions (4–8).
The induction of two host genes has been shown to be specifically regulated by the TAL effectors AvrXa27 and PthXo1 from X. oryzae pv oryzae strain PXO99A (3, 6). Expression of the dominant rice R gene Xa27 is induced by AvrXa27 and results in a hypersensitive resistance reaction. Susceptible plants harbor an allele of Xa27 that is not expressed in the presence of AvrXa27. The second TAL effector/host gene combination involves the TAL effector PthXo1 and the rice gene Os8N3, which are required for full strain virulence and increased host disease susceptibility, respectively (3). Recessive alleles of Os8N3, collectively known as xa13, interfere with Os8N3 induction during infection by PXO99A and result in resistance to strains that rely on PthXo1 for virulence (3, 9). Os8N3, which encodes a member of the MtN3 family, is also required for normal pollen development in uninfected plants, indicating that X. oryzae pv. oryzae commandeers an otherwise developmental pathway to induce a state of susceptibility (3, 9). How the host proteins Xa27 and Os8N3 mediate the respective resistant and susceptible states is unknown.
Individual strains of X. oryzae pv. oryzae and several related species encode multiple TAL effector genes, although many of the genes within a given strain have no known phenotypic effects (8). Extra copies of TAL effector genes in other species of Xanthomonas have been attributed to redundant virulence functions and as sources of substrates for the enhanced generation of potentially new virulence genes through recombination. One study indicated that additional TAL effector genes within a given strain of X. oryzae pv. oryzae may contribute independently to virulence (10). In strain PXO99A, only pthXo1 and avrXa27 have known associated phenotypes, whereas the strain is predicted to harbor upwards of 22 TAL effector genes (3, 6, 8). We are using microarray hybridization analysis of host gene expression during bacterial blight to identify host genes that are potential targets of the TAL effector genes of X. oryzae pv. oryzae. In the process, we identified two TAL effector genes of PXO99A that are linked to the induction of two host genes other than Os8N3 and characterized the genes with regards to their involvement in strain virulence and host susceptibility, respectively.
Results
OsTFX1 and OsTFIIAγ1 Are Differentially Expressed upon Pathogen Challenge.
Microarray analysis was performed with RNA from plants challenged with either virulent bacteria of strain PXO99A or water (mock inoculation) and a custom-designed Affymetrix GeneChip microarray representing ≈23,000 rice genes (11). Nine probe sequences were identified with an arbitrarily assigned cut-off of 5-fold or greater change in expression values and maximum expression value of 400, and eight probe sets had perfect matches to cDNA clones in NCBI or the KOME full-length rice cDNA databases. The genes represented by the top two probe sets in terms of fold change were examined in more detail. Probe set 1 represented a gene (CB097192) that encoded the γ, or small, subunit of the general transcription factor IIA on chromosome 1, which was named OsTFIIAγ1. Probe set 2 represented a member of the bZIP transcription factor gene family (AK108319) and is encoded on chromosome 9. The probe set 2 locus was named OsTFX1. Induction of expression was confirmed for both genes by using Northern hybridization with gene-specific probes that were designed from the respective gene sequences (data not shown), and a time course of induction was subsequently performed for each gene (Fig. 1). Expression levels of both OsTFX1 and OsTFIIAγ1 were undetectable in untreated leaves (0 h) or after water-inoculations and evident within 6 h after inoculation with bacteria (Fig. 1 Top and Middle). A slight increase in OsTFX1 was observed by 24 h during water-inoculation (Fig. 1 Middle). Data regarding the identity and sequences of all nine probe sets, the coding sequences for OsTFIIAγ1 and OsTFX1, gene-specific primers and alignment to related proteins, respectively, are provided in supporting information (SI) Tables 1–3 and SI Figs. 8 and 9.
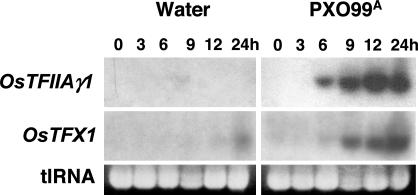
Fig. 1.
Effect of X. oryzae pv. oryzae infection on the expression of rice genes OsTFIIAγ1 and OsTFX1. Fourteen-day-old rice seedlings were inoculated with water (mock inoculation) or X. oryzae pv. oryzae strain PXO99A. Total RNA was extracted from leaves at the indicated times after inoculation and subjected to the Northern blot hybridization analysis. Treatments and times are indicated at the top of the results for OsTFIIAγ1. Gene specific probes are indicated at left. Ethidium bromide stained gels of total RNA are shown at the bottom of each treatment to assess relative total RNA (tlRNA) loading. Times are indicated in hours (h).
OsTFX1 and OsTFIIAγ1 Are Differentially Expressed in a T3S-Dependent Manner.
The requirement for T3S function of OsTFX1 and OsTFIIAγ1 induction was examined by Northern blot analysis of total leaf RNA from plants inoculated with bacterial strains harboring mutations in genes encoding different components of the type III system (Fig. 2A). Mock-inoculation with water resulted in no water-soaking, a symptom of bacterial infection, at the inoculation site compared with full symptoms after inoculation with PXO99A (Fig. 2A, Water and WT, respectively). PXO99AME7 (hereafter ME7) has a mutation in an operon (hrpC/hrcU) encoding conserved and critical components of the T3S apparatus, is incapable of type III effector secretion, and essentially nonpathogenic [Fig. 2A, ME7 (T3S−); (12)]. Inoculation with ME7 resulted in no induction of OsTFX1 or OsTFIIAγ1 [Fig. 2B, ME7 (T3S−)] in comparison with PXO99A (Fig. 2B, WT). ASX41 is a PXO99A mutant of hrpF, whose product is proposed to increase the efficiency of the entry of the T3S effectors into the host cells, yet retains a limited ability to deliver T3S effectors and induces reduced disease symptoms in comparison with WT [Fig. 2A, hrpF−; (13)]. Low, but detectable levels of induction for OsTFX1 and OsTFIIAγ1 were observed with ASX41 (Fig. 2B, hrpF−). Finally, PXO99AME2 (hereafter, ME2) has a transposon insertion in pthXo1, and lacks the major TAL effector gene for virulence and the ability to induce Os8N3 (8). ME2 remains fully competent to secrete T3S effectors, induces considerable water-soaking, yet is reduced in virulence [Fig. 2A, pthXo1−; (8)]. ME2 retained the ability to induce both OsTFX1 and OsTFIIAγ1 (Fig. 2B, pthXo1−). The results, therefore, provided evidence that the changes in expression of both genes depended on a functioning T3S but were not dependent on the major TAL effector PthXo1.
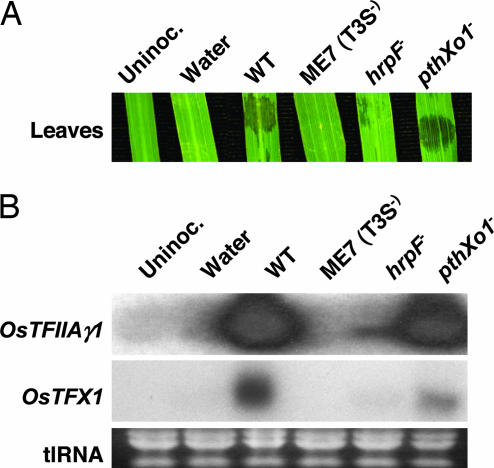
Fig. 2.
Effect of T3S mutations on host gene expression. (A) Phenotypes of strains on rice leaves inoculated with each strain in the Northern blot hybridization analysis. Strain descriptions are as follows: Uninoc., uninoculated; water, mock inoculated; WT, wild type PXO99A; ME7 (T3S−), hrpC− mutant incapable of type III-mediated secretion; hrpF−, ASX41 with mutation in hrpF and severely reduced in type III secretion; pthXo1−, ME2 lacking TAL effector gene pthXo1, severely reduced in virulence, and proficient in type III secretion. Photographs were taken 3 days after inoculation. (B) Northern blot hybridization analysis of host gene expression after inoculation with T3S mutant strains. RNA samples were prepared 24 h after inoculation with T3S mutants that vary in strain virulence and subjected to Northern blot analysis. Ethidium bromide stained total RNAs (tlRNA) are shown in Bottom. Strain descriptions are identical to A.
TAL Effectors Are Responsible for OsTFX1 and OsTFIIAγ1 Induction.
The T3S dependence of OsTFX1 and OsTFIIAγ1 expression prompted us to determine whether OsTFX1 and OsTFIIAγ1 expression depended on members of the TAL effector gene family of PXO99A. Candidate TAL effector genes were identified first by assaying host gene expression after individual plant inoculations with 28 candidate TAL effector gene mutants of strain PXO99A, which were generated in a previous study by maker-exchange mutagenesis (8). Inoculations with two mutants, PXO99AME1 and PXO99AME10 (hereafter, ME1 and ME10, respectively), resulted in the loss of OsTFX1 or OsTFIIAγ1 induction, respectively (Fig. 3 A and B, respectively). ME1 and ME10 were then used as recipients for complementation by using cosmid genomic clones from PXO99A containing TAL effector genes (8). A single cosmid, and, upon further subcloning, a single TAL effector gene was recovered that directed the induction of each host gene, respectively. pthXo6 (pathogenicity Xo6) and pthXo7, when introduced into ME1 and ME10, respectively, restored the ability to induce OsTFX1 and OsTFIIAγ1 (Fig. 3 A and B, respectively). The central BamHI fragment of each gene was sequenced (NCBI accessions DQ233317 and DQ231560), and each gene consisted of the typical TAL gene structure with 22.5 and 21.5 repeat units, respectively. Based on alignment of the repeats to those of other family members, pthXo6 and pthXo7 represented previously undescribed genes of the family. The alignments are presented in SI Fig. 10.
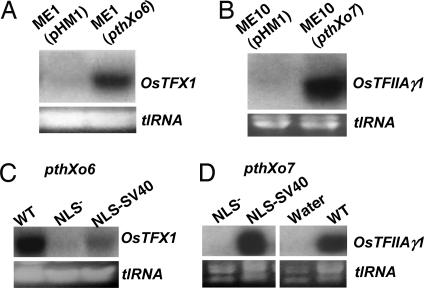
Fig. 3.
Identification and characterization of the TAL effector genes pthXo6 and pthXo7. (A) Northern blot hybridization analysis of OsTFX1 induction by ME1 (pHM1) (empty vector) and ME1 (pthXo6). Induction of OsTFX1 was examined 24 h after treatment of rice leaves. Lane assignments are as follows: ME1 (pHM1), empty vector; ME1 (pthXo6), single pthXo6 gene in pHM1 vector. (B) Northern blot hybridization analysis of OsTFIIAγ1 induction by ME10 (pHM1) and ME10 (pthXo7). Induction of OsTFIIAγ1 was examined 24 h after treatment of rice leaves. Lane assignments are as follows: ME10 (pHM1), empty vector; ME10 (pthXo7), single pthXo7 gene in pHM1 vector. Ethidium bromide-stained total RNA (tlRNA) is shown Lower. (C and D) Substitutions in the NLS and AD mutations of PthXo6 or PthXo7, respectively, result in loss of OsTFX1 and OsTFIIAγ1 induction. Northern blot hybridization analysis was performed at 24 h after inoculation as described in Fig. 1. Strain assignments in C are ME1 with the following genes: WT, wild-type pthXo6; NLS−, pthXo6M123 lacking consensus NLS sequences; NLS-SV40, pthXo6M123 with SV40 NSL sequence. Assignments in D are ME10 with the following: NLS−, pthXo7M123 lacking NLS sequences; NLS-SV40, pthXo7M123 with SV40 NLS sequences; Water, mock-inoculation; WT, wild-type pthXo7.
Previous results have shown that the NLS motifs of the TAL effectors avrXa7 and avrXa27 are required for activity, reflecting their nuclear localization requirement (6, 14). To determine whether PthXo6 and PthXo7 have similar requirements for the induction of OsTFX1 and OsTFIIAγ1, respectively, mutations that were previously generated for the NLS motifs of avrXa7 were introduced into pthXo6 and pthXo7 by replacing the central SphI fragment of avrXa7 and the various mutant versions of avrXa7 with the SphI fragment from pthXo6 or pthXo7. The exchange is possible because of the highly conserved nature of the TAL effector genes, which differ, with a few exceptions, only in the repetitive region (8). These alterations included replacing all three NLS motifs with non-consensus sequences in both genes. The altered versions of pthXo6 were introduced into ME1, and those for pthXo7 were introduced into ME10, and the ability of the strains to mediate induction of the respective host genes was examined by Northern blot hybridization analysis (Fig. 3 C and D, respectively). Removal of all three NLS motifs from both genes resulted in the loss of host gene induction (Fig. 3 C and D, NLS−, respectively, for pthXo6 and pthXo7) compared with the intact genes (Fig. 3 C and D, WT). Replacement of the NLS motifs with the single SV40 NLS sequence (Fig. 3 C and D, NLS-SV40) restored induction activity to both pthXo6 and pthXo7, although the activity of pthXo6 was considerably weaker than the WT gene based on the signal strength (Fig. 3 C and D, NLS-SV40). Thus, both PthXo6 and PthXo7 require the NLS motifs for induction of the respective host genes, and the NLS requirement can be met by the heterologous SV40 NLS motif, indicating that nuclear localization of either TAL effector is necessary for host gene induction.
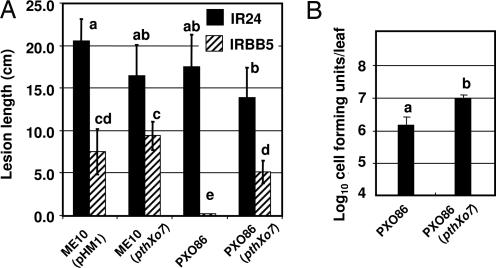
pthXo7-Mediated Induction of OsTFIIAγ1 Is Strain-Specific and Contributes to Virulence on Rice Containing xa5.
A collection of strains was tested for the ability of different X. oryzae pv. oryzae field isolates to induce either OsTFX1 or OsTFIIAγ1. Infections by all of the strains tested were found to cause the induction of OsTFX1, whereas induction of OsTFIIAγ1 was limited to PXO99A (Fig. 4 A and B, respectively). The ability to induce OsTFIIAγ1 was not suppressed by coinoculation of PXO99A and noninducing strains (data not shown), and the transfer of pthXo7 to PXO86, an otherwise OsTFIIAγ1 noninducing strain, resulted in the acquisition of the ability to induce OsTFIIAγ1 expression (Fig. 4C). The potential contribution of pthXo7 to virulence was assessed by the comparison of the abilities of ME10, the pthXo7-deficient derivative of PXO99A, and the complemented strain ME10 (pthXo7) to cause lesions on plants. An allele of OsTFIIAγ5 is known as xa5, which is a recessive resistance gene for bacterial blight and has a glutamic acid codon substitution for valine at codon position 39 (15, 16). With the idea that OsTFIIAγ1 expression may complement to some extent the xa5 allele of OsTFIIAγ5, virulence assays of ME10 and ME10 (pthXo7) were conducted in plants with (IRBB5) and without the xa5 allele (IR24). Regardless of the host, ME10 and ME10 (pthXo7) did not display demonstrable differences in lesion formation [Fig. 5A, ME10 (pHM1) and ME10 (pthXo7), respectively]. The effect of pthXo7 was then tested on strain PXO86, which does not grow well in plants with the xa5 allele, with the idea that a virulence effect of pthXo7, if any, might be more apparent in a strain with less overall virulence. Strains of PXO86 with and without pthXo7 are virulent on IR24 (Fig. 5A, strains 3 and 4, respectively, solid columns). However, PXO86 caused very short lesions on IRBB5, averaging <1 cm (Fig. 5A, PXO86, hatched column). PXO86 (pthXo7), on the other hand, formed lesions with a statistically significant average length of lesions >5 cm [Fig. 5A, PXO86 and PXO86 (pthXo7), hatched column]. The difference in lesion lengths was also reflected in changes in bacterial leaf populations in IRBB5 (at 17 days after inoculation), which were higher for PXO86 with pthXo7 than without the gene (Fig. 5B). Thus, pthXo7 may be an adaptation of the strain PXO99A to multiply in the plants that possess xa5. However, the virulence effect might be masked by other virulence factors that also allow ME10 to grow in the presence of the xa5 mutation.
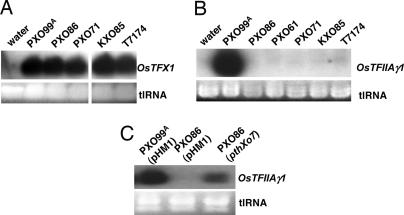
Fig. 4.
Strain-specific regulation of OsTFX1 and OsTFIIAγ1 expression. (A) Northern blot hybridization analysis of OsTFX1expression after challenge by four additional strains of X. oryzae pv. oryzae. The expression for OsTFX1 was examined by Northern blot analysis 24 h after inoculation of 14-day-old plants with needless syringe at multiple sites in each leaf. A gene-specific fragment was generated by PCR and used as a probe. Ethidium bromide-stained total RNA (tlRNA) is shown in Lower. (B) Northern blot analysis of OsTFIIAγ1 expression 24 h after challenge with five additional strains of X. oryzae pv. oryzae. (C) OsTFIIAγ1 expression by a noninducing field strain of X. oryzae pv. oryzae after transfer of pthXo7.
Fig. 5.
pthXo7 converts PXO86 from an incompatible to a compatible strain on IRBB5. (A) Lesion formation is enhanced by pthXo7 in PXO86 on 50-day-old IRBB5 (xa5/xa5) plants. Strains as indicated in the graph were inoculated by the leaf-clip method on IR24 (filled columns) and IRBB5 (hatched columns), and lesions were measured from tip at 17 days after inoculation. The values represent the means of eight plants for each treatment. Columns with the same lowercase letters do not differ from each other at the significance level of P < 0.01 using the Tukey test for post-ANOVA analysis. The experiment was repeated twice. (B) Bacterial leaf populations of PXO86 are enhanced by pthXo7. Rices leaves inoculated with the indicated strains were excised 17 days after inoculation, ground in water, and plated after serial dilution for counting colony forming units, which were measured on a per leaf basis. Small case letters indicate significant differences in values at P < 0.01.\.
pthXo6 Contributes to the Virulence of PXO99A and Ectopic OsTFX1 Expression Contributes to Host Susceptibility.
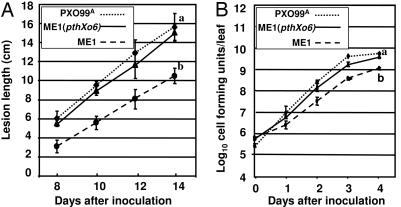
The potential contribution of pthXo6 to the virulence of the pathogen was also determined by comparing the abilities of the PXO99A, ME1 and ME1 (pthXo6) to elicit disease symptoms and grow within the host. By both measures, ME1 (pHM1), containing the empty vector, showed a decrease in virulence. Using lesion length as an indicator, ME1 (pHM1) caused lesions with an ≈35% decrease in length by day 14 using the leaf clip assay on young seedlings in comparison with the parental strain PXO99A and the complemented strain ME1 (pthXo6) (10.6 ± 0.8 vs. 15.7 ± 1.4 and 15.1 ± 0.8 cm, respectively; Fig. 6A). Similarly, ME1 population levels were also significantly lower, as measured here during the first four days, in comparison with the populations for PXO99A and ME1 (pthXo6) (Fig. 6B).
Fig. 6.
pthXo6 is a virulence factor of PXO99A. (A) Loss of pthXo6 in ME1 resulted in reduction in lesion forming ability of the pathogen. The ability of PXO99A, ME1, and ME1 (pthXo6) was assayed by the leaf clip method, clipping the leaf tips of 28-day-old plants with scissors contaminated with the appropriate strain at a concentration of 0.5 OD at 600 nm. The lengths of lesions extending from the tips were then measured each day over a 14-day period and starting on day 8 after inoculation in cultivar IR24. The curves represent the average lesion length for 10 plants and are labeled according to the legend superimposed on the graph. Lesion measurements are in centimeters (cm). Significance between treatments at final time points was assessed at a P value of <0.05 by using the Tukey test for post-ANOVA analysis. The lesion length experiment was repeated three times, and the population measurements were preformed twice with similar results. (B) ME1 had reduced bacterial leaf populations. Bacterial populations of PXO99A, ME1, and ME1 (pthXo6) were measured in cultivar IR24 over a 5-day period starting at day 1. The curves represent the average leaf population over the 5-day period and are labeled according to the legend superimposed on the graph. Inoculation and statistical analysis are the same as for A.
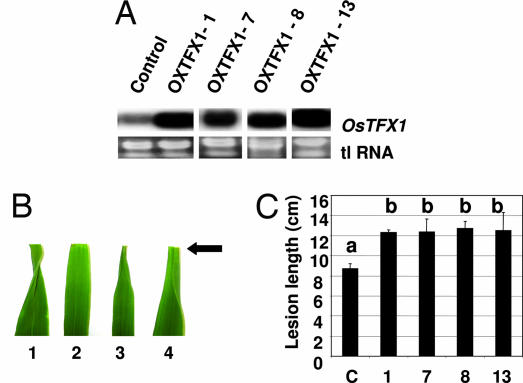
To determine whether OsTFX1 expression specifically contributes to virulence effect of pthXo6, OsTFX1 was expressed by using the Ubi1 promoter (OsTFX1Ubi1) with the prediction that, if involved in susceptibility, constitutive expression might alleviate the need for pthXo6 in the pathogen and increase host susceptibility to infection by strain ME1. OsTFX1Ubi1 was introduced into plants by using Agrobacterium-mediated transformation, and four homozygous T1 lines with constitutive expression of OsTFX1 were selected for further analysis along with a vector-only control line (Fig. 7A, plants OXTFX1-1, -7, -8, and -13). Upon inoculation with ME1, phenotypic differences were readily apparent in the OsTFX1Ubi1 lines within 5 days compared with control plants on the basis of lesion progression after inoculation of the leaf tips with the bacteria, which at five days is evident by leaf curling at the tip (Fig. 7B, results shown from plant OXTFX1-8). Leaves of control plants showed typical leaf-tip curling at 5 days after inoculation with PXO99A (Fig. 7B, leaf 1) and no curling response after inoculation with the pthXo6 mutant ME1 (Fig. 7B, leaf 2). At the same time, OXTFX1-8, as an example, showed the same degree of leaf curling upon inoculation with PXO99A (Fig. 7B, leaf 3) and ME1 (Fig. 7B, leaf 4). Changes in susceptibility to infection of the mutant ME1 compared with control plants analysis was conducted on 15 homozygous (for the T-DNA insert) T2 progeny from each line [T-DNA: portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells]. Based on lesion length, all four lines with OsTFX1Ubi1 expression were more susceptible to the mutant in comparison with a control line (Fig. 7C).
Fig. 7.
Ectopic expression of OsTFX1 increases host susceptibility to ME1. (A) Northern blot hybridization analysis of four independent T0 (primary transgenic) plants expressing OsTFX1Ubi1. Total RNA was extracted from leaves of plants selected from hygromycin-resistant calli after Agrobacterium-mediated gene transfer. Four independent lines with elevated levels of OsTFX1 expression were chosen for further study and are labeled OXTFX1-1, -7, -8, and -13. Total RNA (tlRNA) stained with ethidium bromide is shown in Lower to indicate relative loading. (B) Leaf phenotype of F1 progeny from transgenic plant OXTFX1-8 as an example showing increased susceptibility to pathogen strain ME1 compared with control plants. Leaf 1, Control plant inoculated with PXO99A; leaf 2, control plant with ME1; leaf 3, plant OXTFX1-8 with PXO99A; and leaf 4, OXTFX1-8 with ME1. Arrow indicates position of clip-inoculation of the leaf tips. Note leaf curling in leaves 1, 3, and 4 and lack of curling in leaf 2. Leaves were photographed 5 days after inoculation. (C) Measurements of susceptibility in four independent OXTFX1 lines by lesion length after inoculation with ME1. Values with same lowercase letters above the column are not significantly different at the P < 0.05 level using the Tukey test.
Discussion
Microarray hybridization analysis represents an excellent tool for the analysis of bacterial blight because of the reliance of the pathogen on hijacking the expression of host genes for virulence. On the chance that the most highly induced genes might represent targets of the TAL effectors of PXO99A, we examined whether the two genes with the highest fold change induction could be linked to specific TAL genes of the pathogen. Indeed, upon screening the pool of mutants, the two type III effector genes pthXo6 and pthXo7 were shown to be required for the expression of OsTFX1 and OsTFIIAγ1, respectively. Furthermore, functional analysis of the effector genes revealed that each has a potential contribution to virulence and fitness. On the host side, the functional analyses of OsTFX1 and OsTFIIAγ1 represent good candidates for genes targeted by the TAL effectors and involved in mediating host susceptibility. At the same time, the actual mechanism underlying host gene induction as mediated by TAL effectors function is unknown, and, therefore, we cannot conclude that induction of the targeted host genes is result of a direct interaction of an effector and host gene promoter or an indirect effect of interference with an upstream regulatory process.
The results provide corroborative evidence that pthXo6 and pthXo7 have potential contributions to fitness based on the loss of virulence by the pathogen in our experimental setting and that the associated host genes in question are involved in susceptibility in the infection process. The evidence for the involvement of OsTFX1 in host susceptibility is more direct. Ectopic expression of OsTFX1 indicates that OsTFX1 represents a host susceptibility gene that is targeted by PthXo6. The gene is induced in both indica and japonica varieties of rice (data not shown) and by all of the strains of X. oryzae pv. oryzae examined, which represent a wide variety of geographical locations. In fact, we have yet to identify a field isolate that does not induced OsTFX1 and, as a consequence, could not test the effect of OsTFX1 ectopic expression on field isolates of X. oryzae pv. oryzae lacking pthXo6. The ability to induce OsTFX1 is wide-spread and seems to be a disease-specific phenomenon of bacterial blight. The overexpression of OsTFX1 only shows the ability of OsTFX1 ectopic expression is sufficient to compensate for loss of pthXo6 in the pathogen and does not demonstrate that OsTFX1 is necessary. We know of no recessive resistance alleles of OsTFX1 among the many resistance genes in rice to X. oryzae pv. oryzae, and, therefore, are generating plants that suppress the expression of OsTFX1 by RNA silencing to test the requirement of OsTFX1 in disease susceptibility. On the other hand, the broad ability to induce OsTFX1 by many strains of X. oryzae pv. oryzae indicates that induction of a resistance response using the OsTFX1 induction signals would be useful for broad resistance to bacterial blight disease. Further exploitation of OsTFX1 induction will require a better understanding of the elements involved in the developmental and disease-associated expression of OsTFX1.
Whereas we have not demonstrated a requirement for OsTFIIAγ1 induction in host susceptibility directly, the relatedness to OsTFIIAγ5 is intriguing and provided an opportunity to compare the effects of OsTFIIAγ1 expression on host susceptibility in the xa5 and OsTFIIAγ5 (dominant allele) backgrounds. OsTFIIAγ5 has been shown previously to be constitutively expressed in rice leaves, and, therefore, seems to represent the housekeeping form of TFIIAγ in leaves (15, 16). The constitutive role for OsTFIIAγ5 is consistent with the effect of xa5 allele on the disease. Although PXO99A is categorized as a compatible strain with IRBB5, the virulence of this strain is also reduced in IRBB5. Therefore, we used the xa5 allele for TFIIAγ as a pseudomutant for OsTFIIAγ1 with the assumption that xa5/xa5 homozygous plants (IRBB5) have a preponderance of defective TFIIAγ (TFIIAγxa5). PXO86, which is otherwise incompatible on IRBB5 becomes compatible on xa5 upon the introduction of pthXo7. In our model, the elevation of OsTFIIAγ1 expression is not likely an inconsequential side effect of disease, but that PthXo7 would serve to induce OsTFIIAγ1 expression and increase the amount of TFIIAγ available without the xa5 mutation. We presume that pthXo7 behaves similar to a number of T3S effector genes, including avrPto and avrRpt2, whose contribution to virulence is only measurable in certain strains or contexts (17, 18) and that pthXo7 has also a contribution to the fitness of PXO99A in the field but the contribution is difficult to measure given the relative high virulence of PXO99A.
Whereas different TAL effectors within a given strain have qualitatively different effects, the results do not preclude cooperation between the effectors. In fact, we proposed a model whereby PthXo7 could enhance the functioning of other TAL effectors by influencing the availability of a host transcription component TFIIAγ. TFIIA is a part of the RNA polymerase II preinitiation complex for transcription, the basic components of which are conserved among eukaryotic organisms (19). TFIIAγ is reported also to interact with the transcription activator VP16, which can substitute for the AD of TAL effectors under some circumstances (15, 20, 21). Similarly, OsTFX1 could function to assist in the expression of other TAL effector targets. A number of bZIP transcription factors act in concert with other transcription factors to enhance expression of developmentally regulated genes. A search of related genes in other species has provided few clues as to the involvement of OsTFX1 in development or defense, and no specific function has been ascribed to the closely related homologs in other species. bZIP transcription factors, as a class, represent an ancient evolutionary line of transcription factors and regulate the expression of genes in a wide variety of developmental and physiological response pathways in a wide range of organisms including plants and animals (22, 23).
Our data here and other unpublished results have important implications for the function of TFIIAγ in the pathogenesis of X. oryzae pv. oryzae and important implications for the mechanism of the recessive resistance gene xa5. In the classical view, PXO86 is considered as an incompatible strain because of the presence of an avirulence protein (e.g., Avrxa5) (24). We do not presume the presence of an avirulence protein in PXO86 but propose that resistance is due, in part, to the lack of appropriate virulence factors, of which pthXo7 is an example. In this model, xa5 does not confer resistance because of recognition of a corresponding avirulence protein. Rather, xa5, a mutant version of OsTFIIAγ5, functions primarily to disrupt effector function through modification in TFIIAγ, severely reducing the ability of the pathogen to alter host gene expression. Similar to the addition of avrXa7 to PXO99A and the gain of virulence on xa13, the addition of pthXo7 converted PXO86 into a virulent strain on plants with xa5 (3). However, the OsTFIIAγ1 subunit does not seem to support the level of susceptibility conferred by the normal OsTFIIAγ5 subunit, nor does the presence of pthXo7 fully explain the ability of PXO99A to grow in plants with xa5 allele as evidenced by the ability of ME10 to grow in the IRBB5 line. An alternative explanation is that PthXo7 may suppress host defense functions elicited by PXO86. Future experiments will be conducted to distinguish these possibilities.
We have provided an evidence that X. oryzae pv. oryzae utilizes a virulence strategy involving the alteration in the expression of key host genes. Here, we demonstrated that, in addition to affecting the regulation of Os8N3, the pathogen alters the regulation of additional host genes via effectors of the same family. Structurally, the effectors are highly similar with differences in the central repetitive region. Strikingly, both OsTFX1 and OsTFIIAγ1 encode components of the host transcription system. Thus, X. oryzae pv. oryzae, as represented by strain PXO99A, may not only facilitate colonization by induction of specific major target genes but attempt to alter transcription pathways and the transcription process itself.
Materials and Methods
Bacterial Strains and Plasmids.
The bacterial strains and plasmids used in the experiments are listed in SI Table 4. Escherichia coli was grown in terrific broth (TB) or on Luria agar plates at 37°C with the appropriate antibiotics. Xanthomonas strains were cultured on tryptone sucrose agar at 28°C (12). Carbenicillin, spectinomycin, and kanamycin were used at 100 μg/ml.
Recombinant DNA Techniques.
DNA manipulations were performed by standard procedures. Amino acid alignments were constructed by using ClustalW (25). pthXo6 and pthXo7 indicate the pHM1 version of each gene. NLS mutants of pthXo6 and pthXo7 were generated by using previously constructed NLS mutants of avrXa7 (14). Further details of the mutants are provided in SI Fig. 11.
DNA Amplification.
The primer sequences and parameters used for preparing gene-specific probes for Northern blot analysis are provided in SI Table 3.
Inoculation, RNA Preparation, and Hybridization.
Rice IR24 plants were grown in growth chambers at 28°C with a 12 h photoperiod. The second leaf of 14 days old rice plants was inoculated at multiple sites with bacterial suspensions of optical density of 0.5 at 600 nm (≈5 × 107 cell-forming units per ml) by using a needless syringe. RNA extraction and Northern blot analysis were performed as described (3). For microarray hybridization, quality control of RNA, complementary RNA labeling, and hybridization to a GeneChip were performed as described (26). Three biological replicates were analyzed.
Microarray Data Acquisition and Manipulation.
The GeneChip image is acquired by using Affymetrix scanner 3000 and digitized by MAS 5.0. The CEL files were then processed by using Robust Multiarray Average (RMA) algorithm (27). More details are provided in SI Table 2.
Plant Assays.
Lesion lengths and bacterial populations were assayed by using the leaf clip inoculation (28). Bacterial inocula were prepared at an optical density of 0.5 at 600 nm (≈5 × 107 cell-forming units per ml). Scores are the mean of eight leaves. Measurements were taken at the times indicated in the respective figure legends. Significance between treatments was assessed on the basis of a P value of <0.05 by using the Tukey test for post-ANOVA analysis. All virulence assays were repeated three times with similar results.
Ectopic Gene Expression Construction and Stable Rice Transformation.
For OsTFX1 construction, OverTFX1-F, the forward primer matching the ATG start codon region, and OverTFX1-R, the reverse primer complementary to 3′ end of cDNA, were used to amplify the OsTFX1 from a cDNA pool of the reverse transcribed mRNA extracted from PXO99A infected leaves (primer sequences can be found in SI Table 3), and the gene was cloned behind the maize Ubi1 promoter (29). The expression cassettes were inserted into binary vector pCAMBIA 1300 (CAMBIA), and the resulting binary vectors were electroporated into Agrobacterium strain EHA105. Calli derived from immature embryos of rice cultivar Kitake were transformed as described (30). Significance was assessed on the basis of a P value of <0.05 by using the Tukey test for post-ANOVA analysis.
Supplementary Material
Acknowledgments
We thank Sherman Chang, Makoto Ono, and Charles Chilcott for conducting the GeneChip experiments, Dr. Jianfa Bai for assistance and use of the Kansas State University (KSU) Gene Expression Facility, Dr. John Fellers of the KSU Facility, Dr. A. Robert-Seilaniantz for critical reading of the manuscript, and Ms. Miranda Leathers for excellent technical assistance. This research was supported by funds from National Science Foundation Award 0544269, the Kansas Agricultural Experiment Station (KAES), and the Biology of Plant-Microbe Associations Program of the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Services National Research Initiative. This publication is number 07-282A of the KAES.
Abbreviations
- T3S
type III secretion system
- TAL
transcription activator-like
- NLS
nuclear localization signal
- AD
activator-like domain
- ME1
-2, -7, and -10, PXO99AME1, -2, -7, and -10.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ233317 and DQ231560).
This article contains supporting information online at www.pnas.org/cgi/content/full/0701742104/DC1.
References
- 1.He SY, Nomura K, Whittam TS. Biochim Biophys Acta. 2004;1694:181–206. doi: 10.1016/j.bbamcr.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Schornack S, Meyer A, Romer P, Jordan T, Lahaye T. J Plant Physiol. 2006;163:256–272. doi: 10.1016/j.jplph.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Yang B, Sugio A, White FF. Proc Natl Acad Sci USA. 2006;103:10503–10508. doi: 10.1073/pnas.0604088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herbers K, Conrads-Strauch J, Bonas U. Nature. 1992;356:172–174. [Google Scholar]
- 5.Hopkins CM, White FF, Choi SH, Guo A, Leach JE. Mol Plant–Microbe Interact. 1992;5:451–459. doi: 10.1094/mpmi-5-451. [DOI] [PubMed] [Google Scholar]
- 6.Gu K, Yang B, Tian D, Wu L, Wang D, Sreekala C, Yang F, Chu Z, Wang GL, White FF, et al. Nature. 2005;435:1122–1125. doi: 10.1038/nature03630. [DOI] [PubMed] [Google Scholar]
- 7.Yang B, Sugio A, White FF. Mol Plant–Microbe Interact. 2005;18:142–149. doi: 10.1094/MPMI-18-0142. [DOI] [PubMed] [Google Scholar]
- 8.Yang B, White FF. Mol Plant–Microbe Interact. 2004;17:1192–1200. doi: 10.1094/MPMI.2004.17.11.1192. [DOI] [PubMed] [Google Scholar]
- 9.Chu Z, Yuan M, Yao J, Ge X, Yuan B, Xu C, Li X, Fu B, Li Z, Bennetzen JL, et al. Genes Dev. 2006;20:1250–1255. doi: 10.1101/gad.1416306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bai J, Choi SH, Ponciano G, Leung H, Leach JE. Mol Plant–Microbe Interact. 2000;13:1322–1329. doi: 10.1094/MPMI.2000.13.12.1322. [DOI] [PubMed] [Google Scholar]
- 11.Zhu T. Curr Opin Plant Biol. 2003;6:418–425. doi: 10.1016/s1369-5266(03)00083-9. [DOI] [PubMed] [Google Scholar]
- 12.Zhu W, Magbanua MM, White FF. J Bacteriol. 2000;182:1844–1853. doi: 10.1128/jb.182.7.1844-1853.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugio A, Yang B, White FF. Mol Plant–Microbe Interact. 2005;18:546–554. doi: 10.1094/MPMI-18-0546. [DOI] [PubMed] [Google Scholar]
- 14.Yang B, Zhu W, Johnson LB, White FF. Proc Natl Acad Sci USA. 2000;97:9807–9812. doi: 10.1073/pnas.170286897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iyer AS, McCouch SR. Mol Plant–Microbe Interact. 2004;17:1348–1354. doi: 10.1094/MPMI.2004.17.12.1348. [DOI] [PubMed] [Google Scholar]
- 16.Jiang GH, Xia ZH, Zhou YL, Wan J, Li DY, Chen RS, Zhai WX, Zhu LH. Mol Genet Genomics. 2006;275:354–366. doi: 10.1007/s00438-005-0091-7. [DOI] [PubMed] [Google Scholar]
- 17.Shan L, He P, Zhou JM, Tang X. Mol Plant–Microbe Interact. 2000;13:592–598. doi: 10.1094/MPMI.2000.13.6.592. [DOI] [PubMed] [Google Scholar]
- 18.Chen Z, Kloek AP, Boch J, Katagirl F, Kunkel BN. Mol Plant–Microbe Interact. 2000;13:1312–1321. doi: 10.1094/MPMI.2000.13.12.1312. [DOI] [PubMed] [Google Scholar]
- 19.Reinberg D, Orphanides G, Ebright R, Akoulitchev S, Carcamo J, Cho H, Cortes P, Drapkin R, Flores O, Ha I, et al. Cold Spring Harbor Symp Quant Biol. 1998;63:83–103. doi: 10.1101/sqb.1998.63.83. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi N, Boyer TG, Berk AJ. Mol Cell Biol. 1995;15:6465–6473. doi: 10.1128/mcb.15.11.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez E. Plant Mol Biol. 2002;50:925–947. doi: 10.1023/a:1021258713850. [DOI] [PubMed] [Google Scholar]
- 22.Siberil Y, Doireau P, Gantet P. Eur J Biochem. 2001;268:5655–5666. doi: 10.1046/j.0014-2956.2001.02552.x. [DOI] [PubMed] [Google Scholar]
- 23.Dong X. Curr Opin Plant Biol. 2004;7:547–552. doi: 10.1016/j.pbi.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Niño-Liu DO, Ronald PC, Bogdanove A. Mol Plant Pathol. 2006;7:303–324. doi: 10.1111/j.1364-3703.2006.00344.x. [DOI] [PubMed] [Google Scholar]
- 25.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu T, Budworth P, Chen W, Provart N, Chang H, Guimil S, Su W, Estes B, Zou G, Wang X. Plant Biotech J. 2003;1:59–70. doi: 10.1046/j.1467-7652.2003.00006.x. [DOI] [PubMed] [Google Scholar]
- 27.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 28.Kauffman HE, Reddy APK, Hsiek SPV, Marca SD. Plant Dis Rep. 1973;57:537–541. [Google Scholar]
- 29.Christensen AH, Quail PH. Transgenic Res. 1996;5:213–218. doi: 10.1007/BF01969712. [DOI] [PubMed] [Google Scholar]
- 30.Hiei Y, Komari T, Kubo T. Plant Mol Biol. 1997;35:205–218. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.