Abstract
This study was undertaken to investigate the role of mouse double minute 2 (MDM2) oncogene in prostate cancer growth and the potential of MDM2 as a target for prostate cancer therapy. An antisense anti-human–MDM2 mixed-backbone oligonucleotide was tested in human prostate cancer models with various p53 statuses, LNCaP (p53wt/wt), DU145 (p53mt/mt), and PC3 (p53null). In a dose- and time-dependent manner, it specifically inhibited MDM2 expression and modified expression of several genes, at both mRNA and protein levels. In LNCaP cells, p53, p21, Bax, and hypophosphorylated retinoblastoma tumor suppressor protein (pRb) levels increased, whereas Bcl2, pRb protein, and E2F transcription factor 1 (E2F1) levels decreased. In DU145 cells, p21 levels were elevated and E2F1 levels decreased, although mutant p53, Rb, and Bax levels remained unchanged. In PC3 cells, MDM2 inhibition resulted in elevated p21, Bax, and pRb levels and decreased ppRb and E2F1 levels. In all three cell lines, MDM2 inhibition reduced cell proliferation, induced apoptosis, and potentiated the effects of the chemotherapeutic agents 10-hydroxycamptothecin and paclitaxel. The anti-MDM2 oligonucleotide showed antitumor activity and increased therapeutic effectiveness of paclitaxel in both LNCaP and PC3 xenografts, causing changes in gene expression similar to those seen in vitro. In summary, this study demonstrates that MDM2 has a role in prostate cancer growth via p53-dependent and p53-independent mechanisms and that multiple genes are involved in the process. MDM2 inhibitors such as second-generation antisense oligonucleotides have a broad spectrum of antitumor activities in human cancers regardless of p53 status, providing novel approaches to therapy of human prostate cancer.
Keywords: p53, p21, Bax, E2F1, oligonucleotides
Antisense therapy is designed to deliver to the target cells antisense molecules that target to mRNA with which they can hybridize and specifically inhibit the expression of pathogenic genes (1), thus offering the possibility of specific, rational, genetic-based therapy (1, 2). With encouraging results from preclinical and clinical studies in the past decade, significant progress has been made in the field (3–6). Although the first-generation phosphorothioate antisense oligonucleotides (oligo) are in clinical trials, a number of factors, including in vivo biostability and sequence motifs that could lead to unwanted effects, have been identified (4). The second-generation antisense oligos are designed to overcome these limitations (7, 8). Recently, we have developed several mixed-backbone antisense oligos (MBO) that are being tested for antitumor activity in vitro and in vivo (8–11).
Prostate cancer poses a major public health problem in the United States and worldwide. It has the highest incidence and is the second most common cause of cancer deaths in North American men, with estimates of 220,900 new cases and 28,900 deaths in 2003 (12). There is a need to develop novel therapeutic approaches. The molecular mechanisms of development and progression of prostate cancer are complicated and likely to involve multiple factors, such as tumor suppressor genes, oncogenes, growth factors, signal transduction molecules, adhesion molecules, and angiogenesis (13). Abnormal p53 expression occurs in prostate cancer patients (14–16). Mouse double minute 2 (MDM2) is amplified or overexpressed in a number of human tumors, including prostate cancer (17–21). The expression of MDM2 is induced by p53 and MDM2 oncoprotein functions as a negative regulator of p53. MDM2 also interacts with pRB, E2F transcription factor 1 (E2F1), and RNA, suggesting that MDM2 has p53-independent activities (21). Overexpression of MDM2 is associated with resistance to G1/M growth arrest in prostate cancer DU145 cells (p53mt/mt) (22) and with the androgen-independent phenotype (23), a characteristic of advanced prostate cancer. Therefore, we hypothesized that MDM2 may be a rational target for prostate cancer therapy.
Cancer chemotherapeutic agents and radiation often exert their cytotoxic effects through activation of wild-type p53, which may be limited in cancers with MDM2 expression. Therefore, inactivation of the MDM2–p53 negative feedback loop may increase the magnitude of p53 activation after DNA damaging treatments, thus enhancing therapeutic effectiveness. In addition, we further hypothesized that the role of MDM2 in tumor development and progression is not only associated with p53 pathway, but also with other p53-independent pathways. To that end, in the present study, we used in vitro and in vivo human prostate cancer models with distinct p53 genetic statuses, i.e., LNCaP (p53 wild type), DU145 (p53 mutant), and PC3 (p53 null), to demonstrate that the MDM2 antisense inhibitor has a broad spectrum of antitumor activity in human cancers regardless of p53 status. More importantly, in previous studies, we demonstrated the effect of MDM2 antisense oligo in human cancer cells with MDM2 overexpression (9); we now present data supporting the effect of second-generation antisense MDM2 inhibitor in cell lines that are not MDM2 overexpressors, implying that the application of the specific antisense anti-MDM2 oligo can be widened.
Materials and Methods
Antisense Oligonucleotides. Oligo AS, a 20-mer anti-human-MDM2 MBO (5′-UGACACCTGTTCTCACUCAC-3′) and its 5-base mismatch control, Oligo ASM (5′-UGTCACCCTTTTTCATUCAC-3′) were synthesized, purified, and analyzed as described (7, 10). Two nucleosides at the 5′ end and four nucleosides at the 3′ end are 2′-O-methylribonucleosides (represented by boldface letters); the remaining are deoxynucleosides. The underlined nucleosides of Oligo ASM are the sites of the mismatch controls compared with Oligo AS. All internucleotide linkages are phosphorothioate.
Cell Lines and Culture. The cell lines LNCaP, DU145, and PC3 were obtained from the American Type Tissue Culture Collection. LNCaP cells were cultured in RPMI medium 1640 supplemented with 4.5 g/liter glucose, 1% l-glutamine, 1.5 g/liter sodium bicarbonate, 1% Hepes buffer, and 1% sodium pyruvate. DU145 cells were cultured in EME medium (24). Media for PC3 cells consisted of Ham's F-12 medium supplemented with 1.5 g/liter sodium bicarbonate and 1% l-glutamine. All media contained 10% FBS and 1% penicillin/streptomycin. For in vitro experiments, cells were transfected with oligos in the presence of Lipofectin (7 μg/ml; Life Technologies, Gaithersburg, MD) and 1% FBS for various times before analysis of mRNA and protein levels, apoptosis, and cell proliferation. In combination treatments, oligo-transfected cells were incubated for an additional 12–36 h after adding chemotherapeutic agents, paclitaxel (Mead Johnson Oncology Products, Princeton, NJ) or 10-hydroxycamptothecin (HCPT) (Midwest, Beijing).
BrdUrd Cell Proliferation Assay. BrdUrd incorporation into cells was accomplished by using a BrdUrd cell proliferation assay kit from Oncogene Science. Cells were seeded in 96-well plates (8 × 103 to 1.2 × 104 cells per well) and transfected with oligos for 24 h (12 h for LNCaP). In combination treatments, cells were then exposed to paclitaxel for 36 h (12 h for LNCaP) or HCPT for 24 h (12 h for LNCaP). BrdUrd was added to the medium 10 h before treatment termination. The levels of BrdUrd incorporated into cells were quantified by anti-BrdUrd antibody, measuring absorbance at dual wavelengths of 450/540 nm with an OPTImax microplate reader (Molecular Devices, Sunnyvale, CA).
Detection of Apoptosis. Following a similar treatment protocol as above, cells in early and late stages of apoptosis were detected with an annexin V-FITC apoptosis detection kit from BioVision (Mountain View, CA) as described (24).
Western Blot Analysis. The protein levels of MDM2, p53, p21, hypophosphorylated retinoblastoma tumor suppressor protein (pRb), hyperphosphorylated pRb protein (ppRb), Bcl2, Bax, E2F1, and β-actin in cultured cells and xenografts were analyzed by using previously described methods, including densitometry measurement of protein bands (10, 11, 24). The monoclonal antibodies against MDM2 (Ab-1), p53 (Ab-6), p21 (Ab-1), Rb (Ab-5), Bcl2 (Ab-1), and Bax (Ab-2) were obtained from Oncogene Research Products (Boston, MA). Anti-E2F1 (KH95) monoclonal antibody was purchased from Santa Cruz Biotechnology. Anti-β-actin (SC-15) monoclonal antibody was obtained from Sigma.
Quantitation of mRNA. The mRNA levels in cells treated with Oligo AS or ASM were analyzed by RT-PCR. Cells were transfected with oligos as described above, and total RNA was extracted by using the Trizol reagent from Invitrogen, quantified by UV spectrophotometry, and used to create cDNA by using the SuperScript RT-PCR kit from Invitrogen. The PCR coamplifications of MDM2 (25), P53 (26), BAX (27), E2F1 (28), WAF1, RB, and BCL2 (29) with β-actin (29) were accomplished. The detailed methods are described in Supporting Text and Table 1, which are published as supporting information on the PNAS web site, www.pnas.org.
Xenograft Models. The models of LNCaP and PC3 xenografts were established by using the methods described previously (10, 11, 24). Male, 4- to 6-week-old severe combined immunodeficient mice (for LNCaP) and athymic nude mice (nu/nu) (for PC3), were obtained from Frederick Cancer Research and Development Center (Frederick, MD). Cultured cells were washed with and resuspended in serum-free media. The suspension (5 × 106 cells in 0.2 ml per mouse) mixed with Matrigel at a ratio of 1:1 (LNCaP) or 5:1 (PC3) was then injected into the left inguinal area of the mice. The mice were monitored by measuring tumor growth and body weight and by general clinical observation (24). Tumor-bearing mice were randomly divided into multiple treatment and control groups (five mice per group). All oligos, dissolved in physiological saline (0.9% NaCl), were given by i.p. injection at doses of 10 or 25 mg/kg per day, 5 days/week for 4 weeks. The control group received saline only. Paclitaxel (10 mg/kg) was given i.p., twice per week for 4 weeks. In combination therapy, oligos were given at 25 mg/kg level and paclitaxel as described above.
Results
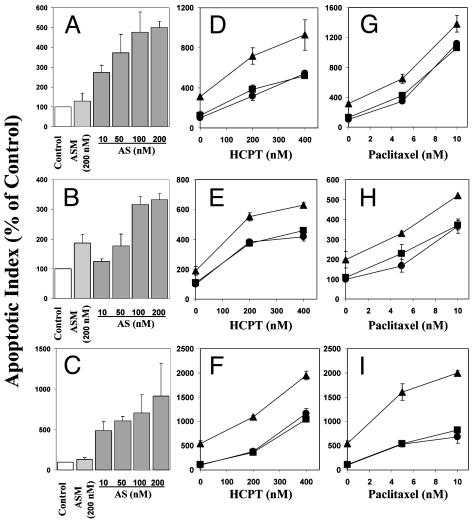
In Vitro MDM2 Antisense Inhibition Resulted in Induction of Apoptosis, Inhibition of Cell Proliferation, and Chemosensitization in Prostate Cancer Cells, Regardless of p53 Status. Induction of apoptosis. In a dose-dependent manner, Oligo AS induced apoptosis in all three prostate cancer cell lines, regardless of p53 status (Fig. 1). After treatment with 200 nM Oligo AS, the apoptotic index increased by 400% in LNCaP cells (Fig. 1 A), 230% in DU145 cells (Fig. 1B), and 820% in PC3 cells (Fig. 1C). The mismatch control Oligo ASM had no or minimal effects. Pretreatment with 50 nM Oligo AS, but not the control Oligo ASM, sensitized all of the three cell lines to the cancer chemotherapeutic agents HCPT (Fig. 1 D–F) and paclitaxel (Fig. 2 G–I).
Fig. 1.
Effects of Oligo AS alone or in combination with HCPT or paclitaxel on apoptosis in prostate cancer cells. The cells were transfected with Oligo AS or ASM at various concentrations for 12 h (A, LNCaP) or 24 h (B, DU145; C, PC3). In combination treatment with HCPT, the cells were transfected with 50 nM Oligo AS or ASM for 12 h (D, LNCaP) or 24 h (E, DU145; F, PC3) and then exposed to HCPT for an additional 12 h (D and E) or 24 h (F). In combination treatment with paclitaxel, the cells were transfected with 50 nM Oligo AS or ASM for 12 h (G, LNCaP) or 24 h (H, DU145; I, PC3) and then exposed to paclitaxel for an additional 12 h (G) or 36 h (H and I). Cells that stained positive for annexin V-FITC (early apoptosis) and positive for FITC and propidium iodide (late apoptosis) were counted. Relative levels of apoptotic indices were expressed as percentage of Lipofectin control. (D–I) Filled circles, Lipofectin control; filled squares, Oligo ASM 50 nM; filled triangles, Oligo AS 50 nM.
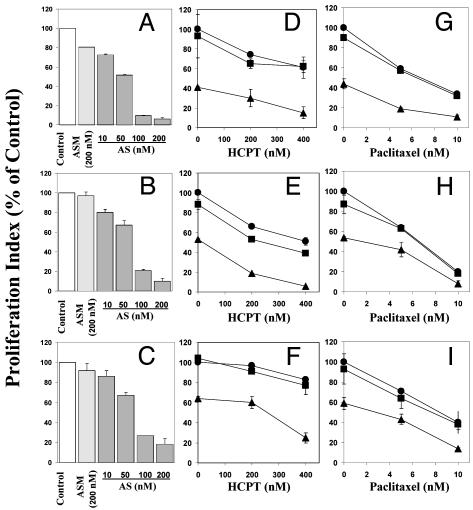
Fig. 2.
Effects of Oligo AS on the proliferation of prostate cancer cells, analyzed by BrdUrd incorporation. By using the same treatment protocol as for Fig. 1, LNCaP (A, D, and G), DU145 (B, E, and H), and PC3 (C, F, and I) cells were treated with oligos alone (A–C) or in combination with HCPT (D–F) or paclitaxel (G–I). Relative levels of BrdUrd incorporations were expressed as percentage of Lipofectin control. (D–I) Filled circles, Lipofectin control; filled squares, Oligo ASM 50 nM; filled triangles, Oligo AS 50 nM.
Inhibition of cell proliferation. In a dose-dependent manner, Oligo AS inhibited proliferation, as measured by BrdUrd incorporation, in all three prostate cancer cell lines, regardless of p53 status (Fig. 2). After treatment with 200 nM Oligo AS, the proliferation index decreased by 94% in LNCaP cells (Fig. 2 A), 90% in DU145 cells (Fig. 2B), and 82% in PC3 cells (Fig. 2C), respectively. The mismatch control Oligo ASM had no or minimal effects. Pretreatment with 50 nM of Oligo AS, but not ASM, sensitized all of the three cell lines to HCPT (Fig. 2 D–F) and paclitaxel (Fig. 2 G–I).
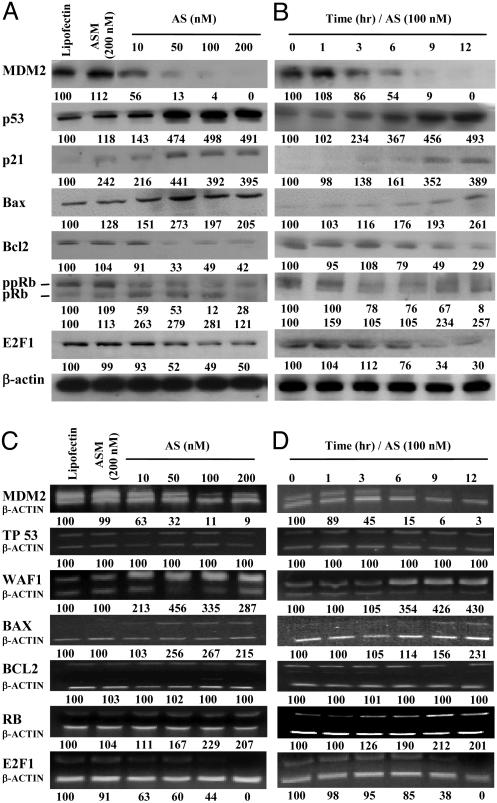
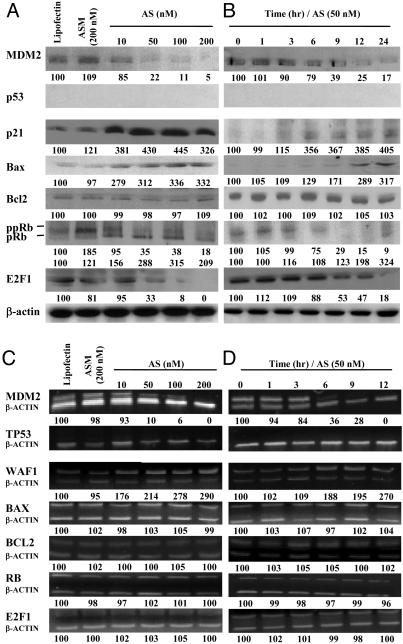
In Vitro MDM2 Antisense Inhibition Modified Multiple Gene Expression. LNCaP cells. Both MDM2 protein and mRNA were inhibited by Oligo AS in a dose-, time-, and sequence-dependent manner (Fig. 3). The MDM2 knockdown resulted in p53 protein elevation in a dose- and time-dependent fashion (Fig. 3 A and B). However, P53 mRNA levels did not change (Fig. 3 C and D), confirming that inactivation of p53 by MDM2 is mainly via the p53 degradation pathway. The levels of p21 and Bax proteins and mRNAs were also elevated. There were also increases in pRb and RB mRNA and decreases in ppRb. Both E2F1 protein and mRNA levels decreased. The Bcl2 protein level decreased (Fig. 3 A and B), but its mRNA level was not changed (Fig. 3 C and D).
Fig. 3.
Protein (A and B) and mRNA (C and D) levels of various genes in LNCaP cells treated with Oligo AS or controls. (A) The cells were transfected with Oligo AS (10–200 nM) or ASM (200 nM) for 24 h. Proteins were analyzed by Western blotting. The number under each band is expressed as a percentage of Lipofectin control, normalized by the corresponding β-actin level. (B) Cells were incubated with Oligo AS (100 nM) for various times. (C) Cells were transfected as in A. Target mRNAs were analyzed by RT-PCR and coamplified with β-actin mRNA. The number under each band is expressed as a percentage of Lipofectin control, normalized by the corresponding coamplified β-actin level. (D) Cells were incubated with Oligo AS (100 nM) for various times.
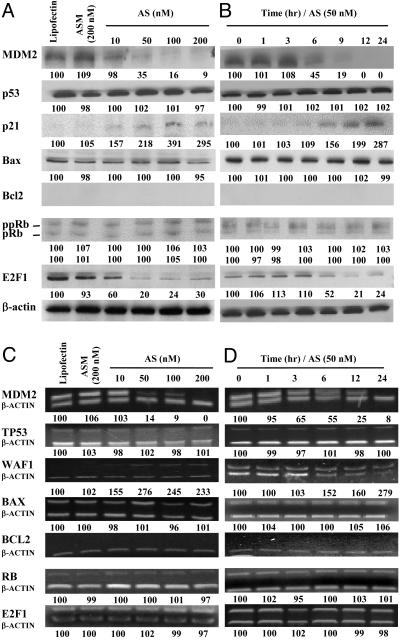
DU145 cells. In a dose-, time-, and sequence-dependent manner, MDM2 protein and mRNA levels were inhibited by Oligo AS (Fig. 4). No changes in mutant p53 protein and mRNA were observed. Interestingly, both p21 protein and mRNA levels were elevated, which is independent of p53. E2F1 protein levels were decreased (Fig. 4 A and B) without remarkable changes in mRNA levels (Fig. 4 C and D). DU145 cells are Bcl2 null and express mutant Bax and Rb, which were not changed after MDM2 inhibition.
Fig. 4.
Protein (A and B) and mRNA (C and D) levels of various genes in DU145 cells treated with Oligo AS or controls. The assay conditions were the same conditions as described for Fig. 3, except for the concentration of Oligo AS used in B and D (50 nM).
PC3 cells. After antisense MDM2 inhibition, p21 mRNA and protein levels were significantly elevated (Fig. 5). Bax protein levels were also elevated (Fig. 5 A and B) without remarkable changes in mRNA levels (Fig. 5 C and D). The pRb protein levels increased and ppRb levels decreased (Fig. 5 A and B) without significant changes in RB mRNA levels (Fig. 5 C and D). E2F1 protein levels decreased (Fig. 5 A and B) without changes in its mRNA levels (Fig. 5 C and D). No changes in Bcl2 protein or mRNA levels were found (Fig. 5).
Fig. 5.
Protein (A and B) and mRNA (C and D) levels of various genes in PC3 cells treated with Oligo AS or controls. The assay conditions were the same conditions as described for Fig. 3, except for the concentration of Oligo AS used in B and D (50 nM).
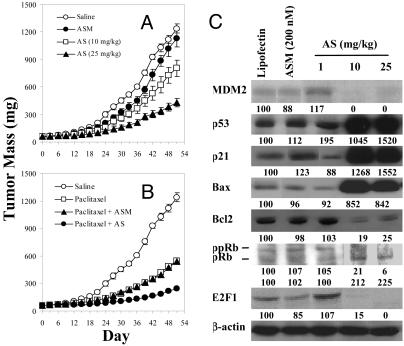
In Vivo MDM2 Antisense Inhibition Showed Dose-Dependent Antitumor Activity and Chemosensitization and Modified Multiple Gene Expression in Prostate Cancer Xenografts. LNCaP xenografts. Oligo AS showed significant antitumor activity in a dose-dependent manner (Fig. 6A) and increased therapeutic effectiveness of paclitaxel in severe combined immunodeficient mice bearing LNCaP xenografts (Fig. 6B). Oligo ASM showed minimal effects on tumor growth or on paclitaxel efficacy, demonstrating the specificity of Oligo AS (Fig. 6 A and B). To confirm the antisense mechanism in vivo, a separate study was accomplished to analyze protein levels of MDM2 and related genes, by Western blot analysis of pooled LNCaP xenografts. As shown in Fig. 6C, Oligo AS specifically inhibited MDM2 expression in a dose-dependent manner, resulting in elevation of p53, p21, Bax, and pRb and reduction of ppRb, Bcl2, and E2F1. The control Oligo ASM had no or minimal effects.
Fig. 6.
In vivo antitumor activity of Oligo AS administered alone (A) or in combination with paclitaxel (B) in severe combined immunodeficient (SCID) mice bearing LNCaP xenografts and the protein expression profile of xenografts (C). (A) Oligo AS (10 or 25 mg/kg per day) or ASM (25 mg/kg per day) treatments were initiated on day 0 and given i.p., 5 days/week for 4 weeks. (B) Oligos were given i.p. at 25 mg/kg per day, 5 days/week for 4 weeks, and paclitaxel was given i.p. at 10 mg/kg per day, twice per week for 4 weeks. (C) The SCID mice bearing LNCaP xenografts (60–70 mg) were treated i.p. with Oligo AS (10 or 25 mg/kg) or ASM (25 mg/kg) for 7 consecutive days. Proteins from tumor homogenates were analyzed by Western blotting. The number under each band is expressed as a percentage of saline control, normalized by the corresponding β-actin level.
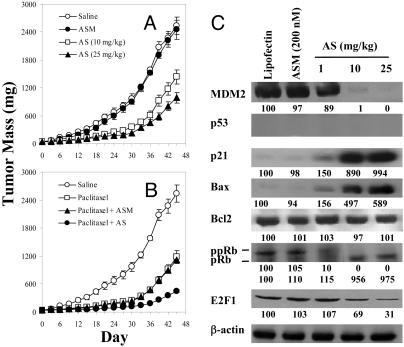
PC3 xenograft model. The antitumor activity of Oligo AS was further shown in the PC3 model, which is p53 null, providing direct evidence for p53-independent activity of MDM2 (Fig. 7A). In this model, Oligo AS also increased therapeutic effectiveness of paclitaxel (Fig. 7B). Oligo ASM showed no effect (Fig. 7 A and B). The protein levels of MDM2 were specifically decreased by Oligo AS in a dose-dependent manner, resulting in elevation of p21, Bax, and pRb and reduction of ppRb and E2F1 (Fig. 7C). Further quantitative analysis of the mode of chemosensitization effects in both LNCaP and PC3 models is presented in Supporting Text and Table 2, which is published as supporting information on the PNAS web site.
Fig. 7.
In vivo antitumor activity of Oligo AS administered alone (A) or in combination with paclitaxel (B) in nude mice bearing PC3 xenografts and the protein expression profile (C). The treatment and analytical procedures were the same as described for Fig. 6.
Discussion
Antisense oligonucleotides have been extensively studied as a research tool in determining gene function and as a novel therapeutic approach to treatment of various human diseases (1–6). However, there still are many questions regarding their specificity and efficacy. For the first-generation antisense oligos, there are several nonspecific effects that detract from their clinical efficacy and safety profiles (3, 4). In the present study, we used various in vitro and in vivo models to determine the specific antitumor activity and the underlying mechanism of action of the anti-human-MDM2 MBO, a new generation of antisense oligos. Unlike most reported sequences of first-generation antisense oligos under clinical investigation (4), our test and control oligos do not contain any CpG motifs or other sequences known to have nonspecific, non-antisense effects.
In the present study, we have demonstrated at least six noteworthy results. First, the novel anti-MDM2 MBO, Oligo AS, specifically knocked-down MDM2 expression, as demonstrated at both mRNA and protein levels, in a dose- and time-dependent manner, regardless of p53 status. Second, MDM2 inhibition resulted in enhanced apoptosis and decreased cell proliferation in a dose-dependent, sequence-specific manner, regardless of p53 status. Third, in a dose-dependent manner, Oligo AS demonstrated sequence-specific in vivo antitumor activity in both LNCaP (p53wt/wt) and PC3 (p53null) xenografts. Fourth, MDM2 inhibition resulted in chemosensitization in vitro and in vivo, regardless of p53 status. Fifth, in a dose- and time-dependent manner, Oligo AS modified the expression of several genes, with varying profiles in cells with different p53 status. In the LNCaP cells (p53wt/wt), MDM2 inhibition resulted in significant elevation of the protein levels of p53, p21, pRb, and Bax and reduction of ppRb, Bcl2, and E2F1. The changes in mRNA levels did not always correspond to those in protein levels, suggesting that there are multiple mechanisms of action for MDM2 inhibition. In DU145 cells (p53mt/mt), MDM2 inhibition also resulted in elevation of p21 at both mRNA and protein levels, and decreases in E2F1 protein levels. In PC3 cells, significant elevation of the protein levels of p21, Bax, and pRb and reduction of ppRb and E2F1 were observed. Finally, in vivo dose-dependent modulation of expression in several genes after MDM2 inhibition was demonstrated in both LNCaP and PC3 xenograft models, consistent with the above in vitro findings. Our results provide direct evidence supporting sequence-specific in vitro and in vivo antisense efficacy of the anti-MDM2 oligo, which should be considered as a proof of principle of antisense technology.
Thus far, most investigations of MDM2 functions focus on its interaction with p53. As a negative regulator of p53, MDM2 plays an important role in tumor formation and growth. In tumors with wild-type p53 expression, MDM2 interacts with p53 via various mechanisms such as inhibition of its transactivation activity (30) and facilitation of its degradation (21). Therefore, inhibition of MDM2 is a promising means of restoring p53 function, including p21 and Bax induction, resulting in cell growth arrest and/or apoptosis. Our results with the in vitro and in vivo LNCaP models provide direct evidence for p53–MDM2 interaction.
The tumorigenic properties of MDM2, however, may be independent of p53 (21). Approximately one-third to one-half of human prostate cancers have MDM2 overexpression with or without p53 expression (20, 21), suggesting that MDM2 may be involved directly in the development of prostate cancer. Moreover, the induction of MDM2 may be associated with resistance to chemotherapy and radiation therapy in prostate cancer patients (31, 32). Our results with the DU145 and PC3 models suggest a p53-independent activity of MDM2, which may have therapeutic applications, because the majority of advanced prostate cancers contain no p53 expression or mutant p53 (13–20) and because MDM2 is associated with the androgen-independent phenotype (23).
Unlike other published reports, the present study demonstrated changes in expression of multiple genes, not only the targeted gene, an observation that is important to understanding the mechanism of action for antisense therapy. Furthermore, the expression profiles were analyzed at both protein and mRNA levels. After MDM2 inhibition, the expression of p21 and Bax was observed in vitro and in vivo, regardless of p53 status. Although WAF1 (p21) gene is a reporter gene of p53 activity, several reports demonstrate that p53-independent p21 induction is mediated by various stimuli, including epidermal growth factor and fibroblast growth factor (20), silibinin (31), zinc (33), and genistein (34). The lack of p21 expression is correlated with poorer clinical outcome in bladder, colon, and hepatocellular carcinomas, and down-regulation of p21 is involved in the development of androgen-independence in prostate cancer (20). Our results from the present study support the concepts that induction of p21 is both p53-dependent and p53-independent and that MDM2 inhibition affects both p21 protein and mRNA levels.
The expression of Bax may also be controlled by p53 (35) or p53-independent mechanisms such as proteosome inhibition (36). In the present study, we demonstrated Bax induction in both LNCaP (p53wt/wt) and PC3 (p53null) cell lines, after MDM2 inhibition. Because Bax degradation by proteosome may be an important regulation mechanism in advanced prostate cancer (36) and because MDM2 has ubiquitin ligase activity (21), MDM2 may have a direct role in Bax regulation. The ratio of Bcl2/Bax is important in determining which cells undergo apoptosis and which survive after DNA damage (37). Bcl2 up-regulation is associated with androgen independence and resistance to chemotherapy (38). In the present study, we report that MDM2 inhibition results in simultaneous Bcl2 reduction and Bax elevation in LNCaP cells and xenografts, explaining partly the significant antitumor and chemosensitization effects of the antisense therapy.
The RB tumor suppressor is involved in cell cycle control and differentiation by modulating the activity of transcription factors such as E2F protein family (39); its mutations or deletions are evident in human prostate cancer (40). ppRb promotes cell progression (39). pRb regulates the G2/M transition (41), inhibiting androgen-independent development (42) and inducing p53-dependent (43) or p53-independent apoptosis (41). The elevated p21 after MDM2 inhibition may lead to Rb hypophosphorylation. However, the RB mRNA increase in LNCaP cells may be p53-dependent, because no changes in RB mRNA were observed in PC3 (p53null) cells.
E2F1 stimulates cellular proliferation and cell cycle progression from G1 to S phase (44–46) and correlates with increased tumor cell invasiveness and metastasis (45). In addition to being negatively regulated by pRb, it was activated by MDM2 directly (21). Results from the present study further demonstrate that MDM2 inhibition results in decrease of E2F1, regardless of p53 status, indicating a role for E2F1 in the antitumor activity and chemosensitization effects of the anti-MDM2 oligo.
In summary, although the role of MDM2 in the development and progression of human cancer was originally thought to be associated with p53 inactivation, we conclude, based on our results from in vitro and in vivo MDM2 antisense treatment in various prostate cancer models, that MDM2 has both p53-dependent and p53-independent activities. MDM2 inhibitors, such as second-generation antisense oligos described in this report, may have a broad spectrum of antitumor activity in human cancers, regardless of p53 status, providing a basis for future development of this novel approach to the therapy of human cancers.
Supplementary Material
Acknowledgments
We thank Dr. D. Yu and G. Prasad for excellent technical assistance. This study was supported by National Institutes of Health/National Cancer Institute Grant R01CA80698 (to R.Z.).
Abbreviations: MDM2, mouse double minute 2; oligo, oligonucleotide; MBO, mixed-backbone oligos; HCPT, 10-hydroxycamptothecin; Rb, retinoblastoma tumor suppressor protein; pRb, hypophosphorylated Rb; ppRb, hyperphosphorylated Rb; E2F1, E2F transcription factor 1.
References
- 1.Zamecnik, P. C. (1996) in Antisense Therapeutics, ed. Agrawal, S. (Humana, Totowa, NJ), pp. 1–11.
- 2.Harth, G., Horwitz, M. A., Tabatadze, D. & Zamecnik, P. C. (2002) Proc. Natl. Acad. Sci. USA 99, 15614–15619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang, H., Prasad, G., Buolamwini, J. K. & Zhang, R. (2001) Curr. Cancer Drug Targets 1, 177–196. [DOI] [PubMed] [Google Scholar]
- 4.Agrawal, S. & Kandimalla, E. R. (2001) Curr. Cancer Drug Targets 1, 197–209. [DOI] [PubMed] [Google Scholar]
- 5.Crooke, S. T. (2000) Oncogene 19, 6651–6659. [DOI] [PubMed] [Google Scholar]
- 6.Opalinska, J. B. & Gewirtz, A. M. (2002) Nat. Rev. Drug Discovery 1, 503–514. [DOI] [PubMed] [Google Scholar]
- 7.Agrawal, S., Jiang, Z., Zhao, Q., Shaw, D., Cai, Q., Roskey, A., Channavajjala, L., Saxinger, C. & Zhang, R. (1997) Proc. Natl. Acad. Sci. USA 94, 2620–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang, H., Cai, Q., Zeng, X., Yu, D., Agrawal, S. & Zhang, R. (1999) Proc. Natl. Acad. Sci. USA 96, 13989–13994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, L., Agrawal, S., Zhou, W., Zhang, R. & Chen, J. (1998) Proc. Natl. Acad. Sci. USA 95, 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang, H., Nan, L., Yu, D., Agrawal, S. & Zhang, R. (2001) Clin. Cancer Res. 7, 3613–3624. [PubMed] [Google Scholar]
- 11.Wang, H., Nan, L., Yu, D., Lindsey, J. R., Agrawal, S. & Zhang, R. (2002) Mol. Med. 8, 184–198. [PMC free article] [PubMed] [Google Scholar]
- 12.Jemal, A., Murray, T., Samuels, A., Ghafoor, A., Ward, E. & Thun, M. J. (2003) CA Cancer J. Clin. 53, 5–26. [DOI] [PubMed] [Google Scholar]
- 13.Ross, J. S., Sheehan, C. E., Dolen, E. M. & Kallakury, B. V. (2002) Adv. Anat. Pathol. 9, 115–128. [DOI] [PubMed] [Google Scholar]
- 14.Tamboli, P., Amin, M. B., Xu, H. J. & Linden, M. D. (1998) Mod. Pathol. 11, 247–252. [PubMed] [Google Scholar]
- 15.Stackhouse, G. B., Sesterhenn, I. A., Bauer, J. J., Mostofi, F. K., Connelly, R. R., Srivastava, S. K. & Moul, J. W. (1999) J. Urol. 162, 2040–2045. [DOI] [PubMed] [Google Scholar]
- 16.Takayama, H., Shin, M., Nonomura, N., Okuyama, A. & Aozasa, K. (2000) Jpn. J. Cancer Res. 91, 941–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ittmann, M., Wieczorek, R., Heller, P., Dave, A., Provet, J. & Krolewski, J. (1994) Am. J. Pathol. 145, 287–293. [PMC free article] [PubMed] [Google Scholar]
- 18.Gao, X., Porter, A. T. & Honn, K. V. (1997) Adv. Exp. Med. Biol. 407, 41–53. [DOI] [PubMed] [Google Scholar]
- 19.Osman, I., Drobnjak, M., Fazzari, M., Ferrara, J., Scher, H. I. & Cordon-Cardo, C. (1999) Clin. Cancer Res. 5, 2082–2088. [PubMed] [Google Scholar]
- 20.Leite, K. R., Franco, M. F., Srougi, M., Nesrallah, L. J., Nesrallah, A., Bevilacqua, R. G., Darini, E., Carvalho, C. M., Meirelles, M. I., Santana, I. & Camara-Lopes, L. H. (2001) Mod. Pathol. 14, 428–436. [DOI] [PubMed] [Google Scholar]
- 21.Zhang, R. & Wang, H. (2000) Curr. Pharm. Des. 6, 393–416. [DOI] [PubMed] [Google Scholar]
- 22.Taj, M. M., Tawil, R. J., Engstrom, L. D., Zeng, Z., Hwang, C., Sanda, M. G. & Wechsler, D. S. (2001) Prostate 47, 194–204. [DOI] [PubMed] [Google Scholar]
- 23.Agus, D. B., Cordon-Cardo, C., Fox, W., Drobnjak, M., Koff, A., Golde, D. W. & Scher, H. I. (1999) J. Natl. Cancer Inst. 91, 1869–1876. [DOI] [PubMed] [Google Scholar]
- 24.Wang, H., Yu, D., Agrawal, S. & Zhang, R. (2003) Prostate 54, 194–205. [DOI] [PubMed] [Google Scholar]
- 25.Ko, J. L., Cheng, Y. W., Chang, S. L., Su, J. M., Chen, C. Y. & Lee, H. (2000) Int. J. Cancer 89, 265–270. [DOI] [PubMed] [Google Scholar]
- 26.Shinohara, T., Miki, T., Nishimura, N., Nokihara, H., Hamada, H., Mukaida, N. & Sone, S. (2001) Cancer Res. 61, 673–678. [PubMed] [Google Scholar]
- 27.Leiter, U., Schmid, R. M., Kaskel, P., Peter, R. U. & Krahn, G. (2000) Arch. Dermatol. Res. 292, 225–232. [DOI] [PubMed] [Google Scholar]
- 28.Kanazawa, S., Fujiwara, Y. & Mizuno, K. (1994) J. Med. Sci. 40, 165–174. [PubMed] [Google Scholar]
- 29.Shen, J. C., Klein, R. D., Wei, Q., Guan, Y., Contois, J. H., Wang, T. T., Chang, S. & Hursting, S. D. (2000) Mol. Carcinog. 29, 92–102. [DOI] [PubMed] [Google Scholar]
- 30.Kokontis, J. M., Wagner, A. J., O'Leary, M., Liao, S. & Hay, N. (2000) Oncogene 20, 659–668. [DOI] [PubMed] [Google Scholar]
- 31.Liang, J. Y., Liu, Y. Y., Zou, J., Franklin, R. B., Costello, L. C. & Feng, P. (1999) Prostate 40, 200–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi, Y. H., Lee, W. H., Park, K. Y. & Zhang, L. (2000) Jpn. J. Cancer Res. 91, 164–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gariboldi, M. B., Rimoldi, V., Supino, R., Favini, E. & Monti, E. (2000) Free Radical Biol. Med. 29, 633–641. [DOI] [PubMed] [Google Scholar]
- 34.Therrien, J. P., Loignon, M., Drouin, R. & Drobetsky, E. A. (2001) Cancer Res. 61, 3781–3786. [PubMed] [Google Scholar]
- 35.Stros, M., Ozaki, T., Bacikova, A., Kageyama, H. & Nakagawara, A. (2002) J. Biol. Chem. 277, 7157–7164. [DOI] [PubMed] [Google Scholar]
- 36.Li, B. & Dou, Q. P. (2000) Proc. Natl. Acad. Sci. USA 97, 3850–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaudhary, K. S., Abel, P. D., Stamp, G. W. & Lalani, E. (2001) J. Pathol. 193, 522–529. [DOI] [PubMed] [Google Scholar]
- 38.Finnegan, N. M., Curtin, J. F., Prevost, G., Morgan, B. & Cotter, T. G. (2001) Br. J. Cancer 85, 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fribourg, A. F., Knudsen, K. E., Strobeck, M. W., Lindhorst, C. M. & Knudsen, E. S. (2000) Cell Growth Differ. 11, 361–372. [PubMed] [Google Scholar]
- 40.Bowen, C., Spiegel, S. & Gelmann, E. P. (1998) Cancer Res. 58, 3275–3281. [PubMed] [Google Scholar]
- 41.Zhao, X. & Day, M. L. (2001) Urology 57, 860–865. [DOI] [PubMed] [Google Scholar]
- 42.Wallace, M., Coates, P. J., Wright, E. G. & Ball, K. L. (2001) Oncogene 20, 3597–3608. [DOI] [PubMed] [Google Scholar]
- 43.Hsieh, J. K., Chan, F. S., O'Connor, D. J., Mittnacht, S., Zhong, S. & Lu, X. (1999) Mol. Cell 3, 181–193. [DOI] [PubMed] [Google Scholar]
- 44.Stanelle, J., Stiewe, T., Theseling, C. C., Peter, M. & Putzer, B. M. (2002) Nucleic Acids Res. 30, 1859–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsieh, J. K., Yap, D., O'Connor, D. J., Fogal, V., Fallis, L., Chan, F., Zhong, S. & Lu, X. (2002) Mol. Cell. Biol. 22, 78–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ookawa, K., Tsuchida, S., Kohno, T. & Yokota, J. (2001) FEBS Lett. 500, 25–30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.