Abstract
Despite the well established role of the frontal and posterior perisylvian cortices in many facets of human-cognitive specializations, including language, little is known about the developmental patterning of these regions in the human brain. We performed a genome-wide analysis of human cerebral patterning during midgestation, a critical epoch in cortical regionalization. A total of 345 genes were identified as differentially expressed between superior temporal gyrus (STG) and the remaining cerebral cortex. Gene ontology categories representing transcription factors were enriched in STG, whereas cell-adhesion and extracellular matrix molecules were enriched in the other cortical regions. Quantitative RT-PCR or in situ hybridization was performed to validate differential expression in a subset of 32 genes, most of which were confirmed. LIM domain-binding 1 (LDB1), which we show to be enriched in the STG, is a recently identified interactor of LIM domain only 4 (LMO4), a gene known to be involved in the asymmetric pattering of the perisylvian region in the developing human brain. Protocadherin 17 (PCDH17), a neuronal cell adhesion molecule, was highly enriched in focal regions of the human prefrontal cortex. Contactin associated protein-like 2 (CNTNAP2), in which mutations are known to cause autism, epilepsy, and language delay, showed a remarkable pattern of anterior-enriched cortical expression in human that was not observed in mouse or rat. These data highlight the importance of expression analysis of human brain and the utility of cross-species comparisons of gene expression. Genes identified here provide a foundation for understanding molecular aspects of human-cognitive specializations and the disorders that disrupt them.
Keywords: cortex, microarray, gene expression, evolution, CASPR2
The capacity of the human brain for tasks involving abstract thought, creativity, and language is a key feature of our species' evolution (1). Although the gross anatomical substrates for some of these features are at least partially known, the underlying circuitry and its molecular basis remain obscure. More is known about regions that subserve language, especially frontal and posterior perisylvian cortex (2–4), but few of the molecules involved in patterning these areas in humans have been identified. Because aspects of language are asymmetrically distributed in humans (5), we and others have worked to identify the molecular basis of cerebral asymmetry (6), demonstrating gene expression asymmetries in perisylvian cortex during human development (7). The development of human cognition also builds on the significant bilateral (8, 9) expansion of densely connected circuits involving frontal, temporal, and parietal association areas in non-human primates, as well as the likely elaboration of novel functionality in humans (10–13).
Although some aspects of human-elaborated circuits are present in other mammals (14), the homologous structures, even when present, are less developed (15) and are unable to support the functionality typical of our species. Thus, it is important to understand what aspects of cortical patterning are conserved in mammals, and which may involve human-specific specializations. Because differences in gene regulation are thought to drive phenotypic divergence among species (16), finding genes for which transcript distribution in human is distinct from other species would inform our understanding of brain evolution and disorders involving higher cognition.
To identify genes that may contribute to the structure and function of the anterior and posterior areas of the perisylvian cortex, we performed an in-depth analysis of regional gene expression in human fetal brain. We used two whole-genome microarray platforms to extensively survey midgestation cortical gene expression to identify genes enriched in frontal cortex and superior temporal regions. This period was chosen because it encompasses the peak of neurogenesis and neuronal migration, a stage that is critical for regional and functional specialization of the cerebral cortex (CTX) (17–23). We also performed detailed analysis of several genes by in situ hybridization, which revealed the marked anterior cortical enrichment of CNTNAP2 and PCDH17 in humans. Strikingly, the CNTNAP2 expression pattern observed in humans was not present in mouse or rat. Such focally expressed genes represent key candidates for understanding the development of human-cognitive specializations,
Results
Whole-Genome Microarray Experiments Identify Genes Differentially Expressed (DE) Between Superior Temporal Gyrus (STG) and CTX.
To identify genes that were DE during cortical regionalization, we compared eight STG samples to CTX by using Agilent (Palo Alto, CA) G4110A two-color arrays. These analyses identified 343 DE genes, with 44 enriched and 299 reduced in STG vs. CTX at a false-discovery rate of <5%. Unsupervised hierarchical clustering of expression values grouped samples according to tissue type (Fig. 1) but not gestational age, sex, postmortem interval, or side of origin, showing that these secondary variables [see supporting information (SI) Table 1] were not significant factors contributing to differential expression. Although background levels are typically low with Agilent microarrays, we further limited this set of genes by also requiring expression at least twice background. This filtering resulted in 172 DE genes, 25 up and 147 down in STG.
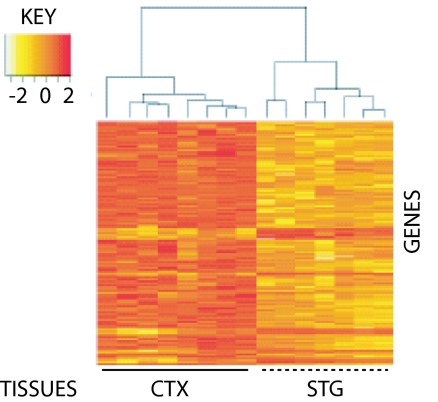
Fig. 1.
Regional differences in gene expression discriminate between STG and remaining CTX. RNA from midgestation human STG and CTX (average of 18.6 weeks old) were labeled with Cy3 and Cy5, respectively, and pairs of samples hybridized against Agilent G4110A arrays. After normalization of expression levels with Agilent Feature Extraction software (Ver. 7.5.1), single-channel values were analyzed for >1.5-fold differences using the Limma package from Bioconductor/R. An unsupervised clustering algorithm, based on Euclidian Distance, was then used to calculate intersample relationships using expression values for these DE genes (n = 343; 44 up- and 299 down-regulated). In contrast to regional clustering, no effect was observed for sex, side of origin (Left or Right), or developmental age (data not shown).
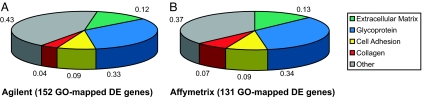
DAVID, an online tool for identification of enriched functional groups within gene lists (24), was then used to analyze DE genes. Nucleus-localized DNA-binding proteins (including FLJ12228, H1F0, HOXB13, LDB1, PIASXβ, and NR4A2) were overrepresented among STG up-regulated genes (6 of 24; P < 0.04), consistent with the known role of transcription factors in CNS patterning. In contrast, extracellular matrix constituents (18 of 152; P < 1e-16), glycoproteins (50 of 152; P < 1e-8), cell-adhesion molecules (13 of 152; P < 7e-6), and collagens (6 of 152; P < 2e-5) were evident among genes down in STG relative to CTX (Fig. 2a). It is well established that matrix constituents are important modulators of neurite outgrowth and pathfinding in model systems (25, 26); our data highlight regional expression differences for these molecules in the developing human cortex.
Fig. 2.
GO analysis of frontal circuit-enriched genes highlights close correspondence for Agilent and Affymetrix array platforms. GO terms genes identified as DE for Agilent (A) and Affymetrix (B) were extracted by using DAVID and analyzed for enrichment of functional groups. Both platforms revealed an overrepresentation of matrix related proteins involved in neuronal pathfinding.
Validation by Gene Ontology (GO) and Overlap Analysis.
We performed a second array experiment using HGU133A Affymetrix (Santa Clara, CA) single-color arrays to validate the Agilent data and provide a broader basis for the genomic screen. These analyses identified 186 DE genes, 36 of which were increased, and 150 of which were decreased in STG vs. CTX. As with Agilent, unsupervised hierarchical clustering of Affymetrix expression data grouped samples according to tissue type (not shown). Likewise, multiple STG-enriched genes identified by Affymetrix arrays (n = 5; PAK7, RAB7L1, RASL11B, RRP22, and TAX1BP3) were annotated as molecules that bind nucleic acids. Finally, GO term analysis of Affymetrix genes down in STG gave results virtually identical to those for Agilent (Fig. 2b). The tight correspondence between Agilent and Affymetrix data validates results obtained by each platform independently.
To assess genic overlap between Agilent and Affymetrix arrays, we mapped probe names to UniGene IDs and identified IDs common to both lists. These analyses highlighted 13 genes identified as DE by both platforms (SI Table 2), all of which showed decreased expression in STG vs. CTX. Independent identification of targets by both single-channel Affymetrix and two-color Agilent microarrays is notable given differences in probe type, probe inclusion, and the stringent statistical cutoffs used. The overlap between the two platforms was also highly significant; the exact hypergeometric probability of obtaining such a result by chance alone is P < 3.3 × 10−13. We also carried out 100,000 permutation trials in which genes were selected from each array platform randomly and then compared for overlap. In contrast to our experimental overlap of 13 genes, the mean overlap from these simulations was under 2, yielding an empiric P < 10−5 (SI Fig. 6). Between the two platforms, a total of 345 genes were identified as DE, with 61 enriched and 284 down in STG, respectively.
Quantitative RT-PCR (Q-PCR) Confirms Differential Expression.
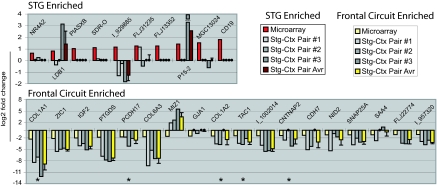
Although 13 genes were identified as DE by both array platforms, the majority of genes that met criteria (332 of 345) did so on only one. To test directly whether additional single-platform genes were DE, we used Q-PCR (see SI Tables 1 and 3) to compare levels of five two-platform genes and an additional 30 single-platform genes, obtaining informative data for 28 of 35 targets (80%). We chose a cross section of genes with >1.5-fold changes, to be within the more reliable detection range of SYBR green-based Q-PCR, also focusing on genes with known roles in brain patterning or disease. Q-PCR results for all two-platform genes confirmed array data (Fig. 3), with Q-PCR based estimates of fold change exceeding the corresponding array values in all cases. Nineteen of 23 single-platform genes showed changes in the same direction as the microarray, and approximately half had fold changes ≥1.5 in the same direction as observed by microarray (0.58 on the log2 scale).
Fig. 3.
Q-PCR validates a subset of microarray-identified genes as DE between STG and remaining CTX. Q-PCR was carried out in triplicate on cDNA prepared from three STG and three CTX samples. Illustrated here (from left to right for individual genes) are the log2-transformed fold change values obtained by (i) averaging across arrays (red and pale yellow for STG and frontal circuit enriched, respectively), (ii) plotting individual Q-PCR-determined STG-CTX comparisons (shades of gray), and (iii) averaging across pairs of Q-PCR comparisons (maroon and bright yellow for STG and frontal circuit enriched, respectively). Q-PCR fold change values between −1 and +1 were not converted to log2 but rather plotted as large black circles on the x axis. Q-PCR results agreed with array data for each of five genes identified as DE by both Agilent and Affymetrix arrays (*), exceeding array fold change values in all cases. Approximately half (11 of 23) of the “single-platform” genes tested showed an average fold change of ≥1.5 (0.58 on the log2 scale) in the same direction as observed by microarray. Several targets not validated by Q-PCR were later confirmed by in situ hybridization (e.g., LBD1 and NR4A2; Fig. 6). The lowest fold change we were able to confirm by Q-PCR was 2.2 (=1.1 on the log2 scale).
In Situ Hybridization Highlights Regionally Restricted Expression in Perisylvian Language-Related Structures and Circuits.
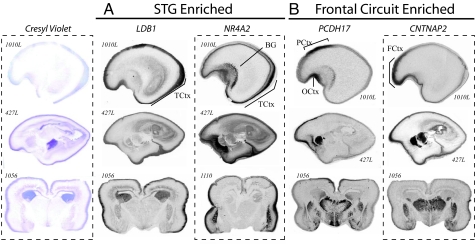
Given cellular and regional heterogeneity within brain and between fetuses (27), we reasoned that a second method, in which data are not averaged across individuals, might be useful to assay patterns of gene expression. We therefore used in situ hybridization to characterize transcript distribution of 13 genes in an additional set of human fetal brains (see SI Table 4). Informative data were obtained for 12 of 13 targets, and support for microarray results was obtained for 11 of 12, including several STG-enriched genes not confirmed by Q-PCR (e.g., LDB1, MAB21L1, MGC13024, NR4A2, and PIASXβ). Consistent enrichment in STG and posterior cortex (Fig. 4A) was observed for LIM domain-binding 1 (LDB1) and nuclear receptor 4A2 (NR4A2), whereas Protocadherin 17 (PCDH17) and contactin-associated protein-like 2 (CNTNAP2) showed consistent frontal and anterior enrichment (Fig. 4B).
Fig. 4.
In situ hybridization validates array data and highlights regionally restricted transcript distribution. Transcript distribution was assayed in 19- and 20-week-old human brains sectioned in either the sagittal (ID no. 1010L and 427L shown here) or coronal orientation (ID no. 1056 and 1110 shown here). LBD1 and NR4A2 are enriched in posterior temporal cortex (A), whereas PCDH17 and CNTNAP2 are enriched in frontal cortex (B). Whereas LDB1 shows a broad-intensity signal across multiple layers within the STG anlage, a high-intensity signal for NR4A2 was restricted to a subset of the later-born neurons from more superficial cortical plate. Similar expression is seen for PCDH17 and CNTNAP2; autoradiograms for both show strong enrichment in frontal gray matter. PCDH17 is restricted to paracentral and orbitofrontal cortex. CNTNAP2 is expressed within the cortex between the orbital gyrus and superior frontal anlage, spanning the inferior and middle frontal gyruses. Sense controls tested on adjacent sections (not shown) gave no signal. TCtx, temporal cortex; BG, basal ganglia; FCtx, frontal cortex; PCtx, paracentral cortex; and OCtx, olfactory cortex.
As illustrated in Fig. 4, both LDB1 and NR4A2 showed enrichment in perisylvian cortex. Both genes also showed similar subcortical expression patterns at the level of the STG, with signal in subventricular zone, striatal neuroepithelium, and portions of the lateral basal ganglia. In contrast, PCDH17 and CNTNAP2 overlapped tightly with corticostriatothalamic circuitry involved in diverse higher cognitive processes including language (4, 28). PCDH17 was present at high levels in the exterior margins of the thalamus, ventromedial striatal neuroepithelium, and anterior cingulate. In subcortical structures, CNTNAP2 was expressed at high levels in dorsal thalamus, caudate, putamen, and amygdala.
Marked CNTNAP2 Expression Differences in the CTX of Rodents and Humans.
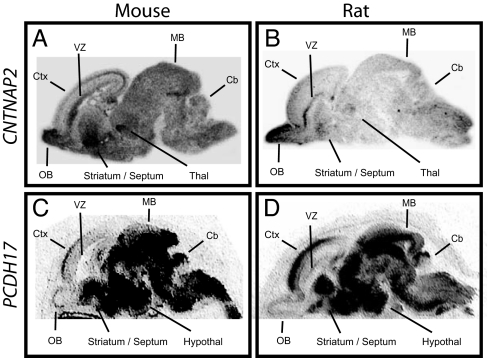
To test the hypothesis that genes enriched in circuits involved in higher cognitive function may show expression differences between humans and other species, we analyzed expression in rodents and compared results to those from humans. We were particularly interested in CNTNAP2 and PCDH17, because variation in CNTNAP2 has been linked to language and autism spectrum disorders (29, 30), and PCDH17 expression was recently suggested to be increased in the prefrontal cortex in a cohort of schizophrenic subjects (31). In sharp contrast to the regionally restricted distribution observed for human CNTNAP2, expression in the developing mouse brain was broad (Fig. 5A). Similarly, in contrast to the frontal enrichment observed in human frontal cortex, the corresponding developmental period in mouse [embryonic day 17 (E17)] showed only limited expression in the cortical plate, with highest levels of expression posteriorly. Highest levels of Cntnap2 expression were in olfactory bulb, ventricular zones, striatum, and thalamus. Expression in the developing rat brain was remarkably similar to that observed for mouse (Fig. 5B). Again, the signal was broadly distributed throughout the brain and uniformly low or absent in the cortical plate. The diffuse Cntnap2 signal observed in rodent is not an artifact of tissue preparation as adjacent sections processed for Pcdh17 showed tightly regionalized expression (Fig. 5 C and D). Moreover, unlike rodent Cntnap2, Pcdh17 showed very similar expression in humans and rodents, including prominent enrichment in the anterior cerebral cortical anlage in both embryonic mouse and rat.
Fig. 5.
In situ hybridization reveals striking differences between human and rodent CNTNAP2 expression in the developing brain. In situ hybridizations for CNTNAP2 and PCDH17 were carried out on sagittal brain sections prepared from E17 mice and E21 rats. In both rodent species, CNTNAP2 was low in cortex and lacking the gradient observed in human (A and B). The diffuse signal observed in rodent is not an artifact of tissue preparation; adjacent sections hybridized with an antisense probe for Pcdh17 show tightly regionalized expression. Moreover, like human Pcdh17, prominent enrichment in anterior cortex was seen in both rodent species (C and D). Sense controls on adjacent sections gave no signal (not shown). Cb, cerebellum; Ctx, cortex; hypothal, hypothalamus; MB, midbrain; OB, olfactory bulb; Thal, thalamus; VZ, ventricular zone.
Although the timepoints examined for mouse, rat, and human approximate one another in terms of cortical regionalization, the formal possibility remains that a more restricted Cntnap2 distribution might be observed in rodent at earlier or later developmental stages. Three lines of evidence argue that this is unlikely. First, expression analyses at multiple points in mouse development (E14, E16, and postnatal day 1) indicate a broad transcript distribution similar to data outlined in Fig. 5; no prefrontal enrichment was observed at any timepoint. Second, analyses at multiple points over an extended period of human development (14, 16, 18, 20, and 22 gestational weeks) determined that expression remain restricted to a core set of structures; prefrontal enrichment was observed in every case. Third, analysis of large-scale expression data for adult human and mouse CNTNAP2 (32) revealed prefrontal enrichment by multiple probes in human, but not with any of 10 in mouse (SI Fig. 7). Together, these data suggest that differences between species are unrelated to sample staging.
To ensure that CNTNAP2 expression differences were indeed the result of distinct gene regulation, we generated an additional isoform-specific in situ probe for each species. Hybridization of these isoform-specific probes (pBSA83 or 98; SI Table 4) showed characteristic frontal enrichment only in human tissue. Reciprocal experiments using the human probe on rodent tissue, and vice versa, gave the expected species-specific result. These results demonstrate that mouse–human expression differences for CNTNAP2 are not attributable to distinct isoforms being assayed in each species.
To explore possible mechanisms underlying the distinct patterns of expression, we compared the human, mouse, and rat CNTNAP2 loci for obvious structural differences. Gene structure was conserved across the three species with each ortholog having 24 coding exons distributed across >2 Mb of genomic DNA. Similarly, protein motif structure (100% identity among human, mouse, and rat), amino acid composition (94% human vs. mouse, 92% human vs. rat), and coding DNA (87% human vs. mouse, 87% human vs. rat) were highly conserved between the three species. Our data suggest that, in addition to differential regulation of transcript abundance (33–35), disparate transcript distribution is likely to be an important driver of cerebral cortical evolution.
Discussion
We hypothesized that genes important for the patterning of perisylvian regions that underlie human higher cognitive functions would show focal enrichment during human development in the posterior perisylvian (STG) or frontal cortex (CTX). Because few such genes had previously been characterized, we used a two-stage microarray design to identify a subset of genes with most robust differences in expression. Expression-based hierarchical clustering was successful in distinguishing between cortical regions (Fig. 1). A total of 345 DE genes were identified, with 61 enriched and 284 down-regulated in STG) across two microarray platforms, with 13 genes identified by both array platforms (SI Table 2). Because of experimental differences among array platforms, genes identified by only one platform should be seen as potentially interesting data awaiting confirmation by a second technique. In this regard, it is reassuring that analysis of functional annotations for Affymetrix genes gave results virtually identical to those observed for Agilent (Fig. 2). Q-PCR (Fig. 3) was performed for a significant cross section (nearly 10%) of the 345 genes identified as DE. These experiments confirmed array results for all two-platform genes tested (five of five) and provided support for 83% of informative single-platform genes (19/23). Further support for microarray data and detailed characterization of transcript distribution were obtained for 11 of 12 genes evaluated by in situ hybridization (Fig. 4). A total of 23 of 32 targets were confirmed by either PCR or in situ hybridization, and four of eight genes tested, CNTNAP2, COL1A1, PI5–2 and PCDH17, were validated using both methods (SI Table 6). These Q-PCR and in situ results, across a significant cross section of targets, suggest that, in addition to the 13 genes identified on both array platforms, many but not all of the single-platform genes with fold changes >1.5 are likely to be DE (SI Table 6).
Identification of LDB1, a Regulator of Asymmetrically Expressed LMO4.
LMO4 is one of the few genes known to be asymmetrically expressed during fetal human brain development (7). That LDB1, an essential cofactor and key regulator of LMO4 (36), was among STG-enriched genes confirmed by in situ hybridization suggests that the combined actions of multiple LMO4 signaling partners may combine to specify language-related structures (3, 37, 38). These results also predict that additional LMO4 interactors may be important for the functional specification of perisylvian regions.
A Previously Uncharacterized Developmental Role for CNTNAP2 in Human Brain Patterning.
CNTNAP2 was identified as DE on both array platforms by Q-PCR and by in situ hybridization. Analyses of CNTNAP2, a gene for which both rare and common variants appear related to autism (29, 30, 57–59), showed marked differences in the pattern of cerebral cortical expression between human and rodent (Fig. 5). The anterior temporal and prefrontal regions, in which CNTNAP2 expression is high in humans and low or absent in rodents, are much more highly developed in human and non-human primates, consisting of numerous anatomical structures and circuits such as the pars triangularis in the inferior frontal gyrus (39) and spindle projection neurons in the cingulate (10). Thus, more detailed studies of CNTNAP2 in human and non-human primates may inform our understanding of the evolution and conservation of these structures and the circuits to which they belong.
Published work (40, 41) demonstrates a role for CNTNAP2 in the clustering of potassium channels to juxtaparanodes on axons. Because myelination is not observed in humans until 28 weeks of gestation (42), our data suggest that CNTNAP2 may have additional unappreciated functionality. The regional transcript distribution for human CNTNAP2 detailed here, together with neuronal migration defects in patients homozygous for truncating mutations (30), supports involvement in the developmental patterning of cells comprising corticostriatothalamic circuitry.
Relationship of Gene Expression to Neurodevelopmental Disorders in Humans.
The frontal enrichment observed for each of CNTNAP2 and PCDH17, together with restricted expression in basal ganglia and dorsal thalamus, is remarkable. That both genes recapitulate well characterized circuits known to be involved in many higher cognitive functions in humans, ranging from complex motor planning, imitation, joint attention, and implicit language learning (4, 43, 44), leads to simple hypotheses regarding how allelic variation in each may contribute to neurodevelopmental disability. Genetic variation in human PCDH17 has yet to be studied in the context of disease but is likely warranted, given that loci for each of dyslexia (45) and specific language impairment (46) include this gene, and changes in its expression have also been documented for schizophrenia (31). The dramatic expression differences we observed are also of immediate practical importance to the use of model systems for the study of CNTNAP2 in human disease.
Finally, much of our knowledge of mammalian brain development comes from work in rodent (47, 48). Because many core features of brain patterning, structure, and circuit composition are conserved between rodents and humans (22, 49–51), these data have proven invaluable to our understanding of human neurobiology in health and disease. Looking forward, studies in the developing human brain, followed by detailed interspecies comparisons, will be necessary for an improved understanding of how regions that are highly developed in humans, such as the prefrontal cortex and interconnected regions in temporal and parietal cortex (44, 52), develop the capacity to support cognition and behavior.
Materials and Methods
Sample Preparation.
Fresh-frozen human midgestation brains (see SI Table 1) were obtained from the National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders (University of Maryland, Baltimore, MD). After separation of the two hemispheres, tissue from STG was extracted. RNA from STG and remaining CTX was prepared by using TRIzol (Invitrogen, Carlsbad, CA) and evaluated by Nanodrop (A260/280 >1.8) and Agilent Bio-Analyzer (28s:18s >2).
Microarrays.
For both G4110A (Agilent) and HGU133A (Affymetrix) arrays, labeling, hybridization, and scanning were carried out according to the manufacturer's instructions. Whereas the Agilent G4110A array is a two-color system with one 60-bp probe for each feature (20,173 features mapping to an estimated 18,716 genes), the Affymetrix HGU133A array is a one-color system with 11 oligonucleotide match-mismatch probe pairs of 25 bp each (22,000 features interrogating an estimated 14,500 distinct genes). For Agilent arrays, STG and CTX samples were labeled with Cy5 and Cy3, respectively. The number of genes common to the two platforms (as determined by the proportion of shared Unigene IDs) is 10,836.
Array Analysis.
For Agilent arrays, normalized expression values were obtained from the Agilent Feature Extraction software (Ver. 7.5.1). We then used Limma (60), a package in Bioconductor (53), to determine statistical significance. To identify DE genes, we first limited the data to the 500 probes with the best statistical support (false-discovery-adjusted P values <0.001). Data were then subset further to include only those probes with a 1.5-fold difference between STG and CTX and expression values at least twice background. Data from Affymetrix CEL files were processed with Probefilter (in R/Bioconductor) to exclude nonspecific and misannotated probes, log2-transformed, and then corrected for batch effects with ComBat (54). CyberT was used (54) to identify genes for which the posterior probability of differential expression was at least 0.99.
Q-PCR.
SYBR green-based Q-PCR (SI Tables 1, 3, and 6) was performed in a 384-well plate format using iTAQ SYBR Green Supermix with ROX (Bio-Rad, Hercules, CA) on an ABI 7900HT (Applied Biosystems, Foster City, CA).
In Situ Hybridization.
In situ hybridization on fresh frozen tissues (SI Tables 4 and 5) was performed as described (55, 56). Each antisense probe was hybridized against multiple sections from at least three separate brains.
Supplementary Material
Acknowledgments
We thank Eric Wexler, Gena Konopka, and Jamee Bomar for insightful discussion; Amir Abu Khalil for technical help, and Lauren Kawaguchi for superb laboratory and administrative support. We thank the National Institutes of Health-funded Developmental Brain and Tissue Bank at University of Maryland for access to the tissues used in these studies (National Institute of Child Health and Human Development Contract no. NO1-HD-4-3368 and NO1-HD-4-3383). We also acknowledge funding awarded from the National Institute of Mental Health (Grant R01 MH 64547, to D.H.G.), the Cure Autism Now Foundation, and Autism Speaks. B.S.A. was supported by a Postdoctoral Fellowship from the Tourette Syndrome Association.
Abbreviations
- DE
differentially expressed
- STG
superior temporal gyrus
- CTX
cerebral cortex
- GO
gene ontology
- Q-PCR
quantitative RT-PCR
- En
embryonic day n.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Our array data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus repository, www.ncbi.nlm.nih.gov/geo (accession no. GSE9335).
This article contains supporting information online at www.pnas.org/cgi/content/full/0706128104/DC1.
References
- 1.Hauser MD, Chomsky N, Fitch WT. Science. 2002;298:1569–1579. doi: 10.1126/science.298.5598.1569. [DOI] [PubMed] [Google Scholar]
- 2.Geschwind N. Science. 1970;170:940–944. doi: 10.1126/science.170.3961.940. [DOI] [PubMed] [Google Scholar]
- 3.Galaburda AM, LeMay M, Kemper TL, Geschwind N. Science. 1978;199:852–856. doi: 10.1126/science.341314. [DOI] [PubMed] [Google Scholar]
- 4.Lieberman P. Am J Phys Anthropol. 2002;35(Suppl):36–62. doi: 10.1002/ajpa.10171. [DOI] [PubMed] [Google Scholar]
- 5.Baynes K, Eliassen JC, Lutsep HL, Gazzaniga MS. Science. 1998;280:902–905. doi: 10.1126/science.280.5365.902. [DOI] [PubMed] [Google Scholar]
- 6.Geschwind DH, Miller BL. Am J Med Genet. 2001;101:370–381. [PubMed] [Google Scholar]
- 7.Sun T, Patoine C, Abu-Khalil A, Visvader J, Sum E, Cherry TJ, Orkin SH, Geschwind DH, Walsh CA. Science. 2005;308:1794–1798. doi: 10.1126/science.1110324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jung-Beeman M. Trends Cognit Sci. 2005;9:512–518. doi: 10.1016/j.tics.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Vargha-Khadem F, Carr LJ, Isaacs E, Brett E, Adams C, Mishkin M. Brain. 1997;120:159–182. doi: 10.1093/brain/120.1.159. [DOI] [PubMed] [Google Scholar]
- 10.Nimchinsky EA, Gilissen E, Allman JM, Perl DP, Erwin JM, Hof PR. Proc Natl Acad Sci USA. 1999;96:5268–5273. doi: 10.1073/pnas.96.9.5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Preuss TM, Qi H, Kaas JH. Proc Natl Acad Sci USA. 1999;96:11601–11606. doi: 10.1073/pnas.96.20.11601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Astafiev SV, Shulman GL, Stanley CM, Snyder AZ, Van Essen DC, Corbetta M. J Neurosci. 2003;23:4689–4699. doi: 10.1523/JNEUROSCI.23-11-04689.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caceres M, Suwyn C, Maddox M, Thomas JW, Preuss TM. Cereb Cortex. 2006;17:2312–2321. doi: 10.1093/cercor/bhl140. [DOI] [PubMed] [Google Scholar]
- 14.Robbins TW. J Comp Neurol. 2005;493:140–146. doi: 10.1002/cne.20717. [DOI] [PubMed] [Google Scholar]
- 15.Cantalupo C, Hopkins WD. Nature. 2001;414:505. doi: 10.1038/35107134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King MC, Wilson AC. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 17.McConnell SK. Neuron. 1995;15:761–768. doi: 10.1016/0896-6273(95)90168-x. [DOI] [PubMed] [Google Scholar]
- 18.Fishell G. Curr Opin Neurobiol. 1997;7:62–69. doi: 10.1016/s0959-4388(97)80121-3. [DOI] [PubMed] [Google Scholar]
- 19.Rakic P, Lombroso PJ. J Am Acad Child Adolesc Psychiatry. 1998;37:116–117. doi: 10.1097/00004583-199801000-00026. [DOI] [PubMed] [Google Scholar]
- 20.Rubenstein JL, Beachy PA. Curr Opin Neurobiol. 1998;8:18–26. doi: 10.1016/s0959-4388(98)80004-4. [DOI] [PubMed] [Google Scholar]
- 21.Rakic S, Zecevic N. Eur J Neurosci. 2000;12:2721–2734. doi: 10.1046/j.1460-9568.2000.00153.x. [DOI] [PubMed] [Google Scholar]
- 22.Monuki ES, Walsh CA. Nat Neurosci. 2001;4:1199–1206. doi: 10.1038/nn752. [DOI] [PubMed] [Google Scholar]
- 23.Ross ME, Walsh CA. Annu Rev Neurosci. 2001;24:1041–1070. doi: 10.1146/annurev.neuro.24.1.1041. [DOI] [PubMed] [Google Scholar]
- 24.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 25.Hoang B, Chiba A. J Neurosci. 1998;18:7847–7855. doi: 10.1523/JNEUROSCI.18-19-07847.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonner J, O'Connor TP. J Neurosci. 2001;21:9782–9791. doi: 10.1523/JNEUROSCI.21-24-09782.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geschwind DH. Proc Natl Acad Sci USA. 2000;97:10676–10678. doi: 10.1073/pnas.97.20.10676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diamond A. Child Dev. 2000;71:44–56. doi: 10.1111/1467-8624.00117. [DOI] [PubMed] [Google Scholar]
- 29.Geschwind DH. World Congress on Psychiatric Genetics. Cagliari, Italy: International Society of Psychiatric Genetics; 2006. [PubMed] [Google Scholar]
- 30.Strauss KA, Puffenberger EG, Huentelman MJ, Gottlieb S, Dobrin SE, Parod JM, Stephan DA, Morton DH. N Engl J Med. 2006;354:1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 31.Dean B, Keriakous D, Scarr E, Thomas EA. Aust N Z J Psychiatry. 2007;41:308–320. doi: 10.1080/00048670701213245. [DOI] [PubMed] [Google Scholar]
- 32.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, et al. Proc Natl Acad Sci USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khaitovich P, Muetzel B, She X, Lachmann M, Hellmann I, Dietzsch J, Steigele S, Do HH, Weiss G, Enard W, et al. Genome Res. 2004;14:1462–1473. doi: 10.1101/gr.2538704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Preuss TM, Caceres M, Oldham MC, Geschwind DH. Nat Rev Genet. 2004;5:850–860. doi: 10.1038/nrg1469. [DOI] [PubMed] [Google Scholar]
- 35.Caceres M, Lachuer J, Zapala MA, Redmond JC, Kudo L, Geschwind DH, Lockhart DJ, Preuss TM, Barlow C. Proc Natl Acad Sci USA. 2003;100:13030–13035. doi: 10.1073/pnas.2135499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xing G, Zhang L, Russell S, Post R. Schizophr Res. 2006;84:36–56. doi: 10.1016/j.schres.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 37.Chi JG, Dooling EC, Gilles FH. Arch Neurol. 1977;34:346–348. doi: 10.1001/archneur.1977.00500180040008. [DOI] [PubMed] [Google Scholar]
- 38.Geschwind N. Sci Am. 1979;241:180–199. doi: 10.1038/scientificamerican0979-180. [DOI] [PubMed] [Google Scholar]
- 39.Desmond JE, Sum JM, Wagner AD, Demb JB, Shear PK, Glover GH, Gabrieli JD, Morrell MJ. Brain. 1995;118:1411–1419. doi: 10.1093/brain/118.6.1411. [DOI] [PubMed] [Google Scholar]
- 40.Poliak S, Gollan L, Martinez R, Custer A, Einheber S, Salzer J, Trimmer J, Shrager P, Peles E. Neuron. 1999;24:1037–1047. doi: 10.1016/s0896-6273(00)81049-1. [DOI] [PubMed] [Google Scholar]
- 41.Poliak S, Salomon D, Elhanany H, Sabanay H, Kiernan B, Pevny L, Stewart CL, Xu X, Chiu SY, Shrager P, et al. J Cell Biol. 2003;162:1149–1160. doi: 10.1083/jcb.200305018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levitt P. J Pediatr. 2003;143:S35–S45. doi: 10.1067/s0022-3476(03)00400-1. [DOI] [PubMed] [Google Scholar]
- 43.Mundy P. J Child Psychol Psychiatry. 2003;44:793–809. doi: 10.1111/1469-7610.00165. [DOI] [PubMed] [Google Scholar]
- 44.Geschwind D, Iacoboni M. In: The Human Frontal Lobes. 2nd Ed. Miller BL, Cummings JL, editors. New York: Guilford; 2007. [Google Scholar]
- 45.Fisher SE, Francks C, Marlow AJ, MacPhie IL, Newbury DF, Cardon LR, Ishikawa-Brush Y, Richardson AJ, Talcott JB, Gayan J, et al. Nat Genet. 2002;30:86–91. doi: 10.1038/ng792. [DOI] [PubMed] [Google Scholar]
- 46.Bartlett CW, Flax JF, Logue MW, Smith BJ, Vieland VJ, Tallal P, Brzustowicz LM. Hum Hered. 2004;57:10–20. doi: 10.1159/000077385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caviness VS, Jr, Rakic P. Annu Rev Neurosci. 1978;1:297–326. doi: 10.1146/annurev.ne.01.030178.001501. [DOI] [PubMed] [Google Scholar]
- 48.Puelles L, Rubenstein JL. Trends Neurosci. 1993;16:472–479. doi: 10.1016/0166-2236(93)90080-6. [DOI] [PubMed] [Google Scholar]
- 49.Jiang YH, Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, Sweatt JD, Beaudet AL. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 50.Skradski SL, Clark AM, Jiang H, White HS, Fu YH, Ptacek LJ. Neuron. 2001;31:537–544. doi: 10.1016/s0896-6273(01)00397-x. [DOI] [PubMed] [Google Scholar]
- 51.Abrahams BS, Kwok MC, Trinh E, Budaghzadeh S, Hossain SM, Simpson EM. J Neurosci. 2005;25:6263–6270. doi: 10.1523/JNEUROSCI.4757-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goldman-Rakic PS. Philos Trans R Soc London B. 1996;351:1445–1453. doi: 10.1098/rstb.1996.0129. [DOI] [PubMed] [Google Scholar]
- 53.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson WE, Li C, Rabinovic A. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 55.Easterday MC, Dougherty JD, Jackson RL, Ou J, Nakano I, Paucar AA, Roobini B, Dianati M, Irvin DK, Weissman IL, et al. Dev Biol. 2003;264:309–322. doi: 10.1016/j.ydbio.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 56.Abu-Khalil A, Fu L, Grove EA, Zecevic N, Geschwind DH. J Comp Neurol. 2004;474:276–288. doi: 10.1002/cne.20112. [DOI] [PubMed] [Google Scholar]
- 57.Alarcón M, Abrahams BS, Stone JL, Duvall JD, Perederiy JV, Bomar JM, Sebat J, Wigler M, Martin CL, Ledbetter DH, et al. Am J Hum Genet. 2007 doi: 10.1016/j.ajhg.2007.09.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bakkaloglu B, O'Roak BJ, Louvi A, Gupta AR, Abelson JF, Morgan TM, Chawarska K, Klin A, Ercan-Sencicek G, Stillman AA, et al. Am J Hum Genet. 2007 doi: 10.1016/j.ajhg.2007.09.017. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, Rea A, Guy G, Lin S, Cook EHJ, et al. Am J Hum Genet. 2007 doi: 10.1016/j.ajhg.2007.09.015. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smyth G. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.