Abstract
RNA folding in the cell occurs during transcription. Expedient RNA folding must avoid the formation of undesirable structures as the nascent RNA emerges from the RNA polymerase. We show that efficient folding during transcription of three conserved noncoding RNAs from Escherichia coli, RNase P RNA, signal-recognition particle RNA, and tmRNA is facilitated by their cognate polymerase pausing at specific locations. These pause sites are located between the upstream and downstream portions of all of the native long-range helices in these noncoding RNAs. In the paused complexes, the nascent RNAs form labile structures that sequester these upstream portions in a manner to possibly guide folding. Both the pause sites and the secondary structure of the nonnative portions of the paused complexes are phylogenetically conserved among γ-proteobacteria. We propose that specific pausing-induced structural formation is a general strategy to facilitate the folding of long-range helices. This polymerase-based mechanism may result in portions of noncoding RNA sequences being evolutionarily conserved for efficient folding during transcription.
Keywords: RNase P, tmRNA, long-range helix
Because of the degeneracy and stability of base-pairing, RNA has a high propensity to form long-lived undesirable intermediates along their folding pathways (1–3). Particularly challenging for RNA folding during transcription is the formation of duplexes composed of two strands located far apart in sequence. In the time before the transcription of the downstream strand, the upstream strand can form stable yet nonnative base pairs with other regions, which could hinder folding. This undesirable possibility can be circumvented by sequestering the upstream strand in specific but labile structures. Such structures can guide the formation of the native long-range helices by avoiding the formation of long-lived unproductive species.
When compared with Mg2+-induced refolding of full-length RNAs, two obvious properties that influence RNA folding during transcription are the 5′ to 3′ polarity of RNA synthesis and the transcriptional speed of the RNA polymerase (4–8). In addition, bacterial RNA polymerases pause during transcriptional elongation (9, 10). The location and/or duration of the pause sites can alter folding (7, 11). A previous work on the folding during transcription of a circularly permuted version of the Bacillus subtilis RNase P RNA demonstrated that pausing at a specific site alters its folding during transcription by Escherichia coli RNA polymerase (11). Only the pause at this site was required for the accelerated folding of one of the two domains of P RNA; varying the transcriptional speed had no effect. Pausing at this site altered the nascent RNA so that an undesirable structure involving a region downstream to the pause site was avoided.
Although the above result demonstrated that pausing can influence folding, it did not address the question of biological relevancy, because (i) circularly permuted RNA transcripts are unnatural, having changed the order of nucleotide synthesis; and (ii) the study mismatches the RNA (from B. subtilis) with a noncognate RNA polymerase (from E. coli). In light of the result that E. coli and B. subtilis RNA polymerase have very distinct pausing properties (9), a coevolutionary relationship between the RNA polymerase and folding during transcription remains unanswered. Transcriptional pausing at a specific location also affects the folding of the FMN-responsive riboswitch by providing FMN with a greater time window to bind the RNA during transcription (7). This case was different from the circularly permuted P RNA study, in that pausing did not alter the structure of the RNA in the paused complex. Here, decreased transcriptional speed also led to the same result.
This current work addresses two questions by studying the folding of three conserved noncoding RNAs from E. coli during transcription by their cognate E. coli RNA polymerase: (i) For naturally evolved RNAs, how do pause sites affect folding during transcription by their cognate RNA polymerase? (ii) What principle(s) underlies the placement of these pause sites for structured noncoding RNAs? All three RNAs, RNase P RNA,signal-recognition particle (SRP) RNA, and tmRNA, contain numerous long-range helices. We show that pausing after the transcription of the 5′ strands of all long-range helices allows for the formation of nonnative structures in the paused complexes. These nonnative structures sequester these upstream portions, possibly preventing the formation of stable nonnative structures that would later hinder the correct formation of the native helices. Our results suggest that one mechanism to achieve efficient folding during transcription is for the polymerase to pause after the transcription of the upstream portions of long-range helices, allowing for the formation of an appropriately labile nonnative intermediate.
Results and Discussion
The catalytic RNA component of ribonuclease P (P RNA) is an essential enzyme required for the 5′ maturation of all tRNAs and several other noncoding RNAs (12, 13). P RNAs have elaborate tertiary structures required for function (14, 15). Mg2+-initiated refolding of B. subtilis and E. coli P RNA involves the rapid formation of unproductive and long-lived kinetic intermediates that result in slow folding (16–18). The native structure of E. coli P RNA has six long-range helices, defined here as helices whose 5′ and 3′ portions are separated by >50 residues (up to 300 residues in E. coli P RNA).
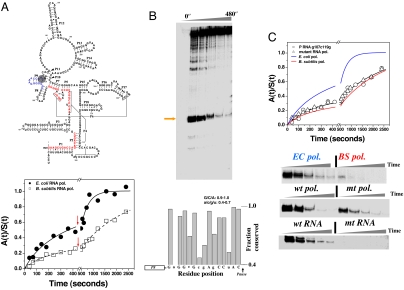
To evaluate whether RNA polymerase-induced pausing influences P RNA folding, we investigated the folding of E. coli P RNA during transcription by its cognate E. coli RNA polymerase and a noncognate B. subtilis polymerase (Fig. 1A). Both polymerases carry out transcription with similar speeds but recognize different pausing signals (9). The formation of native ribozyme was monitored by the recovery of catalytic activity. Fitting the data in Fig. 1A with Eqs. 1 and 2 showed that folding of E. coli P RNA during transcription by B. subtilis RNA polymerase had a fast phase with ≈25% (0.015 s−1, t1/2 ∼ 0.8 min) and a slow phase with ≈75% of the RNA (0.0005 s−1, t1/2 ∼ 23 min). The overall folding profile is only slightly improved when compared with the folding during transcription by T7 polymerase [less pausing, faster speed (19)] or during Mg2+-initated refolding [supporting information (SI) Fig. 6]. Thus, the 5′ to 3′ polarity of RNA during transcription and a variation in transcriptional speed do not significantly alter the folding kinetics of P RNA. Fitting the Fig. 1A data for transcription by E. coli RNA polymerase showed that the fast minor folding population remained the same, but the folding of the major population was ≈6-fold faster (0.003 s−1, t1/2 ∼ 4 min), resulting in significantly more folded molecules at the early time points.
Fig. 1.
Folding and pausing of E. coli P RNA during transcription. (A) At the top, E. coli P RNA. Upstream regions of six long-range helices before the 119 pause (shaded) are in red. At the bottom, folding of P RNA when transcribed by E. coli polymerase [k1 = (15 ± 4) × 10−3·s−1 and f1 = 0.23; k2 = (3 ± 1) × 10−3·s−1 and f2 = 0.77] or by B. subtilis polymerase [k1 = (13 ± 4) × 10−3·s−1 and f1 = 0.21; k2 = (0.5 ± 0.1) × 10−3·s−1 and f2 = 0.79]. Red arrows: +rifampicin to halt transcription. (B Upper) The C119 pause site (orange arrow). (Lower) Fifteen nucleotides preceding the 119 pause manually aligned to show conservation among γ-proteobacteria (53 sequences). (C Upper) The effect on P RNA folding during transcription by the mutant E. coli polymerase [k1 = (12 ± 1)×10−3·s−1 and f1 = 0.30; k2 = (0.5 ± 0.1) ×10−3·s−1 and f2 = 0.70] or by the wild-type polymerase of the C119→G/G107→C mutant [k1 = (10 ± 1)×10−3·s−1 and f1 = 0.33; k2 = (0.5 ± 0.1)×10−3·s−1 and f2 = 0.67]. (Lower) Comparisons of 119 pause during transcription by E. coli and B. subtilis polymerase by wild-type and the mutant E. coli polymerase, and by wild-type polymerase by using the C119→G/G107→C mutant template.
We next investigated whether this fast folding behavior of E. coli P RNA is determined by the pausing properties of its cognate polymerase. A very long-lived pause at nucleotide 119 was observed when transcription was carried out by E. coli RNA polymerase (Fig. 1B; see SI Fig. 6B for pause-site mapping). This pause site had been seen (20) and was present under both pausing (low NTP concentration) and folding (high NTP concentration) conditions (SI Fig. 6). During transcription by B. subtilis RNA polymerase, the strength and duration of this pause were significantly reduced (Fig. 1C and SI Fig. 6). After synthesis up to the pause site at 119, ≈107 nucleotides have emerged from the polymerase exit channel. Forty of these nucleotides belong to the 5′ portions of six long-range helices, P1, P2, P4, P5, P6, and P7 (shown in red in Fig. 1A). Because the downstream portions of these helices have yet to be transcribed, the strong pause at 119 may be an appropriate location to influence the formation of nonnative interactions involving these residues.
To investigate whether pausing at 119 alters folding behavior, we examined the effect of a pausing-defective E. coli RNA polymerase mutant on folding (21) (β: P560S, T563I, Fig. 1C Lower Middle). This mutant polymerase paused at the same site for about half as long as the wild-type polymerase but significantly longer than the B. subtilis RNA polymerase (SI Fig. 6). Nevertheless, P RNA folding during transcription by the mutant polymerase was very similar to that observed for the B. subtilis polymerase (Fig. 1C Upper).
The effect of the 119 pause site on folding during transcription was further investigated with a mutant P RNA that did not pause at this position (Fig. 1C Lower Bottom). The 119 pause site does not resemble either a typical class I or II pause site (10). However, the native P8 helix (Fig. 1A) can form outside of the exit channel of the polymerase when pausing occurs at position 119. We hypothesized that the bacterial family to which E. coli belongs (γ-proteobacteria) may share a common P RNA folding behavior including pausing at this position. Indeed, many of the nucleotides preceding the 119 pause site are highly conserved in this family (Fig. 1B Lower). This conservation includes C119 and its base pairing partner G107. Because C119 is the nucleotide at the pause site, and G107 is in the middle of seven consecutive G residues, we hypothesized that these two residues may be important in pausing at position 119. Therefore, a double-compensating mutant of the E. coli P RNA, C119→G and G107→C, was made that maintained Watson–Crick pairing and the ribozyme's catalytic activity (data not shown) but completely eliminated the 119 pause site (Fig. 1C Lower Bottom). As anticipated, folding of this G107C/C119G mutant P RNA during transcription by the wild-type E. coli polymerase was essentially identical to the folding of wild-type P RNA during transcription by either the B. subtilis or the mutant E. coli RNA polymerase, which also lack the 119 pause site (Fig. 1C Upper).
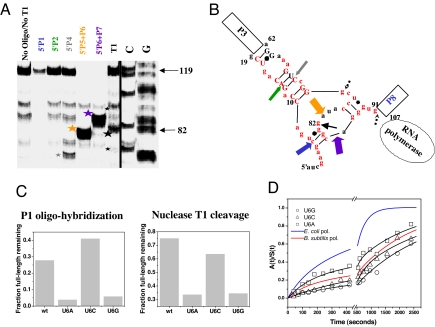
To characterize the nascent RNA structure in the 119-paused P RNA-RNA polymerase complex, structural mapping was performed by using oligonucleotide hybridization/RNase H cleavage (22), and partial nuclease T1 digestion (Fig. 2A). The two native helices P3 and P8 were fully formed, as judged by the absence of nuclease T1 cleavage bands derived from these regions. Strong protection against oligohybridization was observed for the P2 and P4 residues. In contrast, P1, P5, P6, and P7 residues were minimally protected from oligohybridization. However, nuclease T1 primarily cleaved at residue 82 in the P6 region, whereas cleavage at other sites (i.e., residues 74 and 91) were significantly weaker. This result suggests that the 5′ portions of P1, P5, P6, and P7 are structured in the paused complex. However, the oligonucleotide protection results suggest that these structures involving 5′P1/P5/P6/P7 are more labile as compared with the structures involving 5′ P2/P4.
Fig. 2.
Structural analysis of the nascent RNA in the 119-paused complex. (A) Structural mapping of 5′ 32P-labeled 119-paused complex by oligohybridization by using probes complementary to different regions in P RNA and by partial nuclease T1 digestion. The cleavage products are indicated by stars. (B) Proposed structure of the nascent RNA in the 119-paused complex. Colored block arrows show the approximate location and relative intensity of RNase H cleavage upon oligohybridization with the color matching the oligo probe used in A. A black arrow shows the location of the major T1 cleavage product (large star in A). Minor T1 cleavage products are dashed (small stars in A). Red nucleotides indicate the 5′ nucleotides of the long-range helices P1, P2, P4, P5, P6, and P7. Phylogenetically supported base pairs are shown in capital letters. (C) Structural mapping of U6 mutants by T1 nuclease cleavage and by P1 oligohybridization. Mapping with the P2, P4, P5-P6, and P6-P7 oligoprobes produced results similar to wild type (not shown). (D) Folding of U6 mutants when transcribed by E. coli polymerase.
A nonnative structure can be proposed based on the structural mapping results of the 119 paused complex (Fig. 2B). The salient feature of this structure is the sequestration of the 5′ portion of all six long-range helices. The core consists of the P2 and P4 residues forming a helical structure extending from P3. The periphery of this structure consists of portions of P1 and P6 base pairing with each other with some residues in P5 base pairing with P7. The program RNAalifold (23) was used to examine the likelihood of similar nonnative structures being present in bacteria closely related to E. coli (e.g., 39 P RNAs from γ-proteobacteria with an available 5′ P1 sequence; SI Fig. 7). Residues equivalent to 9–19 and 62–73 in E. coli P RNA were given a high probability to base pair with each other, suggesting a conservation of the structural core of this intermediate in the paused complex. The conservation of the peripheral structures is less certain.
To examine the consequences of perturbing this nonnative structure on P RNA folding, residue U6 was mutated to A, G, or C (Fig. 2 C and D). U6 was proposed to pair with G83, and hence, U6A and U6G were expected to decrease, whereas U6C was expected to increase the stability of this intermediate structure. Consistent with these expectations, the U6A and U6G mutants exhibited increased accessibility to the P1 oligo-probe with more T1 cleavage at G82 compared with the wild-type P RNA. Conversely, the U6C mutant had slightly less accessibility to the P1 oligo-probe with similar levels of T1 cleavage at G82 (Fig. 2C). All three U6 mutants folded significantly slower than the wild-type P RNA (Fig. 2D), even though the mutations did not appreciably alter their catalytic activity (data not shown). This result suggests that the structure (the presence and absence of the U6-G83 base pair) plays a role in facilitating P RNA folding during transcription. However, the structure formation alone may be insufficient. The energetics of the P1-P6 helix also may play a role, because the U6C paused intermediate structure may not be labile enough to allow for efficient folding once the downstream portions are transcribed. We could not readily test this possibility with compensatory mutations for residue G83, because it is located in the P6 pseudoknot crucial for the native structure of RNase P RNA (24, 25). Mutating G83 requires additional mutations elsewhere to restore the native structure, and folding results from such complex mutants would have been difficult to interpret.
The specific sequestration of the upstream portions of long-range helices may be a common phenomenon, affecting the folding of a number of noncoding RNAs. We analyzed the folding and structural formation during transcription of two other highly structured and conserved RNAs (Figs. 3 and 4). SRP RNA is essential for the cotranslational insertion of membrane proteins (26, 27). tmRNA is responsible for rescuing stalled ribosomes derived from truncated mRNAs or at codons for which the corresponding charged tRNAs are in limited supply (28, 29). Long-range helices are prominent features in the native structures of both RNAs. The 113 residue E. coli SRP RNA contains two long-range helices, whereas the 363 residue E. coli tmRNA contains five long-range helices.
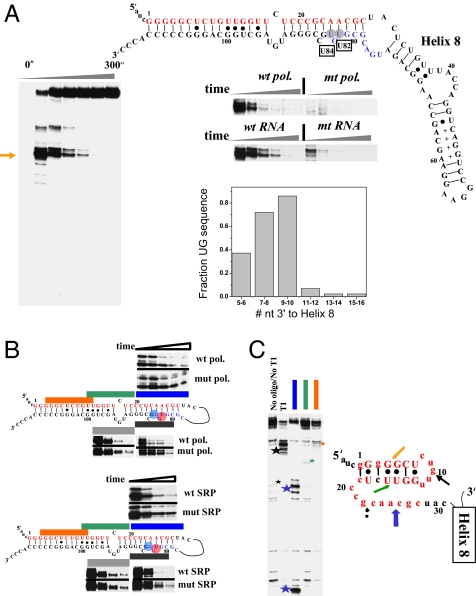
Fig. 3.
Pausing, folding, and structural analysis of E. coli SRP RNA. (A Left) Two pause sites (U82 and U84) are present during transcription by wild-type polymerase. (Right) These pause sites are greatly reduced in the transcription by the mutant polymerase, or upon mutating U82 and its base pairing partner A25 to A82-U25. The graph shows the presence of 5′ UG 7–10 nucleotides downstream to the native helix-8 (manual alignment of 43 sequences). Upstream portions of long-range helices are in red, and the U82/U84 pauses are boxed. (B) Folding of SRP RNA monitored by oligohybridization. (Upper) Transcription by wild-type and mutant polymerase. (Lower) Transcription of wild-type and U25-A82 mutant. The time points are from 15–300 seconds. (C Left) Structural mapping of the body-labeled U82/U84 paused complex by oligohybridization and partial nuclease T1 digestion. The three oligoprobes are complementary to the upstream portions of long-range helices (matching colors as those in B). The cleavage products are indicated by stars. (Right) Proposed structure of the nascent RNA in paused complex. Phylogenetically supported base pairs are shown in capital letters. Colored block arrows, relative intensity and location of RNase H/oligohybridization with the color matching the oligo probe. Black arrows, location of major T1 cleavage (large star in gel on the left) with dashed arrows indicating minor cleavage product (small star in gel on the left).
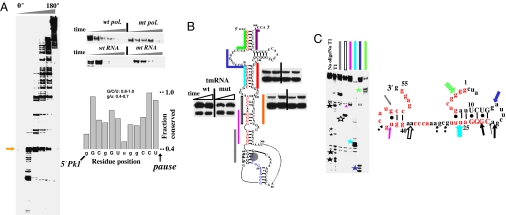
Fig. 4.
Pausing, folding, and structural analysis of E. coli tmRNA. (A Left and Upper Right) Many pause sites are present during transcription by wild-type polymerase. The first major pause (arrow) at U65 is less pronounced in the transcription by the mutant polymerase or through mutation of A51-U65 to U51-A65. Many pause sites are present in the pseudoknot region (residues 200–280). (Lower Right) Sequence preceding the U65 pause is conserved among γ-proteobacteria (manual alignment of 69 sequences). Nucleotides exhibiting especially high levels of conservation are capitalized. (B) Folding monitored by oligohybridization during transcription of wild-type and U25-A82 pausing deficient tmRNA by wild-type polymerase. Upstream portions of the long-range helices are in red and the U65 pause site is shaded. The relative protection from 15–30 seconds is shown. (C Left) Structural mapping of the body-labeled U65 paused complex by oligohybridization and partial nuclease T1 digestion. The six oligos used are complementary to the upstream portions of the long-range helices (matching colors as those in B). The cleavage products are indicated by stars. (Right) Proposed structure of the nascent RNA in paused complex. Phylogenetically supported base pairs are shown in capital letters. Colored block arrows, relative intensity and location of RNase H/oligohybridization with color matching the oligo probe. Black arrows, location of the major T1 cleavage (large stars in gel on the left) with dashed arrows indicating minor cleavage products (small stars in gels on the left).
Transcription of E. coli SRP RNA by E. coli polymerase is characterized by prominent pauses at residues U82 and U84 (Fig. 3A). These pause sites occur just upstream to the 3′ portions of its two long-range helices. Both sites can be tentatively assigned as type I hairpin-dependent pause sites (10). Among γ proteobacteria, 72% have the first, whereas 86% have the second 5′UG signature sequence at this location (Fig. 3A, graph).
Because a functional in vitro folding assay for SRP RNA is unavailable, oligo-probes complementary to the regions encompassing the 5′ and the 3′ portions of these two long range helices were used to assay folding (Fig. 3B). Similar assays have been used extensively in folding studies of the group I ribozyme (30). In wild-type polymerase transcription, the extent of protection increased over time as more SRP RNA molecules presumably folded into their native structures (SI Fig. 8). Both pause sites were absent when transcription was performed by using the same mutant E. coli RNA polymerase (β: P560S, T563I) used in the P RNA folding studies (Fig. 3A Right, top gel). The relative rate of protection was generally greater when transcription was carried out by the wild type compared with the mutant polymerase (Fig. 3B Top). The U82 pause was eliminated by mutating A25-U82 to its complementary U25-A82 base pair (31) (Fig. 3A Right, bottom gel). Once again, the relative rate of protection was greater when the wild-type RNA was transcribed compared with the U82-pausing deficient mutant (Fig. 3B Bottom). These results suggest that the U82 (and presumably U84) pause site influences the folding pathway of SRP RNA during transcription.
We carried out structural mapping of the U82/U84 paused complexes by oligo-hybridization and nuclease T1 cleavage (Fig. 3C). Upon transcription up to helix 8, only 31 residues were available to form a nascent structure involving the upstream portions of the long-range helices. In the paused complex, the protection against oligo-hybridization was strong for residues 3–20 and less so for residues 21–30. The major nuclease T1 cleavage product was at residue 10. Weaker T1 cleavage was also observed at residue 22. This result was consistent with a proposed intermediate structure in which a hairpin core formed within the 5′ portions of both helices, leaving residue 10 particularly exposed (Fig. 3C). RNAalifold analysis of 43 SRP RNAs from γ-proteobacteria indicated a conserved structure in the paused complex at the proposed hairpin (SI Fig. 7). The relative lack of T1 cleavage at residue 27 suggests that residues 20–31 also were involved in the complex, although in a more labile and less phylogenetically conserved manner.
A large number of pause sites were observed when E. coli tmRNA was transcribed by E. coli polymerase, suggesting that transcriptional pausing is important for the proper folding of this molecule as well (Fig. 4A). A major pause site at U65 was located just downstream of the 5′ portions of its five long-range helices. Most of the 10 nucleotides immediately preceding this pause site are highly conserved in γ proteobacteria (Fig. 4A, graph), suggesting that the pause site is conserved in this bacterial branch.
As with SRP RNA, oligo-probes against the long-range helices were used to monitor tmRNA folding. In wild-type polymerase transcription, the absolute protection generally increased over time as more tmRNA molecules fold into their presumed native structures (SI Fig. 9). The same mutant (β: P560S, T563I) E. coli polymerase decreased pausing at U65 (Fig. 4A Right, top gel), but its effect on the pausing of many downstream sites was much stronger. A comparative folding analysis with mutant and wild-type polymerase transcription therefore was unlikely to be informative on the role of U65 pausing on tmRNA folding (data not shown). However, the strength of the U65 pause was reduced when A51-U65 was mutated to U51-A65 (Fig. 4A Right, bottom gel). During transcription of the U65-pausing deficient mutant tmRNA, patterns of protection against oligohybridization (Fig. 4B) were distinct from those produced during transcription of the wild-type tmRNA. Although the protection levels for the downstream probes were similar, the mutant showed decreased protection for an upstream probe when compared with the wild-type tmRNA.
Structural mapping of the U65 paused complex indicated that the nascent tmRNA intermediate is structured (Fig. 4C). The nascent RNA was well protected against hybridization by four of the six probes, but, the protection patterns between this nonnative intermediate and the native structure (SI Fig. 9) were very different. Noticeably, residues 11–20 were very exposed in the native, but relatively well protected in the nonnative structure. Conversely, residues 1–10 and 21–30 were protected in the native, but quite exposed in the nonnative structure. Nuclease T1 cleavage primarily generated cleavage products around nucleotides 18–21, suggesting that this region is particularly exposed to nuclease digestion. Other less prominent T1 cleavage products are also visible. The relatively strong T1 cleavage observed at G21 suggests that the proposed 5′ helix is particularly labile in this structure (Fig. 4C). Alternatively, multiple structures may be present in the paused complex.
Phylogenetic analysis of 69 tmRNAs from γ-proteobacteria indicated that 52 (75%) can have a nonnative structure in this region consistent with our model for the paused complex of E. coli tmRNA (Fig. 4C and SI Fig. 7). Although the U65 pause site appears to be well conserved among γ-proteobacteria, the nonnative intermediate is less well conserved, and there appear to be a number of species-specific interactions. For example, in E. coli, the cytosine residues at residues 35–37 could also interact with the guanine residues toward the 5′ or 3′ end of the structure. In any case, as with the paused complexes of the other two RNAs, the tmRNA paused complex sequesters the upstream portions of all long-range helices, presumably in a marginally stable structure that permits more expedient folding when the downstream portions are transcribed.
Conclusions
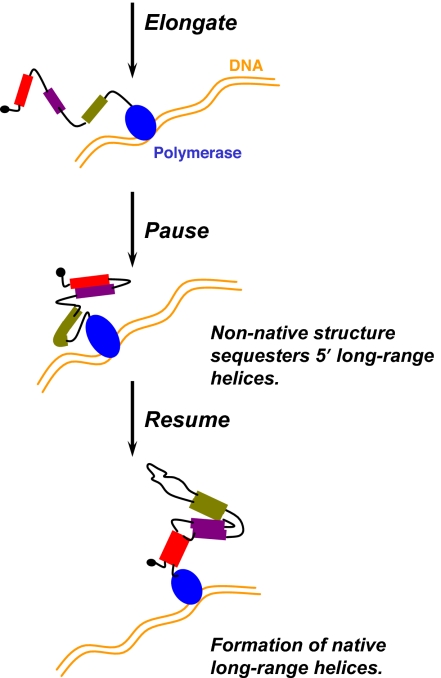
We have shown that a common feature among the folding during transcription of three noncoding RNAs is the formation of defined RNA structures in specifically paused transcription complexes (Fig. 5). These pauses are located downstream to the 5′ portions, but upstream to the 3′ portions of all long-range helices in the native structures of these RNAs. The nascent RNA structures in these paused complexes sequester the upstream portions of the long-range helices in a manner that likely enables more efficient folding when the downstream portions of these helices are transcribed. Indeed, perturbation of these pause sites leads to less efficient folding. Both the pause site and the salient features of the nonnative structure are phylogenetically conserved among γ-proteobacteria.
Fig. 5.
Pausing, long-range helices, and nonnative structure during folding during transcription. DNA template is in gold, and RNA polymerase is in blue. RNA residues involved in long-range helices are shown as filled bars (different colors represent different helices), and other regions of the nascent RNA transcript are shown as black lines. Interaction between regions in the RNA transcript is indicated by proximity.
We hypothesize that site-specific pausing at strategic locations may be a common strategy that allows for the efficient folding of noncoding RNAs during transcription, especially those containing long-range helices in their native structures. For the purpose of efficient folding, the sequence of noncoding RNAs containing long-range helices likely has coevolved with the pausing properties of their cognate RNA polymerases.
Materials and Methods
RNA Polymerases.
The wild-type E. coli RNA polymerase was purchased from USB (Cleveland, OH). The mutant E. coli RNA polymerase (β: P560S, T563I) was a generous gift from Robert Landick (University of Wisconsin, Madison, WI) (21). Polyhis-tagged B. subtilis RNA polymerase was purified in a slightly modified protocol, as described (32).
Transcription Reactions.
Templates for transcription of all RNAs were generated by PCR by using plasmid DNA as templates and confirmed by sequencing. Multiple-round transcription for folding assays was performed as described (ref. 11 and SI Text 1).
Single-round transcription for E. coli RNA polymerase for pausing and structural mapping assays was performed under standard conditions with minor modification (ref. 33 and SI Text 2).
Folding Monitored by Catalytic Activity and Oligohybridization.
Catalysis assays for P RNA were performed as described (ref. 11 and SI Text 4) The fraction of the cleavage product vs. time was fit to a single exponential equation to obtain the reaction rate that determined the relative amount of active ribozyme [A(t)].
To account for RNA synthesis, the data before the addition of rifampicin were fit to the biphasic Eq. 1, where k1 and k2 are the folding rates, and f is the fraction of the population folding at rate k1.
The data after the addition of rifampicin were fit to Eq. 2, where f1 (f2) is the population folding at the same rate k1 (k2) as before the addition of rifampicin still remaining to be folded.
The oligohybridization assay to monitor folding of SRP RNA and tmRNA was a modified version of the method developed by Zarrinkar et al. (ref. 22 and SI Text 3).
Isolation of Polymerase-Paused Complex.
Isolation of the paused complexes was performed similarly to single-round transcription with minor modifications. For P RNA, stalled complexes were formed at 25, 25, and 31 μM ATP, CTP, and GTP, respectively. With internal 32P-labeled paused complexes by using α-32P-GTP, the initiating dinucleotide was unlabeled. For 5′ 32P-labeled paused complexes, the initiating dinucleotide ApU was labeled with γ-[32]ATP. Elongation was resumed upon the addition of UTP to a final concentration of 10 μM. Transcription proceeded at 37°C for 45 seconds (when most of the elongation complexes remain stalled at C119). The reaction was then transferred to ice, and the NTPs were removed through MicroSpin G-25 columns (GE Healthcare, Buckinghamshire, U.K.).
The U82/U84 SRP RNA and U65 tmRNA paused complexes were obtained similarly (SI Text 5).
Structural Mapping of the Paused Complexes.
The oligohybridization assay was a modified version of the method developed by Williamson and coworkers (ref. 22 and SI Text 6). Nuclease T1 mapping was performed as follows. Aliquots of the transcription reaction were mixed with an equal volume of RNase T1 mixture consisting of 20 mM Tris·HCl, pH 7.5/20 mM KCl/14 mM MgCl2/0.1 mM EDTA/0.1 mM DTT/4 μM rifampicin/1.2 mM CaCl2/0.3 μg/μl DNase I/0.24 units/μl RNase T1. The reaction proceeded at 37°C for 45–90 seconds. Aliquots were mixed with an equal volume of 2 mg/ml Proteinase K in 20 mM Tris·HCl, pH 8.0/20 mM NaCl/14 mM MgCl2/14 mM 2-mercaptoethanol/0.1 mM EDTA, and incubated at 55°C for 1 hour.
Phylogenetic Analysis for the Conservation of Nonnative Structures in the Paused Complex.
P RNA, SRP RNA, and tmRNA sequences from numerous γ-proteobacteria were aligned either manually or with ClustalX 1.8.3 (34). In the alignment, the P RNA intermediates were assumed to contain the fully formed P3 native helix. 39/39 P RNA sequences, 43/43 SRP RNA sequences, and 52/69 tmRNA sequences were used in the alignment. Potentially conserved base pairs were identified via the RNAalifold program (http://rna.tbi.univie.ac.at/cgi-bin/alifold.cgi) (23).
Supplementary Material
Acknowledgments
We thank Dr. R. Landick for the generous gift of the mutant E. coli RNA polymerase. We thank Drs. M. Fedor, D. Herschlag, and P. Cluzel for comments on the manuscript and Drs. S. Eddy and R. Landick for insightful discussions. This work was supported by National Institutes of Health Grant GM57880. T.W. was supported by the Medical Scientist Training Program (Grant GM07281).
Abbreviation
- SRP
signal-recognition particle.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705038104/DC1.
References
- 1.Treiber DK, Williamson JR. Curr Opin Struct Biol. 2001;11:309–314. doi: 10.1016/s0959-440x(00)00206-2. [DOI] [PubMed] [Google Scholar]
- 2.Woodson SA. Biochem Soc Trans. 2002;30:1166–1169. doi: 10.1042/bst0301166. [DOI] [PubMed] [Google Scholar]
- 3.Sosnick TR, Pan T. Curr Opin Struct Biol. 2003;13:309–316. doi: 10.1016/s0959-440x(03)00066-6. [DOI] [PubMed] [Google Scholar]
- 4.Isambert H, Siggia ED. Proc Natl Acad Sci USA. 2000;97:6515–6520. doi: 10.1073/pnas.110533697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woodson SA. Cell Mol Life Sci. 2000;57:796–808. doi: 10.1007/s000180050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahen EM, Harger JW, Calderon EM, Fedor MJ. Mol Cell. 2005;19:27–37. doi: 10.1016/j.molcel.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 7.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. Mol Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 8.Pan T, Sosnick T. Annu Rev Biophys Biomol Struct. 2006;35:161–175. doi: 10.1146/annurev.biophys.35.040405.102053. [DOI] [PubMed] [Google Scholar]
- 9.Artsimovitch I, Svetlov V, Anthony L, Burgess RR, Landick R. J Bacteriol. 2000;182:6027–6035. doi: 10.1128/jb.182.21.6027-6035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Artsimovitch I, Landick R. Proc Natl Acad Sci USA. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan T, Artsimovitch I, Fang X, Landick R, Sosnick TR. Proc Natl Acad Sci USA. 1999;96:9545–9550. doi: 10.1073/pnas.96.17.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank DN, Pace NR. Annu Rev Biochem. 1998;67:153–180. doi: 10.1146/annurev.biochem.67.1.153. [DOI] [PubMed] [Google Scholar]
- 13.Altman S, Kirsebom L. In: The RNA World, Second Edition. Gesteland RF, Cech TR, Atkins JF, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1999. pp. 351–380. [Google Scholar]
- 14.Torres-Larios A, Swinger KK, Pan T, Mondragon A. Curr Opin Struct Biol. 2006;16:327–335. doi: 10.1016/j.sbi.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 15.Kazantsev AV, Pace NR. Nat Rev Microbiol. 2006;4:729–740. doi: 10.1038/nrmicro1491. [DOI] [PubMed] [Google Scholar]
- 16.Zarrinkar PP, Wang J, Williamson JR. RNA. 1996;2:564–573. [PMC free article] [PubMed] [Google Scholar]
- 17.Pan T, Sosnick TR. Nat Struct Biol. 1997;4:931–938. doi: 10.1038/nsb1197-931. [DOI] [PubMed] [Google Scholar]
- 18.Kent O, Chaulk SG, MacMillan AM. J Mol Biol. 2000;304:699–705. doi: 10.1006/jmbi.2000.4263. [DOI] [PubMed] [Google Scholar]
- 19.Sousa R, Mukherjee S. Prog Nucleic Acid Res Mol Biol. 2003;73:1–41. doi: 10.1016/s0079-6603(03)01001-8. [DOI] [PubMed] [Google Scholar]
- 20.Lee Y, Ramamoorthy R, Park CU, Schmidt FJ. J Biol Chem. 1989;264:5098–5103. [PubMed] [Google Scholar]
- 21.Landick R, Stewart J, Lee DN. Genes Dev. 1990;4:1623–1636. doi: 10.1101/gad.4.9.1623. [DOI] [PubMed] [Google Scholar]
- 22.Zarrinkar PP, Williamson JR. Science. 1994;265:918–924. doi: 10.1126/science.8052848. [DOI] [PubMed] [Google Scholar]
- 23.Hofacker IL, Fekete M, Stadler PF. J Mol Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 24.Haas ES, Morse DP, Brown JW, Schmidt FJ, Pace NR. Science. 1991;254:853–856. doi: 10.1126/science.1719634. [DOI] [PubMed] [Google Scholar]
- 25.Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A. Nature. 2005;437:584–587. doi: 10.1038/nature04074. [DOI] [PubMed] [Google Scholar]
- 26.Keenan RJ, Freymann DM, Stroud RM, Walter P. Annu Rev Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 27.Doudna JA, Batey RT. Annu Rev Biochem. 2004;73:539–557. doi: 10.1146/annurev.biochem.73.011303.074048. [DOI] [PubMed] [Google Scholar]
- 28.Karzai AW, Roche ED, Sauer RT. Nat Struct Biol. 2000;7:449–455. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- 29.Saguy M, Gillet R, Metzinger L, Felden B. Biochimie. 2005;87:897–903. doi: 10.1016/j.biochi.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 30.Treiber DK, Williamson JR. Methods Enzymol. 2000;317:330–353. doi: 10.1016/s0076-6879(00)17023-5. [DOI] [PubMed] [Google Scholar]
- 31.Mooney RA, Artsimovitch I, Landick R. J Bacteriol. 1998;180:3265–3275. doi: 10.1128/jb.180.13.3265-3275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anthony LC, Artsimovitch I, Svetlov V, Landick R, Burgess RR. Protein Expr Purif. 2000;19:350–354. doi: 10.1006/prep.2000.1272. [DOI] [PubMed] [Google Scholar]
- 33.Landick R, Wang D, Chan CL. Methods Enzymol. 1996;274:334–353. doi: 10.1016/s0076-6879(96)74029-6. [DOI] [PubMed] [Google Scholar]
- 34.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.