Abstract
Can unique protein structures arise from a limited set of amino acids present on the prebiotic Earth? To address this question, we have determined the stability and structure of KIA7, a 20-residue polypeptide containing chiefly Lys, Ile, and Ala. NMR methods reveal that KIA7 tetramerizes and folds on the millisecond time scale to adopt a four-helix X-bundle structure with a tightly and specifically packed core. Denaturation studies and hydrogen exchange measurements of KIA7 and several variants demonstrate that ridges-into-grooves packing of Ala and Ile side chains and the packing of a C-terminal aromatic group into the hydrophobic core are sufficient to give rise to a rather stable, well folded protein structure, with no favorable electrostatic interactions or tertiary or quaternary hydrogen bonds. Both modern proteins and RNAs can adopt specific structures, but RNAs do so with a limited “alphabet” of residues and types of stabilizing interactions. The results reported here show that specific, well folded protein structures can also arise from a highly reduced set of stabilizing interactions and amino acids that are thought to have been present on the prebiotic Earth.
Keywords: four-helix bundle, NMR spectroscopy, protein stability, chemical evolution, protein folding
In the prebiotic Earth, hydrophobic and charged amino acids were thought to have been rather common, whereas polar amino acids were rare (1, 2). Could such a limited set of amino acids have given rise to unique well folded proteins? To gain insight into this question, we have studied the conformational stability and structure of KIA7, a 20-residue polypeptide containing chiefly Lys, Ile, and Ala, plus a –Gly-Gly-Tyr extension for concentration determination, which self-associates to form a well packed, four-helix bundle protein.
KIA7 was previously identified from a combinatorial peptide library as having a highly helical structure (as gauged by CD), a well packed hydrophobic core (1-anilino-8-naphthalene sulfonate fluorescence), and a specific tetrameric quaternary structure (analytical ultracentrifugation) (3, 4). The positively charged lysine side chains prevent the formation of higher-ordered oligomers. Because no tertiary hydrogen bonds, higher-order oligomerization or crystal packing interactions, or favorable electrostatic interactions are present in KIA7, this study constitutes a test of whether or not a specific, unique structure can arise from a highly limited set of stabilizing interactions, namely, the hydrophobic effect and van der Waals interactions.
In addition to KIA7, two other members of the KIA series of peptides, KIA5 and KIA10, were studied (Table 1). These peptides are variants of the KIA7 sequence and possess lesser degrees of ordered structure (3). A stabilized KIA7 variant with a disulfide cross-link, CG3KIA7, was also examined, and additional peptides truncating the –Gly-Gly-Tyr C-terminal extension, KIA7Δ, or carrying Tyr to Phe, KIA7F, or Tyr to Ile, KIA7I, substitutions were studied to probe the possible stabilizing or structural contributions of this extension (Table 1). An additional variant, +KIA7−, was characterized to determined whether and how terminal charges affect stability and structure formation. The results show that the KIA7 tetrameric structure is as well packed as natural four-helix bundle proteins and is stabilized by the hydrophobic effect and van der Waals interactions, with no favorable charge–charge interactions and, in the case of KIA7F, no tertiary hydrogen bonds. Helix-capping interactions that are present in natural proteins also stabilize KIA7.
Table 1.
Sequence of KIA molecules
| Molecule | Sequence | ||||
|---|---|---|---|---|---|
| 1 | 5 | 10 | 15 | 20 | |
| KIA7 | Ac-AKAAA AAIKA IAAII KAGGY-NH2* | ||||
| KIA5 | Ac-AKAIA AIAKA AAAII KAGGY-NH2 | ||||
| KIA10 | Ac-AKAAA AIIKA IAAAI KAGGY-NH2 | ||||
| KIA7Δ | Ac-AKAAA AAIKA IAAII KAGG-NH2 | ||||
| KIA7I | Ac-AKAAA AAIKA IAAII KAGGI-NH2 | ||||
| KIA7F | Ac-AKAAA AAIKA IAAII KAGGF-NH2 | ||||
| CG3KIA7 | [Ac-CGGGAKAAA AAIKA IAAII KAGGY-NH2]2† | ||||
| +KIA7− | +AKAAA AAIKA IAAII KAGGY− | ||||
*The N-terminal amine and C-terminal carboxylate group are acetylated and amidated, respectively, and are uncharged in all the peptides, except +KIA7−. Therefore, apart from +KIA7−, no favorable charge–charge interactions can be formed, as the only charged residues present are Lys.
†In solution, CG3KIA7 dimerizes via disulfide bond formation. Two dimers then associate to form a tetramer.
Results
The Sequence of Ala and Ile and a C-Terminal Aromatic Are Key for Structure Formation.
The KIA peptides were first examined by CD, a probe of secondary structure. The CD spectra at 25°C of KIA5, KIA7Δ, and KIA7I in 0.2 M NaCl, pH 5 show deep minima at 202 nm and a shallow one at 220 nm [supporting information (SI) Fig. 5], which indicate low helix contents. The CD spectra of KIA10 at 25°C in 0.2 M NaCl is typical of a mostly unstructured peptide, but upon lowering the temperature to 5°C and increasing the peptide concentration the spectra becomes characteristically α-helical (data not shown). KIA7, KIA7F, and CG3KIA in 0.2 M NaCl show strong minima at 221 and 208 nm and intense maxima <200 nm, which indicates that they adopt highly helical structures. The variant with terminal charges, +KIA7−, behaves like the corresponding uncharged peptide, KIA7, except higher concentrations of +KIA7− are needed for full helix formation.
The variation of the helix content of the KIA peptides (0.120 mM) upon heating is shown in SI Fig. 5. KIA5, KIA7Δ, KIA7I, and KIA10 in 0.2 M NaCl show small to moderate losses of helix content at elevated temperatures. In 0.2 M NaCl, KIA7, +KIA7−, KIA7F, and CG3KIA7 show broad sigmoidal transitions with apparent denaturation midpoints of 33°C, 40°C, 41°C, and 71°C, respectively, which resemble the thermal unfolding transitions of small, natural proteins. The thermodynamic parameters for KIA7, +KIA7−, KIA7F, and CG3KIA7 unfolding were obtained by applying a two-state model in which dissociation and denaturation are coupled and are shown in SI Table 2 and SI Fig. 6).
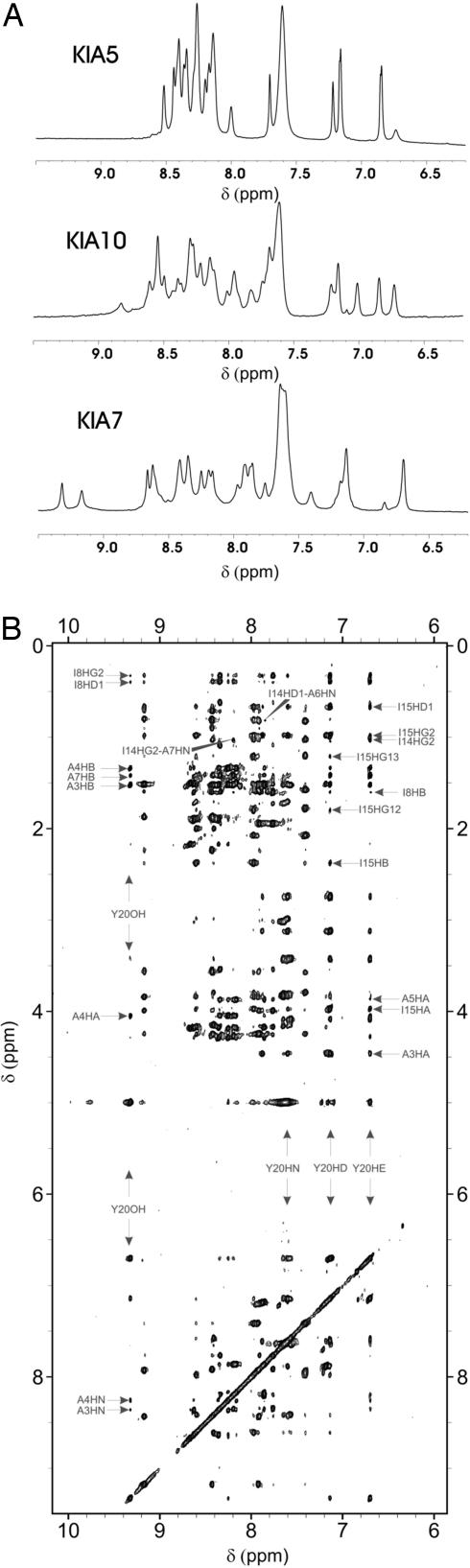
NMR spectroscopy confirms that KIA5, KIA10, KIA7Δ, and KIA7I are not well folded in 0.2 M NaCl at 5°C. The 1D 1H NMR spectrum of KIA5 in 0.2 M NaCl revealed sharp lines with little or no dispersion, which is characteristic of an unfolded protein (Fig. 1A). NOESY spectra confirmed the lack of significant conformational preferences and nonlocal contacts (SI Fig. 7). Similar results were observed for KIA7Δ, KIA7I, and +KIA7I− (data not shown). KIA10 yielded a 1D 1H NMR spectrum with broadened peaks typical of a partly ordered, but fluctuating, structure (Fig. 1A).
Fig. 1.
NMR spectroscopic characterization of KIA molecules in 200 mM NaCl, 10 mM Na/HAc, pH 5.0, 5°C. (A) 1D 1H NMR spectra of KIA5 (Top), KIA10 (Middle), and KIA7 (Bottom). (B) 2D 1H NOESY spectrum of KIA7. Some assignments are labeled.
KIA7 Forms a Stable, Well Ordered Four-Helix Bundle Protein.
KIA7 (Fig. 1A) and +KIA7−, KIA7F, and CG3KIA7 (data not shown) in 0.2 M NaCl gave rise to highly dispersed 1D 1H NMR spectra, which is a hallmark of a well ordered protein. Taking advantage of this dispersion, the 1H and 13C resonances of KIA7, KIA7F, and CG3KIA7 were completely assigned despite the highly repetitive sequence. The three peptides show strong sequential HN-HN NOEs extending along the sequence from the N terminus to Ala-17, and HN-Hα(i, i+3, i+4) NOEs and Hα and Cα conformational chemical shifts, which are all indicative of a fully helical structure. Numerous side-chain NOEs, such as those between Ile-8 and Tyr-20, for example, corroborate an antiparallel orientation for the KIA7 tetramer in solution (Fig. 1B).
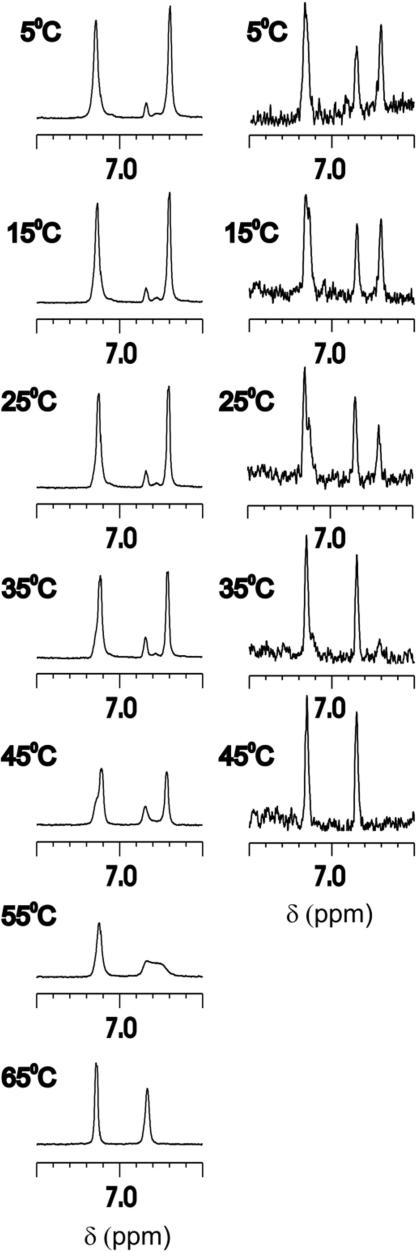
To study the heat-induced unfolding of KIA7, a series of 1D 1H NMR spectra were recorded at different temperatures (Fig. 2). Exchange between the structured and denatured forms is on the slow–intermediate time scale. Disperse peaks are seen until 50°C; >60°C, the sharp lines characteristic of unstructured polypeptides become dominant. Interestingly, the Tyr ring protons also undergo a heat-induced transition. The Tyr ring resonances of the folded state disappear >55°C. From the largest chemical shift difference (δdenatured − δfolded = 0.12 ppm ≡ 96 Hz at 800 MHz), the kinetics of the folding ↔ unfolding reaction is estimated to be <600 s−1, with a kinetic lifetime of >1.7 ms, which is equivalent to a >1.1-ms half-life. Very similar results are observed for +KIA7−, except the apparent midpoint of the transition occurs near 45°C (SI Fig. 8).
Fig. 2.
Thermal denaturation of KIA7 monitored by 1D 1H NMR at 1.2 mM (Left) and 50 μM (Right).
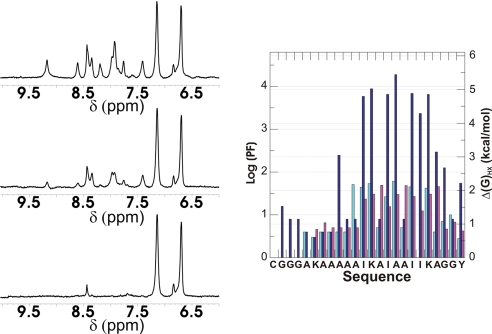
The stability of KIA7, KIA7F, and CG3KIA7 at individual amide hydrogens was probed by NMR-monitored H/D exchange in 0.2 M NaCl (Fig. 3). Little or no protection against exchange was observed in KIA7I. The exchange of hydrogen-bonded amide protons was found to be slowed 20- to 80-fold in KIA7 and KIA7F and 1,000- to 10,000-fold in CG3KIA7 relative to unstructured peptides. These protection factors correspond to free energies of stability (ΔGHX) of ≈2 kcal/mol for KIA7 and KIA7F and ≈5 kcal/mol for CG3KIA7. Chemical denaturation experiments were used to determine the free energy of conformational stability, ΔG°, and the m-value of KIA7, KIA7F, and +KIA7−. ΔG° was found to range from 14 kcal/mol for +KIA7− to 20 kcal/mol for KIA7. The urea m-value was ≈2,400 cal/mol·M for all these variants (SI Table 3, SI Text, and SI Fig. 9).
Fig. 3.
Hydrogen exchange of KIA7 monitored by 1D 1H NMR. (Left) Protonated KIA7 dissolved in D2O after 3 (Top), 113 (Middle), and 840 (Bottom) min. (Right) Left axis, protection factors; right axis, ΔGHX of KIA7 (light blue bars), KIA7F (violet bars), and CG3KIA7 (blue bars).
KIA7I Adopts Ordered Helical Structures in Stabilizing Conditions.
The results described above indicate that KIA7, +KIA7−, KIA7F, and CG3KIA7 are folded in 0.2 M NaCl, but KIA7I is not. To test whether this difference is caused by a decreased stability or a breakdown of tight hydrophobic core packing, additional experiments were performed in 0.5 M sodium sulfate, a well known stabilizer (SI Table 4 and SI Fig. 10). In these conditions, KIA7I adopts a highly helical, tetrameric structure, as gauged by CD and NMR T2 relaxation measurements. Moreover, KIA7I shows a cooperative thermal unfolding transition, induces a moderate enhancement in 1-anilino-8-naphthalene sulfonate fluorescence, and has significant protection against amide H/D exchange. These results suggest that in stabilizing conditions KIA7I adopts an ordered structure whose tight packing is maintained in the center but has loosened near the termini.
The Solution Structure of KIA7 Reveals a Well Ordered Structure Sharing Many Features of Natural Four-Helix Bundles.
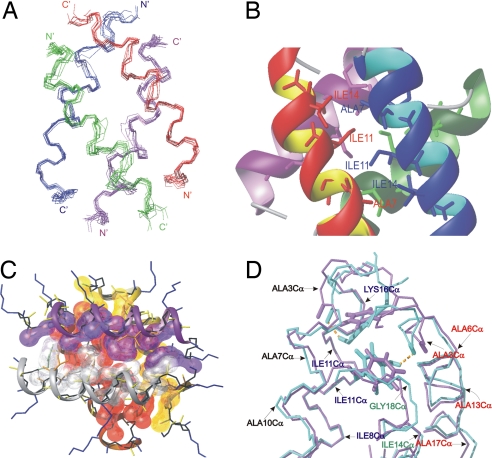
The tetrameric solution structure of KIA7 was performed on the basis of 1,516 NOE-derived distance constraints, 232 of which are interhelical. Ten structures were calculated with restrained molecular dynamics methods following the protocols described in Methods. The final structures exhibit low AMBER energy values, and the residual distance constraints violations were few and small. No distance constraint violation is >0.5 Å. A summary of the input distance constraints and the statistics of the final structures are shown in SI Table 5. The 10 final structures show a good convergence with an average pairwise rmsd of 0.9 Å for the backbone atoms. Most of the side chains of the core residues are well defined (Ile, Ala, and Tyr), although the side chains of the exposed Lys residues are mainly disordered, as they generally are in natural proteins. Similar results and structures were obtained for KIA7F. The NOESY spectrum of +KIA7− is very similar to that of KIA7, which indicates that it adopts a very similar structure (data not shown).
The superposition of the family of KIA7 structures is shown in Fig. 4A. The solution structure confirms that the tetramer consists of an antiparallel arrangement of four helices with relative interhelical angles of ≈110° and 180° in a “ridges-into-grooves” packing (5). All of the Lys side chains are solvent-exposed, forming a highly positively charged surface that prevents higher-order oligomerization. In the first turn of the helix, the residues are in the 3/10 conformation, as detected by their backbone angles and hydrogen-bonding pattern. A turn of 3/10 helix is commonly observed at the termini of helices in natural proteins. Whereas for residues 4–17, the backbone is in the α-helical conformation, the main chain of Gly-18, Gly-19, and Tyr-20 bends back to adopt a Gly helix termination motif, which is also frequently observed in helices in natural proteins (6). In KIA7F, the C-terminal Phe aromatic ring adopts a similar buried conformation and has a similar χ1 torsion angle as the Tyr ring in KIA7. There is a greater difference between the χ2 torsion angle of the Tyr and Phe rings, which reflects an alteration in the packing of these aromatic rings. This difference is likely caused by the Tyr hydroxyl group, which approaches the geometry necessary to form a hydrogen bond with the carbonyl oxygen of Ala-3.
Fig. 4.
Views of KIA7 and KIA7F NMR structures. (A) Superposition of the backbones of the 10 structures. (B and C) Two views of the packing of the Ala and Ile side chains. In C, lysine side chains are drawn as blue lines. (D) Superposition of the KIA7 (light blue) and KIA7F (violet) backbone structures and aromatic residues. The hydrogen bonds between the Tyr-20 hydroxyl and the Ala-3 carbonyl groups are shown as broken yellow lines.
The interaction between the helices is driven by the burial and packing of hydrophobic side chains, as illustrated in Fig. 4 B and C, where the close interaction between Ala-7, Ile-11, and Ile-14 can be observed. In the case of KIA7, the only putative tertiary or quaternary hydrogen bond present in the tetramer is between the hydroxyl group of Tyr-20 and the carbonyl group of the Ala-3 in the adjacent helix (Fig. 4D). In the case of KIA7F, no tertiary or quaternary hydrogen bonds are present.
Discussion
The Stability and Folding Kinetics of KIA7 Are Similar to Those of Natural Proteins.
The conformational stability of KIA7 and KIA7F, as determined by hydrogen exchange at 5°C, yields values of ≈2 kcal/mol. These stability values are modest; they are slightly higher than those determined for the lac repressor DNA binding domain (7) but less than the corresponding values for phage 434 Cro protein (8) and most other small, all-helical proteins. The conformational stability of CG3KIA7 is significantly higher, ≈5 kcal/mol, and similar to variants of IL-4, which also contain two disulfide bonds (9). It is remarkable that the stabilizing interactions in KIA7 are able to pay the substantial entropic penalty of oligomerization (4.5 kcal/mol) (10) and still be as stable as some single-chain proteins, which do not pay this penalty. The cost of tetramerization is also reflected in the remarkable differences in the stability values determined by hydrogen exchange near 1 mM peptide concentration and the standard free energies of stability obtained by denaturation experiments. These ΔG° values and the urea m-values are similar to those obtained for other oligomeric proteins (11). The lower stability of +KIA7− is probably caused by an unfavorable charge-helix macrodipole interaction at the N termini of the helices and the unfavorable desolvation energy of partially buried charges. Favorable charge–charge interactions between the charged groups at the N and C termini also probably contribute to the observed difference in stability between KIA7 and +KIA7−.
KIA7 and +KIA7− fold on the low millisecond timescale at 45°C to 55°C, as judged by the NMR chemical shift difference between the folded and unfolded states of the Tyr ring protons. This value is comparable to the folding rate of phage 434 Cro protein (12). Structural transitions in partly folded peptides are much faster; for example, the slow nucleation step in helix formation by poly l-ornithine is 105 times faster (13).
In addition to reporting on the folding rate, the Tyr ring protons give rise to a large number of NOEs, which indicates that the ring is ordered and plays an important role in the structure. The conformation adopted by the C-terminal extension, –Gly-Gly-Tyr is that of a Gly-terminal loop seen in natural proteins (6). In contrast, this same C-terminal extension has been shown to be flexible and lack a preferred structure in monomeric helical peptides (14).
The Presence of an Aromatic Ring at the C Terminus Is Required for Stability and Structure Formation in KIA7.
The solution structures show that the aromatic rings of Tyr-20 (KIA7) or Phe-20 (KIA7F) are well packed in the protein core, whereas peptides lacking the C-terminal aromatic group, such as KIA7I, appear to be largely unfolded in 0.2 M NaCl. The inability of KIA7I to adopt a well folded structure is probably caused by the loss of stabilizing contributions, ≈0.6 kcal/mol per aromatic >C-H group (15), from the hydrophobic effect when four Tyr (Phe) are replaced by four Ile. Nevertheless, under stabilizing conditions, i.e., 0.5 M sodium sulfate, KIA7I adopts a highly helical, tetrameric structure with hallmarks of a well packed hydrophobic center. The hydroxyl group of Tyr-20 in KIA7 forms a hydrogen bond with the carbonyl oxygen of Ala-3. The contribution of this hydrogen bond is not necessary to maintain the structure and stability of KIA7, because the KIA7F variant has essentially equivalent NMR spectra, a very similar 3D solution structure and a similar (hydrogen exchange and urea denaturation) or superior (CD-monitored thermal denaturation) conformational stability.
Structural Comparison with Other Four-Helical Bundles.
The tertiary structure adopted by KIA7 belongs to a class of four-helix bundles described previously as the “X-bundle” (16), and it has been observed as a structural motif within natural proteins such as the globins or thermolysin (see SI Table 6 for a more complete list), but not as an independent protein, as no close matches were reported in the above cited work or in a recent searches using the Dali and BLAST programs. Therefore, to our knowledge, KIA7 appears to be the first stable isolated X-bundle structure. The stability of KIA7 is striking, because Cohen and coworkers (16) predicted the X-bundle structure to be unstable on its own, because this structure leaves many open “holes” and unsatisfied “knobs” at the helix ends. In KIA7, these open holes at the helix ends are satisfied, at least in part, by the Gly termination motif, in which the C-terminal aromatic group caps the hydrophobic core.
The Origin of Specificity and Stability in Designed Proteins.
Many proteins that were designed by optimizing hydrophobic interactions do adopt a preferred tertiary structure but have mobile, loosely packed side chains. The ground-breaking work of DeGrado and coworkers (17–20) has led to the development of well packed helix bundle proteins with natural-like sequences and stabilizing interactions. In their work, and pioneering work from other laboratories, a specific fold has been achieved via the formation of tertiary hydrogen bonds (20–23), crystal contacts (18, 24), disulfide bonds (25, 26) or favorable charge–charge interactions (19, 20, 23, 24, 26, 27). Previous work toward simplifying proteins has shown that a native-like Src homology 3 fold can be achieved with chiefly five different residues (although 13 in total) (28), and a four-helix bundle protein can be formed from only nine different residues (29). In the latter case, from the Hecht laboratory (29), a high-resolution NMR solution structure revealed that the protein is stabilized mainly by hydrophobic interactions and van der Waals packing, although some stabilizing tertiary electrostatic interactions and hydrogen bonds are also present. Other work has demonstrated the importance of hydrophobic packing (30), particularly of Ile (31–33), in endowing proteins with a unique structure. Nakamura and colleagues (31) observed that replacing Ile by Leu produced a loss of tight packing in a small DNA binding protein domain. Moreover, Hu and collaborators (32) showed that substituting four Ile residues endows the GCN4 leucine zipper with a unique quaternary structure. Harbury and coworkers (33) designed a protein with a novel backbone fold, taking advantage of the specific packing of Ile. The ability of Ile to afford stability and structural uniqueness stems undoubtedly from its large, hydrophobic side chain, which has few allowed rotamers caused by β-branching (34). In the case of KIA7F, it is shown here that ridges-into-grooves packing of nonpolar residues, Ala and Ile, and a capping C-terminal aromatic group are sufficient to give rise to a stable, unique, native-like protein structure without any favorable charge–charge interactions, crystal packing interactions, or tertiary hydrogen bonds.
Modern proteins are endowed with a large set of amino acid residues and a variety of different stabilizing interactions. However, modern RNA ribozymes adopt specific conformations and achieve efficient and selective catalysis despite being restricted to a limited “alphabet” of residues and functional groups. On the basis of the present findings, we propose that prebiotic proteins could have also adopted unique and functional structures using a much more reduced set of amino acids and stabilizing interactions.
Implications for Prebiotic Biochemistry.
It is believed that amino acids, nucleotides, and other organic monomers emerged on Earth through either abiotic synthesis or transport by comets and meteorites (35). Nonenzymatic synthesis of oligopeptides and oligonucleotides has been accomplished under abiotic conditions (36). Although it is likely that the sequences of prebiotic peptides were generated at random, a hypothesis has been forwarded that the physicochemical environment of these peptides can exert “prebiological” selection pressure for sequences that fold into compact soluble structures (37). This hypothesis assumes that the prebiotic environment was hydrolytic toward the amide bond; hence, peptides that sequester their amide bonds avoid hydrolysis. Two methods of sequestering amide bonds are compaction of the chain into close-packed (including secondary structure formation) structure and aggregation. The concentration of the peptides in the primordial soup likely determined whether aggregated peptides or soluble folded peptides were preserved. Our results with the KIA7 series demonstrate the existence of sequences that fold into compact soluble structures using a highly reduced amino acid alphabet that is restricted to a subset of those present in the prebiotic Earth.
Methods
Peptide Synthesis and Purification.
Peptides were synthesized by using the Merrifield solid-phase method on a 9050 Plus PepSynthesizer (PerSeptive Biosystems, Framingham, MA) and later deprotected and purified by RP-HPLC as described (3). +KIA7− was purchased from the Caslo Laboratory (Lyngby, Denmark). The purity and identity of the peptides was further confirmed by MALDI-TOF mass spectroscopy. Peptide concentrations were estimated by UV absorbance for KIA peptides containing Tyr and ε(280 nm) = 1,390 cm−1·M−1 (38) or using a bicinchoninic acid (BCA) assay for KIA peptides lacking Tyr. Tyrosine-containing KIA peptides were used as standards in the BCA assays.
NMR Spectroscopy.
The KIA samples for NMR were dissolved in 90% H2O/10% D2O containing 0.20 M NaCl to reduce electrostatic repulsion between the charged lysine side chains, 10 mM NaAc/HAc (deuterated at the methyl group) to buffer the solution to pH 5, and 50 μM sodium 2,2-dimethyl-2-silapentane-5-sulfonate as the internal chemical shift standard. In some experiments, 0.50 M sodium sulfate was used instead of 0.20 M sodium chloride. The final peptide concentration was 1.2 mM for KIA7, CG3KIA7, +KIA7−, and KIA5, 4.5 mM for KIA7F, 2.8 mM for KIA7Δ, and 0.46 mM for KIA10. An additional sample of KIA7 containing only 50 μM peptide with the same buffer solution was also studied. NMR spectra were acquired on 800- and 600-MHz AV spectrometers (Bruker, Billerica, MA), the latter equipped with a cryoprobe. NOESY (39) spectra for structure determination were acquired in 0.2 M NaCl at pH 5, 5.0°C with mixing times of 75 and 150 ms. TOCSY (40) spectra were recorded with the standard MLEV-17 spin-lock sequence and a mixing time of 80 ms. In general, water suppression was achieved by including a WATERGATE (41) module in the pulse sequence before acquisition. Spectra were processed with Bruker software and analyzed with the program SPARKY (42). This program was used to aid the manual assignment of the NOESY cross-peaks and the quantitative evaluation of the NOE intensities. The formula k = 2 πΔδ, where k is the rate constant and Δδ is the chemical shift difference in Hz, was used to calculate the folding rate. NMR T2 relaxation measurements to gauge the oligomeric state of KIA molecules were performed by using a spin-echo pulse sequence (43).
Spectral Assignments and Structure Determination.
NMR assignment was performed by standard sequence-specific methods. A large number of distance constraints could be assigned despite the intrinsic ambiguity between intermolecular and intramolecular distances in multimeric structures. Most cross-peaks of the KIA7 and KIA7F peptides could be directly assigned on the basis of a preliminary model of a tetramer consisting of four antiparallel α-helices. Trial assignments were made for the few remaining cross-peaks and used in preliminary structure calculations. When a distance constraint was consistently violated in all of the resulting structures, an alternative assignment was considered. After several cycles of assignment and structure calculation, a consistent set of constraints was obtained. Any remaining cross-peaks that were still ambiguous were not included in the subsequent calculations. For KIA7, a total of 1,516 constraints, of which 232 were interhelical, were used.
Structures were calculated with the program DYANA 1.5 (44) and further refined with the molecular dynamics package AMBER 7.0 (45). The AMBER refinement consisted of an annealing protocol in vacuo followed by long trajectories where explicit solvent molecules were included. The temperature and the relative weights of the experimental constraints were varied during the in vacuo simulations according to standard annealing protocols used in our group (46, 47). The final structures were then hydrated with ≈4,000 water molecules and 12 chloride counter ions. The structures were first equilibrated for 130 ps using our standard equilibration process (48), and then subjected to 10 runs of 100-ps MD simulations in water using NMR restraints, periodic boundary conditions, and the Particle-Mesh-Ewald method (49). Simulations in water were performed in the isothermal/isobaric ensemble [P = 1 atm (1 atm = 101.3 kPa), T = 298 K] using SHAKE in all atoms and a 2-fs time step for integration of Newton's equations. MD-averaged structures were obtained by averaging the last 250 ps of individual trajectories and further relaxation of the structure. The same procedure was used to obtain a global MD-averaged structure, resulting from averaging 10 ns of independent MD trajectories.
The programs Procheck/NMR (50) and MOLMOL (51) were used in structure analysis, molecular graphic manipulations, and detection of possible hydrogen bonds. The criteria for hydrogen bond formation were: (i) donor-acceptor distance <3.0 Å and (ii) the donor-proton-acceptor angle <35°. Protein structure comparisons were performed by using the Dali server (52).
Hydrogen Exchange.
Hydrogen/deuterium exchange was initiated by adding a precooled D2O solution containing 0.20 M NaCl, 10 mM sodium acetate/acetic acid (pH 5.3), and 50 μM sodium 2,2-dimethyl-2-silapentane-5-sulfonate to lyophilized KIA peptides. Exchange was monitored by recording a series of 1D 1H spectra (16 transients with two dummy scans per spectrum, acquisition time = 45 s) at 5.0°C. The spectra were transformed, and the NH peaks were integrated by using Bruker Topspin software. The observed exchange rates of the NH protons were determined by fitting a single exponential decay function to the data by using a least-squares fitting algorithm.
The protection factor for each amide proton was calculated as the ratio of the observed exchange rate to the intrinsic exchange rate of hydrogen exchange, in short unstructured peptides using the parameters determined by Englander and coworkers (53). The free energy of stability is calculated by using the Gibbs equation, ΔG = −RT ln 1/P, where R is the gas constant, T is the absolute temperature, and P is the protection factor (= krc/kex), which is the inverse of the equilibrium constant, Kop (= ko/kc), between the open and closed states (54). This equation implicitly assumes that hydrogen exchange proceeds via the EXII mechanism (54), which has been frequently verified for proteins <pH 7. Moreover, simulation of KIA7 hydrogen exchange at pH 5.3, 5°C using reasonable values for the opening, closing, and intrinsic rate constants are consistent with the EXII mechanism.
Supplementary Material
Acknowledgments
We thank Dr. J. M. Pérez Cañadillas and Prof. J. Santoro for assistance with NMR pulse sequences; Prof. J. Santoro for additional support (Ministerio de Ciencia y Tecnología Grant BIO2002-00720); Prof. R. M. M. Brito and Prof. M. H. Hecht for helpful discussion and unpublished data; and Drs. M. A. Jiménez, J. L. Neira, and S. Padmanabhan and Profs. R. L. Baldwin, M. Bruix, M. Levitt, M. Libonati, C. N. Pace and M. Rico for critical comments. This work was supported by Spanish Ministry of Science and Education Grant CTQ2004-08275, Alzheimer's Association Grant NIRG-04-1083, Consejo Superior de Investigaciones Científicas Grant 200580F0131, and a grant from the National Science and Engineering Research Council of Canada.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2jo4 and 2jo5).
This article contains supporting information online at www.pnas.org/cgi/content/full/0706876104/DC1.
References
- 1.Miller SL. Science. 1953;117:528–529. doi: 10.1126/science.117.3046.528. [DOI] [PubMed] [Google Scholar]
- 2.Matthews CN, Moser RE. Nature. 1967;215:1230–1234. doi: 10.1038/2151230a0. [DOI] [PubMed] [Google Scholar]
- 3.Boon CL, Frost D, Chakrabartty A. Biopolymers. 2004;76:244–257. doi: 10.1002/bip.20074. [DOI] [PubMed] [Google Scholar]
- 4.Frost DWH, Yip CM, Chakrabartty A. Biopolymers. 2005;80:26–33. doi: 10.1002/bip.20188. [DOI] [PubMed] [Google Scholar]
- 5.Chothia C, Levitt M, Richardson D. J Mol Biol. 1981;145:215–250. doi: 10.1016/0022-2836(81)90341-7. [DOI] [PubMed] [Google Scholar]
- 6.Aurora R, Srinivasan R, Rose GD. Science. 1994;264:1126–1130. doi: 10.1126/science.8178170. [DOI] [PubMed] [Google Scholar]
- 7.Felitsky DJ, Record MT. Biochemistry. 2003;42:2202–2217. doi: 10.1021/bi0270992. [DOI] [PubMed] [Google Scholar]
- 8.Padmanabhan S, Laurents DV, Fernández AM, Elias-Arnanz M, Ruiz-Sanz J, Mateo PL, Rico M, Filimonov VV. Biochemistry. 1999;38:15536–15547. doi: 10.1021/bi991757+. [DOI] [PubMed] [Google Scholar]
- 9.Vaz DC, Rodrigues JR, Sebald W, Dobson CM, Brito RMM. Protein Sci. 2006;15:33–44. doi: 10.1110/ps.051593306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamura A, Privolov PL. J Mol Biol. 1997;273:1048–1060. doi: 10.1006/jmbi.1997.1368. [DOI] [PubMed] [Google Scholar]
- 11.Panse VG, Swaminathan CP, Aloor JJ, Surolia A, Varadarajan R. Biochemistry. 2000;39:2362–2369. doi: 10.1021/bi992484l. [DOI] [PubMed] [Google Scholar]
- 12.Laurents DV, Corrales S, Elias-Arnanz M, Sevilla P, Rico M, Padmanabhan S. Biochemistry. 2000;39:13963–13973. doi: 10.1021/bi001388d. [DOI] [PubMed] [Google Scholar]
- 13.Hammes GG, Roberts PB. J Am Chem Soc. 1969;91:1812–1816. doi: 10.1021/ja01035a036. [DOI] [PubMed] [Google Scholar]
- 14.Chakrabartty A, Kortemme T, Padmanabhan S, Baldwin RL. Biochemistry. 1993;32:5560–5565. doi: 10.1021/bi00072a010. [DOI] [PubMed] [Google Scholar]
- 15.Pace CN. J Mol Biol. 1992;226:29–35. doi: 10.1016/0022-2836(92)90121-y. [DOI] [PubMed] [Google Scholar]
- 16.Harris NL, Presnell SR, Cohen FE. J Mol Biol. 1994;236:1356–1368. doi: 10.1016/0022-2836(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 17.DeGrado WF, Wasserman ZR, Lear JD. Science. 1989;243:622–628. doi: 10.1126/science.2464850. [DOI] [PubMed] [Google Scholar]
- 18.Hill CP, Anderson DH, Wesson L, DeGrado WF, Eisenberg D. Science. 1990;249:543–546. doi: 10.1126/science.2382133. [DOI] [PubMed] [Google Scholar]
- 19.Walsh STR, Cheng H, Bryson JW, Roder H, DeGrado WF. Proc Natl Acad Sci USA. 1999;96:5486–5491. doi: 10.1073/pnas.96.10.5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geremia S, Di Costanzo L, Randaccio L, Engel DE, Lombardi A, Nastri F, DeGrado WF. J Am Chem Soc. 2005;127:17266–17276. doi: 10.1021/ja054199x. [DOI] [PubMed] [Google Scholar]
- 21.Lumb KJ, Kim PS. Biochemistry. 1995;34:8642–8648. doi: 10.1021/bi00027a013. [DOI] [PubMed] [Google Scholar]
- 22.Oakley MG, Kim PS. Biochemistry. 1998;37:12603–12610. doi: 10.1021/bi981269m. [DOI] [PubMed] [Google Scholar]
- 23.Rudresh U, Ramagopal A, Inai Y, Goel S, Sahal D, Chauhan VS. Structure (London) 2004;12:389–396. doi: 10.1016/j.str.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 24.Kramer RZ, Venugopal MG, Bella J, Mayville P, Brodsky B, Berman HM. J Mol Biol. 2000;301:1191–1205. doi: 10.1006/jmbi.2000.4017. [DOI] [PubMed] [Google Scholar]
- 25.Yan Y, Erickson BW. Protein Sci. 1994;3:1069–1073. doi: 10.1002/pro.5560030709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Domingues H, Cregut D, Sebald W, Oschkinat H, Serrano L. Nat Struct Biol. 1999;6:652–656. doi: 10.1038/10706. [DOI] [PubMed] [Google Scholar]
- 27.Dolphin GT, Baltzer L. Folding Des. 1997;2:319–330. doi: 10.1016/S1359-0278(97)00043-6. [DOI] [PubMed] [Google Scholar]
- 28.Riddle DS, Santiago JV, Bray-Hall ST, Doshi N, Grantcharova VP, Yi Q, Baker D. Nat Struct Biol. 1997;4:805–809. doi: 10.1038/nsb1097-805. [DOI] [PubMed] [Google Scholar]
- 29.Wei Y, Kim S, Fela D, Baum J, Hecht MH. Proc Natl Acad Sci USA. 2003;100:13270–13273. doi: 10.1073/pnas.1835644100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dahiyat BI, Mayo SL. Proc Natl Acad Sci USA. 1997;94:10172–10177. doi: 10.1073/pnas.94.19.10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Furukawa K, Oda M, Nakamura H. Proc Natl Acad Sci USA. 1996;93:13583–13588. doi: 10.1073/pnas.93.24.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu H, Celinski SA, Scholtz JM, Hu JC. Protein Sci. 2001;10:24–33. doi: 10.1110/ps.30901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harbury PB, Plecs JJ, Tidor B, Alber T, Kim PS. Science. 1998;282:1462–1467. doi: 10.1126/science.282.5393.1462. [DOI] [PubMed] [Google Scholar]
- 34.Dunbrack RLJ, Karplus M. Nat Struct Biol. 1994;1:334–339. doi: 10.1038/nsb0594-334. [DOI] [PubMed] [Google Scholar]
- 35.Oro J, Miller SL, Lazcano A. Annu Rev Earth Planet Sci. 1990;18:317–356. doi: 10.1146/annurev.ea.18.050190.001533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oro J, Stephen-Sherwood E. Orig Life. 1974;5:159–172. [PubMed] [Google Scholar]
- 37.Abkevich VI, Gutin AM, Shakhnovich EI. Proc Natl Acad Sci USA. 1996;93:839–844. doi: 10.1073/pnas.93.2.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edelhoch H. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 39.Kumar A, Ernst RR, Wüthrich K. Biochem Biophys Res Commun. 1980;95:1–6. doi: 10.1016/0006-291x(80)90695-6. [DOI] [PubMed] [Google Scholar]
- 40.Bax A, Davis DG. J Magn Reson. 1985;65:355–360. [Google Scholar]
- 41.Piotto M, Saudek V, Sklenar V. J Biomol NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 42.Goddard TD, Kneller DG. SPARKY 3. San Francisco: University of California; 2007. [Google Scholar]
- 43.Sklenar V, Bax A. J Magn Reson. 1987;74:469–479. [Google Scholar]
- 44.Güntert P, Mumenthaler C, Wüthrich K. J Mol Biol. 1997;273:282–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 45.Ferguson G, Seibel GL, Kollman PA. Comput Phys Commun. 1995;91:1–41. [Google Scholar]
- 46.Escaja N, Pedroso E, Rico M, González C. J Am Chem Soc. 2000;122:12732–12742. [Google Scholar]
- 47.Soliva R, Monaco V, Gómez-Pinto I, Meeuwenoord NJ, Marel GA, Boom JH, González C, Orozco M. Nucleic Acids Res. 2001;29:2973–2985. doi: 10.1093/nar/29.14.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shields GC, Laughton CA, Orozco M. J Am Chem Soc. 1997;119:7463–7469. [Google Scholar]
- 49.Darden T, York D, Pedersen L. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 50.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 51.Koradi R, Billeter M, Wüthrich K. J Mol Graphics. 1996;14:51–52. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 52.Holm L, Sander C. Trends Biochem Sci. 1995;20:206–209. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 53.Bai Y, Milne JS, Mayne L, Englander SW. Proteins Struct Funct Genet. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hvidt A, Nielsen SO. Adv Protein Chem. 1966;21:287–386. doi: 10.1016/s0065-3233(08)60129-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.