Abstract
The Neolithic Revolution began 11,000 years ago in the Near East and preceded a westward migration into Europe of distinctive cultural groups and their agricultural economies, including domesticated animals and plants. Despite decades of research, no consensus has emerged about the extent of admixture between the indigenous and exotic populations or the degree to which the appearance of specific components of the “Neolithic cultural package” in Europe reflects truly independent development. Here, through the use of mitochondrial DNA from 323 modern and 221 ancient pig specimens sampled across western Eurasia, we demonstrate that domestic pigs of Near Eastern ancestry were definitely introduced into Europe during the Neolithic (potentially along two separate routes), reaching the Paris Basin by at least the early 4th millennium B.C. Local European wild boar were also domesticated by this time, possibly as a direct consequence of the introduction of Near Eastern domestic pigs. Once domesticated, European pigs rapidly replaced the introduced domestic pigs of Near Eastern origin throughout Europe. Domestic pigs formed a key component of the Neolithic Revolution, and this detailed genetic record of their origins reveals a complex set of interactions and processes during the spread of early farmers into Europe.
Keywords: European colonization, mtDNA, phylogeography
The transition from hunting to husbandry and the spread of economic and cultural elements associated with Neolithic farming across Europe between the 7th and 4th millenniums B.C. remain poorly understood despite decades of research. The question of how the Neolithic “package” first arrived in Europe has focused on three primary hypotheses that assert that the European Neolithic resulted from (i) the migration of immigrant farmers from the Near East [demic diffusion (1–3)], (ii) the transmission of ideas through established trade and exchange networks (cultural diffusion), or (iii) the independent development of agriculture (including the domestication of some animals such as pigs and cattle) by indigenous European Mesolithic cultures (4, 5), although it has been pointed out that these explanations are not mutually exclusive (6). A separate but related debate has focused on the degree and nature of the interaction between proposed incoming Near Eastern populations and indigenous cultures. Linguistic evidence (7) and modern human genetic evidence (8) have advocated admixture between indigenous and incoming cultures, but the issue of whether Near Eastern (9) or indigenous European populations (10) have played the more significant role in the genesis of the genomes of modern Europeans remains contentious.
Parallel debates also exist with respect to the domestic animals associated with the Neolithic revolution, although some cases are easier to interpret than others. For example, because neither wild sheep (Ovis orientalis) nor wild goats (Capra aegagrus) were naturally distributed across Europe during the Holocene (11–13), the recovery of their remains from European Neolithic archeological sites directly implies their introduction from the Near East (14).
The wild ancestors of domestic cows (Bos primigenius), dogs (Canis lupus), and pigs (Sus scrofa), however, were widely distributed throughout Europe during the early and mid Holocene, leaving open the possibility of their indigenous domestication within Europe. Additionally, the genetic legacy of modern domestic animal stocks may have been further complicated by the interbreeding of independently domesticated European and Near Eastern lineages, just as humans from Near Eastern Neolithic and European Mesolithic populations may have done. Deciphering whether these interactions occurred and what their ramifications may have been, however, has proven as difficult to unravel for domestic animals as it has for humans. Genetic data from modern and ancient European cattle, for example, have revealed that although cattle domesticated in the Near East were subsequently introduced into Europe (14, 15), modern European cattle also possess genes derived from wild aurochsen indigenous to Europe. Although this phenomenon has been shown to be the result of hybridization between European aurochsen and introduced Near Eastern domestics (16), in some cases it has been argued that native European wild cattle were also independently domesticated (17), a conclusion that remains contentious (18).
Zooarcheological research at sites in eastern Turkey [Hallam Çemi (19), Çayönü Tepesi (20), and Neval Çori (21)] and on Cyprus (22) reveals an intensification of the relationship between humans and pigs during the second half of the 9th millennium B.C. Once domesticated, pigs formed a key component of the Neolithic package, as evidenced by their presence in numerous Near Eastern and European Neolithic contexts, although, like cows, their endemic or exotic status in European Neolithic contexts remains unresolved (23).
A previous genetic study (24) of modern wild boar and domestic pigs revealed a robust phylogeographic pattern from which two key conclusions were drawn. First, genetic sequences from modern European domestic pigs indicated that indigenous European wild boar were domesticated at some point in the past. Second, because Near Eastern genetic signals were absent from all modern European samples (save one feral pig from Corsica), Near Eastern domestic pigs were either never introduced into Europe, or, if they were, their progeny has all but disappeared.
The purpose of this study was to identify the geographic origin of pigs found in European Neolithic contexts, to assess the timing and nature of the domestication of European wild boar, and to elucidate the dispersal pattern of Neolithic domestic pigs in western Eurasia. To do this we used appropriately rigorous ancient DNA methods (25, 26) to analyze 478 ancient pig samples representing >140 archeological sites across the Near East and Europe, spanning 29 countries and 13,000 years. An 80-bp diagnostic fragment of the mitochondrial control region that allowed samples to be assigned to specific geographical regions was designed, tested, and successfully amplified in 221 of the 478 ancient samples. In addition, we amplified 663 bp of mitochondrial DNA derived from 42 museum specimens and combined the data with 280 wild and domestic pig sequences in GenBank. Phylogenetic analyses were performed by using a Bayesian Monte Carlo–Markov chain (27) methodology and median-joining networks (28). A range of standard metrical and nonmetrical analyses (29) were carried out on the zooarcheological remains to determine their wild or domestic status [see supporting information (SI) Discussion]. Archeological material was dated through direct or associated 14C analyses and by stratigraphic or cultural association.
Results and Discussion
Endogenous or Exotic? Establishing the True Status of Neolithic Pigs in Europe.
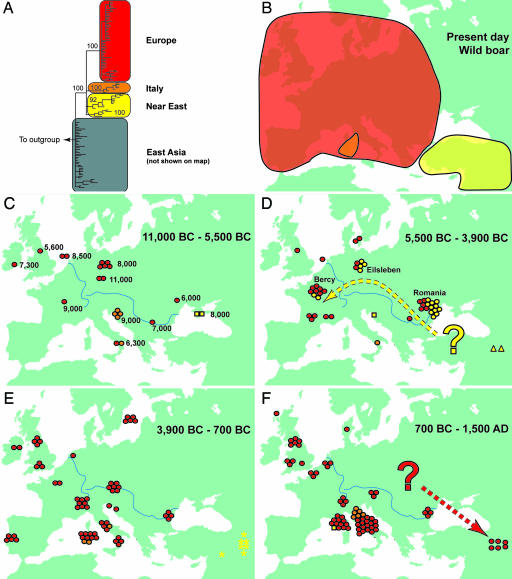
The genetic sequences obtained from the ancient specimens support (24) a modern phylogeographic boundary between Near Eastern and European wild boar haplotypes. Phylogenetic analyses clearly separate four clades with posterior probabilities >0.92. These are geographically distributed with two clades (labeled yellow) predominating in the Near East, another (labeled red) as the major feature of European diversity, and a final minor (orange) clade encountered only in Italy (Fig. 1 A and B).
Fig. 1.
A series of maps depicting the shifting geographic positions of European and Near Eastern pig haplotypes over the past 13,000 years. (A and B) (A) Bayesian (Monte Carlo–Markov chain) consensus tree of 112 modern wild Sus mtDNA control region haplotypes rooted by a common warthog (Phacochoerus aethiopicus). Red, orange, and yellow represent three clusters on the tree that correspond to specific regions on the map in B (Europe, Italy, and the Near East, respectively), where the majority of pigs possess haplotypes within that cluster. Posterior probabilities of the major nodes are listed for each of the branches. (C–F) Time series of maps identifying the locations of ancient pig samples from which DNA haplotypes were generated within Europe. Each symbol corresponds to a single sample, and the colors correspond to those used in A and represent the cluster on the tree to which the samples belong. The four Near Eastern haplotypes discussed in Results and Discussion (Y1, Y2, A1, and A2) are represented by yellow circles, squares, asterisks, and triangles, respectively. Numbers to the right of sample locations in C represent approximate sample ages (in calibrated years B.C.).“Bercy” and “Eilsleben” in D refer to specific sites, and “Romania” refers to several sites discussed in Results and Discussion. Clustered symbols represent multiple samples from the same or geographically proximate sites. The upper and lower blue lines represent the Rhine and Danube rivers, respectively. The dotted yellow arrow in D depicts the hypothesized Danubian trajectory along which the Y1 haplotype was transported, and the dotted red arrow in F highlights the movement of European domestic pigs transported into Armenia. The question marks at the origins of the arrows reflect the uncertainty regarding the precise locations from where the dispersal routes began. C–F very broadly represent the European Mesolithic, the European Neolithic, the Bronze Age, and all subsequent ages to the medieval period, respectively. Additional details regarding all of the modern and ancient samples used in this study can be found in SI Discussion.
To examine the temporal changes in these phylogeographic patterns, we grouped the results into four time periods (Fig. 1 C–F). Data from ancient specimens showed that none of the 20 pre-Neolithic (11000–5500 B.C.) wild boar samples from Europe possessed Near Eastern haplotypes (Fig. 1C). Additionally, four wild individuals within the second temporal stratum (5500–3900 B.C.; Fig. 1D) from Neolithic and post-Neolithic (Chalcolithic) sites in Romania close to the phylogeographic boundary also only possessed European haplotypes. The consistency of the regional restriction of haplotypes through time suggests that the modern phylogeographic boundary between European and Near Eastern wild Sus populations (24) has been intact since at least the early Holocene. Although the exact location of this boundary is difficult to establish, the appearance of a Near Eastern pig haplotype (Y2) in two wild specimens from Mesolithic and Neolithic sites in the Crimea (Fig. 1C) indicates that the distribution of Near Eastern haplotypes probably extended to the north shore of the Black Sea. Given the apparent long-term integrity of the boundary, it follows that samples found on either side that possess haplotypes not matching those of local wild boar most likely represent domestic pigs (derived from exotic wild boar lineages) that have been introduced by humans.
The data clearly show that pigs with Near Eastern haplotypes crossed this boundary during the Neolithic and began to appear in European contexts. Eleven pig specimens, identified as domestic, from four Neolithic Romanian sites dating to 5500 B.C. (Fig. 1D) possess an 80-bp sequence (the Near Eastern Y1 haplotype) identical to that found in a single recent modern boar from Turkey and two from Iran. However, five wild specimens from the same sites possess European haplotypes. The same Y1 haplotype was also identified in four specimens from the 6th-millennium B.C. Linearbandkeramik (LBK) site of Eilsleben in northern Germany, and in two samples from the mid Neolithic (very early 4th-millennium B.C. Chasséen culture) site of Bercy (30) in the Paris Basin. This evidence clearly demonstrates that Near Eastern-derived domestic pigs were dispersed westward into central and western Europe during the Neolithic (Fig. 1D).
Importantly, six additional pigs identified as domestic from Bercy possessed European haplotypes. Bercy is therefore not only the earliest site in our study at which domestic pigs derived from native European wild boar were identified but is also the only site so far to possess definitive domestic pigs of both Near Eastern and European ancestry and the latest site to possess pigs of Near Eastern ancestry. All mainland and British domestic specimens dating from the mid 4th millennium B.C. to the present (including those from sites in Portugal, Switzerland, the Czech Republic, Britain, Croatia, and Romania) possess European haplotypes (Fig. 1E).
The period between the initial incorporation of European wild boar into domestic stocks and the near total replacement of the introduced Near Eastern pig lineages appears relatively brief. A logistic curve fit to data points representing the relative proportion of Near Eastern and European haplotypes in domestic samples through time suggests that European haplotypes increased from 5% to 95% in as little as 500 years. Although this estimate is bracketed by large confidence intervals (see Materials and Methods), all of the derived logistic curves demonstrated a rapid increase in the percentage of European domestic pigs at the expense of pigs with Near Eastern ancestry.
The evidence indicates that by at least the 4th millennium B.C., European wild boar had been domesticated and spread throughout Europe, replacing pigs of Near Eastern origin. Interestingly, although pigs are comparatively scarce relative to other domestic animals from Neolithic Romanian sites (31) and from most LBK sites in central and western Europe (32), they increase in abundance (33) and size (34) from the first part of the 5th millennium B.C., well after the initial introduction of Near Eastern domestic pigs. In the Paris Basin for example, pigs constitute <15% of the total identifiable faunal specimens in late LBK contexts but >30% in post-LBK sites, and in eastern Germany, pigs account for <10% in early LBK contexts but >15% in late LBK sites (33). Metrical evidence also indicates that the proportion of small pigs is greater at LBK sites in eastern Germany and Alsace (35, 36) than at the later site of Bercy, thus suggesting a general increase in the size of pigs through time.
These observations, in combination with the genetic data, suggest that the domestication of European wild boar may not have been truly independent but instead occurred only after the introduction of Near Eastern domestic pigs during the early Neolithic and perhaps as a direct consequence. However, an earlier timeframe and greater degree of independence of the domestication of European wild boar cannot be excluded, because it is possible that domestic pigs possessing European signatures before the introduction of Near Eastern pigs may yet be discovered.
Inferring Separate European Neolithic Dispersal Routes from Pig Ancient DNA.
Previous studies have discussed the possibility that Near Eastern Neolithic cultures entered Europe along at least two separate routes: a northern route along the so-called Danubian Corridor, which followed the Danube and Rhine River valleys into northwest Europe and was associated with the adoption and preferential cultivation of the two hulled wheat [emmer and einkorn (12, 37)], and a southern maritime route that traversed the north shore of the Mediterranean and was characterized instead by a predominance of naked wheat varieties (37–39).
Although based on only a few samples, it appears that the chronology and geographic distribution of the Y1 Near Eastern ancient pig haplotypes mirrors the northern route (Fig. 1D). The presence of Y1 haplotypes within the mid-6th millennium B.C., and more recent contexts in Romania, Germany, and France, suggests that domestic pigs of this lineage were transported along a northern trajectory similar to that of emmer and einkorn wheat. A second Near Eastern haplotype (Y2, found in two modern Turkish wild boar) was identified in two archeological samples: a late Neolithic pig in Croatia (at Pupicina cave; Fig. 1D), and a 15th-century A.D. specimen from Corsica (Fig. 1F), although additional sampling is required to establish a correlation between this haplotype and a Mediterranean dispersal. Curiously, however, the Y2 haplotype was identified in a modern Corsican feral pig, making it the only modern European specimen to possess a Near Eastern haplotype and suggesting that this pig's lineage is descended from the first domestic pigs to arrive on Corsica with the initial Neolithic settlers of the island.
Origins and Subsequent Dispersal of European Domestic Pigs.
The reported (24) presence of Italian (Fig. 1 A and B, orange clade) haplotypes in modern “wild” pigs of Sardinia [like Corsica, an island devoid of pigs before the arrival of Neolithic settlers (40)] supports the possibility that indigenous Italian wild boar were also separately domesticated (41). In this study, the Italian haplotype was identified in numerous ancient samples, including: Mesolithic wild boar from Pupicina cave in Croatia, early and mid Neolithic pigs from Grotta della Madonna cave in Southwestern Italy, a 2nd-millennium B.C. (middle Bronze Age) Sardinian site, and numerous medieval wild boar from Tuscany and Rome (Fig. 1 C–F). These samples indicate that not only were indigenous Italian wild boar distributed beyond their current restricted region of Maremma in Northwest Italy, but the presence of the Italian haplotype in Bronze Age central Sardinia (Fig. 1E) also suggests either an independent domestication of native Italian wild boar or the incorporation of female Italian wild boar into domestic stocks that were subsequently imported to Sardinia by at least the end of the 2nd millennium B.C.
DNA preservation in the Near East is generally poor (42, 43). This is borne out by our analyses in which only 1 of 57 archeological pigs from the Near East (excluding Armenia) yielded DNA. However, for reasons possibly related to altitude and/or local climate, the majority of Armenian samples did produce sequences. Two samples dating from the 5th to the 3rd millennium B.C. (one of which is certainly wild) possessed a Near Eastern haplotype (A2) also found in a modern Armenian wild boar (Fig. 1D). Six samples (four of which were probably domestic) dating from the 2nd millennium B.C. through the early Iron Age all possessed a different Near Eastern haplotype (A1; Fig. 1E) found both in a modern Syrian wild boar and in a 2nd-millennium B.C. domestic pig from Çhagar Bazar (northern Syria). Lastly, a total of six Armenian pigs (two of which are clearly domestic) from five sites dating from the 7th century B.C. to the 13th century A.D. all possess the same European haplotype (Fig. 1F).
These data suggest that European domestic pigs spread eastward to Armenia by the 7th century B.C., apparently completely replacing the earlier domestic pigs of Near Eastern ancestry (Fig. 1F). This transition likely reflects the major reorganization that occurred in the various Neo-Hittite polities during the 1st millennium B.C. whereby large-scale shifts in the economic and political dynamics, linked with the contemporaneous expansion of, for example, the Urartian and Phrygian territorial empires within Armenia and Anatolia, respectively, likely involved the large-scale movement of peoples and the expansion of trade and exchange networks during the later Iron Age (44).
Conclusions
The ancient genetic records reveal a complex temporal and geographical pattern of changes in pig haplotype distributions in Holocene Europe. These data provide unique insights and answers to specific questions about the nature of the European Neolithic revolution. First, the presence of Near Eastern haplotypes in Neolithic contexts in Romania, Germany, France, and Croatia demonstrates that Near Eastern pigs were definitely introduced by people into Europe and that they may have traveled along at least two distinct routes. Second, given the timeframe of the initial introduction of Near Eastern domestics and the first appearance of domestic pigs derived from European wild boar, it appears, at least at the broad spatial and temporal scale of these data, that European pig domestication may not have been a truly independent event but rather a direct consequence of the introduction of Near Eastern domestic pigs (and other animals) into Europe by early farmers. If true, the process of pig domestication in Europe (and possibly the degree of intention among early farmers) could have been fundamentally different from that in the Near East. Regardless of the specific cause or progression of European pig domestication, what is clear is that once European wild boar were domesticated, they rapidly became the predominant lineage within European domestic swine.
The Bronze and Iron Age genetic sequences reveal two additional movements of separate pig haplotypes. First, domestic pigs with Italian wild boar signatures were transported to Sardinia. Second, European domestic pigs, having already displaced Near Eastern pigs in Europe, began replacing indigenous Near Eastern pigs in Armenia during later prehistory (Fig. 1F).
The conclusions of this study are based on a mitochondrial locus that solely reflects maternal inheritance. Although these results likely differ from the paternal history, the maternal pattern is more likely to reflect human movement, given the greater likelihood of humans keeping piglets born to domestic sows relative to wild-born piglets sired by domestic boars. An investigation of nuclear markers (including the Y chromosome) would undoubtedly reveal an even more complex pattern than that demonstrated by the mitochondrial data alone.
Two general conclusions can be drawn from this study. First, the significant differences in the geographical range of wild and domestic haplotypes through time demonstrates that inferences regarding ancient biogeographical ranges of wild and domestic animals based solely only on their modern distribution may not be valid. Second, this study illustrates the effectiveness of ancient genetic data from domestic species in revealing detailed insights into the patterns of past human migration as well as trade and exchange networks around the globe.
Materials and Methods
Ancient, Modern, and Museum Samples.
Of 52 modern and museum specimens (SI Table 3) for which DNA was extracted, 42 samples yielded enough DNA to be combined with 280 GenBank entries (SI Table 4) to generate a dataset comprising 322 individuals. The remaining 10 samples were sufficiently degraded that they only yielded ≈80 bp, using a primer pair (discussed below) applied to ancient samples.
Bones and teeth (478) from ancient pigs from >140 sites in 29 countries were subjected to DNA extraction techniques. Of those, 221 yielded amplifiable DNA (SI Tables 5 and 6).
Extraction, DNA Amplification, and Sequencing of Museum and Ancient Specimens.
Because the preservation of the museum samples varied significantly, all museum and ancient specimens were treated as ancient and were subjected to the same extraction procedures [including the use of multiple extraction and PCR blanks, independent replication of PCRs, and replication (25) of extractions in Adelaide, Australia; Basel, Switzerland; and Dublin, Ireland]. The extraction, amplification, and sequencing protocols for all of the specimens analyzed at the Henry Wellcome Ancient Biomolecules Centre (HWABC) followed Shapiro et al. (45).
Cloning reactions were performed at the HWABC using the Invitrogen (Paisley, U.K.) Topo-TA cloning kit according to the manufacturer's instructions and were amplified by using the primers T7 and M13R (Invitrogen). Eight sequences from each cloning reaction were sequenced to evaluate template damage and check for the presence of contaminating sequences and/or nuclear copies of mitochondrial sequences.
A 663-bp fragment of the mitochondrial control region was targeted for the 42 museum samples analyzed at the HWABC, but not all of the samples yielded the entire fragment. A list of haplotype codes and the samples that are represented by each haplotype are found in SI Table 7. A variety of primer combinations were used (SI Table 8), depending on the nature of the museum samples and stringent ancient DNA protocols.
The full complement of ancient samples was analyzed in three separate facilities. A total of 208 of 409 samples were successfully amplified at the HWABC following the same protocols referenced above.
Of the 19 S. scrofa bones analyzed at Trinity College in Dublin by C.J.E., eight generated a reproducible sequence. Four bones were sourced from four sites in Israel by Liora Kolska Horwitz (The Hebrew University, Jerusalem, Israel). Seven bones from Carsington Pasture Cave in Derbyshire were sourced by Andrew Chamberlain (University of Sheffield, Sheffield, U.K.). Seven of the eight bones from Ireland were sourced by Finbar McCormick (Queens University, Belfast, Ireland), and the County Waterford specimen was provided by the Natural History Museum Dublin, through Nigel Monaghan, Keeper. Details of these bones/sites can be found in refs. 46–48.
Before extraction, each bone was sandblasted to remove external contamination that can out-compete endogenous DNA. The bone samples were all extracted by using a silica-based methodology (49), except that 200 μg/ml proteinase K (rather than 100 μg/ml) was added to the extraction buffer. Varying numbers of DNA extractions were performed for each specimen. In the cases where only one extract was attempted, this was because no amplification products were obtained from several amplification attempts using the first extract.
Nested primers (SI Table 9) were designed to amplify the 5′ end of the mtDNA control region. The resulting amplification products were sent away for direct sequencing to either MWG-Biotech (Ebersberg, Germany) or Macrogen (Rockville, MD). Second-round PCR was not undertaken on any samples that did not amplify in the first round.
Six samples were analyzed in Basel, Switzerland, by A.S., three of which were simultaneously analyzed in Oxford. Bone powder was generated from each bone according to methods published elsewhere, and DNA was extracted by using a modified silica-based method (50).
PCRs in Basel were carried out by using the same methodology as used in Oxford except that AmpliTaq Gold was used as the enzyme. PCR products were viewed on NuSieve Agarose gel and purified with Qiagen (Valencia, CA) PCR product purification kit. Products were directly sequenced by Microsynth (Balgach, Switzerland).
At the HWABC, a highly variable ≈80-bp fragment (that included numerous indels) was identified within the 663-bp alignment. A single primer pair (SI Table 10) was used to amplify the 80-bp fragment in the ancient samples, and this same primer pair was used in Oxford, Basel, and Dublin. The results of each PCR and the haplotype associations of each successful sample are listed in SI Table 5.
Analysis of Sequence Data.
Sequences obtained from museum specimens were aligned by eye with 280 sequences from previous published studies deposited on GenBank, using Se-Al (51). Phylogenetic analysis was performed by using MrBayes software, Version 3 (27) and model parameters identified by ModelTest (52). Under the HKY85+G+I model, parameter estimates (including posterior probabilities) and consensus trees, resulting from five MrBayes runs of at least 5 million (but <30 million) generations each, were recorded and contrasted. The skeleton of the tree is presented in Fig. 1, and a more detailed version of the same tree is presented in SI Fig. 2. The posterior probabilities listed on both trees represent the lowest recorded values amongst all runs.
A neighbor-joining tree (SI Fig. 3) was drawn using only the 80-bp fragment to ascertain how many and which haplotypes derived from the full 663-bp alignment matched those amplified in the ancient samples. The resulting neighbor-joining tree possessed the same cladistic structure as the tree based on the 663 bp (SI Fig. 2), and numerous SNPs were 100% correlated with specific clades. Because of the strong phylogeographic correlation evident within the dataset, the SNPs were then associated with specific geographical regions, and, in some cases, matched unique, individual, modern samples. The relative positions of the 18 identified haplotypes (each of which begin with “ANC”) derived from the 80-bp fragment are depicted in SI Fig. 3. The polymorphic sites differentiating each of the 18 haplotypes are presented in SI Table 11. A more in-depth discussion of the haplotypes based on the 80-bp fragment can be found in the SI Discussion.
Lastly, the entire alignment derived by using the 663-bp fragment was used to draw a median-joining network (28) (shown in SI Fig. 4) to further elucidate the differences between the major clades.
Morphological and Biometric Analysis.
Although the most commonly accepted method for determining the wild or domestic status of animal remains excavated from archeological sites has been an analysis of the relative sizes of teeth and postcranial bones, the reliability of these kinds of markers remains contentious. For the purposes of this article, the wild or domestic attributions of the majority of the specimens listed in SI Tables 5 and 6 were assigned by U.A., K.D., and P.R.-C., although J.-D.V., J.S., A. Tresset, and P.M. also analyzed some material.
The principles guiding the status designations used in this study have been published elsewhere (53–57), although in general, the morphological determination of wild and domestic status involves a wide range of criteria, some of which can be considered more definitive than others. A full discussion of these issues can be found in SI Discussion, and SI Table 6 outlines the specific criteria used to assign wild or domestic status to each individual specimen and provides details used to support these conclusions. In cases where we felt unable to assign a definitive status, we have suggested what is the most likely determination (i.e., domestic or wild) and why, and in circumstances where the evidence is entirely ambiguous, we assigned an “undetermined” status. Lastly, a map shown in SI Fig. 5 depicts the locations of all sample sites.
Regression Analysis.
A nonlinear regression analysis was carried out to assess the time frame over which domestic pigs of European ancestry replaced those from the Near East. Modern and ancient domestic pig samples possessing European and Near Eastern haplotypes were assigned to four separate time bins listed in Table 1. The time frames for the bins were determined by: (i) the appearance of Near Eastern haplotypes in Europe, (ii) the first appearance of domestic samples with European haplotypes, (iii) all additional ancient samples through the 16th century A.D., and (iv) all modern day samples.
Table 1.
Percentages of European and Near Eastern domestic samples within four timeframes
| Time range | Avg. | Euro, n | N.E., n | Euro, % | Binomial C.I., % |
|---|---|---|---|---|---|
| 5500–4000 B.C. | 4750 B.C. | 0 | 16 | 0 | 0–17.1 |
| 4000–3500 B.C. | 3750 B.C. | 6 | 2 | 75 | 34.8–96.8 |
| 3500 B.C.–1500 A.D. | 1500 B.C. | 59 | 0 | 100 | 95.1–100 |
| Present | 2000 A.D. | 140 | 0 | 100 | 97.9–100 |
Avg., average; Euro, European; N.E., Near Eastern; C.I., confidence interval.
Binomial confidence intervals were calculated for each percentage of European pigs based on the total number of samples in each bin. Three logistic curves were then fit to the data using extremes of the confidence interval (34.8%, 96.8%) and the strict ratio (75%) within the 4000–3500 B.C. bin. Using the logistic equation: y = 1/(1 + be−cx), values were generated for b and c and are listed Table 2. To assess the total time over which the ratio of European pigs increased from 5% to 95% for each equation, y values of 0.05 and 0.95 were placed into the equation yielding total times listed in Table 2.
Table 2.
Parameters and results of the regression analysis carried out to estimate the timeframe over which European domestic pigs replaced Near Eastern pigs
| Values from the 4000–3500 B.C. bin, % | b | c | Total time, years |
|---|---|---|---|
| 34.8 | 211.65 | 4.73 | 1,245 |
| 75 | 14,507.59 | 10.96 | 477 |
| 96.8 | 2,375.86 | 11.45 | 514 |
b and c, parameters of the logistic equation discussed in Materials and Methods.
Supplementary Material
Acknowledgments
We thank Simon Ho, Andrew Rambaut, and the institutions and individuals listed in SI Discussion that provided access to collections. This study was supported by Wellcome Trust Bioarchaeology Fellowships (K.D. and A.C.), the Arts and Humanities Research Council (P.R.-C. and U.A.), the Leverhulme Trust (G.L. and A.C.), Smithsonian Short-Term Fellowships (K.D.), and the Irish Research Council for Science, Engineering and Technology (C.J.E.).
Abbreviation
- LBK
Linearbandkeramik.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. DQ872931–DQ873203).
This article contains supporting information online at www.pnas.org/cgi/content/full/0703411104/DC1.
References
- 1.Pinhasi R, Fort J, Ammerman AJ. PLoS Biol. 2005;3:e410. doi: 10.1371/journal.pbio.0030410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Childe VG. The Dawn of European Civilization. St. Albans, UK: Paladin; 1957. [Google Scholar]
- 3.Sampietro ML, Lao O, Caramelli D, Lari M, Pou R, Marti M, Bertranpetit J, Lalueza-Fox C. Proc Biol Sci. 2007;274:2161–2167. doi: 10.1098/rspb.2007.0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark JGD. Antiquity. 1966;40:172–189. [Google Scholar]
- 5.Renfrew C. Before Civilization. The Radiocarbon Revolution and Prehistoric Europe. London: Penguin; 1972. [Google Scholar]
- 6.Zvelebil M. In: Archaeogenetics: DNA and the Population History of Europe. Boyle K, editor. Cambridge, UK: McDonald Inst for Archaeol Res; 2000. pp. 57–79. [Google Scholar]
- 7.Renfrew C. In: Examining the Farming/Language Dispersal Hypothesis. Bellwood P, Renfrew C, editors. Cambridge, UK: McDonald Inst for Archaeol Res; 2002. pp. 3–16. [Google Scholar]
- 8.Cavalli-Sforza LL. Genes, People, and Languages. Berkeley, CA: Univ of California Press; 2001. [Google Scholar]
- 9.Belle EM, Landry PA, Barbujani G. Proc Biol Sci. 2006;273:1595–1602. doi: 10.1098/rspb.2006.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haak W, Forster P, Bramanti B, Matsumura S, Brandt G, Tanzer M, Villems R, Renfrew C, Gronenborn D, Alt KW, Burger J. Science. 2005;310:1016–1018. doi: 10.1126/science.1118725. [DOI] [PubMed] [Google Scholar]
- 11.Luikart G, Gielly L, Excoffier L, Vigne JD, Bouvet J, Taberlet P. Proc Natl Acad Sci USA. 2001;98:5927–5932. doi: 10.1073/pnas.091591198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uerpmann HP. Probleme der Neolithisierung des Mittelmeerraumes. Beihefte zum Tübinger Atlas des Vorderen Orients, Reihe B. Freiburg, Germany: Geisteswissenschaften Band 28; 1979. [Google Scholar]
- 13.Davis SJM. The Archaeology of Animals. London: Batsford; 1987. [Google Scholar]
- 14.Fernandez H, Hughes S, Vigne JD, Helmer D, Hodgins G, Miquel C, Hanni C, Luikart G, Taberlet P. Proc Natl Acad Sci USA. 2006;103:15375–15379. doi: 10.1073/pnas.0602753103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bollongino R, Edwards CJ, Alt KW, Burger J, Bradley DG. Biol Lett. 2006;2:155–159. doi: 10.1098/rsbl.2005.0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gotherstrom A, Anderung C, Hellborg L, Elburg R, Smith C, Bradley DG, Ellegren H. Proc Biol Sci. 2005;272:2345–2350. doi: 10.1098/rspb.2005.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beja-Pereira A, Caramelli D, Lalueza-Fox C, Vernesi C, Ferrand N, Casoli A, Goyache F, Royo LJ, Conti S, Lari M, et al. Proc Natl Acad Sci USA. 2006;103:8113–8118. doi: 10.1073/pnas.0509210103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards CJ, Bollongino R, Scheu A, Chamberlain A, Tresset A, Vigne JD, Baird JF, Larson G, Ho SY, Heupink TH, et al. Proc Biol Sci. 2007;274:1377–1385. doi: 10.1098/rspb.2007.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redding R, Rosenberg M. In: Ancestors for the Pigs: Pigs in Prehistory. Nelson S, editor. Vol 15. Philadelphia: Univ of Pennsylvania MASCA Researcher Papers in Science and Archaeology; 1998. pp. 65–76. [Google Scholar]
- 20.Ervynck A, Dobney K, Hongo H, Meadow R. Paléorient. 2002;27:47–73. [Google Scholar]
- 21.Peters J, von den Driesch A, Helmer D. In: First Steps of Animal Domestication, New Archaeozoological Approaches. Vigne J-D, Peters J, Helmer D, editors. Oxford: Oxbow Books; 2005. pp. 96–124. [Google Scholar]
- 22.Vigne JD, Carrère I, Guilaine J. In: Néolithique de Chypre (Actes Coll Int Nicosie, 17–19 mai 2001) Guilaine J, Brun AL, editors. Vol suppl 43. Athens, Greece: Ecole Française d'Athènes; 2003. pp. 239–251. [Google Scholar]
- 23.Albarella U, Dobney K, Rowley-Conwy P. In: Documenting Domestication: New Genetic and Archaeological Paradigms. Zeder MA, Decker-Walters D, Bradley D, Smith BD, editors. Berkeley, CA: Univ of California Press; 2006. pp. 209–227. [Google Scholar]
- 24.Larson G, Dobney K, Albarella U, Fang MY, Matisoo-Smith E, Robins J, Lowden S, Finlayson H, Brand T, Willerslev E, et al. Science. 2005;307:1618–1621. doi: 10.1126/science.1106927. [DOI] [PubMed] [Google Scholar]
- 25.Cooper A, Poinar HN. Science. 2000;289:1139. doi: 10.1126/science.289.5482.1139b. [DOI] [PubMed] [Google Scholar]
- 26.Gilbert MTP, Bandelt HJ, Hofreiter M, Barnes I. Trend Ecol Evol. 2005;20:541–544. doi: 10.1016/j.tree.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 27.Ronquist F, Huelsenbeck JP. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 28.Bandelt HJ, Forster P, Rohl A. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 29.Vigne JD, Helmer D, Peters J. In: The First Steps of Animal Domestication. Vigne JD, Peters J, Helmer D, editors. Oxford: Oxbow Books; 2005. pp. 1–16. [Google Scholar]
- 30.Balasse M, Tresset A. J Archaeol Sci. 2002;29:853–859. [Google Scholar]
- 31.Bălăçsescu A, Moise D, Radu V. Cultură çsi civilizaţie la Dunărea de Jos XXII: In Honorem Silvia Marinescu Bâlcu – 70 ani. Bucharest, Romania: Daim; 2005. pp. 167–206. [Google Scholar]
- 32.Tresset A, Vigne JD. In: Role et statut de la chasse dans le Neolithique qncien Danubien (5500–4900 av JC) Arbogast RM, Jeunesse C, Schibler J, editors. Radhen/Westfalen, Germany: Marie Leidorf; 2004. pp. 129–151. [Google Scholar]
- 33.Tresset A. In: Ancient Europe 8000 B.C. to A.D. 1000: Encyclopedia of the Barbarian World. Bogucki P, Crabtree P, editors. Farmington Hill, MI: Charles Scribner's Sons; 2003. [Google Scholar]
- 34.Tresset A, Vigne JD. In: Whittle A, editor. Going Over: The Mesolithic-Neolithic Transition in North-west Europe; Proceedings of the Conference Held at Cardiff; May 2005; Oxford: Oxbow Books; 2007. in press. [Google Scholar]
- 35.Müller H. Die Haustiere der mitteldeutschen Bandkeramiker. Berlin: Akademie; 1964. [Google Scholar]
- 36.Arbogast R-M. Premiers élevages néolithiques du Nord-Est de la France, Études et recerches archeologiques de l'Université de Liège 67. Liège, Belgium: Université de Liège; 1994. [Google Scholar]
- 37.Colledge S, Connolly J, Shennan S. Eur J Archaeol. 2006;8:137–156. [Google Scholar]
- 38.Maier U. Vegetat Hist Archaeobot. 1996;5:39–55. [Google Scholar]
- 39.Colledge S, Connolly J, Shennan S. Curr Anthropol. 2004;45:s35–s38. [Google Scholar]
- 40.Vigne JD. Mammal Rev. 1992;22:87–96. [Google Scholar]
- 41.Albarella U, Tagliacozzo A, Dobney K, Rowley-Conwy P. Proc Prehist Soc. 2006;72:193–227. [Google Scholar]
- 42.Edwards CJ, MacHugh DE, Dobney KM, Martin L, Russell N, Horwitz LK, McIntosh SK, MacDonald KC, Helmer D, Tresset A, et al. J Archaeol Sci. 2004;31:695–710. [Google Scholar]
- 43.Smith CI, Chamberlain AT, Riley MS, Stringer C, Collins M. J Human Evol. 2003;45:203–217. doi: 10.1016/s0047-2484(03)00106-4. [DOI] [PubMed] [Google Scholar]
- 44.Smith AT. The Political Landscape: Constellations of Authority in Early Complex Polities. Berkeley, CA: Univ of California Press; 2003. [Google Scholar]
- 45.Shapiro B, Drummond AJ, Rambaut A, Wilson MC, Matheus PE, Sher AV, Pybus OG, Gilbert MTP, Barnes I, Binladen J, et al. Science. 2004;306:1561–1565. doi: 10.1126/science.1101074. [DOI] [PubMed] [Google Scholar]
- 46.Chamberlain AT. [Accessed April 2006];Cave Archaeol PalaeontolRes Archive. 1999 Available at http://capra.group.shef.ac.uk/1/carsabs.html.
- 47.McCormick F. In: From Megaliths to Metals. Essays in Honour of George Eogan. Roche H, Gorgan E, Bradley J, Coles J, Raferty B, editors. Oxford: Oxbow; 2004. pp. 1–5. [Google Scholar]
- 48.Woodman P, McCarthy M, Monaghan N. Quaternary Sci Rev. 1997;16:129–159. [Google Scholar]
- 49.MacHugh DE, Edwards CJ, Bailey JF, Bancroft DR, Bradley DG. Anc Biomol. 2000;3:81–102. [Google Scholar]
- 50.Schlumbaum A, Turgay M, Schibler J. Anim Genet. 2006;37:373–375. doi: 10.1111/j.1365-2052.2006.01435.x. [DOI] [PubMed] [Google Scholar]
- 51.Rambaut A. Se-Al: Sequence Alignment Editor. 1996 Available at http://evolve.200.ox.ac.uk.
- 52.Posada D, Crandall KA. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 53.Rowley-Conwy P, Dobney K. In: Pigs and Humans: 10,000 Years of Interactions. Albarella U, Dobney K, Ervynck A, Rowley-Conwy P, editors. Oxford: Oxford Univ Press; 2007. [Google Scholar]
- 54.Albarella U, Payne S. J Archaeol Sci. 2005;32:589–599. [Google Scholar]
- 55.Albarella U, Tagliacozzo A, Dobney K, Rowley-Conwy P. Proc Prehist Soc. 2006;72:193–227. [Google Scholar]
- 56.Albarella U, Manconi F, Rowley-Conwy P, Vigne JD. In: Archaeozoological Studies in Honour of Alfredo Riedel. Tecchiati U, Sala B, editors. Bolzano, Italy: Province of Bolzano; 2006. pp. 285–302. [Google Scholar]
- 57.Albarella U, Davis S, Detry C, Rowley-Conwy P. Anthropozoologica. 2006;40:27–54. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.