Abstract
Posttranslational modification by the ubiquitin-like protein SUMO (small ubiquitin-like modifier) is emerging as an important regulator in many cellular processes, including genome integrity. In this study, we show that the kinetochore proteins Ndc10, Bir1, Ndc80, and Cep3, which mediate the attachment of chromosomes to spindle microtubules, are sumoylated substrates in budding yeast. Furthermore, we show that Ndc10, Bir1, and Cep3 but not Ndc80 are desumoylated upon exposure to nocodazole, highlighting the possibility of distinct roles for sumoylation in modulating kinetochore protein function and of a potential link between the sumoylation of kinetochore proteins and mitotic checkpoint function. We find that lysine to arginine mutations that eliminate the sumoylation of Ndc10 cause chromosome instability, mislocalization of Ndc10 from the mitotic spindle, abnormal anaphase spindles, and a loss of Bir1 sumoylation. These data suggest that sumoylation of Ndc10 and other kinetochore proteins play a critical role during the mitotic process.
Introduction
Sumoylation plays a key role in many cellular processes, including nuclear transport, signal transduction, transcriptional regulation, and maintenance of genome integrity (Seeler and Dejean, 2003; Johnson, 2004; Hay, 2005). Sumoylation is a process by which a small ubiquitin-like modifier (SUMO) protein (Smt3 in yeast) is conjugated to a target protein at a lysine residue. The consequences of sumoylation are substrate specific and can involve altering a substrate's interaction with other macromolecules (e.g., proteins and DNA) or blocking lysine residues on target proteins from being modified by other lysine-targeted modifications such as ubiquitin (Desterro et al., 1998; Hoege et al., 2002). The addition of SUMO to a protein is reversible through the action of ubiquitin-like proteases (Ulps) that are able to cleave SUMO from its substrate (Li and Hochstrasser, 1999, 2000). Ulps are also required for the posttranslational maturation of SUMO through cleavage at its C terminus to reveal the diglycine motif used to form the isopeptide bond with its substrate.
In the budding yeast Saccharomyces cerevisiae, sumoylation is essential for viability and is required for proper chromosome segregation, with sumoylation-deficient cells arresting in G2/M with short spindles and replicated DNA (Seufert et al., 1995; Li and Hochstrasser, 1999, 2000). In mammalian cells, defects in sumoylation cause abnormal nuclear architecture, chromosome missegregation, and embryonic lethality (Nacerddine et al., 2005). The critical protein targets that lead to these phenotypes when SUMO conjugation is disrupted remain unknown. In S. cerevisiae, proteomic approaches have identified >400 potential sumoylation targets; thus, pinpointing the biologically relevant sumoylation events is a challenge (Panse et al., 2004; Wohlschlegel et al., 2004; Zhou et al., 2004; Denison et al., 2005; Hannich et al., 2005; Wykoff and O'Shea, 2005). The phenotypes associated with sumoylation-defective mutants point to proteins involved in chromosome segregation as critical targets of SUMO modification. Indeed, many proteins involved in chromosome segregation, including kinetochore proteins that mediate the attachment of chromosomes to microtubules, have been identified as sumoylation substrates (Bachant et al., 2002; Hoege et al., 2002; Stead et al., 2003; Panse et al., 2004; Wohlschlegel et al., 2004; Zhou et al., 2004; Denison et al., 2005; Hannich et al., 2005; Wykoff and O'Shea, 2005). However, the functional roles of sumoylation on these proteins are not understood.
The kinetochore is a supramolecular protein complex that assembles on centromere DNA to perform two functions that are critical for genome stability. First, the kinetochore functions in a structural manner by facilitating the connection between sister chromatids and spindle microtubules, allowing microtubule-dependent forces to separate duplicated DNA during mitosis and meiosis (Kapoor and Compton, 2002; Biggins and Walczak, 2003). In budding yeast, this connection is mediated by ∼60 proteins that bridge sister chromatids to the microtubule interface of the mitotic spindle (McAinsh et al., 2003). Second, the kinetochore functions in a regulatory manner by providing a framework for the generation of signals used by cell cycle checkpoints to gauge the progress of mitosis, through which the timing of mitotic events are coordinated. The kinetochore does this by producing a wait anaphase signal until the proper bipolar attachment of sister chromatids to opposite spindle poles and the generation of tension has occurred (Nasmyth, 2005; Pinsky and Biggins, 2005). The Ndc80 kinetochore complex is required to localize spindle checkpoint proteins to the kinetochore, which is critical for the generation of the wait anaphase signal (Gillett et al., 2004; Maiato et al., 2004). Correct attachment and subsequent silencing of the checkpoint ultimately leads to the progression of cells into anaphase and separation of sister chromatids to daughter cells (Wolfe and Gould, 2005).
A subset of kinetochore proteins are relocalized to the mitotic spindle in anaphase and to the spindle midzone before cytokinesis, reflecting the localization of chromosome passenger proteins in higher eukaryotes (Adams et al., 2001; Vagnarelli and Earnshaw, 2004). In yeast, the set of chromosome passenger-like proteins includes the Aurora kinase Ipl1, the inhibitor of apoptosis (IAP) protein Bir1, and the inner kinetochore proteins Cep3 and Ndc10 (Goh and Kilmartin, 1993; Biggins et al., 1999; Buvelot et al., 2003; Huh et al., 2003; Bouck and Bloom, 2005; Widlund et al., 2006). The localization of these proteins to the mitotic spindle is thought to be important for regulating mitotic spindle dynamics and for cytokinesis (Adams et al., 2001; Buvelot et al., 2003). Indeed, temperature-sensitive alleles of ndc10-1 or bir1-33 cause cytokinesis defects at the restrictive temperature (Bouck and Bloom, 2005; Gillis et al., 2005). However, mutations in Bir1 that result in the loss of Ndc10 from the mitotic spindle cause defects in proper spindle elongation, not in cytokinesis (Widlund et al., 2006).
To gain insight into the function of Ndc10 on the mitotic spindle, we began by identifying protein-interacting partners of Ndc10 with the goal of discovering proteins that are required for Ndc10's spindle localization. Using a genome-wide two-hybrid screen, we identified multiple interactions between Ndc10 and the sumoylation machinery of budding yeast, and subsequent analysis demonstrated that Ndc10 is a target for sumoylation in vivo, as are other kinetochore proteins (Bir1, Cep3, and Ndc80). We also found that sumoylation of these proteins is differentially regulated in response to checkpoint activation, suggesting that sumoylation has distinct roles in modulating the function of these kinetochore proteins. Importantly, lysine residues required for Ndc10's sumoylation are necessary for Ndc10's proper localization to the mitotic spindle, suggesting that sumoylation plays a direct role in facilitating Ndc10's interaction with the spindle apparatus. As is the case when Bir1 is mutated, the mislocalization of Ndc10 is not associated with cytokinesis defects, suggesting that the spindle-bound form of Ndc10 is not responsible for Ndc10's cytokinesis-related functions. Furthermore, the loss of Ndc10's mitotic spindle association results in anaphase spindles of abnormal length, highlighting a role for Ndc10 in controlling mitotic spindle dynamics.
Results
Ndc10 interacts with multiple components of the sumoylation machinery
In two independent genome-wide two-hybrid screens using Ndc10 as bait with the Gal4 DNA-binding domain fused to either the N or C terminus, 10 proteins were identified as putative Ndc10 protein interactors (Table I). Three of these interactions occurred with only one of the baits, suggesting that the presence of the Gal4 DNA-binding domain on the N or C terminus may be interfering with specific protein–protein interactions, as has been previously reported (Millson et al., 2003). Of these 10 interacting proteins, Bir1 and Ubc9 were previously identified in two-hybrid screens with Ndc10 (Jiang and Koltin, 1996; Yoon and Carbon, 1999); Bir1 has also recently been shown to play a role in the localization of Ndc10 to the mitotic spindle (Bouck and Bloom, 2005; Widlund et al., 2006). Within this set of two-hybrid interacting proteins, three components of the sumoylation machinery were also identified (Ubc9, Smt3, and Nfi1). SMT3, which was originally identified as a high copy suppressor of a mutation in the kinetochore protein Mif2 (Meluh and Koshland, 1995), encodes the ubiquitin-like protein SUMO.
Table I.
Ndc10 two-hybrid interactions
| Prey | pOBD2-Ndc10a b | pBDC-Ndc10b c | Biological processd |
|---|---|---|---|
| BIR1 | +++++ | +++++ | Chromosome segregation and mitotic spindle elongation |
| CTF13 | − | ++ | Chromosome segregation and kinetochore assembly |
| FIR1 | +++++ | +++++ | mRNA polyadenylylation |
| HEX3 | +++ | +++ | DNA recombination |
| NFI1 | +++ | +++ | Protein sumoylation |
| NIS1 | +++ | +++ | Regulation of mitosis |
| SAP1 | ++++ | − | Unknown |
| SLI15 | + | + | Chromosome segregation |
| SMT3 | + | + | Protein sumoylation |
| UBC9 | − | ++++ | Protein sumoylation |
Encodes Ndc10 with the GAL DNA-binding domain fused to the N terminus.
Two-hybrid positive colony growth from weak (+) to strong (+++++).
Encodes Ndc10 with the GAL DNA-binding domain fused to the C terminus.
Obtained from the Saccharomyces Genome Database (http://www.yeastgenome.org/).
Ndc10 is sumoylated in vitro and in vivo
Given SMT3's genetic interaction with the kinetochore and the number of two-hybrid interactions between Ndc10 and the sumoylation machinery, we hypothesized that these interactions occur because Ndc10 is a target for sumoylation. To address this possibility, we tested whether bacterially expressed GST-Ndc10 is a substrate for sumoylation in an in vitro sumoylation reaction. Western blot analysis of the reaction products revealed two SUMO-modified forms of Ndc10 that were generated in an ATP-dependent manner, indicating that Ndc10 is sumoylated in vitro (Fig. 1 A). To determine whether Ndc10 is modified by SUMO in vivo, we immunoprecipitated Ndc10 from a yeast cell lysate and detected SUMO-modified proteins by Western blot analysis using an antibody that recognizes the yeast SUMO protein. Two signals were apparent by Western blotting that corresponded to the correct molecular weight to be SUMO-modified forms of Ndc10 (Fig. 1 B). Both of these signals changed electrophoretic mobility upon switching the tag on Ndc10 from 13 to 3 copies of myc, demonstrating that both of these signals represent in vivo SUMO-modified forms of Ndc10 and not a coprecipitated protein (Fig. 1 B). The presence of two modified forms of Ndc10 both in vitro and in vivo suggests that Ndc10 may be sumoylated on at least two lysine residues. However, other posttranslational modifications and/or polysumoylation on a single lysine residue cannot be ruled out as reasons for the appearance of multiple modified forms of Ndc10 from these data alone. We estimate that the fraction of SUMO-conjugated Ndc10 is ∼1% or less, which is consistent with the level of modification of most known SUMO substrates.
Figure 1.
Sumoylation of the kinetochore protein Ndc10. (A) Ndc10 is sumoylated in vitro. ATP-dependent sumoylation reactions were performed with recombinant proteins purified from E. coli without (−) or with (+) ATP. The asterisk and arrows denote unmodified and SUMO-modified forms of Ndc10, respectively. (B) Ndc10 is sumoylated in vivo. (C) The E3 proteins Siz1 and Nfi1 function in Ndc10 sumoylation.
The addition of SUMO is often facilitated by a protein ligase (E3; Johnson, 2004; Hay, 2005), and the observed two-hybrid interaction between the E3 protein Nfi1 and Ndc10 suggests that Nfi1 may act as an E3 for Ndc10 sumoylation. We found that strains carrying deletions (nfi1Δ and siz1Δ) or point mutations (mms21-11) in known E3 proteins individually had no effect on Ndc10 SUMO modification levels (Fig. 1 C). However, in the absence of both NFI1 and SIZ1, Ndc10 sumoylation was reduced (Fig. 1 C), indicating a functional redundancy between Nfi1 and Siz1 in targeting Ndc10 for sumoylation. It is worth noting that Ndc10 has also been implicated as a substrate for ubiquitination (Yoon and Carbon, 1995; Kopski and Huffaker, 1997), raising the possibility of alternative regulation through both sumoylation and ubiquitination. However, in preliminary experiments, we have not been able to identify ubiquitinated forms of Ndc10. During the course of this study, Ndc10 was also identified as a sumoylated protein in a proteomic analysis of sumoylated proteins in yeast (Wohlschlegel et al., 2004).
Dynamics of Ndc10 sumoylation
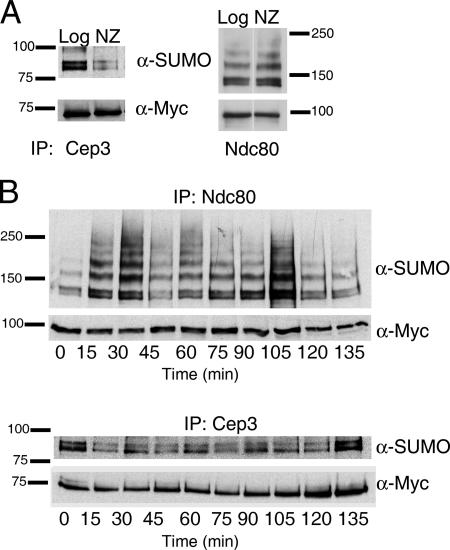
To begin investigating the possible functions of Ndc10 sumoylation, we examined the modification state of Ndc10 at discrete arrest points within the mitotic cell cycle. The mating pheromone α factor (αF) and the microtubule-destabilizing drug nocodazole (NZ) were used to arrest cells in G1 and G2/M, respectively. The terminal arrest of each population after treatment was verified by flow cytometry (unpublished data). Immunoprecipitations (IPs)/Western blotting showed that the sumoylation of Ndc10 was decreased to almost undetectable levels in those cells treated with NZ, whereas it was maintained in αF-treated cells (Fig. 2 A). The loss of sumoylation is not a general consequence of NZ treatment because other sumoylated proteins were not affected (Fig. 3 A, Ndc80) or exhibited increased levels of sumoylation upon NZ treatment (Johnson and Blobel, 1999). Moreover, the loss of sumoylation is not a result of the G2/M arrest because Ndc10 does not become desumoylated in cells that are arrested in G2/M with temperature-sensitive alleles in the anaphase-promoting complex/cyclosome (Fig. 2 A). To further analyze Ndc10 sumoylation during the cell cycle, we assayed the modification state of Ndc10 in a synchronized population of cells as they progressed through mitosis after release from αF arrest (Fig. 2 B). Timing of the progression of cells through the cell cycle was monitored by flow cytometry (Fig. 2 C). The modification state of Ndc10 remained relatively constant throughout the synchronized cell cycle, and, in replicate experiments, the fluctuation observed between individual time points was not consistent, suggesting that Ndc10 sumoylation does not change dramatically over the cell cycle. These results suggest that the loss of Ndc10 sumoylation in NZ-arrested cultures may be a direct consequence of NZ addition relating to checkpoint activation and/or loss of microtubules.
Figure 2.
Dynamics of Ndc10 sumoylation. (A) Ndc10 sumoylation is reduced in NZ-treated cells. IP/Western blotting was performed on protein extracts from logarithmic, G1- (AF), and G2/M (NZ)-arrested yeast cell cultures or cdc26Δ culture arrested in G2/M by shift to the nonpermissive temperature of 37°C for 3 h. (B) Ndc10 sumoylation is constant over an unperturbed cell cycle. An αF-synchronized population was released into rich media at 30°C, and samples for analysis by IP/Western blotting were taken every 15 min. (C) DNA content was analyzed at each 15-min interval by flow cytometry to determine cell cycle progression. (D) The loss of sumoylation in NZ requires Ulp2. Cells that are WT (+) or mutant (Δ) for the SUMO protease Ulp2 were assayed for sumoylation after the treatment of cells with DMSO (−) or DMSO + NZ (+).
Figure 3.
Sumoylation of the kinetochore proteins Cep3 and Ndc80. (A) Cep3 and Ndc80 are sumoylated in vivo. (B) Ndc80 and Cep3 sumoylation is constant over an unperturbed cell cycle. An αF-synchronized population was released into rich media at 30°C, and samples for analysis by IP/Western blotting were taken every 15 min. DNA content was analyzed at each 15-min interval by flow cytometry to determine cell cycle progression (not depicted; see Fig. 2 C).
Specific cysteine proteases are responsible for cleaving SUMO from modified substrates and for maturation of the SUMO protein itself (Johnson, 2004). Of the two SUMO proteases in yeast, Ulp2 localizes to the nucleus, placing it in the proper location to mediate the removal of SUMO from Ndc10 during NZ exposure. We treated ulp2Δ cells with NZ and found that the sumoylated forms of Ndc10 were still present (Fig. 2 D); therefore, Ulp2 is required for the loss of Ndc10 sumoylation in response to NZ treatment.
Kinetochore proteins Cep3 and Ndc80 are sumoylated
To understand whether sumoylation is a common modification on kinetochore proteins, we tested a panel of 14 proteins comprised of inner (Cep3 and Mif2), central (Chl4, Cnn1, Ctf3, Ctf19, Iml3, Ndc80, Nuf2, and Spc24), and outer kinetochore components (Dam1 and Spc34) for modification by SUMO. We also tested the spindle checkpoint protein Bub1 and the kinetochore-associated microtubule-binding protein Bik1. Of all the proteins tested, Cep3 and Ndc80 were found to be sumoylated by IP/Western blotting, with the detection of a doublet signal for Cep3 and a ladder of sumoylated forms of Ndc80 (Fig. 3 A). Like Ndc10, the fraction of SUMO-conjugated Cep3 is 1% or less, whereas we estimate Ndc80 to be modified at slightly higher levels (∼1–5%). Ndc80 and Cep3 were also found to be sumoylated throughout the cell cycle, and, in replicate experiments, the fluctuations observed between individual time points was not consistent, suggesting that Ndc80 and Cep3 sumoylation, as observed for Ndc10, is not altered dramatically over the cell cycle (Fig. 3 B).
Ndc80 was recently identified as a sumoylated protein in a proteomic analysis of sumoylated proteins in yeast by mass spectrometry along with the kinetochore proteins Bir1, Sli15, and Mcm21 (Panse et al., 2004; Wohlschlegel et al., 2004; Zhou et al., 2004; Denison et al., 2005; Hannich et al., 2005; Wykoff and O'Shea, 2005). Our analysis revealed that Cep3, which is a member of the CBF3 complex with Ndc10 that also localizes to the mitotic spindle (Bouck and Bloom, 2005), showed decreased sumoylation in response to NZ, an effect similar to that seen for Ndc10 (Fig. 3 A). For Ndc80, which does not localize to the mitotic spindle, sumoylation does not change in NZ-treated cells, suggesting that desumoylation during the NZ-induced spindle checkpoint arrest (Fig. 3 A) may be associated only with those proteins interacting with the mitotic spindle. Furthermore, the differing response in NZ is indicative of a distinct role for Ndc80 sumoylation relative to that of Ndc10 and Cep3.
Identification of lysine residues required for Ndc10, Ndc80, and Cep3 sumoylation
Sumoylation often occurs on lysine residues found in the consensus motif ΨKxE, where Ψ is any large hydrophobic residue and x is any residue (Johnson, 2004). To identify the possible sites of sumoylation in Ndc10, Ndc80, and Cep3, we mutated select lysine residues in amino acid sequences that resemble the consensus sequence for sumoylation. In total, we individually mutated 11, 14, and 6 potential sumoylation sites in Ndc10, Ndc80, and Cep3, respectively (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200605019/DC1). Strains expressing each K→R mutant were assayed for growth at 37°C, chromosome instability (CIN), and changes in sumoylation by IP/Western blotting. None of the 31 K→R individual mutations in Ndc10, Cep3, or Ndc80 caused a Ts phenotype or CIN. However, lysine residues were identified that when mutated caused an overall decrease in the SUMO modification state of Ndc10 (K651, 652R, and K779R) or abolished one specific SUMO-modified form of Ndc10 (K556R; Fig. 4 A). Given Ndc10's localization to the mitotic spindle, these three Ndc10 K→R mutants were also assayed for proper subcellular localization. In all three cases, the localization of Ndc10 was indistinguishable from that of wild type (WT; unpublished data).
Figure 4.
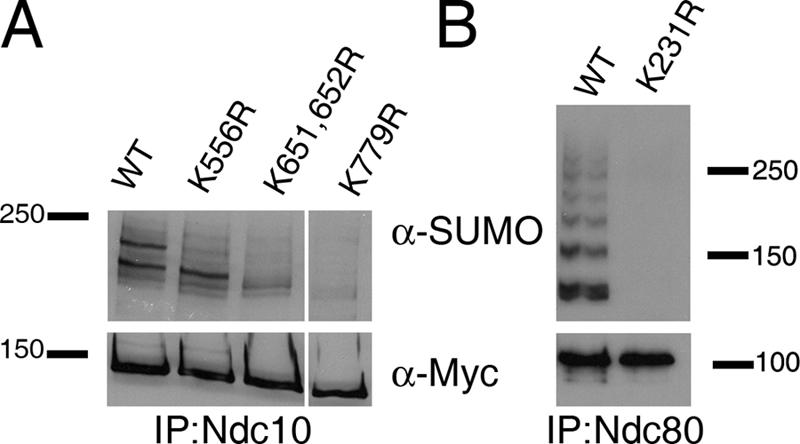
Identification of lysine residues in Ndc10 and Ndc80 that affect sumoylation. (A) Residues K556, K651/652, and K779 affect the sumoylation of Ndc10 in vivo. (B) Residue K231 is required for the sumoylation of Ndc80 in vivo. Mutations were introduced at the endogenous locus for each mutant strain, and protein extracts were generated for assaying sumoylation by IP/Western blotting.
Of the 14 K→R mutations in Ndc80, one lysine mutation (K231R) abolished the majority of Ndc80 sumoylation (Fig. 4 B), whereas none of the six K→R mutations in Cep3 affected its sumoylation state (not depicted). The complete loss of sumoylation in Ndc80 through a single mutation suggests that K231 may be required for SUMO modification of other lysine residues, which leads to the ladder of modified species, or that this site is polysumoylated. To distinguish between these possibilities, we checked Ndc80 sumoylation in a strain that carries a form of SUMO with mutations at positions K11, 15, and 19 that eliminate the formation of polysumoylated chains (Bylebyl et al., 2003). In Ndc80 IPs/Western blots using this strain, the ladder of modified Ndc80 proteins recognized by the SUMO antibody remained present, indicating that the Ndc80 K231 site is not polysumoylated (unpublished data).
Ndc10 mitotic spindle localization is lost in the ndc10-4xK→R mutant
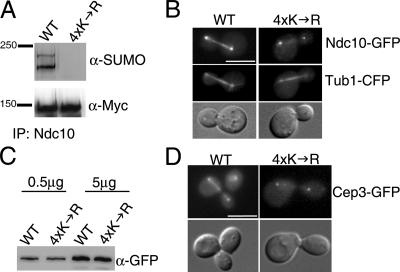
Both in vitro and in vivo Ndc10 sumoylation data showed two modified forms of Ndc10, indicating that Ndc10 may be modified by SUMO on more than one site (Fig. 1). Moreover, lysine mutations in single SUMO consensus sites within Ndc10 did not completely block sumoylation, suggesting that there is more than one sumoylation site in Ndc10 (Fig. 4 A). If each of these lysine residues represents a potential sumoylation site, defects caused by the loss of one site may be masked as a result of sufficient levels of modification at one of the other two sites. To circumvent this problem, we combined all three potential sumoylation site mutations to create Ndc10-4xK→R, which carries four lysine to arginine mutations at residues 556, 651, 652, and 779. By IP/Western blotting, Ndc10-4xK→R lacked any detectable levels of sumoylation, indicating that these three sites are required for the majority of Ndc10 sumoylation (Fig. 5 A).
Figure 5.
Loss of Ndc10 spindle localization. (A) Combining mutations that individually affect Ndc10 sumoylation results in the loss of sumoylation in vivo. NDC10 was mutated at the endogenous locus to encode K556R, K651, K652, and K779R (4xK→R). (B) Ndc10-4xK→R mislocalizes from the mitotic spindle. Mutant and WT protein was tagged with GFP and imaged in live cells. To visualize the mitotic spindle, Tub1-CFP was also introduced into each strain. (C) Ndc10-4xK→R is expressed at normal levels. Whole cell protein extracts were analyzed by Western blotting using α-GFP antibodies (Roche). (D) Cep3 mislocalizes from the mitotic spindle in ndc10-4xK→R strains. Cep3 was tagged with GFP in a WT or ndc10-4xK→R strain and imaged in live cells. Bars, 5 μm.
Combining the mutations at residues 556, 651, 652, and 779 also caused a dramatic reduction in the amount of Ndc10 localized to the anaphase mitotic spindle both along the length of the spindle and at the spindle midzone (Fig. 5 B), which was not caused by a change in the overall protein level of Ndc10-4xK→R (Fig. 5 C). Using a cdc5-10 allele to arrest ndc10-4xK→R strains in anaphase with elongated spindles, we quantified the localization defects associated with these mutations and found that 57% of cells had undetectable levels of Ndc10-4xK→R on the spindle, whereas 43% had faint staining that was observable but well below WT levels (n = 200). This result is in contrast to WT cells, which showed robust spindle staining in 88% of cells and faint staining in only 12% of the population (n = 200). Ndc10 may be on the spindle as part of the CBF3 complex because Cep3 was recently shown to localize to the mitotic spindle (Bouck and Bloom, 2005). We found that Cep3 was also mislocalized from mitotic spindles in ndc10-4xK→R mutants, supporting the idea that Ndc10 and Cep3 are present on the spindle as a complex (Fig. 5 D). The chromosomal passenger protein Sli15 and the microtubule-associated protein Ase1 still localized normally in the ndc10-4xK→R mutant (not depicted) as did Bir1 (see Fig. 7 A), suggesting that the mislocalization of Ndc10 and Cep3 is not caused by a gross defect in spindle structure. We also found that ndc10-4xK→R strains are not Ts and did not show changes in Ndc10's ability to dimerize (Russell et al., 1999), suggesting that these mutations have not altered the structure of the protein (unpublished data).
Figure 7.
Bir1 is sumoylated in an Ndc10-dependent manner independently of CBF3 function. (A) Bir1-GFP remains spindle localized in ndc10-4xK→R anaphase cells. BIR1 was tagged with GFP in a WT and ndc10-4xK→R strain and imaged in live cells. For B–E and G, Bir1 was immunoprecipitated with α-myc–conjugated beads and used for Western blot analysis with α-myc, α-Ndc10, or α-SUMO antibodies as indicated from WT or ndc10-4xK→R strains. (B) Ndc10 and Bir1 interact in vivo independently of Ndc10 sumoylation. (C) The interaction between Ndc10 and Bir1 does not require microtubules (NZ) nor is it specific to mitotic cells (αF). (D) Bir1 is sumoylated in vivo but reduced in response to NZ treatment. (E) Bir1 sumoylation is reduced in an ndc10-4xK→R strain. (F) Cep3 and Ndc80 sumoylation is unchanged in the ndc10-4xK→R mutant strain. (G) Bir1 sumoylation is not affected by mutations in CBF3 components other than Ndc10. Protein extracts from WT, ndc10-1, cep3-1, and ctf13-30 were made after 3 h at 37°C. Bar, 5 μm.
Beyond Ndc10's canonical function at the kinetochore in chromosome segregation, recent studies have described roles for the CBF3 complex and Ndc10 in cytokinesis (Bouck and Bloom, 2005; Gillis et al., 2005). For this reason, we tested ndc10-4xK→R mutants for multibudding, proper septin ring maturation, and defects in the axial pattern of haploid budding. In these assays, we did not observe any differences between WT and ndc10-4xK→R (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200605019/DC1; and not depicted).
ndc10-4xK→R mutants display CIN
In addition to the spindle localization defects, ndc10-4xK→R strains showed increased CIN in a color sector assay (Fig. 6 A). When quantified by half-sector analysis (Koshland and Hieter, 1987; Hyland et al., 1999), ndc10-4xK→R strains had rates of chromosome loss and chromosome nondisjunction 160 and 60× greater, respectively, than WT. In comparison, the loss of checkpoint function in bub1Δ or bub3Δ strains causes a 50× increase in the rate of chromosome loss (Warren et al., 2002). We found that these strains also lose endogenous chromosomes at an increased rate as determined by a diploid bimating assay (Spencer et al., 1990). In this assay, ndc10- 4xK→R homozygous diploid strains formed mating colonies at a rate 10× that of WT diploids, presumably as a result of the loss of chromosome III (2N-1; Fig. 6 B). Although ndc10-4xK→R strains showed increased rates of CIN, we have been unable to observe a delay in the cell cycle progression of a αF-synchronized cell culture (Fig. 6 C). ndc10-4xK→R strains are also G2/M checkpoint proficient in both their ability to arrest and recovery from NZ exposure (unpublished data). These results suggest that the defect causing chromosome missegregation is not eliciting a checkpoint response and frequent repair but is more likely to be either a rare event that always leads to failure or an event that does not trigger a G2/M checkpoint response at all.
Figure 6.
ndc10-4xK→R strains have increased rates of chromosome missegregation and mitotic spindle defects. (A) ndc10-4xK→R strains lose an artificial chromosome fragment at elevated rates. Strains carrying a nonessential chromosome fragment suppressing red pigment formation were plated on nonselective media. The formation of red color indicates the loss of the chromosome fragment. (B) ndc10-4xK→R strains lose endogenous chromosomes. Independent isolates of WT and ndc10-4xK→R diploid strains were mated with haploid tester strains, and mating products were selected. Two representative patches are shown. (C) Cell cycle progression is not delayed in ndc10-4xK→R strains. WT and ndc10-4xK→R strains were arrested in G1 with αF and released into YPD at 30°C. DNA content was analyzed at each 15-min interval by flow cytometry to determine cell cycle progression. (D) ndc10-4xK→R strains are benomyl sensitive. Fivefold serial dilutions of each strain were spotted on YPD plates containing 0, 10, 15, or 20 g/ml benomyl and incubated at 30°C for 2 d. Benomyl-hypersensitive tub1-1 and the resistant strain tub2-104 are included as controls. (E) ndc10-4xK→R strains have mitotic spindles of abnormal length. Anaphase mitotic spindle lengths were measured in asynchronous cultures of WT and ndc10-4xK→R strains containing Tub1-CFP.
ndc10-4xK→R mutants have mitotic spindle defects
A common phenotype associated with mutations affecting kinetochore and/or spindle function is increased sensitivity to the microtubule-destabilizing drug benomyl. ndc10-4xK→R strains displayed benomyl sensitivity (Fig. 6 D), which, taken together with the mitotic spindle mislocalization phenotype, raises the possibility that the loss of Ndc10 sumoylation may be causing specific defects in spindle function. To investigate this possibility, we measured the lengths of mitotic spindles in anaphase-stage cells of an asynchronous culture. In the ndc10-4xK→R mutant, we found that anaphase spindles are of abnormal length (representative images in Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200605019/DC1), with spindles averaging 7.30 ± 0.08 μm (n = 483) compared with 6.62 ± 0.06 μm (n = 443) for WT cells. In comparing the distribution of spindle lengths graphically, we noted that the ndc10-4xK→R mutants had abnormally long anaphase spindles ranging up to 10–12 μm, which are lengths that are never seen in WT cells (Fig. 6 E). The observed spindle defect and CIN, which is associated with no observable G2/M delay, suggests that the defects caused by the loss of Ndc10 sumoylation is related to events in anaphase after the G2/M checkpoint has been silenced (e.g., spindle elongation).
Bir1 and Ndc10-4xK→R physically interact
In anaphase, Ndc10 and Bir1 colocalize on the mitotic spindle in a Bir1-dependent manner, suggesting that Ndc10 and Bir1 are part of a complex on the mitotic spindle (Bouck and Bloom, 2005; Widlund et al., 2006). In the ndc10-4xK→R strain, Ndc10 no longer localized to the mitotic spindle (Fig. 5 B), whereas Bir1 remained spindle bound (Fig. 7 A). Therefore, we hypothesized that the spindle length defect and mislocalization of Ndc10 was caused by a disruption of the Ndc10–Bir1 protein interaction. However, co-IPs demonstrated that the Ndc10-4xK→R interaction with Bir1 was comparable with that seen in WT cells (Fig. 7 B). We also found that the interaction between Ndc10 and Bir1 was not disrupted in cells arrested with αF or NZ, demonstrating that this interaction is not specific to mitosis and is not microtubule dependent (Fig. 7 C). These results suggest that the observed interaction between Ndc10 and Bir1 occurs independently of the mitotic spindle and that the spindle length defect in the ndc10-4xK→R strain is not caused by the failure of Ndc10-4xK→R to interact with Bir1.
Bir1 is sumoylated in an Ndc10-dependent manner
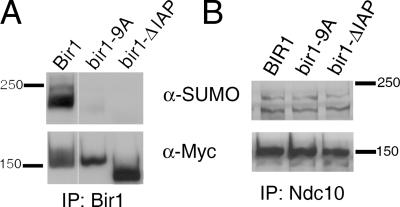
Bir1 is a phosphorylated protein that was also recently identified as a sumoylated protein in proteomic studies (Wohlschlegel et al., 2004; Zhou et al., 2004; Widlund et al., 2006). We were able to confirm that Bir1 is sumoylated by IP/Western blotting, with an estimated 1% of Bir1 being SUMO conjugated in an asynchronous culture. As seen for Ndc10, we also observed that the treatment of cells with NZ resulted in the loss of Bir1 sumoylation (Fig. 7 D). Given that the interaction between Ndc10 and Bir1 is not disrupted in an ndc10-4xK→R strain, we wondered whether the loss of Ndc10 sumoylation and/or spindle localization resulted in a change in Bir1's modification state. Therefore, we assayed both the phosphorylation and sumoylation state of Bir1 in the ndc10-4xK→R mutant strain. Western blots show that the overall appearance of Bir1 phosphorylation (as indicated by the diffuse Western signal representing phosphorylated forms of Bir1) is unchanged in the mutant as compared with WT (Fig. 7 E, α-myc blot) but that the sumoylation of Bir1 is reduced in the ndc10-4xK→R mutant to undetectable levels (Fig. 7 E, α- SUMO blot). We tested Cep3 and Ndc80 for changes in the sumoylation state in the ndc10-4xK→R mutant (Fig. 7 F) but found no significant differences compared with WT; thus, the loss of sumoylation in the ndc10-4xK→R mutant appears to be specific to the modification of Bir1. These data suggest that the sumoylation of Ndc10 is required for Bir1 sumoylation.
Bir1 sumoylation is independent of CBF3 function
Ndc10's effect on Bir1 sumoylation may be dependent on the activity of the CBF3 complex or may occur via a CBF3-independent mechanism. To distinguish these two possibilities, we tested Bir1 sumoylation in strains carrying Ts mutations in the CBF3 component Cep3 (cep3-1) or Ctf13 (ctf13-30). In both cases, when cells were arrested at the nonpermissive temperature, Bir1 sumoylation was detected; however, the loss of sumoylation was observed when the ndc10-1 Ts allele was used to abrogate Ndc10's function. This result indicates that Ndc10's effect on Bir1 sumoylation is not caused by altered CBF3 activity (Fig. 7 G).
Discussion
In this study, we report that the kinetochore proteins Ndc10, Cep3, Ndc80, and Bir1 are substrates for sumoylation. In the case of Ndc10, the effect of lysine mutations on Ndc10's localization demonstrates that sumoylation demarcates a specific subset of Ndc10 that associates with the mitotic spindle. This demarcation most likely occurs through an alteration of Ndc10's binding interactions with microtubules or microtubule-associated proteins, leading to the establishment or maintenance of spindle localization. Our data also assigns a function to the fraction of Ndc10 protein on the mitotic spindle in controlling mitotic spindle dynamics.
Several other kinetochore proteins also localize to the spindle during anaphase, including Bir1, Cep3, Cin8, Dam1, Duo1, Ipl1, Sli15, Slk19, and Stu2, most of which have also been shown to play a role in controlling spindle dynamics (Hoyt et al., 1992; Hofmann et al., 1998; Biggins et al., 1999; Jones et al., 1999; Zeng et al., 1999; Kosco et al., 2001; Bouck and Bloom, 2005; Widlund et al., 2006). The localization of Ipl1, Sli15, and Ndc10 to the spindle requires the function of the phosphatase Cdc14 that is activated in anaphase to regulate mitotic exit (Pereira and Schiebel, 2003; Stegmeier and Amon, 2004; Bouck and Bloom, 2005). Moreover, Ndc10, Bir1, Cep3, and Sli15 are SUMO substrates (Wohlschlegel et al., 2004; and this study), indicating that these spindle-associated kinetochore proteins may be regulated via common mechanisms to control mitotic spindle dynamics during anaphase.
The regulation of spindle dynamics likely includes delivery of these kinetochore proteins to the spindle midzone, which may be used as a signal to coordinate the collapse of the mitotic spindle and to initiate other late-stage mitotic events once chromosome segregation has occurred. Given ndc10-4xK→R's long spindle phenotype, it is possible that the signal to commence spindle disassembly is delayed as a result of the absence of Ndc10 on the mitotic spindle, providing time for additional spindle elongation. It is currently unknown whether the altered spindle dynamics are also responsible for the increase in CIN observed in ndc10-4xK→R mutants, but the lack of an observable cell cycle delay is suggestive of events that are invisible to the mitotic checkpoint machinery or that occur during stages of the cell cycle when the checkpoint has already been satisfied (e.g., anaphase). Intriguingly, Ndc10 also localizes to microtubules in telophase and into G1 of the next cell cycle, raising the possibility that microtubule-associated Ndc10 may be required in nonmitotic stages of the cell cycle (Bouck and Bloom, 2005). This may include S phase when centromere DNA is replicated and kinetochore microtubule attachments are being established.
Ndc10's localization to the mitotic spindle is dependent on sumoylation and Bir1 (Bouck and Bloom, 2005; Widlund et al., 2006; and this study). Our work demonstrates that the interaction between Ndc10 and Bir1 occurs in nonmitotic cells and is independent of microtubules, Ndc10 spindle localization, and sumoylation of Ndc10 or Bir1. How then does Bir1 facilitate Ndc10's spindle association if not by physically interacting with Ndc10 on the mitotic spindle? Based on localization data, the other structure at which these two proteins colocalize is the kinetochore (Widlund et al., 2006). At the kinetochore, Bir1 could direct Ndc10 to microtubules in a manner dependent on the function of the Bir1–Sli15–Ipl1 kinase complex. In support of this possibility, Ndc10 has a weak two-hybrid interaction with Sli15 (Table I). Moreover, Ndc10 has been shown to be an Ipl1 substrate for phosphorylation in vitro, and ipl1 mutants, like ndc10-4xK→R mutants, contain spindles of abnormal length (Biggins et al., 1999; Buvelot et al., 2003). In experiments that use the ipl1-321 temperature-sensitive mutant, Ndc10 shows only slightly lower levels of mitotic spindle localization (Bouck and Bloom, 2005). However, it is known that at the restrictive temperature, ipl1-321 retains a low level of kinase activity, and, thus, a phosphorylation-dependent mechanism is still possible (Pinsky et al., 2006).
In contrast to Ndc10, mutations that abolish Bir1 sumoylation (Fig. 8 A) do not result in a loss of Bir1 mitotic spindle localization (Widlund et al., 2006), implying that Bir1's association with the mitotic spindle does not rely on sumoylation. A clue to the function of Bir1's sumoylation may come from a recent study on the mammalian homologue of Bir1, Survivin. In mammals, the modification of Bir1/Survivin with ubiquitin appears to regulate dynamic protein–protein interactions that are important for chromosome segregation at the centromere (Vong et al., 2005). Intriguingly, these modifications map to the IAP region of mammalian Bir1/Survivin (Vong et al., 2005), the deletion of which results in a loss of sumoylation in yeast Bir1 (Fig. 8 A). Thus, the IAP domain may be used to regulate both yeast and mammalian Bir1/Survivin's function by lysine-directed modifications.
Figure 8.
Bir1 phosphorylation and the IAP domain are required for Bir1 sumoylation. (A) Bir1 sumoylation is undetectable in the bir1-9A and bir1-ΔIAP mutants. Bir1-ΔIAP (lacks IAP) interacts with Ndc10 and the mitotic spindle, whereas Bir1-9XA (lacking nine potential phosphorylation sites) does not interact with Ndc10 but remains spindle bound (Widlund et al., 2006). (B) Ndc10 sumoylation is not dependent on Bir1 sumoylation or on a physical interaction with Bir1.
Once Ndc10 and Bir1 are on the mitotic spindle, it appears that these two proteins function to regulate spindle dynamics in an opposing manner given that the spindle length defect observed in ndc10-4XK→R mutants is in contrast to that seen in Bir1 mutants in which spindles fail to fully elongate and are on average shorter than WT (Widlund et al., 2006; and this study). The interplay between these two proteins is further illustrated by the fact that in the ndc10-4xK→R mutant, Bir1 sumoylation is undetectable even though the Ndc10 mutant protein and Bir1 still interact normally. This dependence is not reciprocal in that Ndc10 sumoylation is not affected by mutations that block Bir1's sumoylation or that disrupt the interaction between Bir1 and Ndc10 (Fig. 8 B). The reliance on Ndc10 being competent for sumoylation suggests that Ndc10 requires prior modification with SUMO before the sumoylation of Bir1 can occur and is indicative of a cascade of SUMO modification events in which Ndc10 functions in trans to facilitate the modification of Bir1 by SUMO.
Although Ndc10 is not conserved in higher eukaryotes, Bir1 and Ndc80 are well conserved. An important question is whether the sumoylation of kinetochore proteins, including Bir1 and Ndc80, function in an analogous manner in these evolutionally diverse organisms. Although neither Bir1 nor Ndc80 homologues have been shown to be sumoylated in higher eukaryotes, there is evidence linking sumoylation to kinetochore and mitotic spindle functions. For example, SUMO-2–modified proteins are enriched at the inner centromere of chromatids in Xenopus laevis egg extracts, and the alteration of SUMO modification of chromosomal substrates by SUMO-2 causes a block in the segregation of sister chromatids in anaphase (Azuma et al., 2003, 2005). Moreover, RanGap1, the first identified sumoylated substrate, is targeted to mitotic spindles and kinetochores, and, like Ndc10, mutations that abolish sumoylation lead to RanGAP1 mislocalization from the spindle (Matunis et al., 1996; Mahajan et al., 1997; Saitoh et al., 1997; Joseph et al., 2002, 2004). Complexed with RanGAP1 is the SUMO E3 ligase RanBP2, which, when depleted in mitotic cells, results in the mislocalization of RanGAP1, the spindle checkpoint proteins Mad1 and Mad2, and the kinetochore proteins CENP-E and -F (Joseph et al., 2004). The phenotypic consequences of these effects include the accumulation of mitotic cells with multipolar spindles and unaligned chromosomes. The localization of RanBP2 to kinetochores places an E3 protein near the kinetochore, highlighting the possibility that there may be many proteins of the kinetochore targeted for sumoylation.
The number of sumoylated substrates in yeast and higher eukaryotes continues to grow rapidly, but the biological functions of the majority of these modifications remains elusive. The data presented here provide evidence for the regulation of chromosomal passenger proteins and kinetochore proteins by SUMO modification, including the localization of Ndc10 to the mitotic spindle during anaphase. Findings in higher eukaryotes provide evidence that the mechanism for localizing proteins to microtubules via sumoylation may be conserved (e.g., RanGAP1). Future work will be required to discern the details and extent of this mechanism in eukaryotic cells and the importance of sumoylation in regulating the mitotic program.
Materials and methods
Yeast strains, plasmids, and microbial techniques
Yeast strains (S288C background) and plasmids used in this study are listed in Tables S2 and S3 (available at http://www.jcb.org/cgi/content/full/jcb.200605019/DC1). Mutant alleles were integrated into the yeast genome at the endogenous locus by cotransformation of a PCR product carrying the desired mutations and a PCR product containing a nutritional marker (URA3). Integration of the desired mutation was confirmed by DNA sequencing. To arrest cell cultures, 1 mg/ml αF (in methanol) or 5 mg/ml NZ (in DMSO) was added, and cultures were incubated for 2 h at 30°C. To assay benomyl sensitivity, benomyl was added at the indicated concentration to YPD media, dimethyl sulfoxide was used in the control plate (0 μg/ml benomyl), and fivefold serial dilutions were spotted on the plates and grown at 30°C for 2 d. Flow cytometry analysis to monitor DNA content was performed as previously described (Haase and Lew, 1997).
Genome-wide two-hybrid screen
NDC10 was cloned into pOBD2 and pBDC using standard techniques (Cagney et al., 2000; Millson et al., 2003). Fusion of the DNA-binding domain to the C terminus (pBDC) but not the N terminus of Ndc10 (pOBD2) resulted in a functional protein as judged by the ability to rescue an ndc10Δ strain. Two independent genome-wide two-hybrid screens were performed using an activation domain array (Hazbun et al., 2003) as described previously (Uetz et al., 2000). The two-hybrid positives from these genome-wide screens were reconfirmed by repeating the two-hybrid assay. The identities of the activation domain fusions were confirmed by rescuing plasmids and sequencing.
In vitro and in vivo sumoylation assays
GST-Ndc10 was expressed in Escherichia coli strain BL21 from a pGex 4T-2 vector (GE Healthcare) and was purified using glutathione beads. In vitro sumoylation reactions were performed using bacterially expressed proteins (expression plasmids were provided by L. McIntosh, University of British Columbia, Vancouver, British Columbia, Canada) as previously described (Macauley et al., 2005), and Western blotting was performed with an Ndc10 antibody (Measday et al., 2005). To detect sumoylation in vivo, yeast cells (100–200 OD600) were lysed by bead beating (10 times for 30 s) in 2.5 ml lysis buffer (50 mM Tris-HCl, pH 8.0, 5 mM EDTA, 150 mM NaCl, 0.2% Triton X-100, complete protease inhibitor [one tablet per 25 mL; Roche], 10 mM N-ethylmaleimide, 2 mM PMSF, and 20 μg each of leupeptin, aprotinin, and pepstatin per milliliter) on ice. Lysates were cleared by centrifugation at 30,000 g for 20 min, and soluble protein concentrations were determined by protein assay (Bio-Rad Laboratories). Equal amounts of protein were incubated with α-myc–conjugated beads for 3 h at 4°C and washed four times with cold lysis buffer for 2 min. Immunoprecipitated protein was then eluted with lysis buffer containing 2% SDS at 42°C for 15 min. 2.5 μL of the eluted protein was used for Western blotting with α-myc antibody to confirm pull down of the tagged protein; 25 μl was used for blotting with α-SUMO antibody to detect sumoylated protein. To detect the Ndc10–Bir1 interaction, 25 μl Bir1 IP was used for Western blotting with α-Ndc10 antibody. α-SUMO polyclonal antibodies were generated in rabbits (Covance Research Products) as previously described (Johnson and Blobel, 1999). Initial experiments also made use of an α-SUMO polyclonal antibody provided by E. Johnson (Thomas Jefferson University, Philadelphia, PA).
Light, fluorescence, and immunofluorescence microscopy
Strains used for microscopy were grown in either YPD (for synchronous culture experiments) or in fluorescent protein medium (minimal medium supplemented with adenine and containing 6.5 g/L sodium citrate). Cells were imaged at room temperature using a microscope (Axioplan 2; Carl Zeiss MicroImaging, Inc.) with a plan-Apochromat 100× NA 1.4 differential interference contrast oil immersion objective (Carl Zeiss MicroImaging, Inc.) with filter sets 38 and 47 (Carl Zeiss MicroImaging, Inc.) and filter 488000 (Chroma Technology Corp.). 3D images (0.25-μm steps) were acquired with a camera (CoolSNAP HQ; Roper Scientific) and analyzed using MetaMorph software (Invitrogen). Images are presented as maximum intensity 2D projections. For fluorescence microscopy, WT and mutant proteins were tagged at the endogenous locus with GFP or a GFP variant (Longtine et al., 1998). Tub1-CFP–containing strains were generated by integrating plasmid pSB375 (a gift from K. Bloom, University of North Carolina, Chapel Hill, NC) digested with StuI at the URA3 locus. Spindle length measurements were performed on asynchronous cultures of Tub1-CFP–containing cells fixed in 70% ethanol and 200 mM Tris-HCl, pH 8.0, which was judged to be in anaphase by the presence of part of the spindle in the daughter cell. To visualize the haploid budding pattern, cells were incubated in PBS with 20 μg/ml Calcofluor white (Fluorescent Brightener 28; Sigma-Aldrich) at 25°C for 5 min, washed in PBS, and visualized for bud scar staining.
CIN assays
Quantitative half-sector analysis was performed as described previously (Koshland and Hieter, 1987; Hyland et al., 1999). To perform the diploid bimater assay, 10 single colonies were patched onto YPD plates, replica plated to both MATa and MATα lawns, and mating products were selected. The median number of mating products from the 10 patches was compared with WT in order to calculate the increase in frequency over mating of a WT diploid strain.
Online supplemental material
Fig. S1 shows that ndc10-4xK→R strains do not have cytokinesis-related defects. Fig. S2 contains representative images of CFP-Tub1–marked anaphase mitotic spindles in WT and ndc10-4xK→R strains. Table S1 lists the potential sumoylation sites targeted by site-directed mutagenesis in Ndc10, Ndc80, and Cep3. Tables S2 and S3 list the yeast strains and plasmids used in this study, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200605019/DC1.
Supplementary Material
Acknowledgments
We would like to thank Kerry Bloom, Erica Johnson, and Lawrence McIntosh for their gifts of reagents. We are grateful to Erica Johnson, Doug Koshland, Vivien Measday, Forrest Spencer, and members of the Hieter laboratory for their support, helpful discussions, and input into this project.
This work was supported by awards to B. Montpetit from the Natural Sciences and Engineering Research Council of Canada and the Michael Smith Foundation for Health Research. S. Fields is supported by the National Center for Research Resources (grant PHS P41 RR11823) and is an investigator of the Howard Hughes Medical Institute. P. Hieter is supported by the Canadian Institutes of Health Research (grant MOP-38096) and the U.S. National Institutes of Health (grant P01-CA0161519).
Abbreviations used in this paper: αF; α factor; CIN, chromosome instability; IAP, inhibitor of apoptosis; IP, immunoprecipitation; NZ, nocodazole; SUMO, small ubiquitin-like modifier; Ulp, ubiquitin-like protease; WT, wild type.
References
- Adams, R.R., M. Carmena, and W.C. Earnshaw. 2001. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 11:49–54. [DOI] [PubMed] [Google Scholar]
- Azuma, Y., A. Arnaoutov, and M. Dasso. 2003. SUMO-2/3 regulates topoisomerase II in mitosis. J. Cell Biol. 163:477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma, Y., A. Arnaoutov, T. Anan, and M. Dasso. 2005. PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. EMBO J. 24:2172–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachant, J., A. Alcasabas, Y. Blat, N. Kleckner, and S.J. Elledge. 2002. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell. 9:1169–1182. [DOI] [PubMed] [Google Scholar]
- Biggins, S., and C.E. Walczak. 2003. Captivating capture: how microtubules attach to kinetochores. Curr. Biol. 13:R449–R460. [DOI] [PubMed] [Google Scholar]
- Biggins, S., F.F. Severin, N. Bhalla, I. Sassoon, A.A. Hyman, and A.W. Murray. 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13:532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck, D.C., and K.S. Bloom. 2005. The kinetochore protein Ndc10p is required for spindle stability and cytokinesis in yeast. Proc. Natl. Acad. Sci. USA. 102:5408–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvelot, S., S.Y. Tatsutani, D. Vermaak, and S. Biggins. 2003. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J. Cell Biol. 160:329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylebyl, G.R., I. Belichenko, and E.S. Johnson. 2003. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J. Biol. Chem. 278:44113–44120. [DOI] [PubMed] [Google Scholar]
- Cagney, G., P. Uetz, and S. Fields. 2000. High-throughput screening for protein-protein interactions using two-hybrid assay. Methods Enzymol. 328:3–14. [DOI] [PubMed] [Google Scholar]
- Denison, C., A.D. Rudner, S.A. Gerber, C.E. Bakalarski, D. Moazed, and S.P. Gygi. 2005. A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol. Cell. Proteomics. 4:246–254. [DOI] [PubMed] [Google Scholar]
- Desterro, J.M., M.S. Rodriguez, and R.T. Hay. 1998. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol. Cell. 2:233–239. [DOI] [PubMed] [Google Scholar]
- Gillett, E.S., C.W. Espelin, and P.K. Sorger. 2004. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J. Cell Biol. 164:535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis, A.N., S. Thomas, S.D. Hansen, and K.B. Kaplan. 2005. A novel role for the CBF3 kinetochore-scaffold complex in regulating septin dynamics and cytokinesis. J. Cell Biol. 171:773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh, P.Y., and J.V. Kilmartin. 1993. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 121:503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase, S.B., and D.J. Lew. 1997. Flow cytometric analysis of DNA content in budding yeast. Methods Enzymol. 283:322–332. [DOI] [PubMed] [Google Scholar]
- Hannich, J.T., A. Lewis, M.B. Kroetz, S.J. Li, H. Heide, A. Emili, and M. Hochstrasser. 2005. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. 280:4102–4110. [DOI] [PubMed] [Google Scholar]
- Hay, R.T. 2005. SUMO: a history of modification. Mol. Cell. 18:1–12. [DOI] [PubMed] [Google Scholar]
- Hazbun, T.R., L. Malmstrom, S. Anderson, B.J. Graczyk, B. Fox, M. Riffle, B.A. Sundin, J.D. Aranda, W.H. McDonald, C.H. Chiu, et al. 2003. Assigning function to yeast proteins by integration of technologies. Mol. Cell. 12:1353–1365. [DOI] [PubMed] [Google Scholar]
- Hoege, C., B. Pfander, G.L. Moldovan, G. Pyrowolakis, and S. Jentsch. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 419:135–141. [DOI] [PubMed] [Google Scholar]
- Hofmann, C., I.M. Cheeseman, B.L. Goode, K.L. McDonald, G. Barnes, and D.G. Drubin. 1998. Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. J. Cell Biol. 143:1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt, M.A., L. He, K.K. Loo, and W.S. Saunders. 1992. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J. Cell Biol. 118:109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, W.K., J.V. Falvo, L.C. Gerke, A.S. Carroll, R.W. Howson, J.S. Weissman, and E.K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691. [DOI] [PubMed] [Google Scholar]
- Hyland, K.M., J. Kingsbury, D. Koshland, and P. Hieter. 1999. Ctf19p: a novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J. Cell Biol. 145:15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W., and Y. Koltin. 1996. Two-hybrid interaction of a human UBC9 homolog with centromere proteins of Saccharomyces cerevisiae. Mol. Gen. Genet. 251:153–160. [DOI] [PubMed] [Google Scholar]
- Johnson, E.S. 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73:355–382. [DOI] [PubMed] [Google Scholar]
- Johnson, E.S., and G. Blobel. 1999. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J. Cell Biol. 147:981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M.H., J.B. Bachant, A.R. Castillo, T.H. Giddings Jr., and M. Winey. 1999. Yeast Dam1p is required to maintain spindle integrity during mitosis and interacts with the Mps1p kinase. Mol. Biol. Cell. 10:2377–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph, J., S.H. Tan, T.S. Karpova, J.G. McNally, and M. Dasso. 2002. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 156:595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph, J., S.T. Liu, S.A. Jablonski, T.J. Yen, and M. Dasso. 2004. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr. Biol. 14:611–617. [DOI] [PubMed] [Google Scholar]
- Kapoor, T.M., and D.A. Compton. 2002. Searching for the middle ground: mechanisms of chromosome alignment during mitosis. J. Cell Biol. 157:551–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopski, K.M., and T.C. Huffaker. 1997. Suppressors of the ndc10-2 mutation: a role for the ubiquitin system in Saccharomyces cerevisiae kinetochore function. Genetics. 147:409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosco, K.A., C.G. Pearson, P.S. Maddox, P.J. Wang, I.R. Adams, E.D. Salmon, K. Bloom, and T.C. Huffaker. 2001. Control of microtubule dynamics by Stu2p is essential for spindle orientation and metaphase chromosome alignment in yeast. Mol. Biol. Cell. 12:2870–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland, D., and P. Hieter. 1987. Visual assay for chromosome ploidy. Methods Enzymol. 155:351–372. [DOI] [PubMed] [Google Scholar]
- Li, S.J., and M. Hochstrasser. 1999. A new protease required for cell-cycle progression in yeast. Nature. 398:246–251. [DOI] [PubMed] [Google Scholar]
- Li, S.J., and M. Hochstrasser. 2000. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 20:2367–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., A. McKenzie III, D.J. Demarini, N.G. Shah, A. Wach, A. Brachat, P. Philippsen, and J.R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Macauley, M.S., W.J. Errington, M. Scharpf, C.D. Mackereth, A.G. Blaszczak, B.J. Graves, and L.P. McIntosh. 2005. Beads-on-a-string: characterization of Ets-1 sumoylated within its flexible N-terminal sequence. J. Biol. Chem. 281:4164–4172. [DOI] [PubMed] [Google Scholar]
- Mahajan, R., C. Delphin, T. Guan, L. Gerace, and F. Melchior. 1997. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 88:97–107. [DOI] [PubMed] [Google Scholar]
- Maiato, H., J. DeLuca, E.D. Salmon, and W.C. Earnshaw. 2004. The dynamic kinetochore-microtubule interface. J. Cell Sci. 117:5461–5477. [DOI] [PubMed] [Google Scholar]
- Matunis, M.J., E. Coutavas, and G. Blobel. 1996. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135:1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh, A.D., J.D. Tytell, and P.K. Sorger. 2003. Structure, function, and regulation of budding yeast kinetochores. Annu. Rev. Cell Dev. Biol. 19:519–539. [DOI] [PubMed] [Google Scholar]
- Measday, V., K. Baetz, J. Guzzo, K. Yuen, T. Kwok, B. Sheikh, H. Ding, R. Ueta, T. Hoac, B. Cheng, et al. 2005. Systematic yeast synthetic lethal and synthetic dosage lethal screens identify genes required for chromosome segregation. Proc. Natl. Acad. Sci. USA. 102:13956–13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P.B., and D. Koshland. 1995. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell. 6:793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millson, S.H., A.W. Truman, and P.W. Piper. 2003. Vectors for N- or C-terminal positioning of the yeast Gal4p DNA binding or activator domains. Biotechniques. 35:60–64. [DOI] [PubMed] [Google Scholar]
- Nacerddine, K., F. Lehembre, M. Bhaumik, J. Artus, M. Cohen-Tannoudji, C. Babinet, P.P. Pandolfi, and A. Dejean. 2005. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell. 9:769–779. [DOI] [PubMed] [Google Scholar]
- Nasmyth, K. 2005. How do so few control so many? Cell. 120:739–746. [DOI] [PubMed] [Google Scholar]
- Panse, V.G., U. Hardeland, T. Werner, B. Kuster, and E. Hurt. 2004. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J. Biol. Chem. 279:41346–41351. [DOI] [PubMed] [Google Scholar]
- Pereira, G., and E. Schiebel. 2003. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science. 302:2120–2124. [DOI] [PubMed] [Google Scholar]
- Pinsky, B.A., and S. Biggins. 2005. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 15:486–493. [DOI] [PubMed] [Google Scholar]
- Pinsky, B.A., C.V. Kotwaliwale, S.Y. Tatsutani, C.A. Breed, and S. Biggins. 2006. Glc7/protein phosphatase 1 regulatory subunits can oppose the Ipl1/aurora protein kinase by redistributing Glc7. Mol. Cell. Biol. 26:2648–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, I.D., A.S. Grancell, and P.K. Sorger. 1999. The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J. Cell Biol. 145:933–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh, H., R. Pu, M. Cavenagh, and M. Dasso. 1997. RanBP2 associates with Ubc9p and a modified form of RanGAP1. Proc. Natl. Acad. Sci. USA. 94:3736–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler, J.S., and A. Dejean. 2003. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 4:690–699. [DOI] [PubMed] [Google Scholar]
- Seufert, W., B. Futcher, and S. Jentsch. 1995. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 373:78–81. [DOI] [PubMed] [Google Scholar]
- Spencer, F., S.L. Gerring, C. Connelly, and P. Hieter. 1990. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 124:237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead, K., C. Aguilar, T. Hartman, M. Drexel, P. Meluh, and V. Guacci. 2003. Pds5p regulates the maintenance of sister chromatid cohesion and is sumoylated to promote the dissolution of cohesion. J. Cell Biol. 163:729–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier, F., and A. Amon. 2004. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38:203–232. [DOI] [PubMed] [Google Scholar]
- Uetz, P., L. Giot, G. Cagney, T.A. Mansfield, R.S. Judson, J.R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 403:623–627. [DOI] [PubMed] [Google Scholar]
- Vagnarelli, P., and W.C. Earnshaw. 2004. Chromosomal passengers: the four-dimensional regulation of mitotic events. Chromosoma. 113:211–222. [DOI] [PubMed] [Google Scholar]
- Vong, Q.P., K. Cao, H.Y. Li, P.A. Iglesias, and Y. Zheng. 2005. Chromosome alignment and segregation regulated by ubiquitination of survivin. Science. 310:1499–1504. [DOI] [PubMed] [Google Scholar]
- Warren, C.D., D.M. Brady, R.C. Johnston, J.S. Hanna, K.G. Hardwick, and F.A. Spencer. 2002. Distinct chromosome segregation roles for spindle checkpoint proteins. Mol. Biol. Cell. 13:3029–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widlund, P.O., J.S. Lyssand, S. Anderson, S. Niessen, J.R. Yates III, and T.N. Davis. 2006. Phosphorylation of the chromosomal passenger protein Bir1 is required for localization of Ndc10 to the spindle during anaphase and full spindle elongation. Mol. Biol. Cell. 17:1065–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel, J.A., E.S. Johnson, S.I. Reed, and J.R. Yates III. 2004. Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 279:45662–45668. [DOI] [PubMed] [Google Scholar]
- Wolfe, B.A., and K.L. Gould. 2005. Split decisions: coordinating cytokinesis in yeast. Trends Cell Biol. 15:10–18. [DOI] [PubMed] [Google Scholar]
- Wykoff, D.D., and E.K. O'Shea. 2005. Identification of sumoylated proteins by systematic immunoprecipitation of the budding yeast proteome. Mol. Cell. Proteomics. 4:73–83. [DOI] [PubMed] [Google Scholar]
- Yoon, H.J., and J. Carbon. 1995. Genetic and biochemical interactions between an essential kinetochore protein, Cbf2p/Ndc10p, and the CDC34 ubiquitin-conjugating enzyme. Mol. Cell. Biol. 15:4835–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, H.J., and J. Carbon. 1999. Participation of Bir1p, a member of the inhibitor of apoptosis family, in yeast chromosome segregation events. Proc. Natl. Acad. Sci. USA. 96:13208–13213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, X., J.A. Kahana, P.A. Silver, M.K. Morphew, J.R. McIntosh, I.T. Fitch, J. Carbon, and W.S. Saunders. 1999. Slk19p is a centromere protein that functions to stabilize mitotic spindles. J. Cell Biol. 146:415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, W., J.J. Ryan, and H. Zhou. 2004. Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J. Biol. Chem. 279:32262–32268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.