Abstract
Salmonella enterica, the cause of food poisoning and typhoid fever, has evolved sophisticated mechanisms to modulate Rho family guanosine triphosphatases (GTPases) to mediate specific cellular responses such as actin remodeling, macropinocytosis, and nuclear responses. These responses are largely the result of the activity of a set of bacterial proteins (SopE, SopE2, and SopB) that, upon delivery into host cells via a type III secretion system, activate specific Rho family GTPases either directly (SopE and SopE2) or indirectly (SopB) through the stimulation of an endogenous exchange factor. We show that different Rho family GTPases play a distinct role in Salmonella-induced cellular responses. In addition, we report that SopB stimulates cellular responses by activating SH3-containing guanine nucleotide exchange factor (SGEF), an exchange factor for RhoG, which we found plays a central role in the actin cytoskeleton remodeling stimulated by Salmonella. These results reveal a remarkable level of complexity in the manipulation of Rho family GTPases by a bacterial pathogen.
Introduction
Salmonella enterica, the cause of food poisoning and typhoid fever, has evolved sophisticated mechanisms to manipulate host cell functions. Central to this ability is a specialized organelle, the type III secretion system (TTSS), which mediates the transfer of a battery of bacterial proteins into host cells (Galán, 2001). Through molecular and functional mimicry of host cell proteins, these bacterial effectors of virulence can stimulate or interfere with a variety of cellular activities (Stebbins and Galán, 2001). For example, a subset of these effectors stimulates Rho family GTPases, triggering actin cytoskeleton rearrangements, macropinocytosis, and bacterial internalization into host cells (Chen et al., 1996a). The stimulation of Rho family GTPases also leads to activation of the MAPK signaling pathways as well as transcription factors, resulting in the reprogramming of host cell gene expression and the production of proinflammatory cytokines (Chen et al., 1996a; Hobbie et al., 1997). Two of these bacterial effectors are SopE and SopE2, which are highly related bacterial proteins with the capacity to catalyze nucleotide exchange on Rho family GTPase members (Hardt et al., 1998; Bakshi et al., 2000; Stender et al., 2000). Another of these effector proteins is SopB, a phosphoinositide phosphatase that can stimulate Rho family GTPase-dependent actin cytoskeleton rearrangements by unknown mechanisms (Norris et al., 1998; Zhou et al., 2001). After bacterial internalization, the activation of Rho family GTPases and the actin cytoskeleton rearrangements are reversed by the TTSS bacterial effector SptP, a GTPase-activating protein for several Rho family members (Fu and Galán, 1999). Therefore, through a carefully orchestrated “yin and yang,” Salmonella reversibly activates Rho family GTPases to induce its own uptake and the production of proinflammatory cytokines.
Previous studies have reported the requirement of Cdc42 and Rac1 for Salmonella-induced actin cytoskeleton remodeling, macropinocytosis, and MAPK activation (Chen et al., 1996a; Hardt et al., 1998). However, it is not known whether these two GTPases act redundantly or whether their activation leads to specific responses. In vitro, SopE and SopE2 exhibit similar specificity, catalyzing exchange on several members of the Rho family of GTPases, including Rac1 and Cdc42 (Hardt et al., 1998; Stender et al., 2000; Friebel et al., 2001). However, the relative contribution of these GTPases to the different responses induced by Salmonella has not been determined. Evidence indicates that the SopB-mediated actin cytoskeleton rearrangements are also dependent on Rho family GTPases (Zhou et al., 2001). The observation that a noncatalytic mutant of SopB is unable to induce actin remodeling suggests that it must exert its function by activating an endogenous exchange factor, presumably through phosphoinositide fluxes. However, the actual GTPases targeted by the SopB activity or the identity of the exchange factor that this bacterial effector stimulates has not been defined. In this study, we describe distinct roles for different Rho family GTPases in Salmonella-induced cellular responses. In addition, we report that SopB stimulates cellular responses by activating SH3-containing guanine nucleotide exchange factor (SGEF), an exchange factor for RhoG, which we found plays a central role in the actin cytoskeleton remodeling stimulated by Salmonella. Together, these results reveal a remarkable level of complexity in the manipulation of Rho family GTPases by a bacterial pathogen.
Results
Differential activation of Cdc42 and Rac1 by Salmonella effector proteins in vivo
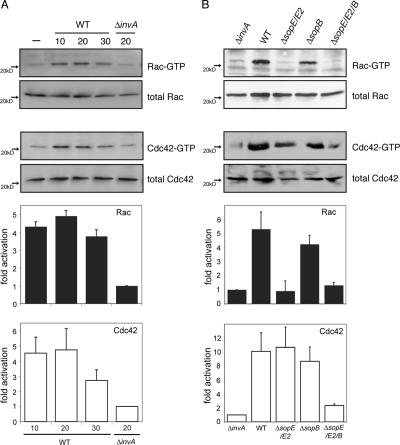
Previous studies have established that the cytoskeletal remodeling driving Salmonella entry into nonphagocytic cells is triggered by three bacterial effector proteins (SopE, SopE2, and SopB) that, upon delivery into host cells, target the Rho family GTPases Cdc42 and Rac1 (Hardt et al., 1998; Bakshi et al., 2000; Stender et al., 2000; Zhou et al., 2001). Despite strong evidence linking these bacterial effectors to Salmonella entry, their specific contribution to the in vivo activation of Cdc42 and/or Rac1 is not known. Therefore, we examined the speci ficity of GTPase activation by S. enterica serovar Typhimurium (S. typhimurium) mutant strains lacking sopE and sopE2 (ΔsopE/ΔsopE2) or sopB (ΔsopB). Cultured COS-2 cells were infected with these different mutant strains, and Cdc42 and Rac1 activation was assessed 20 min after infection, which was the time shown to result in their maximal stimulation by wild-type S. typhimurium (Fig. 1 A). The absence of sopB had little effect on the ability of S. typhimurium to activate Cdc42 or Rac1 because cells infected with a ΔsopB S. typhimurium induced similar stimulation levels of these GTPases as did wild type (Fig. 1 B). In contrast, the ΔsopE/ΔsopE2 S. typhimurium mutant, which solely relies on SopB for Rho family GTPase stimulation, did not activate Rac1 despite its ability to induce Cdc42 activation in a manner similar to wild type (Fig. 1 B). As expected, in the absence of a functional TTSS (Fig. 1 A) or in the combined absence of sopE, sopE2, and sopB (Fig. 1 B), S. typhimurium was unable to stimulate Cdc42 or Rac1. The lack of Rac1 activation by the ΔsopE/ΔsopE2 strain was apparent even up to 30 min after infection (unpublished data), which is much later than the time when the actin cytoskeleton rearrangements induced by this strain are apparent. Therefore, these results suggest that SopB can mediate actin remodeling in a Rac1-independent manner. Collectively, these results show that in vivo, the Salmonella effector proteins SopE, SopE2, and SopB differentially activate Rho family GTPases.
Figure 1.
Differential activation of Cdc42 and Rac1 by Salmonella effector proteins in vivo. (A) COS-2 cells were infected with wild-type (WT) S. typhimurium or its isogenic type III secretion-defective invA mutant (defective in its ability to induce actin cytoskeleton rearrangements and bacterial uptake; Galán et al., 1992) for the indicated times (in minutes), and the relative levels of activated Rac1 or Cdc42 were determined by the amount bound to GST-PAK-CRIB. Activated GTPase levels were normalized to the amount of total Rac1 or Cdc42 in cell lysates as analyzed by Western blotting. The intensity of the bands was quantified using ImageJ software. Values are the mean ± SD (error bars) of three independent experiments normalized to the response of cells infected with the invA mutant. (B) COS-2 cells were infected with different mutant strains of S. typhimurium (as indicated), and, 20 min after infection, the relative levels of activated Rac1 or Cdc42 were determined as indicated in A. Values are the mean ± SD of three independent experiments.
Independent activation of Cdc42 and Rac1 by Salmonella effector proteins in vivo
Wild-type S. typhimurium can robustly activate both Cdc42 and Rac1 in vivo. This is consistent with the observation that its Rho family GTPase exchange factors SopE and SopE2 can catalyze exchange on these GTPases in vitro (Hardt et al., 1998; Friebel et al., 2001). However, it is not known whether the activation of Rac1 in vivo occurs through the direct action of the bacterial proteins on this GTPase or indirectly through the activation of Cdc42. Although there is no evidence that Cdc42 can be activated downstream of Rac1, it is well established that Rac1 activation can also occur subsequent to the activation of Cdc42 through the stimulation of downstream exchange factors (Nobes and Hall, 1995; Hall, 1998; Jaffe and Hall, 2005).
To address the mechanisms of GTPase signaling stimulation by Salmonella in vivo, we examined Cdc42 and Rac1 activation after the selective inhibition of either GTPase. As expected and as previously shown (Chen et al., 1996a), the expression of a dominant-negative mutant of Cdc42 (Cdc42N17) abolished GTP loading of endogenous Cdc42 after wild-type S. typhimurium infection (unpublished data). However, the expression of Cdc42N17 also prevented the Salmonella-induced activation of Rac1 (Fig. 2 A). These results would suggest that Salmonella-induced Rac1 activation in vivo requires Cdc42 and, thus, may not be the consequence of a direct action of the bacterially encoded effectors (e.g., SopE and SopE2) on Rac1. However, the expression of a dominant-negative mutant of Rac1 (RacN17) also inhibited Salmonella-induced Cdc42 activation (unpublished data), suggesting that either of these GTPase mutants sequesters the bacterial exchange factors, thereby preventing the unambiguous interpretation of these results. To address this issue, we used RNAi to selectively inhibit Cdc42 and Rac1 expression. Delivery of specific RNAi constructs caused >90% depletion of Cdc42 or Rac1 in both intestinal Henle-407 and COS-2 cells (Fig. 2, B and C). Cells depleted of Cdc42 and/or Rac1 remained viable with normal cytoskeletal morphology for up to 3 d after treatment (unpublished data). We examined the effect of Cdc42 or Rac1 depletion on S. typhimurium–induced Rac1 and Cdc42 activation, respectively. In contrast to what we observed when expressing dominant-negative mutants, the depletion of one GTPase had no effect on the activation of the other (Fig. 2 A and not depicted). These results indicate that consistent with the in vitro specificity of the exchange factors SopE and SopE2 (Hardt et al., 1998; Friebel et al., 2001), S. typhimurium can independently activate Cdc42 and Rac1 in vivo.
Figure 2.
Independent activation of Cdc42 and Rac1 by Salmonella effector proteins in vivo. (A) COS-2 cells were transfected with plasmids expressing either Cdc42N17, short hairpin RNA directed to Cdc42, or a vector control (pSHAG). 2 d after transfection, cells were infected with either wild-type S. typhimurium (WT) or its isogenic invA mutant, which is defective for type III secretion and bacteria-induced signaling (Galán et al., 1992). 20 min after infection, the relative levels of activated Rac1 were determined by the amount bound to GST-PAK-CRIB that was normalized to the amount of Rac1 in cell lysates analyzed by Western blotting. The intensity of the bands was quantified using ImageJ software. Equivalent results were obtained in several repetitions of this experiment. (B and C) Levels of Cdc42 or Rac in Henle-407 (B) or COS-2 (C) cells transfected with RNAi constructs. GTPase levels were analyzed in cells 2 d after transfection using RT-PCR (B) or Western blotting with antibodies directed to Cdc42, Rac1, or actin (C).
SopE-mediated ruffling and efficient actin-mediated Salmonella internalization into nonphagocytic cells requires Rac1 but not Cdc42
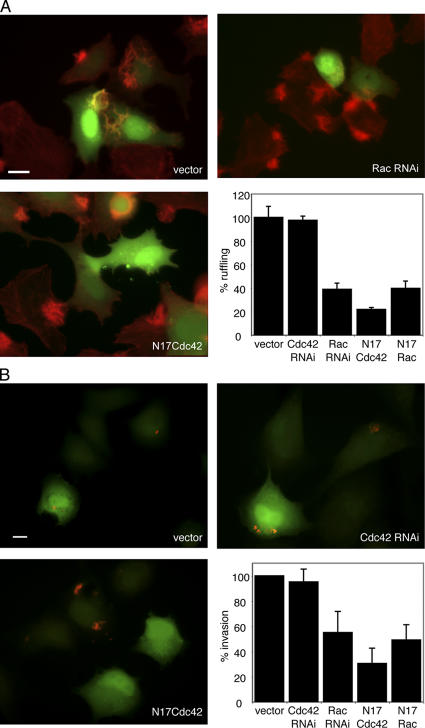
We have previously shown that the transient expression of SopE in cultured cells induces profuse actin rearrangements and membrane ruffling at the cell periphery and that these responses can be completely abrogated after the coexpression of Cdc42N17 or RacN17 (Hardt et al., 1998). However, our observation of the lack of specificity of these dominant-negative constructs (Fig. 2 A) prompted us to reexamine the contribution of Cdc42 and Rac1 to SopE-mediated actin cytoskeleton rearrangements. COS-2 cells were cotransfected with plasmids expressing SopE, GFP (to mark transfected cells), and RNAi constructs to deplete either Cdc42 or Rac1. 2 d after transfection, cells were stained with rhodamine-phalloidin to visualize polymerized actin. Depletion of Rac1 by RNAi effectively abrogated SopE-mediated ruffling (Fig. 3, C and F). The inhibition resulting from Rac1 depletion was equivalent to that observed after the expression of dominant-negative Cdc42 (Cdc42N17; Fig. 3, D and F) or Rac1 (RacN17; Fig. 3, E and F). In contrast, the depletion of Cdc42 by RNAi had no effect on the ability of SopE to induce actin cytoskeleton rearrangements (Fig. 3, B and F). These results indicate that actin rearrangements initiated by the Salmonella effector SopE and presumably its highly related paralogue SopE2 require the GTPase Rac1 but not Cdc42.
Figure 3.
SopE-mediated ruffling requires Rac1 but not Cdc42. (A–F) COS-2 cells were cotransfected with a plasmid expressing SopE, GFP (to detect transfected cells) along with RNAi constructs directed to Cdc42 (B) or Rac1 (C), or plasmids expressing Cdc42N17 (D), Rac1N17 (E), or vector control (A). 2 d after transfection, cells were stained with rhodamine-phalloidin, and the percentage of cells undergoing membrane ruffling as a consequence of SopE expression was enumerated by fluorescence microscopy (F). (F) Numbers indicate the percentage of transfected cells undergoing SopE-mediated actin remodeling (ruffling) and have been standardized considering the number of cells undergoing membrane ruffling in control transfections to be 100%. The results represent the mean ± SD (error bars) of three independent experiments. At least 200 transfected cells were counted for each independent transfection. Bar, 10 μM.
The observation that SopB-mediated actin remodeling and entry proceeds concomitant with the activation of Cdc42 but in the absence of Rac1 activation (Fig. 1 B) suggested that Salmonella must possess alternative mechanisms to induce Rho family GTPase-dependent actin remodeling than those used by SopE or SopE2. Therefore, we examined the ability of wild-type S. typhimurium to induce actin cytoskeleton rearrangements and its uptake into cells depleted of Cdc42 or Rac1. Depletion of Cdc42 had no effect on the induction of actin reorganization (Fig. 4 A) or bacterial entry (Fig. 4 B) in cultured intestinal epithelial cells. In contrast, the depletion of Rac1 markedly reduced but did not completely abrogate bacteria-induced actin cytoskeleton rearrangements and entry into host cells (Fig. 4). Together, these results strongly suggest that Cdc42 is dispensable for the actin remodeling events generated during Salmonella entry. In addition, the failure of Rac1 siRNA to reduce bacteria-induced ruffling and internalization to the same levels observed after the expression of Cdc42N17 coupled with the Rac1-independent actin remodeling seen upon SopB-mediated bacterial entry (Fig. 1 B) indicate that additional host components must contribute to the stimulation of these cellular responses.
Figure 4.
Salmonella-induced actin remodeling and bacterial internalization into nonphagocytic cells requires Rac1 but not Cdc42. (A) Intestinal Henle-407 cells were cotransfected with a plasmid expressing GFP (to detect transfected cells) along with RNAi constructs directed to Cdc42 or Rac1 or plasmids expressing Cdc42N17, Rac1N17, or vector control as indicated. 2 d after transfection, cells were infected with wild-type S. typhimurium for 30 min and stained with rhodamine-phalloidin. The percentage of transfected cells undergoing membrane ruffling as a consequence of bacterial infection was enumerated by fluorescence microscopy. Numbers indicate the percentage of transfected cells undergoing Salmonella-mediated actin remodeling (ruffling) and have been standardized considering the number of transfected cells undergoing membrane ruffling in mock vector control transfections to be 100%. (B) Intestinal Henle-407 cells were transfected as indicated in A. Transfected cells were infected with wild-type S. typhimurium, and the percentage of transfected cells with internalized bacteria was enumerated by fluorescence microscopy as indicated in Materials and methods. Numbers have been standardized considering the number of transfected cells with internalized bacteria in control transfections to be 100%. (A and B) The results represent the mean ± SD (error bars) of three independent experiments. At least 200 transfected cells were counted for each independent transfection. Bars, 10 μm.
Cdc42 is required for the Salmonella-induced nuclear responses in cultured intestinal epithelial cells
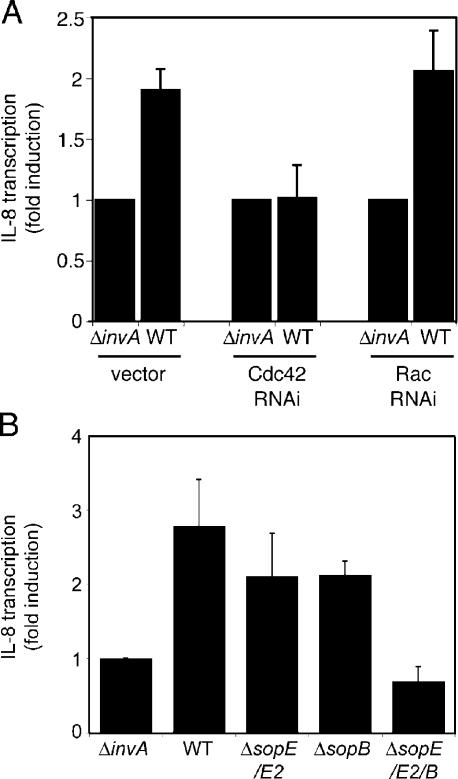
The observation that depletion of Cdc42 did not result in any measurable defect in the ability of S. typhimurium to stimulate actin cytoskeleton rearrangements prompted us to investigate the potential involvement of this GTPase in other cellular responses induced by these bacteria. In addition to inducing bacterial internalization into nonphagocytic cells, the Salmonella TTSS is also essential for the stimulation of a rapid reprogramming of host gene expression, particularly the induction of proinflammatory cytokines such as IL-8 and TNF-α (Hobbie et al., 1997). Stimulation of these nuclear responses is the consequence of the TTSS-dependent activation of MAPK pathways and transcription factors such as AP-1 and NF-κB and can be inhibited by the expression of dominant-negative Cdc42N17 (Chen et al., 1996a; Hobbie et al., 1997). Because the expression of Cdc42N17 nonspecifically blocks the activation of Rac1 (Fig. 2 A), we reevaluated the specific contribution of Cdc42 and Rac1 to the Salmonella-mediated nuclear response using RNAi. Henle-407 cells were cotransfected with a plasmid encoding an IL-8 luciferase reporter and RNAi constructs to deplete either Cdc42 or Rac1. Transfected cells were then infected with wild-type S. typhimurium, and the induction of IL-8 expression was assessed by measuring the luciferase levels in infected cells. Depletion of Rac1 by RNAi had no effect on the ability of S. typhimurium to stimulate IL-8 transcription (Fig. 5 A). In contrast, the depletion of Cdc42 completely abrogated the stimulation of IL-8 expression upon bacterial infection (Fig. 5 A). Similar results were obtained using an NF-κB–dependent reporter cell line (unpublished data). These results indicate that the stimulation of Cdc42 is essential for the bacteria-induced nuclear responses. Furthermore, these results indicate that the different cellular responses induced by Salmonella proceed through the activation of distinct Rho family GTPases: Rac1 for the actin cytoskeleton remodeling and Cdc42 for the nuclear responses.
Figure 5.
Cdc42 is required for the Salmonella-induced nuclear responses in cultured cells. (A) Intestinal Henle-407 cells were cotransfected with an IL-8 transcription firefly luciferase reporter plasmid along with RNAi constructs directed to Cdc42 or Rac1 or vector control. 2 d after transfection, cells were infected with wild-type (WT) S. typhimurium or its isogenic invA mutant, which is defective in type III secretion. The stimulation of IL-8 transcription in transfected cells was assayed by measuring the levels of firefly luciferase. (B) SopE, SopE2, and SopB work redundantly to activate IL-8 expression. Intestinal Henle-407 cells were transfected with an IL-8 transcription firefly luciferase reporter, and, 2 d after transfection, cells were infected with wild-type S. typhimurium or isogenic mutants lacking the indicated effector proteins. The stimulation of IL-8 transcription in transfected cells was assayed by measuring the levels of firefly luciferase. (A and B) Values represent fold induction in cells infected with wild-type S. typhimurium over the value of cells infected with the type III secretion-defective invA mutant strain and are the mean ± SD (error bars) of three independent measurements.
We then examined the specific contribution of the different Salmonella effectors that activate Rho family GTPases (i.e., SopE/SopE2 and SopB) to the stimulation of nuclear responses. Henle-407 cells were transfected with the IL-8 reporter plasmid and were subsequently infected with wild-type S. typhimurium, the isogenic ΔsopE/sopE2, ΔsopB, or a strain lacking the three effectors (ΔsopE, sopE2, and ΔsopB). Both the ΔsopE/sopE2 and the ΔsopB mutant strains stimulated IL-8 expression but at lower levels than wild type (Fig. 5 B). In contrast, the triple mutant did not induce the expression of IL-8. These results indicate that SopE, SopE2, and SopB act redundantly to stimulate nuclear responses, which is consistent with the observation that all of them can activate Cdc42.
SopB-mediated actin remodeling requires RhoG
Infection of cultured epithelial cells with an S. typhimurium ΔsopE/ΔsopE2 mutant, which induces actin remodeling through the activity of the effector protein SopB, resulted in the activation of Cdc42 but not Rac1 (Fig. 1 B). However, Cdc42 does not contribute to bacterial uptake (Fig. 4). These results suggested the possibility that SopB may stimulate actin-cytoskeleton responses through the stimulation of another Rho family GTPase. A potential candidate is RhoG (Vincent et al., 1992), a Rho family GTPase that has been previously shown to mediate dorsal ruffling and macropinocytosis in fibroblasts (Ellerbroek et al., 2004).
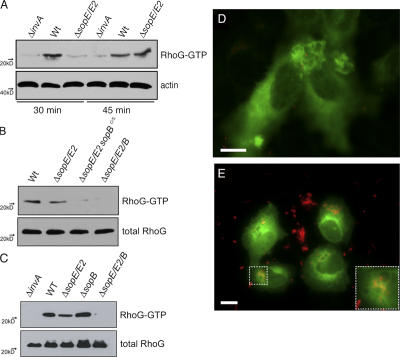
We first investigated whether S. typhimurium infection of cultured epithelial cells resulted in the activation of RhoG. COS-2 cells were infected with wild-type S. typhimurium, and the activation of RhoG was monitored by pull-down assays using the RhoG effector protein ELMO as an affinity probe for activated RhoG (Katoh and Negishi, 2003). RhoG activation by wild-type S. typhimurium was observed as early as 15 min after infection (unpublished data), although maximal activation was not seen until at least 30 min after infection (Fig. 6 A). The activation of RhoG was not observed in cells infected with a TTSS-deficient S. typhimurium strain (Fig. 6 A), indicating that stimulation of this GTPase requires the activity of TTSS effector proteins. Importantly, RhoG stimulation was observed in cells infected with a ΔsopE/ΔsopE2 S. typhimurium mutant strain (Fig. 6 A), which induces signaling exclusively through SopB. SopB-mediated activation of RhoG was dependent on its phosphatase activity because the ΔsopE/ΔsopE2 strain encoding a catalytically inactive SopB (SopBC460S) was unable to stimulate RhoG (Fig. 6 B). A ΔsopB mutant, which signals though SopE and SopE2, was also able to activate RhoG (Fig. 6 C). This is consistent with the observation that SopE can catalyze exchange on RhoG in vitro (Hardt et al., 1998). A strain lacking sopE, sopE2, and sopB was not able to stimulate RhoG activation (Fig. 6, B and C), indicating that these three effector proteins can account for all of the activation of this GTPase by wild-type S. typhimurium. GFP-RhoG was recruited to the site of Salmonella entry shortly after infection (Fig. 6 D and Videos 1–4; available at http://www.jcb.org/cgi/content/full/jcb.200605144/DC1) and remained associated with the Salmonella-containing vacuole as well as with empty vacuoles generated during the stimulation of membrane ruffling (Fig. 6 E, inset; and Videos 1–3). This pattern of recruitment and localization closely resembled the localization of GFP-Rac1 during Salmonella infection (Video 5), although it differs from that of GFP-Cdc42 (Video 6). Collectively, these results indicate that upon infection, Salmonella elicits the transient activation of at least three different Rho family GTPases—Cdc42, Rac1, and RhoG—through the coordinated activities of its TTSS effector proteins SopE, SopE2, and SopB.
Figure 6.
Salmonella infection of cultured epithelial cells results in the activation and recruitment of RhoG. (A–C) COS-2 cells were infected with different mutant strains of S. typhimurium (as indicated) and at the indicated times (A) or, 45 min (B and C) after infection, the relative levels of activated RhoG were determined by the amount bound to GST-ELMO that was normalized to the total amount of RhoG in cell lysates analyzed by Western blotting. Equivalent results were obtained in several repetitions of this experiment. (D and E) Intestinal Henle-407 (D) or COS-2 (E) cells were transfected with a plasmid expressing RhoG-GFP. 24 h after transfection, cells were infected with wild-type S. typhimurium expressing the dsRed protein and imaged by time-lapse fluorescence video microscopy. Images corresponding to 15 (D) and 45 min (E) after infection are shown. Inset shows RhoG-GFP recruited to the Salmonella-containing vacuole. Bars, 10 μm. The entire sequence of time-lapse video microscopy from which the images were obtained is shown in Video 1 (available at http://www.jcb.org/cgi/content/full/jcb.200605144/DC1).
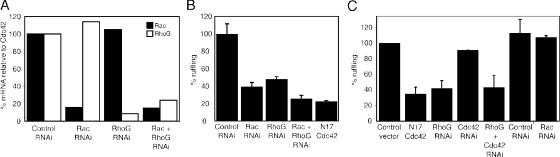
We then examined the potential role of RhoG in Salmonella-induced and, more specifically, SopB-mediated actin remodeling. RhoG was depleted from intestinal Henle-407 cells by the transfection of RNAi constructs designed to target this GTPase (Fig. 7 A), and the ability of S. typhimurium and mutant strains to stimulate actin remodeling in these cells was examined by fluorescence microscopy (Fig. 7, B and C). The depletion of RhoG resulted in a marked inhibition of the ability of wild-type S. typhimurium to induce actin-cytoskeleton rearrangements (Fig. 7 B). However, the inhibition was reproducibly not as pronounced as that observed by the expression of dominant-negative Cdc42 (Fig. 7 B) or Rac1 (Chen et al., 1996a; Hardt et al., 1998), which nonspecifically blocks signaling through several GTPases. However, simultaneous depletion of Rac1 and RhoG achieved the maximum level of inhibition (Fig. 7 B), indicating that S. typhimurium–induced actin remodeling is the result of signaling through both GTPases. Importantly, the depletion of RhoG completely abolished the ability of the ΔsopE/ΔsopE2 S. typhimurium mutant to induce actin remodeling (Fig. 7 C). This mutant strain signals to Rho GTPases exclusively through SopB; therefore, these results indicate that this effector protein stimulates actin remodeling through the activation of RhoG. Consistent with this hypothesis, the depletion of Rac1 had no effect on the ability of the ΔsopE/ΔsopE2 S. typhimurium mutant to induce actin remodeling (Fig. 7 C). Because the expression of SopB results in strong toxicity (Zhou et al., 2001), the effect of RhoG depletion on the actin cytoskeleton rearrangements induced by transiently expressed SopB could not be evaluated. Collectively, these results indicate that S. typhimurium induces actin remodeling by targeting both Rac1 and RhoG via distinct TTSS effector proteins.
Figure 7.
SopB-dependent Salmonella-induced actin remodeling requires RhoG. (A) Levels of Rac1 or RhoG in Henle-407 cells transfected with RNAi constructs that were directed to these GTPases measured by quantitative real-time RT-PCR. Values were standardized relative to the levels of Cdc42 mRNA, which are not affected by these constructs. (B and C) Intestinal Henle-407 cells were cotransfected with a plasmid expressing GFP (to detect transfected cells) along with RNAi constructs directed to Cdc42, Rac1, or RhoG or plasmids expressing Cdc42N17, Rac1N17, and vector control as indicated. Transfected cells were infected with wild-type S. typhimurium for 20 min (B) or its isogenic ΔsopE/ΔsopE2 (which induces actin remodeling solely through SopB) mutant for 40 min (C). After infection, cells were stained with rhodamine-phalloidin, and the percentage of transfected cells undergoing membrane ruffling as a consequence of bacterial infection was enumerated by fluorescence microscopy. Numbers indicate the percentage of transfected cells undergoing Salmonella-mediated actin remodeling (ruffling) and have been standardized considering the number of transfected cells undergoing membrane ruffling in control transfections to be 100%. The results represent the mean ± SD (error bars) of three independent experiments. At least 200 cells were counted for each independent transfection.
SopB-mediated actin cytoskeleton rearrangements requires the cellular RhoG exchange factor SGEF
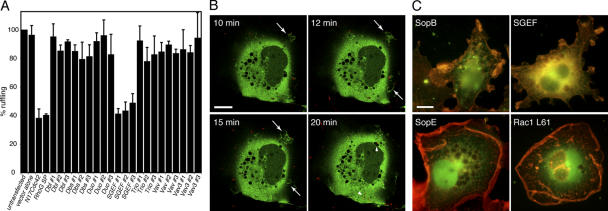
SopB-mediated actin cytoskeleton rearrangements require its phosphoinositide phosphatase activity (Zhou et al., 2001) and are dependent on RhoG. Because SopB lacks measurable in vitro nucleotide exchange activity toward Rho family GTPases (unpublished data), these observations suggest that SopB, presumably through its ability to induce phosphoinositide fluxes, must exert its function by activating an endogenous RhoG exchange factor. Therefore, we investigated the potential involvement of several cellular exchange factors that have been shown to have the ability to catalyze nucleotide exchange on RhoG. Cells were cotransfected with plasmids expressing siRNAs directed to Dbl (May et al., 2002), Dbs (Whitehead et al., 1995), Duo (Kalirin; May et al., 2002; Tse et al., 2005), SGEF (Ellerbroek et al., 2004), Vav, and Vav3 (Schuebel et al., 1998) along with a plasmid expressing GFP to identify transfected cells. Cells were infected with the ΔsopE/ΔsopE2 S. typhimurium mutant strain, which induces actin cytoskeleton rearrangements exclusively through the activity of SopB. Depletion of Dbl, Dbs, Duo (Kalirin), Vav, or Vav3 had little or no effect on the ability of the ΔsopE/ΔsopE2 S. typhimurium mutant strain to induce actin cytoskeleton rearrangements (Fig. 8 A). However, the depletion of SGEF markedly impaired actin cytoskeleton rearrangements induced by the ΔsopE/ΔsopE2 S. typhimurium strain (Fig. 8 A). Furthermore, the depletion of SGEF abolished the ability of the ΔsopE/ΔsopE2 S. typhimurium strain to activate RhoG (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200605144/DC1). The effect of SGEF depletion on the ability of this strain to stimulate actin remodeling was equivalent to the effect of RhoG depletion (Fig. 8 A), suggesting that most of the SopB-mediated signaling to RhoG is transduced through its exchange factor SGEF. Consistent with its involvement in S. typhimurium–induced actin remodeling, SGEF-GFP was efficiently recruited to the site of bacterial entry into host cells (Fig. 8 B and Videos 7 and 8; available at http://www.jcb.org/cgi/content/full/jcb.200605144/DC1).
Figure 8.
SopB-dependent Salmonella-induced actin remodeling requires the RhoG exchange factor SGEF. (A) Intestinal Henle-407 cells were cotransfected with a plasmid expressing GFP (to detect transfected cells), the vector control, a plasmid expressing Cdc42N17, or RNAi constructs directed to RhoG or its exchange factors Dbl, Dbs, Duo (Kalirin), SGEF, Trio, Vav, and Vav3. 2 d after transfection, cells were infected with the S. typhimurium ΔsopE/ΔsopE2 mutant, which induces actin remodeling solely through SopB. 40 min after infection, cells were stained with rhodamine-phalloidin, and the percentage of transfected cells undergoing membrane ruffling as a consequence of bacterial infection was enumerated by fluorescence microscopy. Numbers indicate the percentage of transfected cells undergoing Salmonella-mediated actin remodeling (ruffling) and have been standardized considering the number of transfected cells undergoing membrane ruffling in control transfections to be 100%. The results represent the mean ± SD (error bars) of three independent experiments. At least 200 transfected cells were counted for each independent transfection. (B) COS-2 cells were transfected with a plasmid expressing SGEF-GFP. 24 h after transfection, cells were infected with wild-type S. typhimurium expressing the dsRed protein and were imaged by confocal time-lapse fluorescence video microscopy. Images corresponding to different time points after infection are shown. Arrows indicate membrane ruffles induced by bacterial infection, and arrowheads indicate S. typhimurium. The entire sequence of images is shown in Video 8 (available at http://www.jcb.org/cgi/content/full/jcb.200605144/DC1). (C) COS-2 cells were cotransfected with a plasmid expressing GFP (to detect transfected cells) along with plasmids expressing SopB, SGEF, SopE, or Rac1L61 as indicated. Transfected cells were stained with rhodamine-phalloidin to analyze the morphology of filamentous actin ruffles. Bars, 10 μm.
Previous studies have indicated that the SopB-mediated actin cytoskeleton rearrangements are morphologically distinct from those induced by SopE or SopE2 (Hardt et al., 1998; Zhou et al., 2001). Although the expression of SopE in cultured cells resulted in actin cytoskeleton rearrangements and membrane ruffling throughout the cell (Fig. 8 C) similar to that observed upon the overexpression of constitutively active Rac1 (Fig. 8 C), the expression of SopB within cultured cells, although toxic, induced much more localized membrane ruffles and actin remodeling in the few cells that can be visualized in a standard transfection (Fig. 8 C; Zhou et al., 2001; Hernandez et al., 2004). Consistent with its involvement in SopB-mediated actin remodeling, the overexpression of SGEF induced localized membrane ruffles that closely resembled those induced by SopB (Fig. 8 C; Ellerbroek et al., 2004). Collectively, these results indicate that SGEF is the exchange factor activated by SopB to induce actin cytoskeleton rearrangements and mediate bacterial entry.
Discussion
Many bacterial pathogens target Rho family GTPases with a variety of toxins (Aktories, 1997). These toxins most often introduce covalent modifications on these critical signal transduction molecules, which usually result in their irreversible modification, leading to their constitutive activation, their complete inactivation, or their degradation. On the other hand, S. enterica modulates Rho family GTPases with bacterial proteins that precisely mimic the function of endogenous modulators of this family of small GTP-binding proteins (Galán, 2001; Stebbins and Galán, 2001). Previous studies have defined the precise mechanisms by which two Salmonella-encoded exchange factors, SopE and its highly related paralogue SopE2, catalyze the activation of Rho GTPase family members (Hardt et al., 1998; Friebel et al., 2001; Buchwald et al., 2002). However, the contribution of the specific Rho family members to Salmonella–host cell interactions has remained elusive.
In our studies, we have defined the specific contribution of Rho family GTPases to different Salmonella-induced cellular responses, thus revealing a hitherto unknown complexity in the functional consequences of the modulation of these molecular switches by this bacterial pathogen. Our studies revealed that different Rho GTPases are required for different responses induced by Salmonella during its interaction with intestinal epithelial cells. Previous studies using the dominant-negative mutant of different Rho GTPases have indicated that Cdc42 was central for the ability of Salmonella to induce actin cytoskeleton rearrangements and to enter nonphagocytic cells (Chen et al., 1996a; Hardt et al., 1998). However, using RNAi, we found that Cdc42 is dispensable for these responses. Instead, we found that Rac1 is required for efficient induction of the actin remodeling that leads to bacterial uptake into nonphagocytic cells. Our studies also showed that Cdc42 is essential for the transduction of signals induced by the Salmonella TTSS that lead to nuclear responses, a process in which Rac1 seems to play an unimportant role. Consequently, these studies have uncovered a remarkable mechanism by which a bacterial pathogen has the capacity to generate a rather diverse set of cellular responses that are essential for its pathogenicity through the activity of even a single bacterial effector.
In addition to SopE and SopE2, a previous study has shown that another bacterial effector, SopB, can also induce actin cytoskeleton rearrangements and bacterial uptake into cells (Zhou et al., 2001). Although it has been shown that the phosphoinositide phosphatase activity of SopB is required for the stimulation of these responses (Zhou et al., 2001), the mechanisms by which the signal generated by this bacterial effector protein is transduced to the actin cytoskeleton has remained unknown. In this study, we showed that the stimulation of actin cytoskeleton rearrangements by SopB is dependent on RhoG, a Rho family GTPase not previously implicated in Salmonella–host cell interactions. RhoG has been previously implicated in modulation of the actin cytoskeleton as well as in the induction of membrane ruffling and macropinocytosis (Wennerberg et al., 2002; Ellerbroek et al., 2004). A Salmonella ΔsopE/ΔsopE2 mutant strain, which relies solely on SopB to induce actin cytoskeleton rearrangements, was able to induce the activation of RhoG but not Rac1. More importantly, cells that were depleted of RhoG by RNAi were markedly impaired in their ability to rearrange their actin cytoskeleton upon infection with this S. typhimurium mutant strain. We have previously shown that SopE can catalyze RhoG exchange in vitro (Hardt et al., 1998), and, consistent with this observation, we have shown in this study that SopE and its highly related paralogue SopE2 can activate RhoG in vivo.
The observation that SopB lacks in vitro exchange activity on Rho family GTPases combined with the observation that SopB-mediated actin remodeling requires its phosphoinositide phosphatase activity suggested that this bacterial effector must activate RhoG through the stimulation of an endogenous cellular exchange factor. We found that such an exchange factor is SGEF because its depletion from cells effectively prevented the ability of S. typhimurium to signal to the actin cytoskeleton through the activity of SopB. SGEF is ubiquitously expressed and has been implicated in the induction of localized dorsal membrane ruffles and macropinocytosis (Ellerbroek et al., 2004), which closely resembles the cellular responses induced by S. typhimurium and SopB in particular. We have previously found that the ability of S. typhimurium to induce macropinocytosis is strictly dependent on the phosphoinositide phosphatase activity of SopB (Hernandez et al., 2004). Indeed, an S. typhimurium mutant strain lacking SopB or expressing a catalytically inactive mutant was fully capable of inducing membrane ruffling (through SopE and SopE2) but failed to induce macropinocytosis (Hernandez et al., 2004). The observation that the expression of constitutive active RhoG or its exchange factor SGEF but not Rac1 leads to profuse macropinocytosis (Ellerbroek et al., 2004) is consistent with these observations and with the role of SGEF and RhoG in Salmonella–host cell interactions. It is unknown how SopB may activate SGEF. The requirement of the phosphatase activity of SopB to induce actin rearrangements (Zhou et al., 2001) as well as to activate RhoG (this study) suggests that the phosphoinositide fluxes induced by its catalytic activity may directly activate SGEF. Consistent with this hypothesis, SGEF possesses a phosphoinositide-binding pleckstrin homology domain, which is essential for its membrane localization and activity (Ellerbroek et al., 2004).
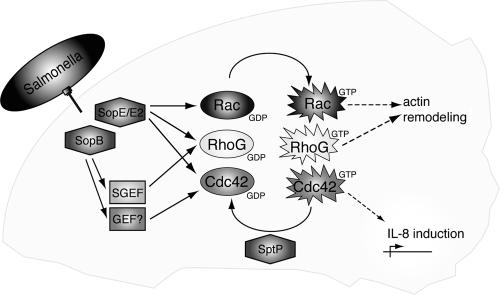
In summary, our study has revealed a remarkable complexity in the stimulation of cellular responses by S. typhimurium through the specific and selective activation of Rho family GTPases (Fig. 9). Both directly through action of the bacterially encoded exchange factors SopE and SopE2 and indirectly through activation of the endogenous RhoG exchange factor SGEF by SopB, this bacterial pathogen orchestrates the coordinated activation of specific Rho family GTPase members to stimulate specific cellular responses. These constitute remarkable examples of pathogen adaptations to modulate specific cellular functions.
Figure 9.
Model for Salmonella signaling to Rho family GTPases. Through its TTSS, Salmonella delivers SopE, SopE2, and SopB, which differentially activate different Rho family GTPase members either directly (SopE and SopE2) or indirectly via the stimulation of the endogenous exchange factor SGEF (SopB). Activation of the different Rho family GTPases leads to specific cellular responses, such as actin remodeling, and the stimulation of nuclear responses, such as the production of IL-8. Responses are subsequently reversed by the action of the TTSS GTPase-activating protein effector protein SptP.
Materials and methods
Bacterial strains and cell lines
The wild-type strain of S. enterica serovar Typhimurium (S. typhimurium) SL1344 and its isogenic derivatives used in this study, ΔsopB (SB1120; Hernandez et al., 2004), ΔsopE ΔsopE2 (SB1301), ΔsopE ΔsopE2 ΔsopB (SB1302; Zhou et al., 2001), and ΔinvA (SB136; Galán et al., 1992), have been previously described. InvA encodes an essential component of the invasion-associated TTSS, and, therefore, strains lacking this gene are completely defective in their ability to stimulate actin cytoskeleton rearrangements, membrane ruffling, bacterial uptake, and nuclear responses (Galán and Curtiss, 1989; Galán et al., 1992; Chen et al., 1996a,b). A ΔsopE ΔsopE2 S. typhimurium derivative expressing the catalytic mutant SopBC460S was constructed by allelic exchange as previously described (Zhou et al., 2001). All bacterial strains were cultured under conditions that stimulate the expression of the Salmonella pathogenicity island-1–encoded TTSS (Galán and Curtiss, 1990). In brief, overnight cultures were diluted 1:25 in L-broth containing 0.3M NaCl, incubated on a rotating wheel for 3 h at 37°C until an OD600nm of ∼0.9, and used immediately for infection. Where appropriate, 0.1% l-arabinose was added to cultures at the early logarithmic phase of growth (OD600nm of 0.4) to induce expression of the dsRed gene under the control of the paraBAD promoter. COS-2 cells were maintained in DME (Invitrogen) supplemented with 10% heat-inactivated FCS (Gemini), 100 U/ml penicillin, and 50 μg/ml streptomycin. Henle-407 cells were grown in DME supplemented with 10% heat-inactivated bovine calf serum (Hyclone) and penicillin/streptomycin. 293T/NF-κB–luc (Panomics) cells were passaged in DME containing 10% FBS (Gemini), penicillin/streptomycin, and 100 μg/ml hygromycin (Sigma-Aldrich) as recommended by the manufacturer. For infection experiments, cells were washed and passaged into media lacking penicillin/streptomycin overnight.
Plasmids and transfections
Eukaryotic expression vectors encoding GFP-tagged wild-type Cdc42 and Rac1 as well as their dominant-negative (Cdc42N17 and Rac1N17) and constitutively active (Cdc42L61 and Rac1L61) mutants have been previously described (Chen et al., 1996a). Wild-type RhoG and SGEF were subcloned into vectors pRK5myc and pEGFP-C1 (CLONTECH Laboratories, Inc.) by PCR using pECEFL-Au5 RhoG (a gift from X. Bustelo, University of Salamanca, Salamanca, Spain) and clone 10624662 (American Type Culture Collection), respectively, as templates. To fluorescently label S. typhimurium, the plasmid vector pdsRed.T3S4T (a gift from D. Bumann, Max-Planck Institute, Berlin, Germany; Sorensen et al., 2003), which encodes the bacterially optimized dsRed protein under the control of an arabinose-inducible promoter, was transformed into the different bacterial strains by electroporation. To measure IL-8 transcription, a firefly luciferase reporter plasmid (pSB2805) was constructed by replacing the CAT gene of pIL-8-CAT (Hobbie et al., 1997) with the firefly luciferase gene (Promega). To standardize transfection experiments, a plasmid was constructed (pSB2806) in which the Renilla luciferase, which was derived from pRL-TK (Promega), was cloned into the eukaryotic expression vector pCDNA3.1 (Invitrogen). Plasmids pSB1141, a bicistronic vector encoding SopE and GFP, and pSB2807 encoding YFP-tagged SopB, have been previously described (Hernandez et al., 2004). Plasmid DNA was purified using the endotoxin-free Maxiprep kit (QIAGEN) and used for the transfection of cells with LipofectAMINE 2000 (Invitrogen) according to the manufacturer's instructions.
Gene silencing by RNAi
Depletion of endogenous Cdc42 was performed using a short hairpin sequence targeting nucleotides 296–318 of human Cdc42. Oligonucleotide pairs A (ACAGTGGTGAGTTATCTCAGGAAGCTTGCTGAGATAACTCACCACTGTCCATTTTTT) and B (GATCAAAAAATGGACAGTGGTGAGTTATCTCAGCAAGCTTCCTGAGATAACTCACCACTGTCG) were annealed and ligated into the vector pSHAG (a gift from G. Hannon, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY; Hannon and Conklin, 2004). Attempts to knockdown endogenous Rac1 and RhoG using a similar vector-based approach proved unsuccessful. Thus, silencing of Rac1 and RhoG gene expression was achieved using synthetic SMARTpools (Dharmacon), each comprising four proprietary siRNA sequences. Three independent RNAi constructs (denoted as 1–3) targeting the RhoG GEFs Dbl, Dbs, Duo (Kalirin), SGEF, Trio, Vav1, and Vav3 comprising short hairpin RNA sequences cloned into pSUPER Retro (Oligoengine) were provided by A. Schmidt and A. Hall (Memorial Sloan Kettering Institute, New York, NY). In all cases, siRNA and short hairpin RNA constructs were transfected into COS-2 and Henle-407 cells using LipofectAMINE 2000 (Invitrogen). Where appropriate, RNAi constructs were cotransfected with pEGFP-C1 at a ratio of 5:1 to mark transfected cells.
The silencing efficiency of these RNAi constructs for their targeted mRNA was tested by quantitative RT-PCR 48 h after transfection on different amounts of total RNA template isolated by TRIzol (Invitrogen) extraction. RNA samples were treated with DNase (Invitrogen) and used to generate cDNA by the iSCRIPT cDNA Synthesis Kit (Bio-Rad Laboratories) according to the manufacturer's protocol. Quantitative RT-PCR was performed using a real-time detection system (iQ iCycler; Bio-Rad Laboratories) with iQ SYBR Green Supermix (Bio-Rad Laboratories) and specific primers for Cdc42 (CCTTTCTTGCTTGTTGGGACTC and CTCCACATACTTGACAGCCTTC), Rac1 (GTCCCAACACTCCCATCATCC and ACAGCACCAATCTCCTTAGCC), and RhoG (TCATCTGCTACACAACTAACG and GCGGTCATACTCCTCCTG). iCycler iQ software (version 3; Bio-Rad Laboratories) was used to analyze threshold cycles. mRNA levels were normalized to GTPase mRNA in untransfected cells. RNAi knockdown efficiency and specificity were also analyzed at the protein level by Western blotting of cell lysates 48 h after transfection. Endogenous Cdc42 and Rac1 levels were probed using rabbit anti-Cdc42 (Santa Cruz Biotechnology, Inc.) and monoclonal anti-Rac1 (clone 23A8; Upstate Biotechnology). Because of a lack of suitable antibodies to test endogenous protein, the knockdown of RhoG and SGEF protein was monitored by blotting for levels of FLAG-tagged versions of these proteins after the cotransfection of relevant expression plasmids and RNAi constructs. In each case, protein depletion by RNAi was normalized to endogenous levels of actin using rabbit anti-actin antibodies (Sigma-Aldrich).
Determination of Rho GTPase activation
Activation of endogenous Cdc42 and Rac1 GTPases was measured by p21-activated kinase (PAK)–Cdc42–Rac1 interaction binding (CRIB) pull-down assays as previously described (Sander et al., 1999). In brief, COS-2 cells transfected as appropriate in 10-cm dishes were washed twice with prewarmed HBSS and infected with S. typhimurium strains at an MOI of 100. At different time points, COS-2 cells were washed twice in cold HBSS and lysed by scraping on ice in cold radioimmunoprecipitation assay buffer (50 mM Tris, pH 8, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 10 mM MgCl2, 0.2 mM PMSF, and Complete protease inhibitor cocktail; Roche). Lysates were cleared by centrifugation at 14,000 rpm for 10 min at 4°C, an aliquot was saved to assess total GTPase levels, and the remaining lysates were divided in two aliquots to assess levels of active Cdc42 and Rac1. Lysates were incubated for 1 h at 4°C with ∼20 μg GST fused to the CRIB domain of PAK (GST-PAK-CRIB) that was prebound to glutathione agarose beads (50% slurry; prepared as previously described by Sander et al., 1999) to precipitate GTPases. Beads were subsequently washed twice in cold wash buffer (50 mM Tris, pH 8, 150 mM NaCl, 1% Triton X-100, 10 mM MgCl2, 0.2 mM PMSF, and protease inhibitor cocktail). Equal amounts of beads and total cell lysate were resolved by SDS-PAGE, transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore), and immunoblotted using anti-Cdc42 or Rac1 antibodies. For RhoG, GTP loading was assessed on exogenously expressed myc-tagged RhoG using GST-ELMO 21-362 (a gift from X. Bustelo) coupled to glutathione agarose beads as bait. The GST fusion protein was expressed and purified as for GST-PAK-CRIB, and the pull-down assays were performed as described for Cdc42 and Rac1 with the exception of the cell lysis buffer (20 mM Tris, pH 7.6, 150 mM NaCl, 5 mM MgCl2, 0.5% Triton X-100, 1 mM DTT, 0.1 mM sodium vanadate, 1 mM PMSF, and protease inhibitor cocktail). Western blots were probed with mouse anti-myc (clone 9E10; Santa Cruz Biotechnology, Inc.) antibody. In all cases, blots were visualized using chemiluminescence reagents (Supersignal chemiluminescence substrate; Pierce Chemical Co.), and, where appropriate, bands were scanned and their intensities were quantified using the public domain ImageJ program (National Institutes of Health; http://rsb.info.nih.gov/nih-image/).
S. typhimurium ruffling and internalization assays
S. typhimurium–induced ruffling assays in COS-2 and Henle-407 cells have been previously described (Chen et al., 1996a). In brief, semiconfluent cells were infected with different S. typhimurium strains for various time intervals (cells were infected with wild-type S. typhimurium for 20 min and with ΔsopE/ΔsopE2 S. typhimurium for 30 min with an MOI of 40 and 100, respectively). After infection, cells were washed twice with HBSS, fixed with 4% PFA for 20 min, permeabilized with 0.05% Triton X-100 for 5 min, and stained with FITC- or rhodamine-labeled phalloidin to visualize the actin cytoskeleton. To visualize S. typhimurium internalization into host cells, we used an assay that takes advantage of the inaccessibility of internalized bacteria to gentamicin, an antibiotic that kills bacteria by inhibiting protein synthesis. In brief, cells were infected with an S. typhimurium strain (MOI of 20) containing a plasmid that expresses the dsRed fluorescent protein under an arabinose-inducible promoter. 45 min after infection, cells were washed twice with warm HBSS, and noninternalized bacteria were killed by the addition of gentamicin (100 μg/ml in DME). 45 min after gentamicin addition, the media was replaced with DME containing 100 μg/ml gentamicin and 0.1% arabinose to induce dsRed expression only in internalized bacteria, as bacterial protein synthesis is inhibited by gentamicin in extracellular bacteria. Internalized S. typhimurium were visualized 2 h after the addition of arabinose by fluorescence microscopy. This assay was validated by carrying out parallel experiments using an antibody-based differential staining of internalized versus external bacteria as previously described (Chen et al., 1996a), obtaining equivalent results. The SopE-mediated ruffling assay has been previously described (Hardt et al., 1998). Immunofluorescence studies used DAPI (Sigma-Aldrich) to detect bacteria (in some experiments), TRITC-conjugated phalloidin (Sigma-Aldrich) to stain the host actin cytoskeleton, AlexaFluor596- and -488–conjugated goat anti–rabbit IgG and goat anti–mouse IgG antisera (Invitrogen), and rabbit anti S. typhimurium lipopolysaccharide (Difco Laboratories). Fluorescence microscopy was performed using a confocal laser-scanning microscope (LSM 510; Carl Zeiss MicroImaging, Inc.) or an inverted microscope (Eclipse TE2000-U; Nikon) equipped with oil immersion plan Apo 60× NA 1.4, 100× NA 1.4, or dry plan Fluor 40× NA 0.6 objectives (Nikon) and a CCD camera (MicroMAX RTE/CCD-1300Y; Princeton Instruments). Acquisition and analysis of still images were performed using MetaMorph Imaging Software (version 6.1; Universal Imaging Corp.). For live cell imaging, cells were maintained at 37°C.
IL-8 and NF-κB gene reporter assay
Henle-407 cells were transiently transfected with the dual luciferase reporter constructs pSB2805 and pSB2806 together with different RNAi constructs. 2 d after transfection (including overnight serum starvation), cells were infected with wild-type S. typhimurium or an isogenic TTSS-defective invA mutant at an MOI of 25 for 40 min. Cells were washed with HBSS and further incubated with DME containing 100 μg/ml gentamicin for 4 h. After passive cell lysis, firefly and Renilla luciferase levels were determined using the Dual Luciferase Reporter assay (Promega) according to the manufacturer's instructions. Transfection efficiency was normalized by the comparison of IL-8–induced firefly luciferase levels with that of constitutively expressed Renilla luciferase. Induction of the IL-8 reporter by S. typhimurium strains was expressed relative to that of an invA isogenic mutant strain, which is defective for type III secretion and the induction of cellular responses. To determine the activation of NF-κB by S. typhimurium, we used a 293T NF-κB luciferase reporter cell line (Panomics). Cells were transiently transfected with different RNAi constructs and assayed as indicated above for IL-8. Induction of the reporter by wild-type S. typhimurium was expressed relative to that of an invA isogenic mutant.
Online supplemental material
Fig. S1 shows that RhoG activation by S. typhimurium strains requires SGEF. Video 1 is a time-lapse video showing RhoG localization during Salmonella infection. Video 2 shows that GFP-RhoG labels vacuoles generated during Salmonella entry. Video 3 is a confocal time-lapse video showing GFP-RhoG localization during Salmonella infection, and Video 4 is a 3D projection showing GFP-RhoG recruitment to a Salmonella-induced ruffle. Video 5 is a time-lapse video showing Rac localization during Salmonella infection. Video 6 is a time-lapse video showing Cdc42 localization during Salmonella infection. Videos 7 (wide field) and 8 (confocal) are time-lapse videos showing GFP-SGEF localization during Salmonella infection. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200605144/DC1.
Supplementary Material
Acknowledgments
We thank Xosé Bustelo and Alan Hall for providing plasmids and reagents and members of the Galán laboratory for critical reading of this manuscript.
This work was supported by Public Health Service grant AI 055472 from the National Institutes of Health to J.E. Galán.
Abbreviations used in this paper: CRIB, Cdc42–Rac1 interaction binding; PAK, p21-activated kinase; SGEF, SH3-containing guanine nucleotide exchange factor; TTSS, type III secretion system.
References
- Aktories, K. 1997. Bacterial toxins that target Rho proteins. J. Clin. Invest. 99:827–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi, C., V. Singh, M. Wood, P. Jones, T. Wallis, and E. Galyov. 2000. Identification of SopE2, a Salmonella secreted protein, which is highly homologous to SopE and involved in bacterial invasion of epithelial cells. J. Bacteriol. 182:2341–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwald, G., A. Friebel, J.E. Galán, W.D. Hardt, A. Wittinghofer, and K. Scheffzek. 2002. Structural basis for the reversible activation of a Rho protein by the bacterial toxin SopE. EMBO J. 21:3286–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L.M., S. Hobbie, and J.E. Galán. 1996. a. Requirement of CDC42 for Salmonella-induced cytoskeletal and nuclear responses. Science. 274:2115–2118. [DOI] [PubMed] [Google Scholar]
- Chen, L.M., K. Kaniga, and J.E. Galán. 1996. b. Salmonella spp. are cytotoxic for cultured macrophages. Mol. Microbiol. 21:1101–1115. [DOI] [PubMed] [Google Scholar]
- Ellerbroek, S.M., K. Wennerberg, W.T. Arthur, J.M. Dunty, D.R. Bowman, K.A. DeMali, C. Der, and K. Burridge. 2004. SGEF, a RhoG guanine nucleotide exchange factor that stimulates macropinocytosis. Mol. Biol. Cell. 15:3309–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebel, A., H. Ilchmann, M. Aepfelbacher, K. Ehrbar, W. Machleidt, and W. Hardt. 2001. SopE and SopE2 from Salmonella typhimurium activate different sets of RhoGTPases of the host cell. J. Biol. Chem. 276:34035–34040. [DOI] [PubMed] [Google Scholar]
- Fu, Y., and J.E. Galán. 1999. A Salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature. 401:293–297. [DOI] [PubMed] [Google Scholar]
- Galán, J.E. 2001. Salmonella interaction with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17:53–86. [DOI] [PubMed] [Google Scholar]
- Galán, J.E., and R. Curtiss III. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA. 86:6383–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán, J.E., and R. Curtiss III. 1990. Expression of Salmonella typhimurium genes required for invasion is regulated by changes in DNA supercoiling. Infect. Immun. 58:1879–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán, J.E., C. Ginocchio, and P. Costeas. 1992. Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J. Bacteriol. 17:4338–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, A. 1998. Rho GTPases and the actin cytoskeleton. Science. 279:509–514. [DOI] [PubMed] [Google Scholar]
- Hannon, G., and D. Conklin. 2004. RNA interference by short hairpin RNAs expressed in vertebrate cells. Methods Mol. Biol. 257:255–266. [DOI] [PubMed] [Google Scholar]
- Hardt, W.-D., L.-M. Chen, K.E. Schuebel, X.R. Bustelo, and J.E. Galán. 1998. Salmonella typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell. 93:815–826. [DOI] [PubMed] [Google Scholar]
- Hernandez, L.D., K. Hueffer, M.R. Wenk, and J.E. Galán. 2004. Salmonella modulates vesicular trafficking by altering phosphoinositide metabolism. Science. 304:1805–1807. [DOI] [PubMed] [Google Scholar]
- Hobbie, S., L.M. Chen, R. Davis, and J.E. Galán. 1997. Involvement of the mitogen-activated protein kinase pathways in the nuclear responses and cytokine production induced by Salmonella typhimurium in cultured intestinal cells. J. Immunol. 159:5550–5559. [PubMed] [Google Scholar]
- Jaffe, A., and A. Hall. 2005. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21:247–269. [DOI] [PubMed] [Google Scholar]
- Katoh, H., and M. Negishi. 2003. RhoG activates Rac1 by direct interaction with the Dock180-binding protein Elmo. Nature. 424:461–464. [DOI] [PubMed] [Google Scholar]
- May, V., M. Schiller, B. Eipper, and R. Mains. 2002. Kalirin Dbl-homology guanine nucleotide exchange factor 1 domain initiates new axon outgrowths via RhoG-mediated mechanisms. J. Neurosci. 22:6980–6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobes, C.D., and A. Hall. 1995. Rho, Rac and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 81:53–62. [DOI] [PubMed] [Google Scholar]
- Norris, F.A., M.P. Wilson, T.S. Wallis, E.E. Galyov, and P.W. Majerus. 1998. SopB, a protein required for virulence of Salmonella dublin, is an inositol phosphate phosphatase. Proc. Natl. Acad. Sci. USA. 95:14057–14059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander, E., J. ten Klooster, S. van Delft, R. van der Kammen, and J. Collard. 1999. Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J. Cell Biol. 147:1009–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuebel, K., N. Movilla, J. Rosa, and X. Bustelo. 1998. Phosphorylation-dependent and constitutive activation of Rho proteins by wild-type and oncogenic Vav-2. EMBO J. 17:6608–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen, M., C. Lippuner, T. Kaiser, A. Misslitz, T. Aebischer, and D. Bumann. 2003. Rapidly maturing red fluorescent protein variants with strongly enhanced brightness in bacteria. FEBS Lett. 552:110–114. [DOI] [PubMed] [Google Scholar]
- Stebbins, C.E., and J.E. Galán. 2001. Structural mimicry in bacterial virulence. Nature. 412:701–705. [DOI] [PubMed] [Google Scholar]
- Stender, S., A. Friebel, S. Linder, M. Rohde, S. Mirold, and W. Hardt. 2000. Identification of SopE2 from Salmonella typhimurium, a conserved guanine nucleotide exchange factor for Cdc42 of the host cell. Mol. Microbiol. 36:1206–1211. [DOI] [PubMed] [Google Scholar]
- Tse, S., J. Broderick, M. Wei, M. Luo, D. Smith, P. McCaffery, S. Stamm, and A. Andreadis. 2005. Identification, expression analysis, genomic organization and cellular location of a novel protein with a RhoGEF domain. Gene. 359:63–72. [DOI] [PubMed] [Google Scholar]
- Vincent, S., P. Jeanteur, and P. Fort. 1992. Growth-regulated expression of rhoG, a new member of the ras homolog gene family. Mol. Cell. Biol. 12:3138–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennerberg, K., S. Ellerbroek, R. Liu, A. Karnoub, K. Burridge, and C. Der. 2002. RhoG signals in parallel with Rac1 and Cdc42. J. Biol. Chem. 277:47810–47817. [DOI] [PubMed] [Google Scholar]
- Whitehead, I., H. Kirk, and R. Kay. 1995. Retroviral transduction and oncogenic selection of a cDNA encoding Dbs, a homolog of the Dbl guanine nucleotide exchange factor. Oncogene. 10:713–721. [PubMed] [Google Scholar]
- Zhou, D., L.M. Chen, L. Hernandez, S.B. Shears, and J.E. Galán. 2001. A Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host cell actin cytoskeleton rearrangements and bacterial internalization. Mol. Microbiol. 39:248–259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.