Abstract
The human intragranulocytic bacterium Anaplasma phagocytophilum promotes variation of P44s, which are surface-exposed proteins encoded by a p44 multigene family. In the present study, the specific p44 gene expression loci in four strains of A. phagocytophilum were identified and it was determined that each consisted of four tandem genes, tr1, omp-1X, omp-1N, and p44. A putative σ70-type promoter was found upstream of tr1. The p44 genes include a central hypervariable region flanked by conserved regions. The hypervariable region sequence in the p44 expression locus was duplicated and, regardless of the expression status, conserved at another locus in both low- and high-passage cell cultures of strain NY-37. No significant differences in the hypervariable region were found when we compared p44 sequences, at the level of cDNA, within the expression locus and within other loci in the genomes of strains NY-37 and HZ. Similarly, in cDNA isolated from patients and from assorted cultures of strains NY-31, NY-36, and NY-37, hypervariable regions of 450 deduced amino acid sequences of various p44s within each strain were found to be identical, as were those of p44 sequences in the genome of strain HZ. These data suggest that variations in p44 sequences at the level of the p44 expression locus occur through unidirectional conversion of the entire (nonsegmental) p44 hypervariable region including flanking regions with a corresponding sequence copied from one of the conserved donor p44 genomic loci. The data suggest that the P44 antigenic repertoire within the hypervariable region is restricted.
Human granulocytic ehrlichiosis (HGE) is an acute systemic and potentially fatal disease that has been reported in the United States and Europe (1, 9, 12, 32, 39). The agent of HGE is Anaplasma phagocytophilum, a gram-negative obligatory intracellular bacterium that infects granulocytes. This pathogen can be transmitted to humans via the bite of an arthropod vector (the Ixodes species of ticks) from wild animal reservoirs (Peromyscus leucopus, a white-footed mouse, or other wild animals) (37). P44s are surface-exposed immunodominant 44-kDa major outer membrane proteins of A. phagocytophilum that are encoded by polymorphic multigene family p44 (43). p44s consist of a central hypervariable region of approximately 280 bp flanked by conserved sequences of 100 to 500 bp (21, 43). Many, but not all, p44s lack ATG start codon (43, 44). The products of hypervariable regions of various p44s have 23 to 80% amino acid identity (21). Although the genome of A. phagocytophilum is relatively small (1.5 Mb), it includes a large number of genes that are paralogous to p44 (43). p44s were found to be differentially expressed in cells cultured at different temperatures and in different host cells, including ticks, experimentally infected mammals, and HGE patients (3, 8, 13, 15, 21, 43, 44).
p44 is homologous to msp2 from Anaplasma marginale, a bovine intraerythrocytic agent (43). It was reported that the segmental gene conversion of multiple msp2s at a single expression locus confers unlimited variation of expressed msp2 chimeras or mosaics (2, 5, 6). Recently, Barbet et al. proposed the same mechanism for the expression of variable p44s in A. phagocytophilum (3). If segmental gene conversion occurs, the sequences of the hypervariable regions of p44 in the expression locus would be chimeras or mosaics derived from more than two donor p44s located elsewhere within the same genome. This is inconsistent with our previous data showing that the hypervariable region sequences in several p44 paralogs cloned from different genomic loci are identical in the corresponding regions of p44 cDNAs (43, 44). In the present study, we tried to resolve this conundrum by testing the msp2 (p44) segmental gene conversion theory in various strains used in our previous studies. Our data suggest that, instead of segmental gene conversion, the entire hypervariable region is replaced to alter p44 expression. Understanding the versatility in the structure and expression of surface component P44s may provide insights into the adaptive capabilities of A. phagocytophilum in mammalian and tick hosts and host clearance mechanisms.
MATERIALS AND METHODS
Cultivation and purification.
Three strains of A. phagocytophilum (NY-31, NY-36, and NY-37) were isolated from patients in the state of New York during the summer of 2000, as described previously (21). An HZ strain was isolated from a patient in New York during the summer of 1995 (35). Three Minnesota strains (MN1, MN2, and MN9) were generously provided by J. Goodman from the University of Minnesota. All A. phagocytophilum strains were cultivated in HL-60 cells at 37°C in 5% CO2-95% air, by using RPMI 1640 medium (Invitrogen, Carlsbad, Calif.) supplemented with 10% heat-inactivated fetal bovine serum (Atlanta Biologicals, Norcross, Ga.). Each strain isolated in the year 2000 was maintained in two cultures: a low-passage cell culture (no more than 10 passages; each passage is 3 to 4 days of culture) and a high-passage cell culture (greater than 20 passages). For the HZ strain, the number of passages exceeded 100. The infection rate was checked daily by Diff-Quik staining of cytocentrifuged cells (Baxter Scientific Products, Obetz, Ohio). After the designated number of passages, the cells were harvested (when the infection rate reached 80 to 100%) and used to prepare total RNA and DNA. A. phagocytophilum organisms purified from infected cells by Sephacryl S-1000 (Amersham Pharmacia Biotech, Piscataway, N.J.) column chromatography were used for the preparation of genomic DNA, as previously described (34).
Reverse transcription-PCR (RT-PCR) analysis.
Total RNA was extracted from 5 × 106 A. phagocytophilum-infected HL-60 cells (80 to 100% infection) with the TRIzol reagent (Invitrogen). The RNA was further purified with the RNeasy minikit (Qiagen, Valencia, Calif.). After DNase I treatment, the isolated RNA (5 μg) was heated at 70°C for 10 min. Samples were then subjected to reverse transcription at 42°C for 50 min in a 20-μl reaction mixture containing 0.5 mM (each) deoxynucleotide triphosphate (dNTP), 200 U of SuperScript II reverse transcriptase (Invitrogen), 200 ng of random hexamers, and 3 mM MgCl2. PCR was performed in a 50-μl reaction mixture containing 4 μl of the cDNA product, 10 pmol of each primer (primer sets 1, 12, 13, 14, and 15, as shown in Table 1), 0.2 mM (each) dNTP, 5 U of Taq DNA polymerase, and 1.5 mM MgCl2. PCR conditions included 3 min of denaturation at 94°C, followed by 35 cycles consisting of 1 min of denaturation at 94°C, 1 min of annealing at 55°C, and 1 min of extension at 72°C. PCR products were purified from a gel and cloned into a pCRII vector (Invitrogen). Twenty to 56 cDNA clones were randomly selected from the transformants and sequenced on an ABI 373XL Stretch DNA sequencer with the ABI PRISM BigDye Terminator Cycle Sequencing Reaction kit.
TABLE 1.
Oligonucleotide primers used in RT-PCR, 5′RACE, and genomic Southern blotting
| Primer set no. | Target | Purpose | Size (bp) | Sequencea (5′-3′) of:
|
||
|---|---|---|---|---|---|---|
| 5′ primer (5′RACE or genome walking primer 2) | 3′ primer (5′RACE or genome walking primer 1) | RT primer | ||||
| 1 | Multiple p44 transcripts | RT-PCR | 420 | CTGCTCTKGCCAARACCTC | CAATAGTYTTAGCTAGTAACC | Random hexamer |
| 2 | p44-1 (transcript) | 5′RACE | TTACTCAAATATTTATGAGCAGCCT | AATGCTATTGCTGTTGTTACTCGA | GTTACTATTAGGTGC | |
| 3 | p44-28 (transcript) | 5′RACE | TTTAAAGACTGCACGGTACCCG | TTTGTGTTAGTATTTCCCGATTTCT | GTTGTCGTTAGGTTG | |
| 4 | p44-18 (transcript) | 5′RACE | TTTTGGTTCTTACCACCACACAAT | GTACCATCACTCAATGTGTTAGAA | TCGATTTAGACGTTTC | |
| 5 | Upstream of tr1 (transcript) | 5′RACE | TTGTACTTGTTGGAATGTAATCCCTAA | ACCTGCTTATCACTATACGGTTAGTA | TTCCTCTAATGACGCTTCAT | |
| 6 | Upstream of p44 (p44 expression locus) | 5′RACE | CGCACATCTTTCTCTAGAACAAATA | ACTAGGGTATTTCTTCTCCGTTATTA | GGATTCAACACTCCCCAAGACA | |
| 7 | Upstream of p44 in p44 expression locus (genomic DNA) | Southern blotting (p44 expression locus) | ATTGGACTTTTGAGCTGTCTT | ACTTTTCAACTTCTCCAAATTC | ||
| 8 | p44-28 (gene) | Southern blotting | 230 | GGACGAAGCGGAAGGCTGGT | AGGTTGAGGCTTTGTGTTAGT | |
| 9 | p44-1 (gene) | Southern blotting | 247 | GAAGGTTTGTAGTGGAAAGCA | ATGCTCCAACTACAATGCTAT | |
| 10 | Upstream of p44 in p44 expression locus (genomic DNA) | Genome walking | CGCACATCTTTCTCTAGAACAAATA | ACTAGGGTATTTCTTCTCCGTTATTA | ||
| 11 | Downstream of p44-28 in p44 expression locus (genomic DNA) | Genome walking | AAGAAGGAATTTGGAGAAGTTGAAAA | GGCTGGAGTGAGGAGACGAAG | ||
| 12 | omp-1N-p44 (transcript) | RT-PCR, DNA PCR | 1,220 | TTGAGGGTGTTGACACTCTGCT | CAATAGTYTTAGCTAGTAACC | Random hexamer |
| 13 | omp-1X-omp-1N (transcript) | RT-PCR | 652 | TGGATTGGAATGTGGTGTTCGAC | CGCAGCGCTAGGGTGAAACT | Random hexamer |
| 14 | tr1-omp-1X (transcript) | RT-PCR | 494 | GTAGATTATGAGACTTCCTATAGG | TGAAAAATCCACAGGTTTTGGCGAA | Random hexamer |
| 15 | ndk-tr1 (transcript) | RT-PCR | 305 | AGAAAGCGTCCTCTCTTGCACTC | CTTCCTATAGTTATGGGGATGGAATTT | Random hexamer |
| 16 | Upstream of p44-28 (gene) | Genome walking | TTTAAAGACTGCACGGTACCCG | TTTGTGTTAGTATTTCCCGATTTCT | ||
| 17 | Downstream of p44-28 (gene) | Genome walking | AGAAATCGGGAAATACTAACACAAAG | CGGGTACCGTGCAGTCTTTAAAAG | ||
| 18 | p44-upstream sequence (p44 expression locus, genomic DNA) | DNA PCR | 1,082 | ATTGGACTTTTGAGCTGTCTT | CAATAGTYTTAGCTAGTAACC | |
K = T and G; R = A and G; Y = C and T.
DNA PCR analysis.
Total DNA was extracted from 5 × 106 A. phagocytophilum-infected HL-60 cells with the QIAamp DNA minikit (Qiagen). PCR was performed (primer sets 12 and 18, as shown in Table 1) under the same conditions as those described above for RT-PCR.
5′RACE.
The experiment involving 5′ rapid amplification of cDNA ends (5′RACE) was performed according to the protocol provided by the manufacturer (Invitrogen). DNase I-treated total RNA (3 μg) was reverse transcribed with Superscript II and gene-specific primers at 42°C for 50 min (primer sets 2, 3, 4, and 5, as shown in Table 1). For cDNA synthesis of long transcripts (>3 kb), the DNase I-treated total RNA (5 μg) was reverse transcribed by using a Thermotranscript kit (Invitrogen) according to the manufacturer's recommended protocol at 54°C for 50 min with a gene-specific primer (primer set 6, as shown in Table 1). The cDNA was tailed by using terminal transferase to add cytidine or adenosine residues at the 3′ end and then amplified by PCR with a second gene-specific primer and either an oligo(dG)- or oligo(dT)-linked amplification primer. The PCR conditions included 35 cycles of 1 min of denaturation at 94°C, 1 min of annealing at 57°C, and 1 min of extension at 72°C. Primary PCR products were further amplified by a nested gene-specific primer and the amplification primer without an oligo(dG) or oligo(dT) anchor. The secondary PCR products were purified and cloned. Inserts of 20 clones were sequenced for each sample.
Genomic Southern blot analysis.
Genomic DNA of A. phagocytophilum was extracted from purified organisms as described elsewhere (29). Briefly, samples were subjected to lysis with sodium dodecyl sulfate, pronase digestion, phenol-chloroform extraction, and ethanol precipitation. The purified genomic DNA was then digested with either EcoRI, PstI, XbaI, or SpeI. PCR products amplified with primer sets 7, 8, and 9 (Table 1) were labeled with [α-32P]dCTP by using Ready-To-Go DNA labeling beads (Amersham Pharmacia Biotech), and the labeled PCR products were then used as DNA probes. Hybridization was carried out under high-stringency conditions (65°C) in Rapid hybridization buffer (Amersham Pharmacia Biotech), as described elsewhere (29). After being washed, the membrane was exposed to Hyperfilm to visualize the locations of the hybridized bands (Amersham Pharmacia Biotech).
Genome walking.
Based on the unique sequences of identified p44 transcripts, unknown 5′- and 3′-end flanking regions were amplified by using a Universal GenomeWalker kit (Clontech Laboratories, Palo Alto, Calif.) or by multiplex restriction site PCR (2, 16, 40). The primers used for genome walking (primer sets 10, 11, 16, and 17) are shown in Table 1. The PCR products were cloned into a pCRII vector or a pCR-XL-TOPO vector and subjected to DNA sequencing.
Western blot analysis.
The P44-28 oligopeptide REKSGNTNTKPQOND, Pep28, was synthesized (Alpha Diagnostic, San Antonio, Tex.). Antiserum against Pep28 was generated by immunizing rabbits with keyhole limpet hemocyanin-conjugated Pep28 (ProSci, Inc., Poway, Calif.). P44-1 and P44-18 oligopeptides and antisera were generated as described elsewhere (43). The monoclonal antibody 5C11 recognizes the N-terminal conserved region of P44 (17), and Western blot analysis was performed as described elsewhere (21, 33).
Sequence analysis.
The protein signal sequence was analyzed with the SignalP program (www.cbs.dtu.dk). Sequence assembling, alignments, and analysis were done with SeqMan, MegAlign, and MapDraw programs in the DNAStar software. The genome of A. phagocytophilum strain HZ was sequenced at The Institute for Genomic Research, and the sequences are available at http://www.tigr.org. The GenBank accession number of the A. phagocytophilum strain HZ unfinished whole genome is NC_004351.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the p44 expression loci in A. phagocytophilum strains are as follows: NY-37, AY137510 and AY319262 (p44-28 and p44-1 in the expression locus, respectively); NY-36, AY319262; NY-31, AY319264; HZ, AY319265 (p44-18 in the expression locus). The accession number for the p44-1/p44-18 gene locus is AY151054, and that for the p44-50/p44-28 pseudogene locus is AY151053.
RESULTS
Characterization of the genetic locus for p44 transcription.
To identify the expression locus for the major p44 transcripts in low-passage cell cultures, the predominant p44 mRNA species were identified by RT-PCR with primers corresponding to the 5′ and 3′ conserved flanking regions (primer set 1 in Table 1). The resulting RT-PCR products were approximately 420 bp in length and contained central hypervariable regions of approximately 280 bp. No amplicons were detected in the absence of reverse transcriptase, indicating that the RNA preparation was not contaminated by genomic DNA. These PCR products were used to isolate cDNAs, and 40 to 50 clones in strains NY-31, NY-36, and NY-37 were subjected to DNA sequencing. We previously demonstrated that relative ratios of p44 cDNA clones isolated are proportional to the levels of transcription of the p44 paralogs (44). In strains NY-31 and NY-36, 47 of 56 and 38 of 42 cDNA clones, respectively, were found to encode P44-28 in the low-passage cell culture. In strain NY-37, 34 of 41 clones were found to encode P44-1.
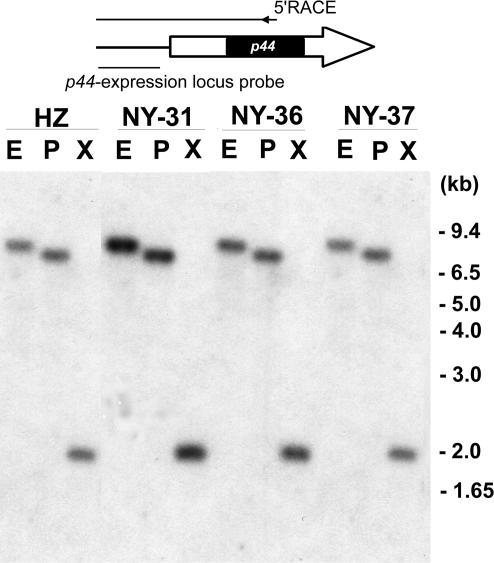
To identify the p44 expression locus, we analyzed the upstream sequences of the major transcript species by 5′RACE in strains NY-31, NY-36, NY-37, and HZ. p44-28 was the major mRNA species in the low-passage cell cultures of strains NY-31 and NY-36 and in the high-passage cell culture of strain NY-37, p44-1 was the major species in the low-passage culture of strain NY-37, and p44-18 was the major species in the high-passage cell culture of strain HZ (passage numbers >100). It was found that all upstream sequences up to 199 bp from the start codon of each transcript species were identical, except for a 1-bp difference in the upstream sequence of p44-18 in strain HZ. These results suggest that all p44 major transcript species in the four A. phagocytophilum strains were expressed at identical genomic loci. To confirm this hypothesis, Southern blot analysis was performed on genomic DNA from each strain using a DNA probe specific to the common p44 upstream sequence (primer set 7; Table 1). The DNA used in these experiments was derived from the same samples from which RNA was extracted for 5′RACE analysis. The genomic DNA from the four strains (HZ, NY-31, NY-36, and NY-37) was digested with either EcoRI, PstI, or XbaI. The relative position of the probe (bp −199 to −42) is shown in Fig. 1. The DNA probe hybridized to a single fragment in samples of genomic DNA treated with each of these restriction enzymes, and hybridization patterns for the four strains were identical (Fig. 1). The hybridization patterns indicated that this genetic locus was conserved in the various human isolates of A. phagocytophilum, including one isolated 5 years earlier (HZ). Since the ank gene sequences of A. phagocytophilum isolates derived from the northeastern United States were shown to be distinct from Minnesota and Wisconsin A. phagocytophilum isolates (25), we examined whether this expression locus was present in the genomes of three A. phagocytophilum strains isolated in Minnesota by DNA PCR using primers specific to the p44 expression locus (primer sets 12 and 18; Table 1 and Fig. 2). The target region corresponding to the p44 expression locus in each of the three Minnesota isolates was amplified (data not shown). Barbet et al. also showed that A. phagocytophilum strains from New York and Wisconsin had the same p44 expression locus (3). These findings strongly suggest that the same p44 expression locus is present in all strains of A. phagocytophilum, regardless of their geographic origins.
FIG. 1.
Genomic Southern blot analysis of a locus for variable p44 transcription in four strains of A. phagocytophilum. A schematic representation of a p44 transcript is shown at the top. Entire arrow, coding region of the transcript; black area within the arrow, hypervariable region of the p44 multigene family; white areas, 5′- and 3′-end conserved regions. Line to the left of the arrow, 5′-end upstream region of the transcript. The relative position of the p44 expression locus-specific DNA probe is shown. A. phagocytophilum strains NY-31, NY-36, NY-37, and HZ were digested with EcoRI (E), PstI (P), and XbaI (X), as indicated. Numbers on the right indicate molecular sizes.
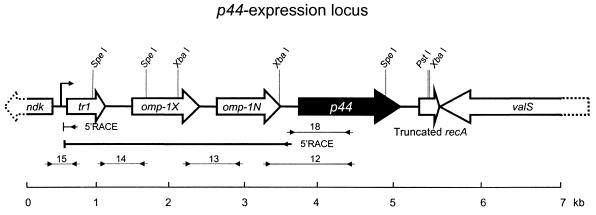
FIG. 2.
Schematic representation of a genetic region including the p44 expression locus of A. phagocytophilum. Black arrow, coding region of p44 transcription (p44 expression locus); white arrows, genes located in close proximity to the expression locus. For both types of arrows, the direction indicates the direction of orientation. Lines with facing arrows at each end, regions of RT-PCR in the transcriptional analysis; bent arrow located upstream of tr1, putative transcriptional start site for polycistronic transcription linked to the expression locus, which was deduced by 5′RACE analysis (arrow labeled 5′RACE); dotted lines, portions of the ndk and valS genes that have not yet been sequenced.
To further characterize this common expression locus, we used genome walking methods based on the unique upstream sequence identified by 5′RACE to analyze unknown genes flanking the expression locus in strains NY-31, NY-36, and NY-37. Since the sequences obtained by the initial genome walking procedure in the NY-31, NY-36, and NY-37 strains were identical, we used strain NY-37 for further analyses. We found a total of six ORFs (ndk, tr1, omp-1X, omp-1N, truncated recA, and valS) flanking the expression locus for p44 paralogs within a 7-kb region of the genome, as illustrated in Fig. 2. The tr1 gene encoded a putative transcriptional regulator which had a characteristic helix-turn-helix DNA binding motif in approximately 30 to 80 amino acid residues at the N-terminal region and was orthologous to Ehrlichia chaffeensis tr1 (BlastP E value of 2e-37), previously designated un1 (27). The omp-1X and omp-1N genes were orthologous to E. chaffeensis omp-1X and omp-1N (BlastP E-values of 9e-25 and 2e-8, respectively). The predicted molecular sizes of the A. phagocytophilum Tr1, OMP-1X, and OMP-1N proteins were 21,000, 33,452, and 29,328 Da, respectively, including putative signal peptides from amino acid positions 1 to 23 for OMP-1X and from 1 to 24 for OMP-1N, as predicted by the SignalP program. Tr1 lacked a signal peptide sequence. The 5′ end of the partially sequenced open reading frame (ORF) which encoded the N-terminal 156 amino acid residues of a nucleoside diphosphate (NDP) kinase (BlastP E value of 4e-43 with Myxococcus xanthus NDP kinase) was located 179 bp upstream of tr1 in an opposite transcriptional orientation. The 3′ end of the partially sequenced ORF encoded the C-terminal 413 amino acid residues of a valine tRNA synthase (BlastP E-value of 7e-40 with Rickettsia prowazekii ValS) and was located downstream of p44 in the expression locus in an opposite transcriptional orientation. There were no ORFs within a 213-bp intergenic region between a p44 paralog and omp-1N, within a 235-bp intergenic region between omp-1N and omp-1X, or within a 339-bp intergenic region between omp-1X and tr1.
The p44 expression locus also included a truncated recA (265 bp) with a start codon (ATG) 294 bp downstream of p44 and a stop codon (TAA). This fragment encoded the N-terminal 88 amino acid residues of the full-length RecA (357 amino acids) and is located elsewhere in the genome. The full-length recA gene was highly expressed by all four A. phagocytophilum strains in HL-60 cell cultures, as determined by RT-PCR (data not shown). The full-length recA in A. phagocytophilum strain HZ was highly homologous to recA in Escherichia coli (BlastP E-value of e-108). The A. phagocytophilum truncated and full-length RecAs each had an ATP binding site consensus sequence GPESSGKT, which corresponds to (G/A)XXXXGK(T/S) of the Walker A box, also referred to as the P loop (24). The putative DNA binding sites were located at amino acid residues 150 to 176 and 190 to 227, which were highly conserved in A. phagocytophilum and E. coli RecA proteins. Because DNA binding activity is essential for the function of RecA proteins (24) and because the recA fragment downstream of the p44 expression locus lacked DNA binding sequences, this RecA fragment is likely to be nonfunctional.
We designated the area consisting of tr1, omp-1X, omp-1N, and a downstream transcriptionally active p44 the p44 expression locus (GenBank accession number AY137510). Barbet et al. (3) independently identified omp-1N, the p44 paralog, and the 3′ terminus recA truncated in A. phagocytophilum strains NY-18, Webster, and HGE2 and in a strain from HGE patient no. 2. The sequence alignment of this region showed that the upstream and downstream sequences of p44 paralogs were 99.6 to 99.9% identical to the respective sequences in strains NY-31, NY-36, NY-37, and HZ.
Transcription at the p44 expression locus.
In the Florida and South Idaho strains of A. marginale, msp2 transcripts were shown to be polycistronically transcribed with ORF4 (OpAG3), ORF3 (OpAG2), and ORF2 (OpAG1) (2, 22). Because tr1, omp-1X, om-1N, and p44 were found in the same transcriptional orientation and because their intergenic spaces were relatively short, we considered the possibility that these genes could be polycistronically transcribed. To examine this possibility, we used RT-PCR to investigate whether transcripts of four sets of two genes adjacent to the p44 expression locus, including their intergenic spaces, were present the NY-31, NY-36, and NY-37 strains of A. phagocytophilum cultivated in HL-60 cells. As shown in Fig. 3A, all transcripts in the tr1-omp-1X, omp-1X-omp-1N, and omp-1N-p44 regions were cotranscribed, including the intergenic spaces. In contrast, no transcript from the ndk-tr1 region was detectable. The major 3-kb fragment was also detected by 5′RACE using specific primers located in the 5′ upstream region of p44 in the expression locus, confirming polycistronic expression from the tr1 promoter (Fig. 3B). These results indicate that the four tandem genes (tr1, omp-1X, omp-1N, and a p44 paralog) are polycistronically transcribed under certain conditions. In addition, several minor fragments, ranging from 1.3 to 2.5 kb, were also detected, suggesting that additional promoters might be present within and upstream of omp-1N. Barbet et al. showed that omp-1N (p44ESup1) and a p44 (msp2) paralog were coexpressed (3).
FIG. 3.
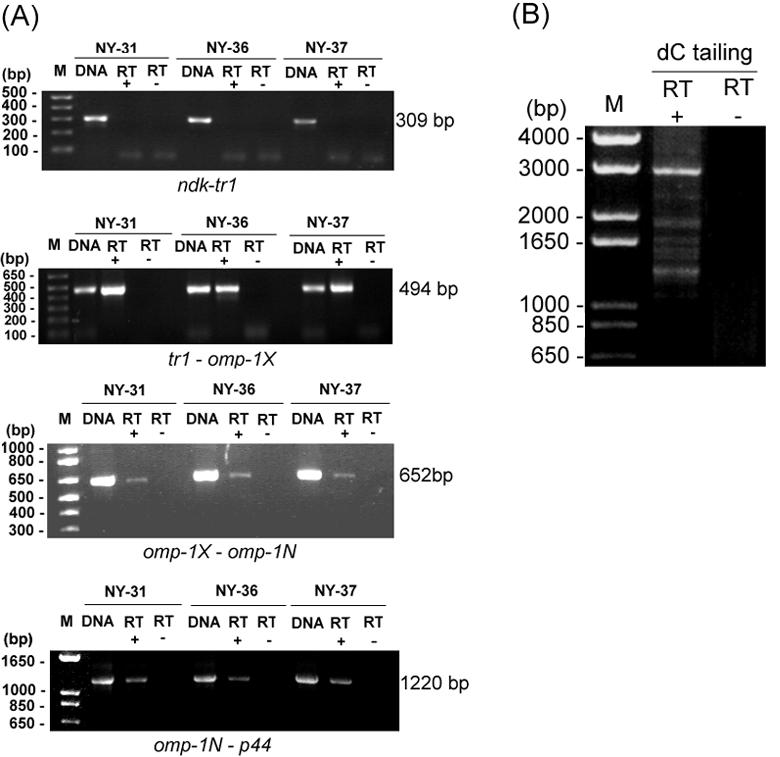
RT-PCR and 5′RACE analysis of polycistronic transcription linked to the p44 expression locus in strains NY-31, NY-36, and NY-37. (A) RT-PCR was used for transcriptional analysis of four sets of two adjacent genes and their intergenic spaces in the p44 expression locus, as indicated below each panel. The DNA template control included 0.1 ng of genomic DNA from the purified HZ strain and shows the intensity and specificity of the bands detected with each pair of primers (lanes DNA). Primers used include sets 12, 13, 14, and 15, as shown in Table 1 and Fig. 2. RT+ and RT−, presence and absence of reverse transcriptase, respectively. (B) 5′RACE analysis of the p44 expression locus operon using primer set 6, as shown in Table 1. The lack of an amplicon in the absence of reverse transcriptase (RT−) shows that there was no genomic DNA contamination in the RNA preparation. The numbers on the left of each panel indicate the respective amplicon sizes.
Characterization of the promoter region for polycistronic transcription of the p44 expression locus.
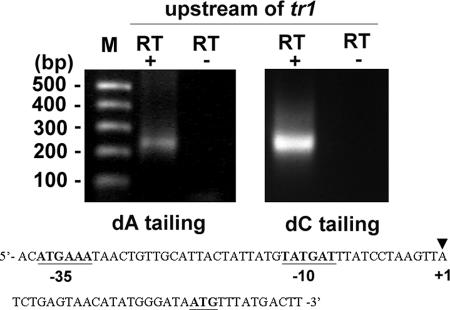
To identify the promoter region for the polycistronic transcription of the p44 expression locus, we analyzed the transcriptional initiation site upstream of tr1 by 5′RACE. With the addition of a polymeric dC tail at the 3′ end of the cDNA, the major 5′RACE product was detected as a single band in strain NY-37 cultivated in HL-60 cells (Fig. 4). Similar results were obtained when we performed 5′RACE analysis with the addition of a poly(dA) tail at the 3′ end of the cDNA. From sequence analysis of these 5′RACE products, the initiation site for polycistronic transcription was determined to be an adenine located 20 bp upstream from the translational start codon of tr1, as shown in Fig. 4. The putative −10 and −35 promoter regions (TATGAT and ATGAAA, respectively), which resemble the σ70-type consensus promoter sequences of E. coli (30), were found upstream of the initiation site. This site differs from the polycistronic transcriptional start site of A. marginale, which is located 74 bp 5′ to the ATG initiation codon of ORF4 (OpAG3) (2). Barbet et al. identified a putative promoter region 80 bp upstream of omp-1N (p44ESup1) (3) that closely resembles the promoter identified in the msp2 expression site of A. marginale.
FIG. 4.
5′RACE analysis of the promoter region located upstream from tr1 in strain NY-37. Primer set 5 was used in 5′RACE, as shown in Table 1. RT+ and RT−, presence and absence of reverse transcriptase, respectively. The putative −10 and −35 regions and the translational start codon are shown in boldface and underlined. Arrowhead (+1), initiation site for polycistronic transcription.
Effect of cell culture passage on expression of p44s at the p44 expression locus of A. phagocytophilum.
Jauron et al. and Barbet et al. demonstrated that P44 proteins and p44 species in the expression locus, respectively, were changed when A. phagocytophilum was passaged in the tick cell line ISE6 and in the human HL-60 cell line under different culture conditions (3, 15). We examined whether the p44 cDNA changes as the passage numbers increase using the same host cell species, HL-60, and culture conditions. cDNA cloning and sequencing showed that, in the low-passage cultures of strain NY-37, p44-1 was the major p44 cDNA species. However, in the high-passage culture (greater than 20 passages), the p44-1 cDNA was undetectable and the p44-28 cDNA was the predominant species (15 out of 20 cDNA clones). We also examined the p44 species at the p44 expression locus in high-passage cultures by expression locus-specific PCR using primer set 18 (Table 1 and Fig. 2); of 19 DNA clones, 15 clones were identified as p44-28. This indicates that, as A. phagocytophilum is progressively passaged in culture, the genetic population of A. phagocytophilum expresses different p44 species at the p44 expression locus, even though culture conditions remain the same. Thus, the major p44 cDNA species correlated with the major genetic population of p44 species at the p44 expression locus, similar to what was shown by Barbet et al. (3). Similar results were obtained by Western blotting with antibodies specific for certain P44 species in NY-37 high-passage cultures (data not shown). These experiments further confirmed that the P44 protein expression profile correlated with the expression profile of p44 transcripts.
To identify the genomic loci of the major p44 cDNA species, we compared the Southern blotting patterns of genomic DNA isolated from low-passage cultures of strain NY-37 (∼10 passages) with those of DNA isolated from high-passage cultures (>20 passages). Three probes, a p44 expression locus probe, a p44-1-specific probe, and a p44-28-specific probe, were used for this purpose. In both low- and high-passage cultures, the expression locus probe hybridized to a single DNA fragment (Fig. 5A), showing that the p44 expression locus was stably located in the genome during the culture passages. In contrast, the p44-1-specific probe hybridized to two fragments in the early culture passages: one corresponding to the p44 expression locus (Fig. 5B) and another corresponding to the p44-1/p44-18 locus that was previously identified in the HZ strain (43). However, in the high-passage culture, the p44-1 band in the p44 expression locus disappeared and a single band in the p44-1/p44-18 locus remained (Fig. 5B). Upon hybridization with the p44-28-specific probe, two DNA fragments were detected in PstI and XbaI digestions of genomic DNA from the low-passage culture: one in the p44 expression locus (weakly hybridized bands in Fig. 5C) and another in an unknown locus (strongly hybridized bands in Fig. 5C). A single overlapping band was detected in the EcoRI digestion of DNA from the low-passage culture (Fig. 5C). In the high-passage culture, the weakly hybridized band in the p44 expression locus became much stronger in relative density (Fig. 5C). The p44-28 paralog within the unknown locus appeared to be transcriptionally inactive, since the p44-28 transcript was not detected despite its presence at an unknown locus within the genome in the low-passage culture. This unknown p44-28 locus was previously detected by genomic Southern blot analysis in five strains of A. phagocytophilum at similar relative densities (21). These results indicate that, during cell culture passages, (i) p44-1 and p44-28 were stably present, regardless of their expression status, and (ii) the genetic population of A. phagocytophilum with p44-1 in the p44 expression locus was replaced with the population of A. phagocytophilum with p44-28 in the expression locus.
FIG. 5.
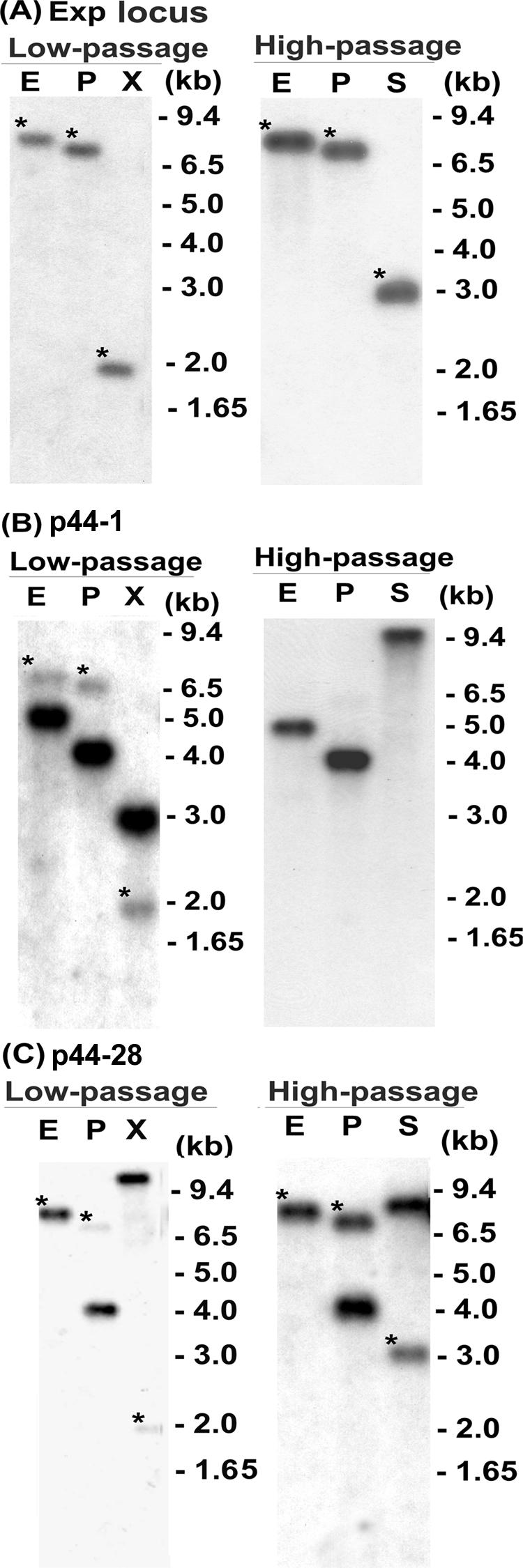
Genomic Southern blot analyses comparing low- and high-passage cultures of A. phagocytophilum strain NY-37. Genomic DNA derived from strain NY-37 was digested with EcoRI (E), PstI (P), and XbaI (X) or SpeI (S). (A) Blots were hybridized with a p44 expression locus probe (primer set 7, shown in Table 1 and Fig. 1) used for Fig. 3. (B) Blots were hybridized with a p44-1-specific probe (primer set 9 in Table 1). (C) Blots were hybridized with a p44-28-specific probe (primer set 8 in Table 1). Low-passage, cells with <10 passages in culture; high-passage, cells with >20 passages in culture. Asterisks, bands corresponding to the p44 expression locus. Numbers on the right indicate molecular sizes.
To characterize the unknown silent locus of p44-28 in strain NY-37, we used genome walking procedures to analyze the upstream and downstream sequences of p44-28 in the high-passage culture of the NY-37 strain. Two pairs of p44-28-specific primers located in the unique central hypervariable region of p44-28 (primer sets 16 and 17 in Table 1) were used for the amplification. We obtained two types of DNA sequences related to p44-28: one corresponded to the p44 expression locus, and another corresponded to a novel locus. The p44-28 paralog in the novel locus lacked an ATG start codon, and its ORF started from an AAG codon at the nucleotide 430 position of the p44-1 paralog (43) and had a TGA at the 3′ end, indicating that this was a pseudogene (p44-28p). We also found a new p44 paralog, p44-50, located 179 bp upstream of the p44-28p, with the genome walking procedure. It is not known whether p44-50 has an AUG start codon, because the sequence was only partially determined. We tentatively designated this locus the p44-50/p44-28 pseudogene locus. We next confirmed that this pseudogene locus corresponds to the unknown locus in Fig. 5C by genomic Southern blot analysis. A DNA probe specific to the pseudogene locus (upstream of p44-28p and downstream of p44-50) hybridized to a DNA band corresponding to the unknown locus in Fig. 5C in strain NY-37. The locus was also detected in strains NY-31, NY-36, and HZ (data not shown). These findings suggest that the p44-50/p44-28 pseudogene locus is conserved and stable among these four strains and that the p44-28 present at the p44 expression locus was generated by unidirectional conversion of p44-28p at this locus.
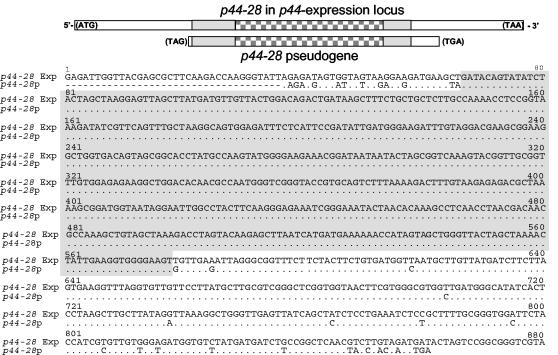
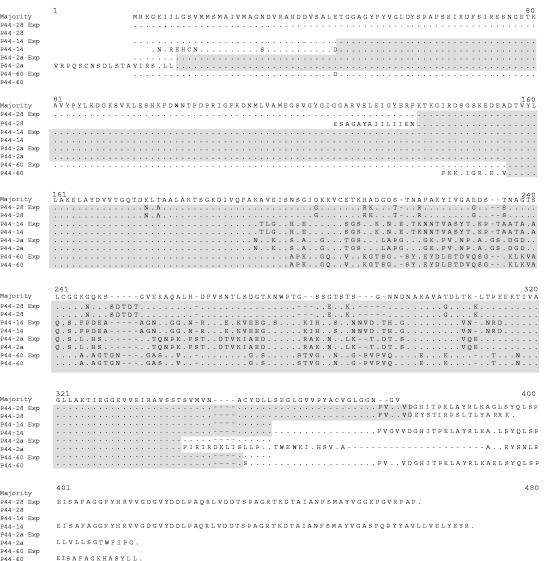
Sequence alignment of the new locus revealed that the sequences of two p44-28 hypervariable regions and flanking regions (411 bp, extending from position 31 to 541 of p44-28p) in the transcriptionally active p44-28 of the p44 expression locus and the inactive p44-28p found in the p44-50/p44-28 pseudogene locus were identical (Fig. 6). Nucleotide sequences external to these regions differed between the expression and pseudogene loci. These results suggest a nonreciprocal and nonsegmental gene conversion, whereby the entire central hypervariable region of p44-28 in the expression locus and a portion of the flanking sequences were derived from p44-28p in the donor locus.
FIG. 6.
Nucleotide sequence alignment of p44-28 in the expression locus and in the silent genomic locus from the same sample of genomic DNA derived from strain NY-37. The upper diagram shows the relative location of p44 in the expression locus and the corresponding p44 in the donor locus. The hypervariable region is shown in a checkerboard pattern. The identical 5′- and 3′-end conserved regions are shown in gray. The lower panel shows the nucleotide sequence alignment of the p44-28 pseudogene (p44-28p) and the p44-28 complete gene in the expression locus. Areas of continuous identical sequence are shaded gray. Dots, identical nucleotides; dashes, sequence gaps.
Sequence comparison of the same p44 species in expression and donor loci and p44 species expressed at different time points in A. phagocytophilum.
Our findings described above are inconsistent with the segmental gene conversion that has been previously proposed for variable expression of p44s in A. phagocytophilum (3). To address this issue, we next compared sequences of (i) p44s in the expression locus and in the multiple donor loci in the genome of strain HZ; (ii) p44 cDNA with its respective copies in the genome of strain HZ; (iii) p44 transcript species within the NY-31, NY-36, and NY-37 strains at three different time points; (iv) all p44 transcript species in strains NY-31, NY-36, and NY-37 (all isolated in 2000) and their respective copies in the genome of strain HZ (isolated in 1995).
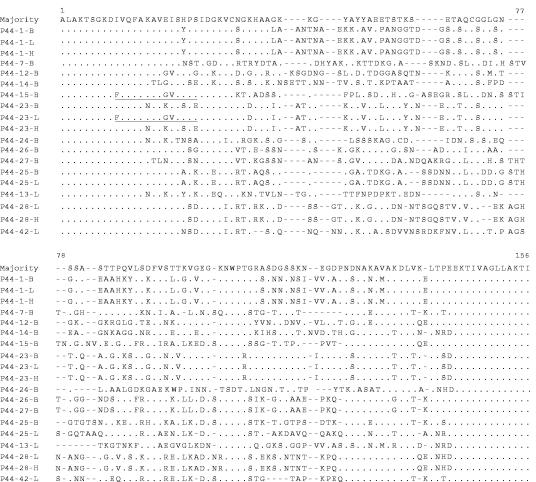
To obtain the sequences of the p44 hypervariable region in the expression locus, DNA PCR was performed with expression locus-specific primers (primer set 18 in Table 1 and Fig. 2). Of a total of 21 clones sequenced, four different p44 species were identified: p44-18, p44-14, p44-2a, and p44-60. Donor p44-2a and donor p44-14 were found to be complete genes, whereas donor p44-18 and p44-60 were pseudogenes. Sequence alignment of four pairs of the p44 species in the expression locus and in their respective donor loci in strain HZ revealed that each pair had identical sequences within the hypervariable region and within a portion of the flanking regions. These identical flanking sequences extended to different lengths in each pair (Fig. 7), and sequence variations were observed within each pair external to the putative recombined sequences (Fig. 7). These results suggest that (i) the entire p44 hypervariable region in the expression locus was derived from the identical region in the donor locus and (ii) the recombination for each p44 species included a part of the flanking 5′ and 3′ conserved regions.
FIG. 7.
Predicted amino acid sequence comparison of p44s in the expression locus (Exp) and the corresponding copies in the genome of strain HZ of A. phagocytophilum. Identical sequences between the expression locus and its corresponding copy in the genome are shaded gray. Dots, identical nucleotides; dashes, sequence gaps.
To further analyze the possibility of nonsegmental gene conversion at the p44 expression locus, we compared the sequences of central hypervariable regions plus partial flanking sequences of 20 different p44 cDNA species previously identified in strain HZ in ticks, mice, horses, and cell cultures (44) and their respective paralogs not in the expression locus. Eighteen of 20 p44 pairs encoded identical amino acid sequences, and 7 of the 20 p44 pairs had identical nucleotide sequences. This further suggests that nonsegmental conversion of the hypervariable p44 region included part of the flanking 5′ and 3′ conserved regions.
We also considered the possibilities that the various p44 species might exhibit minor differences in sequence and/or that they might be differentially affected by temporal factors. Therefore, we compared the predicted amino acid sequences of all p44 clones that were identified by RT-PCR, 5′RACE, and p44 expression locus DNA PCR in the NY-31, NY-36, and NY-37 strains. We made this comparison at three different time points: in patients (when the cultures were isolated), during low passages of cell culture after isolation, and during high passages of cell culture. In strains NY-36 and NY-37, the p44 types had identical sequences in the central hypervariable region and parts of flanking sequences and no chimera sequence was found. In strain NY-31, alignment of 150 predicted amino acid sequences from a total of 13 different p44 species showed that the sequences of all p44 species were identical except for that of p44-23. A total of 15 p44-23 cDNA clones were sequenced, and this species was continuously expressed by strain NY-31 at all three time points. Two of these cDNA clones, one p44-23 cDNA derived from a low-passage culture and one of five p44-23 cDNAs derived from a high-passage culture, included a sequence (encoding FVQFAKAVGVSHPS) which bordered the 5′ conserved region and the central hypervariable region (Fig. 8). The same block of sequence was found in the p44-15 transcript in the blood from the patient. This sequence differed from that encoding IVQFANAVKISSPE found in the same region of 13 other p44-23 cDNA clones derived from the blood of patient NY31 and from high-passage cell cultures. No p44-15 transcript was detected in cell cultures with the NY-31 strain.
FIG. 8.
Predicted protein sequence analysis of p44 expression in strain NY-31 at various times. p44 cDNA clones were derived from blood (B), low-passage cultures (L), and high-passage cultures (H). Dots, identical amino acid residues; dashes, sequence gaps. The underlined sequences are identical among proteins encoded by p44-15, the p44-23 cDNA clone identified in low-passage cultures, and one of five p44-23 cDNA clones identified in high-passage cultures. The sequences of the proteins encoded by the remaining p44-23 cDNA clones are shown as p44-23-B and p44-23-H. p44-15 cDNA was detected only in the blood of patient NY31, and p44-23 cDNA was detected at all three time points.
Finally, by comparing the sequences of cDNA clones from strains NY-31, NY-36, and NY-37 with p44 orthologs from the strain HZ genome sequence database, we found one p44-28 cDNA clone in strain NY-36 that had a short sequence (encoding KAVEISHSD) at the 5′ border of the conserved region and the central hypervariable region. This sequence differed from the sequence encoding NAVKISSPE, found in the other 37 p44-28 cDNA clones derived from low-passage cell culture. The former sequence was identical to the sequence in the corresponding region of p44-24 in the HZ strain. In strain NY-37, there were two p44-1 cDNA clones at 5′ border of the conserved and hypervariable regions encoding the short sequence EISYPS, which differed from the GISHPT sequence encoded by the remaining 33 p44-1 cDNA clones derived from low-passage cultures. The sequence encoding EISYPS was detected in two different p44 paralogs in the HZ strain. Barbet et al. reported an identical short block of sequence at the 3′ border of the conserved and hypervariable regions in two different p44 species each from different strains (3). Nevertheless, neither our present study nor the previous studies of Barbet et al. detected any mosaic sequences that included more than two p44 species within the central hypervariable region.
DISCUSSION
Studies of the p44s and msp2s have generated considerable interest, since so many paralogs of these genes exist (particularly p44s), and many are expressed despite the absence of a start codon. Barbet et al. (2, 3) and Brayton et al. (5, 6) proposed segmental gene conversion and Zhi et al. (42) proposed RNA splicing as mechanisms by which these pseudogenes can be expressed as complete p44 genes. The present study provides new information regarding the mechanisms of p44 expression at the p44 expression locus. Our study revealed that genomic sequences of individual p44 species within a single strain of A. phagocytophilum were identical at the expression locus and donor locus in both the hypervariable region and in parts of the conserved flanking regions. These findings suggest that nonreciprocal and nonsegmental gene conversion could take place at the expression locus, resulting in duplication of each donor gene, including the flanking regions.
Although the 5′ and 3′ regions of p44 paralogs are relatively conserved, they are not identical. Therefore, we propose that heteroduplex formation and resolution occur at the 5′ and 3′ conserved regions in each recombination event. Sequences of p44 hypervariable regions appear to be too diverse to form heteroduplexes in these regions. After heteroduplex resolution, the conserved regions, rather than the hypervariable regions, appear to create chimera or mosaic sequences that include sequences derived from the conserved regions of the new p44 species and the previous sequence in the p44 expression locus. The key finding of this study is that sequences of the hypervariable region, the expression locus, and the corresponding donor locus are conserved within the same A. phagocytophilum strains. Despite extensive analyses, we did not identify any chimera or mosaic sequences within the hypervariable region of p44 cDNA or p44 paralogs in the expression locus. We also identified a short block of shared amino acid sequences encoded by two different p44 species in two different strains of A. phagocytophilum; the coding sequences are outside of the three hypervariable domains in the central hypervariable region (21). This is similar to the findings of Barbet et al., which suggested segmental gene conversion of p44 in A. phagocytophilum (3).
Our extensive comparison of p44 sequences within the same strains revealed a short block of p44-15 sequence within the p44-23 cDNA of the NY-31 strain. One possible explanation for the sequence diversity within this region is the formation of heteroduplexes combined with mismatch base repairing. It is possible that p44-23 recombined at the expression locus to replace p44-15 through homologous recombination. Such a recombination could have included the small DNA fragment within the 5′- and 3′-end conserved region, so that the new p44-23 in the expression locus retained this short block of p44-15. Insertion and deletion of such a short sequence are known to occur during RecA-mediated heteroduplex formation and mismatch base repairing in E. coli, as described by DasGupta and Radding (10) and Bianchi and Radding (4) and in more recent review articles (19, 20). Sequence variations in the msp2 expression locus could potentially also result from heteroduplex formation between two partially homologous sequences (multigene family) with base pair mismatches and bubbles. Since msp2 pseudogenes have short blocks of homologous regions within the hypervariable region (6), the several patterns of bubbles corresponding to the same heteroduplex and their resolution may create complex patchworks, as previously described (18). It is also possible that there is another p44-23 paralog at a different genetic locus that has a hypervariable region sequence identical to that of p44-23 with this block of p44-15 sequence at the border, since we previously found three bands in the genomic Southern blot analysis of NY-31 strain using the p44-23 probe (21). In fact, duplicated p44 paralogs are present in the A. phagocytophilum chromosome. We previously cloned p44-2a and p44-2b, which have identical nucleotide sequences in the central hypervariable region and a part of 5′- and 3′-end conserved regions in two different genomic loci of strain HZ (43, 44). In addition we found that two copies of p44-16, p44-7, and p44-15 are present in the non-p44 expression locus.
Thus, although p44 and msp2 have similar gene structures and expression loci, different mechanisms appear to have evolved to create sufficient antigenic diversity. The reason for the differences may lie in the number and the nature of the hypervariable region sequence of the paralog. The A. phagocytophilum genome includes more than 80 paralogs of p44, whereas only 8 to 10 msp2s are predicted in A. marginale (7). Thus, the number and sequence diversity of p44 paralogs may have been sufficient for A. phagocytophilum to evade host immune surveillance by a simple gene conversion, but the mechanism may have required modification for A. marginale to create sufficient antigenic diversity. In agreement with this hypothesis, to date we have identified 59 different p44 cDNA sequences, all of which have been found in the preliminary A. phagocytophilum genome sequencing database (www.tigr.org).
Interestingly, despite the presence of 5′ and 3′ conserved flanking sequences, no promiscuous recombination was detected within the nonexpressed reserve p44 genes. Since genomic loci of individual p44 paralogs within the same strain were conserved, this suggests that recombination was restricted to the single p44 expression locus. Previous genomic Southern blot analyses with probes specific for each of the p44 paralogs showed identical patterns among A. phagocytophilum strains isolated from HGE patients from New York in 1995 and 2000, suggesting that DNA sequences and genomic loci of individual p44 orthologs are also conserved among different strains and over a time period of at least 5 years in this geographic locus (21). The present study further confirmed that the p44 hypervariable region sequences are conserved among four strains. A few base variations observed within the hypervariable region of p44s may be the result of point mutations. Point mutations and gene conversion are not mutually exclusive and partially account for vlsE variation in Borrelia burgdorferi (36).
Our present results suggest that nonsegmental gene conversion is the primary mechanism for variable p44 gene expression by A. phagocytophilum at the p44 expression site. If this is true, the next question would be what recombination pathway is involved in changing p44s in the expression locus. Our analysis of the preliminary A. phagocytophilum genome sequence data revealed that A. phagocytophilum has genes homologous to E. coli recA, recF, recO, recR, recO, and recJ genes that could be used for the RecA-dependent RecF homologous recombination pathway (14, 19, 23). We did not find recBCD or recE homologs in the A. phagocytophilum genome, indicating that A. phagocytophilum might lack the RecBCD and RecE homologous recombination pathways. We did find the ruvABC and recG genes, which are used to resolve Holliday junctions generated by homologous recombination (19) within the A. phagocytophilum genome. Thus, the mechanism of recombination of A. phagocytophilum may be RecF dependent, similar to antigenic variation in Neisseria gonorrhoeae pilin (26).
No direct evidence for msp2 or p44 recombination has yet been found in either A. marginale or A. phagocytophilum. Unlike the cloning of vlsE in B. burgdorferi (41), the cloning of A. phagocytophilum p44 phenospecies (p44 expression locus genospecies) has been difficult. However, if there is no recombination and all P44 variants observed to date by us and others (3, 8, 13, 21, 43, 44) are due to selection alone, various P44 phenospecies may have been eliminated from each stock of these strains after continuous passages in different environments, and they would not be expected to reappear. In contrast, in one patient's blood (strain NY-37), p44-28 was the major transcript (21), but once this strain was isolated in the HL-60 cell line, the p44-1 transcript became the predominant species in the NY-37 isolate and the p44-28 transcript disappeared. However, in later culture passages, the p44-28 transcript once again became the predominant transcript and p44-1 mRNA disappeared. Similar observations have been reported during the transmission of A. phagocytophilum strain HZ from ticks to horses (44). Without recombination to explain this phenomenon, each isolate of A. phagocytophilum would need to maintain >80 different p44 genotypes at the expression locus, such that >80 different genetic clones would be present in each strain under all circumstances, which would clearly be inefficient. It is more plausible that A. phagocytophilum evolved this mechanism of antigenic variation to maintain the ability to express diverse p44 antigenic repertoires. Of course, differences in growth rate and selection also contribute to the overall dominance of any particular P44 phenotype.
It has been proposed that, in A. marginale, the msp2 variant change is driven by the humoral immune pressure of the cattle (6, 11). However, our present data and those previously reported by others (3, 15) showed that changes in p44 variants occur in cell culture in the absence of antibodies. It will be of considerable interest to determine the factors that influence the dominance of particular phenotypes under identical cell culture conditions.
Other novel findings of the present study are the polycistronic expression of the p44 expression locus from the σ70-type promoter sequence upstream of tr1. In addition, our 5′RACE results suggest that additional promoters may exist within and upstream of omp-1X. Barbet et al. reported that a promoter with significant nucleotide sequence identity to the msp2 expression site promoter of A. marginale exists upstream of omp-1N (p44ESuP1) (3). Taken together, the p44 expression locus appears to be regulated by multiple promoters.
A. phagocytophilum p44s and A. marginale msp2s (31), along with E. chaffeensis (the etiologic agent of human monocytic ehrlichiosis) omp-1s (27, 29) and Ehrlichia canis (the etiologic agent of canine ehrlichiosis) p30s (27, 28, 38), have been identified as immunodominant major outer membrane protein multigene families in the family Anaplasmataceae. In both Ehrlichia spp., 22 paralogous genes are arranged in tandem at a single locus and are regulated by both polycistronic and monocistronic transcription (27, 29). In Anaplasma spp., the paralogs are dispersed throughout the genomes, and their primary transcription seems to primarily depend on a specific expression locus. Previous studies by others (2, 3) revealed that this locus is linked to omp-1 orthologs. Our study showed that the omp-1-linked expression locus of A. phagocytophilum is arranged in tandem downstream from the tr1 gene, similar to those of the Ehrlichia spp. (27, 28). This implies that the expression loci of major outer membrane protein multigene families of pathogenic bacteria in the family Anaplasmataceae evolved from a common locus in a bacterium ancestral to both Ehrlichia and Anaplasma spp. More-detailed analyses of these loci should facilitate our understanding of the evolutionary relationship between these two genera. Understanding the molecular mechanisms underlying regulation of p44 and msp expression represents an important first step toward developing therapies and vaccines against HGE.
Acknowledgments
This research was supported by grant R01AI47407 from the National Institutes of Health. The sequencing project was supported by National Institutes of Health grant R01 AI47885 to Y.R.
Editor: J. T. Barbieri
REFERENCES
- 1.Bakken, J. S., J. Krueth, C. Wilson-Nordskog, R. L. Tilden, K. Asanovich, and J. S. Dumler. 1996. Clinical and laboratory characteristics of human granulocytic ehrlichiosis. JAMA 275:199-205. [PubMed] [Google Scholar]
- 2.Barbet, A. F., A. Lundgren, J. Yi, F. R. Rurangirwa, and G. H. Palmer. 2000. Antigenic variation of Anaplasma marginale by expression of MSP2 mosaics. Infect. Immun. 68:6133-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbet, A. F., P. F. M. Meeus, M. Belanger, M. V. Bowie, J. Yi, A. M. Lundgren, A. R. Alleman, S. J. Wong, F. K. Chu, U. G. Munderloh, and S. D. Jauron. 2003. Expression of multiple outer membrane protein sequence variants from a single genomic locus of Anaplama phagocytophilum. Infect. Immun. 71:1706-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianchi, M. E., and C. M. Radding. 1983. Insertions, deletions and mismatches in heteroduplex DNA made by recA protein. Cell 35:511-520. [DOI] [PubMed] [Google Scholar]
- 5.Brayton, K. A., D. P. Knowles, T. C. McGuire, and G. H. Palmer. 2001. Efficient use of a small genome to generate antigenic diversity in tick-borne ehrlichial pathogens. Proc. Natl. Acad. Sci. USA 98:4130-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brayton, K. A., G. H. Palmer, A. Lundgren, J. Yi, and A. F. Barbet. 2002. Antigenic variation of Anaplasma marginale msp2 occurs by combinatorial gene conversion. Mol. Microbiol. 43:1151-1159. [DOI] [PubMed] [Google Scholar]
- 7.Brown, W. C., K. A. Brayton, C. M. Styer, and G. H. Palmer. 2003. The hypervariable region of Anaplasma marginale major surface protein 2 (MSP2) contains multiple immunodominant CD4+ T lymphocyte epitopes that elicit variant-specific proliferative and IFN-gamma responses in MSP2 vaccinates. J. Immunol. 170:3790-3798. [DOI] [PubMed] [Google Scholar]
- 8.Caspersen, K., J. H. Park, S. Patil, and J. S. Dumler. 2002. Genetic variability and stability of Anaplasma phagocytophila msp2 (p44). Infect. Immun. 70:1230-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, S. M., J. S. Dumler, J. S. Bakken, and D. H. Walker. 1994. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J. Clin. Microbiol. 32:589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DasGupta, C., and C. M. Radding. 1982. Polar branch migration promoted by recA protein: effect of mismatched base pairs. Proc. Natl. Acad. Sci. USA 79:762-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.French, D. M., T. F. McElwain, T. C. McGuire, and G. H. Palmer. 1998. Expression of Anaplasma marginale major surface protein 2 variants during persistent cyclic rickettsemia. Infect. Immun. 66:1200-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardalo, C. J., V. Quagliarello, and J. S. Dumler. 1995. Human granulocytic ehrlichiosis in Connecticut: report of a fatal case. Clin. Infect. Dis. 21:910-914. [DOI] [PubMed] [Google Scholar]
- 13.IJdo, J. W., C. Wu, S. R. Telford III, and E. Fikrig. 2002. Differential expression of the p44 gene family in the agent of human granulocytic ehrlichiosis. Infect. Immun. 70:5295-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivancic-Bace, I., P. Peharec, S. Moslavac, N. Skrobot, E. Salaj-Smic, and K. Brcic-Kostic. 2003. RecFOR function is required for DNA repair and recombination in a RecA loading-deficient recB mutant of Escherichia coli. Genetics 163:485-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jauron, S. D., C. M. Nelson, V. Fingerle, M. D. Ravyn, J. L. Goodman, R. C. Johnson, R. Lobentanzer, B. Wilske, and U. G. Munderloh. 2001. Host cell-specific expression of a p44 epitope by the human granulocytic ehrlichiosis agent. J. Infect. Dis. 184:1445-1450. [DOI] [PubMed] [Google Scholar]
- 16.Johnston, S. L., M. Strausbauch, G. Sarkar, and P. J. Wettstein. 1995. A novel method for sequencing members of multi-gene families. Nucleic Acids Res. 23:3074-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, H. Y., and Y. Rikihisa. 1998. Characterization of monoclonal antibodies to the 44-kilodalton major outer membrane protein of the human granulocytic ehrlichiosis agent. J. Clin. Microbiol. 36:3278-3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kourilsky, P. 1983. Genetic exchanges between partially homologous nucleotide sequences: possible implications for multigene families. Biochimie 65:85-93. [DOI] [PubMed] [Google Scholar]
- 19.Kowalczykowski, S. C., D. A. Dixon, A. K. Eggleston, S. D. Lauder, and W. M. Rehrauer. 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58:401-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 63:751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin, Q., N. Zhi, N. Ohashi, H. W. Horowitz, M. E. Aguero-Rosenfeld, J. Raffalli, G. P. Wormser, and Y. Rikihisa. 2002. Analysis of sequences and loci of p44 homologs expressed by Anaplasma phagocytophila in acutely infected patients. J. Clin. Microbiol. 40:2981-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lohr, C. V., K. A. Brayton, V. Shkap, T. Molad, A. F. Barbet, W. C. Brown, and G. H. Palmer. 2002. Expression of Anaplasma marginale major surface protein 2 operon-associated proteins during mammalian and arthropod infection. Infect. Immun. 70:6005-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lovett, S. T., R. L. Hurley, V. A. Sutera, Jr., R. H. Aubuchon, and M. A. Lebedeva. 2002. Crossing over between regions of limited homology in Escherichia coli. RecA-dependent and RecA-independent pathways. Genetics 160:851-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lusetti, S. L., and M. M. Cox. 2002. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu. Rev. Biochem. 71:71-100. [DOI] [PubMed] [Google Scholar]
- 25.Massung, R. F., J. H. Owens, D. Ross, K. D. Reed, M. Petrovec, A. Bjoersdorff, R. T. Coughlin, G. A. Beltz, and C. I. Murphy. 2000. Sequence analysis of the ank gene of granulocytic ehrlichiae. J. Clin. Microbiol. 38:2917-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehr, I. J., and H. S. Seifert. 1998. Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Mol. Microbiol. 30:697-710. [DOI] [PubMed] [Google Scholar]
- 27.Ohashi, N., Y. Rikihisa, and A. Unver. 2001. Analysis of transcriptionally active gene clusters of major outer membrane protein multigene family in Ehrlichia canis and E. chaffeensis. Infect. Immun. 69:2083-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohashi, N., A. Unver, N. Zhi, and Y. Rikihisa. 1998. Cloning and characterization of multigenes encoding the immunodominant 30-kilodalton major outer membrane proteins of Ehrlichia canis and application of the recombinant protein for serodiagnosis. J. Clin. Microbiol. 36:2671-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohashi, N., N. Zhi, Y. Zhang, and Y. Rikihisa. 1998. Immunodominant major outer membrane proteins of Ehrlichia chaffeensis are encoded by a polymorphic multigene family. Infect. Immun. 66:132-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliver, D. 1985. Protein secretion in Escherichia coli. Annu. Rev. Microbiol. 39:615-648. [DOI] [PubMed] [Google Scholar]
- 31.Palmer, G. H., G. Eid, A. F. Barbet, T. C. McGuire, and T. F. McElwain. 1994. The immunoprotective Anaplasma marginale major surface protein 2 is encoded by a polymorphic multigene family. Infect. Immun. 62:3808-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrovec, M., S. L. Furlan, T. A. Zupanc, F. Strle, P. Brouqui, V. Roux, and J. S. Dumler. 1997. Human disease in Europe caused by a granulocytic Ehrlichia species. J. Clin. Microbiol. 35:1556-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rikihisa, Y., S. A. Ewing, and J. C. Fox. 1994. Western immunoblot analysis of Ehrlichia chaffeensis, E. canis, or E. ewingii infections in dogs and humans. J. Clin. Microbiol. 32:2107-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rikihisa, Y., S. A. Ewing, J. C. Fox, A. G. Siregar, F. H. Pasaribu, and M. B. Malole. 1992. Analyses of Ehrlichia canis and a canine granulocytic Ehrlichia infection. J. Clin. Microbiol. 30:143-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rikihisa, Y., N. Zhi, G. P. Wormser, B. Wen, H. W. Horowitz, and K. E. Hechemy. 1997. Ultrastructural and antigenic characterization of a granulocytic ehrlichiosis agent directly isolated and stably cultivated from a patient in New York State. J. Infect. Dis. 175:210-213. [DOI] [PubMed] [Google Scholar]
- 36.Sung, S. Y., J. V. McDowell, and R. T. Marconi. 2001. Evidence for the contribution of point mutations to vlsE variation and for apparent constraints on the net accumulation of sequence changes in vlsE during infection with Lyme disease spirochetes. J. Bacteriol. 183:5855-5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Telford, S. R., III, J. E. Dawson, P. Katavolos, C. K. Warner, C. P. Kolbert, and D. H. Persing. 1996. Perpetuation of the agent of human granulocytic ehrlichiosis in a deer tick-rodent cycle. Proc. Natl. Acad. Sci. USA 93:6209-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Unver, A., N. Ohashi, T. Tajima, R. W. Stich, D. Grover, and Y. Rikihisa. 2001. Transcriptional analysis of p30 major outer membrane multigene family of Ehrlichia canis in dogs, ticks, and cell culture at different temperatures. Infect. Immun. 69:6172-6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Dobbenburgh, A., A. P. van Dam, and E. Fikrig. 1999. Human granulocytic ehrlichiosis in western Europe. N. Engl. J. Med. 340:1214-1216. [DOI] [PubMed] [Google Scholar]
- 40.Weber, K. L., M. E. Bolander, and G. Sarkab. 1998. Rapid acquisition of unknown DNA sequence adjacent to a known segment by multiplex restriction site PCR. BioTechniques 25:415-419. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, J. R., J. M. Hardham, A. G. Barbour, and S. J. Norris. 1997. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89:275-285. [DOI] [PubMed] [Google Scholar]
- 42.Zhi, N., N. Ohashi, and Y. Rikihisa. 2002. Activation of a p44 pseudogene in Anaplasma phagocytophila by bacterial RNA splicing: a novel mechanism for post-transcriptional regulation of a multigene family encoding immunodominant major outer membrane proteins. Mol. Microbiol. 46:135-145. [DOI] [PubMed] [Google Scholar]
- 43.Zhi, N., N. Ohashi, and Y. Rikihisa. 1999. Multiple p44 genes encoding major outer membrane proteins are expressed in the human granulocytic ehrlichiosis agent. J. Biol. Chem. 274:17828-17836. [DOI] [PubMed] [Google Scholar]
- 44.Zhi, N., N. Ohashi, T. Tajima, J. Mott, R. W. Stich, D. Grover, S. R. Telford III, Q. Lin, and Y. Rikihisa. 2002. Transcript heterogeneity of the p44 multigene family in a human granulocytic ehrlichiosis agent transmitted by ticks. Infect. Immun. 70:1175-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]