Abstract
In all eukaryotes, segregation of mitotic chromosomes requires their interaction with spindle microtubules. To dissect this interaction, we use live and fixed assays in the one-cell stage Caenorhabditis elegans embryo. We compare the consequences of depleting homologues of the centromeric histone CENP-A, the kinetochore structural component CENP-C, and the chromosomal passenger protein INCENP. Depletion of either CeCENP-A or CeCENP-C results in an identical “kinetochore null” phenotype, characterized by complete failure of mitotic chromosome segregation as well as failure to recruit other kinetochore components and to assemble a mechanically stable spindle. The similarity of their depletion phenotypes, combined with a requirement for CeCENP-A to localize CeCENP-C but not vice versa, suggest that a key step in kinetochore assembly is the recruitment of CENP-C by CENP-A–containing chromatin. Parallel analysis of CeINCENP-depleted embryos revealed mitotic chromosome segregation defects different from those observed in the absence of CeCENP-A/C. Defects are observed before and during anaphase, but the chromatin separates into two equivalently sized masses. Mechanically stable spindles assemble that show defects later in anaphase and telophase. Furthermore, kinetochore assembly and the recruitment of CeINCENP to chromosomes are independent. These results suggest distinct roles for the kinetochore and the chromosomal passengers in mitotic chromosome segregation.
Keywords: centromere, CENP, passenger, mitosis, chromosome
Introduction
To segregate the replicated genome, all eukaryotic cells remodel replicated interphase chromatin. This remodeling includes condensation, proper establishment and timely dissolution of cohesion between the sister chromatids, and the assembly of kinetochores (for review see Koshland and Strunnikov 1996; Rieder and Salmon 1998; Maney et al. 2000; Nasmyth et al. 2000). Two kinetochores are assembled per chromosome, one on each of the sister chromatids, to act as chromosome-associated “receptors” for spindle microtubules (MTs). Much recent work has focused on the mechanisms of chromosome condensation and sister chromatid cohesion/separation. In contrast, the mechanisms underlying formation of the interface between centromeric DNA and spindle MTs remain relatively mysterious.
Two major obstacles to understanding kinetochore assembly have been the complex nature of centromeric DNA and the dramatic divergence of centromeric DNA sequences between different eukaryotes (Karpen and Allshire 1997; Tyler-Smith and Floridia 2000). The former has made a “bottom-up” approach starting from centromeric sequences impractical, and the latter has made it difficult to evaluate the general relevance of findings in a given species. A remarkable recent finding resulting from genome sequencing projects is that, despite dramatic variation in centromeric DNA, many protein components of the centromere–kinetochore complex are conserved (Tyler-Smith and Floridia 2000). The most striking conserved components are CENP-A and CENP-C, two proteins that localize to the chromatin-proximal region of the kinetochore. In addition, mitotic checkpoint components that localize to the outer MT binding regions of the kinetochore (Skibbens and Hieter 1998) and three “chromosomal passenger” proteins that localize to the centromeric heterochromatin between the two kinetochores (Adams et al. 2001) are also conserved. The founding member of the chromosomal passengers is INCENP, a protein shown recently to function as a targeting subunit for a second chromosomal passenger, aurora B kinase (Adams et al. 2000; Kaitna et al. 2000). Recent work has also revealed that survivin, originally implicated in inhibition of apoptosis, is a chromosomal passenger (Skoufias et al. 2000; Speliotes et al. 2000; Uren et al. 2000). The centromere-specific histone variant CENP-A, the kinetochore structural component CENP-C, and the chromosomal passengers have been functionally implicated in mitotic chromosome segregation in diverse eukaryotic species, and homologues of CENP-A, CENP-C, and INCENP are essential in mice and budding yeast (Tomkiel et al. 1994; Meluh and Koshland 1995; Kalitsis et al. 1998; Meluh et al. 1998; Cutts et al. 1999; Kim et al. 1999; Howman et al. 2000; Takahashi et al. 2000). These results suggest fundamental conservation among eukaryotes in the mechanisms used to assemble the interface between mitotic chromosomes and spindle MTs.
In vertebrate somatic cells, EM has generated a structural picture of the chromosome–MT interface. In these cells, the kinetochore appears as a trilaminar disk–like structure that assembles on the centromeric region of condensed chromosomes (Rieder and Salmon 1998; Maney et al. 2000). The trilaminar organization of the kinetochore, as well as localization studies of a number of kinetochore–centromere components, suggest a structure composed of distinct subdomains. The region of the kinetochore closest to the centromeric heterochromatin is called the inner plate; both CENP-A and CENP-C localize to this region (Saitoh et al. 1992; Warburton et al. 1997). CENP-A is a histone H3 variant, suggesting a direct role in formation of the specialized chromatin that underlies the kinetochore (Vafa and Sullivan 1997; Meluh et al. 1998; Howman et al. 2000). Like CENP-A, CENP-C may also interact directly with DNA, although how it participates in kinetochore assembly remains unclear (Sugimoto et al. 1994; Yang et al. 1996). Both CENP-A and CENP-C associate with centromeres throughout the cell cycle in vertebrate somatic cells. In contrast, many proteins, such as the MT-destabilizing kinesin MCAK (Wordeman and Mitchison 1995), first localize to the centromeric region early in mitosis, when the kinetochore plate structure is formed. Other proteins that localize specifically during mitosis include the components of the outermost corona regions of the kinetochore that directly interact with spindle MTs. These include the motor proteins CENP-E and cytoplasmic dynein, and proteins such as Bub1 that function in the mitotic checkpoint (Maney et al. 2000). The chromosomal passengers are not kinetochore components per se. Instead, they localize to the centromeric heterochromatin that lies between the inner plates of the two oppositely oriented sister kinetochores. Like Bub1 and MCAK, the passenger proteins INCENP, aurora B, and survivin are recruited to the centromeric region in the early stages of mitosis (Adams et al. 2001).
Dissecting the pathways that contribute to the assembly of the interface between chromosomes and spindle MTs requires an analysis of the dependency relationships that exist between kinetochore–centromere components. For example, does the localization of CENP-C to kinetochores depend on CENP-A or vice versa, or do both proteins target independently of chromosomes? Such an analysis would address whether kinetochore assembly occurs via a linear pathway from the most proximal components near the DNA to the outermost components of the corona, or whether more complex relationships are involved. Similarly, determining if targeting of chromosomal passengers requires assembly of the kinetochore or vice versa would help elucidate the relationship between the kinetochore and this conserved set of proteins. To begin to address these questions we have taken a reverse genetic approach using RNA-mediated interference (RNAi) in the one cell stage Caenorhabditis elegans embryo.
A major difference between C. elegans and other model eukaryotic organisms is that C. elegans, like several nematode, hemipteran insect, and monocotolydenous plant species, has holocentric chromosomes. In these organisms, diffuse kinetochores form along the entire length of the chromosome (Comings and Okada 1972; Albertson and Thomson 1982). Despite this structural difference, the C. elegans genome contains predicted genes homologous to components found at the localized kinetochores of other eukaryotes, suggesting commonalities in molecular composition, assembly mechanisms, and function. Support for this comes from two recent studies that identified a CENP-A–like histone (hcp-3) and two apparently redundant genes with homology to CENP-F (hcp-1 and hcp-2) from C. elegans (Buchwitz et al. 1999; Moore et al. 1999). The CENP-A homologue and one of the CENP-F homologues were shown to localize to two plates on opposing faces of condensed chromosomes. These studies also showed that RNAi of either the CENP-A homologue or of both genes with homology to CENP-F resulted in chromosome segregation defects.
Here, we combine RNAi with live and fixed assays for chromosome dynamics, spindle structure, and assembly of kinetochore–centromere components. Focusing on the first mitotic division of the fertilized C. elegans embryo, we show that depletion of either the CENP-A or the CENP-C homologue results in similar chromosome segregation, kinetochore assembly, and spindle defects that likely represent the “kinetochore null” phenotype. In contrast, depletion of an INCENP homologue results in distinct mitotic chromosome segregation and spindle defects. Consistent with their different phenotypes, we find that kinetochore components and the INCENP homologue target independently to chromosomes. These results lead us to conclude that kinetochores and chromosomal passengers make distinct contributions to mitotic chromosome segregation and allow us to present a preliminary map of dependency relationships during kinetochore assembly in C. elegans.
Materials and Methods
Expression of GFP Fusions
Green fluorescent protein (GFP) fusions were generated in pJH4.52, a plasmid expressing GFP-HIS-11 under control of the pie-1 promoter (a gift of G. Seydoux, Johns Hopkins University, Baltimore, MD). The unspliced genomic locus for the α-tubulin C47B2.3 and the coding region for the γ-tubulin tbg-1 were inserted into pJH4.52 after removal of the his-11 sequence by SpeI digestion. Transformed lines expressing the GFP fusions from complex arrays were established as described (Mello et al. 1991; Kelly et al. 1997). Approximately 20 F2 lines with good transmission frequency were monitored for GFP expression for ∼20 generations by placing individual rollers into 5 μl 20 mM azide in 15-well slides and using a 10×, 0.3 NA objective. Discarding silenced and unstable lines resulted in one reasonably stable line for each injection mixture. All fluorescent worm lines were maintained at 24.5°C.
Live Imaging
Dissected embryos were mounted for filming as described (Gonczy et al. 1999). Widefield microscopy on a motorized microscope (Axioplan II; ZEISS) was used to film GFP-histone and GFP-histone/GFP γ-tubulin strains. The motorized filter turret and focus, external shutters in both light paths, and a 12-bit camera (Orca100; Hamamatsu) were controlled using Metamorph software (Universal Imaging Corp.). Differential interference contrast (DIC)/GFP images were acquired every 6–8 s by sequentially rotating the analyzer and GFP filter set into the light path, resulting in ∼1 s difference between the paired images. Epiillumination intensity was attenuated to 5–10% using neutral density filters. Images were acquired using a 63×, 1.4 NA PlanApochromat objective with 250 ms exposure for GFP and 100 ms for DIC. GFP-tubulin videos were acquired using a spinning disk confocal (QLC100; Visitech International) mounted on a microscope of the type described above. GFP images were collected every 8 s using 500–1,000 ms exposure at ∼40% power on the 50-mW argon laser. Nonconfocal DIC images were collected through the spinning disk.
Quantitative Analysis of Spindle Positioning and Pole Separation Rate
2 s time lapse videos of embryos expressing GFP-histone/GFP–γ-tubulin were analyzed. A line K was drawn through the center of each embryo from anterior to posterior. For each time point, cartesian pixel coordinates for the following were recorded: the intersection of the anterior (A) and posterior (P) cortex and K, the anterior (SPa) and posterior (SPp) spindle poles, and the intersection of the poleward edges of separating chromosome masses with a line connecting the two spindle poles (Ca and Cp). Distances between SPa and SPp (pole-to-pole distance), between Ca and Cp (chromosome mass separation), and the chromosome-to-pole distance (pole-to-pole distance minus the chromosome mass separation divided by 2) were calculated. The “posteriorness” of the spindle within the embryo was represented by the distance between P and the projection of the center of the spindle onto the line K. The projection is necessary because of extensive rocking of the spindle during anaphase. We first calculated the following distances: X = A − P (embryo length); Y = A − SPa; Z = P − SPa; Q = A − SPp; and R = P − SPp. If a = (X 2 + Y 2 − Z 2)/2X and c = (X 2 + Q 2 − R 2)/2X, then the distance between the center of the spindle projected onto K and the posterior of the embryo, expressed as a fraction of egg length, is (2X − a − c)/(2X).
To analyze pole separation in GFP-tubulin videos, pole–pole distance was plotted relative to NEBD. The resulting curves were averaged to reduce noise (5 point running average), and the maximum pole separation rate was calculated by taking a derivative of the curve and finding the inflection point.
RNA-mediated Interference
For production of dsRNA to CeCENP-C (ggaaatgtacggagcgaaaa, acattgttggtgggtccaat) and CeINCENP (ggatgaaagagctcgagaagaa, ttctgacattctcacggacaac) the primers in parentheses with tails containing T3 and T7 promoters were used to amplify regions from genomic N2 DNA and the cDNA yk329a11, respectively. PCR reactions were cleaned (QIAGEN GmbH) and used as templates for 25 μl T3 and T7 transcription reactions (Ambion), which were combined and cleaned using an RNeasy kit (QIAGEN). RNA eluted with 50 μl of H2O was mixed with 25 μl of 3× injection buffer (IX = 20 mM KPO4, pH 7.5, 3 mM K-Citrate, pH 7.5, 2% PEG 6000) and annealed by incubating at 68°C for 10 min followed by 37°C for 30 min. DsRNA for CeCENP-A was made similarly, except cDNA yk325d10 digested with EcoRI (XhoI) was used to template the T7 (T3) reactions. For fixed assays, adult wild-type hermaphrodites injected with dsRNA were placed at 20°C for 24 h before fixation. For live fluorescence assays, young roller adults were injected and kept at 24.5°C for 22–30 h.
Antibody Production and Labeling
To make GST fusion proteins to generate antibodies to CeINCENP (cgcgcgggatccgaaaccgacgaagtgcagac, gcgcgcgaattctcaatcattgaacggaatcacac), CeCENP-A (cgcgcgggatccgccgatgacaccccaattat, gcgcgcgaattctcattcttcgtc-ggagctatcgt), CeCENP-C (cgcgcgggatccacgattgttcctggtcgaaa, gcgcgcgaattc-tcatctctcctcgagaatggttgg), CeMCAK (cgcgcgagatctcagagaaaacgagccgagaa, gcgcgcgaattctcaaggagccatacgaacaggaac), and CeBub1 (gcggaattcgagaaa-acggttgatgatgagga, ggtacgactcgagtggggaggacgcacaagacac), the primers in parentheses were used to amplify fragments of the corresponding genes from cDNAs. Fragments were digested with BamHI-EcoRI (CeINCENP, CeCENP-A, and CeCENP-C), BglII-EcoRI (CeMCAK), or EcoRI-XhoI (CeBub1) and cloned into pGEX6P-1 (Amersham Pharmacia Biotech). Purified GST fusions were injected into rabbits. For affinity purification, fusions were cleaved with Prescission protease (Amersham Pharmacia Biotech) and coupled to 1 ml NHS HiTrap columns (Amersham Pharmacia Biotech). Affinity purification was performed using standard procedures (Harlow and Lane 1988). Direct labeling of antibodies was performed as described previously (Francis-Lang et al. 1999).
Immunofluorescence and Fixed Imaging
Embryos were fixed by freeze cracking and plunging into −20°C methanol as described (Gonczy et al. 1999). Optimal fixation times were: 2 h for CeCENP-A and CeINCENP and 20 min for CeCENP-C, CeMCAK, and CeBub1. Embryos were rehydrated in PBS, blocked in AbDil (PBS plus 2% BSA, 0.1% Triton X-100), incubated overnight at 4°C with 1 μg/ml of each directly labeled antibody and antitubulin monoclonal DM1α (1:500) diluted in AbDil, washed with PBST (PBS plus 0.1% Triton X-100), incubated for 1 h with FITC anti–mouse secondary (Dianova GmbH), washed with PBST, with PBST plus 1 μg/ml Hoechst, and mounted in 0.5% p-phenylenediamine, 20 mM Tris-Cl, pH 8.8, 90% glycerol. Three-dimensional widefield datasets collected using a 63×, 1.4 NA Planapochromat lens on a DeltaVision microscope were computationally deconvolved and projected (Applied Precision). For dependency analysis, one- and two-cell embryos were analyzed. Stages scored for the different markers depended on their wild-type localization and were as follows: CeCENP-A, CeCENP-C, and CeINCENP, pronuclear meeting (midprophase) through late anaphase; CeMCAK, NEBD through late anaphase; CeBub1, pronuclear meeting (midprophase) through metaphase. Although cytokinesis fails, second division CeINCENP-depleted embryos can be recognized by the presence of four asters and are referred to as “two-cell” embryos.
Online Supplemental Material
Quicktime™ videos associated with Fig. 2, Fig. 6, Fig. 7, and Fig. 9A and Fig. B are available at http://www.jcb.org/cgi/content/full/153/6/1209/DC1. In the videos with Fig. 2 (Videos 1–3), a strain expressing GFP-histone H2B was used to visualize chromosome segregation during the first mitotic cell division. Videos are provided for wild-type, CeCENP-A–depleted and CeCENP-C–depleted embryos. The video with Fig. 6 (Video 4) shows an embryo expressing both GFP–γ-tubulin and GFP-histone H2B during the first cell division. Such videos were used to precisely track spindle position within the embryo relative to anaphase onset. In the videos with Fig. 7 (Videos 5–7), the MT cytoskeleton in embryos expressing GFP–α-tubulin was visualized using spinning disk confocal microscopy. Spindle assembly and dynamics were followed in wild-type, CeCENP-A–depleted, and CeCENP-C–depleted embryos. The videos with Fig. 9 A (Videos 8–12) show chromosome segregation in embryos depleted of one of the three chromosomal passengers: CeINCENP, Air-2, or Bir-1. Three different videos of CeINCENP-depleted embryos are shown, one of which (Video 9) provides a fortuitously clear view of the dynamics of sperm-derived paternal chromosomes. Finally, the video associated with Fig. 9 B (Video 13) shows spindle assembly and dynamics in a CeINCENP-depleted embryo.
Figure 2.

Depletion of CeCENP-A or CeCENP-C results in a similar chromosome segregation defect. The panels summarize videos of embryos expressing GFP-histone. Stills from a wild-type embryo (left), a CeCENP-A–depleted embryo (middle), and a CeCENP-C–depleted embryo (right) are shown. The time in the upper right hand corner of each panel is seconds after NEBD. All sequences are oriented with embryo anterior on the left and embryo posterior, determined by the position of sperm entry, on the right. In CeCENP-A– or CeCENP-C–depleted embryos, the maternal and paternal chromosomes form separate compact chromosomal masses that fail to distribute over the spindle equator and fail to segregate at anaphase. The insets show a face on view of metaphase spindles. Insets are magnified 3.4-fold relative to the other panels. Online supplemental videos are available at http://www.jcb.org/cgi/content/full/153/6/1209/DC1. Bar, 5 μm.
Figure 6.
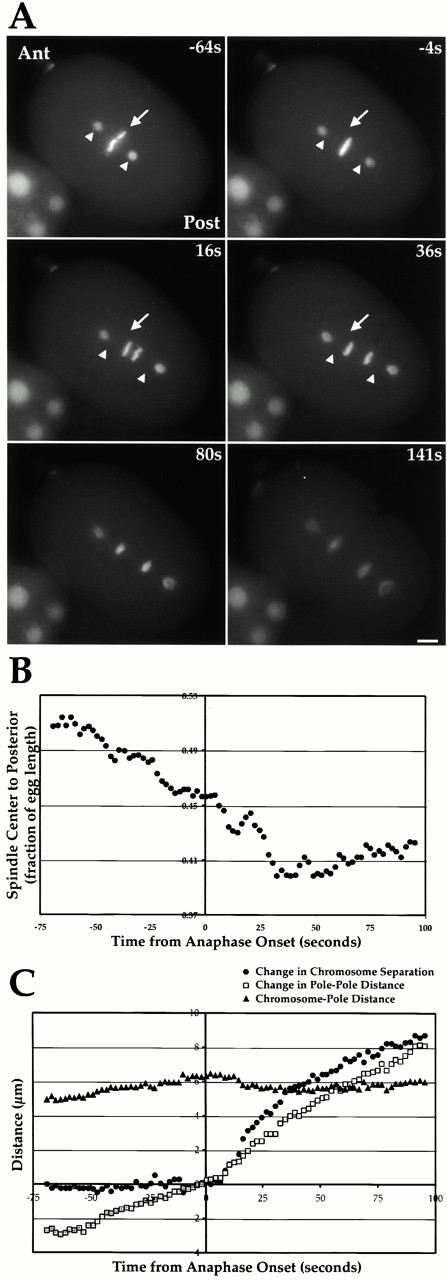
Polarity-cued forces begin moving the spindle towards the embryo posterior before anaphase onset. (A) Stills from a video of a C. elegans embryo expressing GFP-histone to mark the DNA and GFP–γ-tubulin to mark the spindle poles. The embryo anterior (Ant) is on the top left and the embryo posterior (Post) on the lower right. Time in seconds on the upper right of each panel is relative to anaphase onset. For reference, the position of the spindle center (arrow) and the two spindle poles (arrowheads) at the first time point (−64s) are marked on the subsequent three panels. (B) The distance between the spindle center and the embryo posterior (expressed as a fraction of egg length) is plotted. About half of the posterior movement of the spindle occurs before anaphase onset. (C) Chromosome segregation during the first mitotic division of the C. elegans embryo is primarily due to anaphase B. Chromosome-to-pole distance (black triangles), change in chromosome separation (medium gray circles), and change in pole-to-pole distance (light grey squares) are plotted relative to anaphase onset. Chromosome separation is the distance between the polar edges of the separating chromosomes measured along the pole–pole axis. Changes in chromosome separation and pole-to-pole distance were obtained by subtracting the relevant values 4 s before anaphase onset. Online supplemental videos are available at http://www.jcb.org/cgi/content/full/153/6/1209/DC1. Bar, 5 μm.
Figure 7.

Mechanically stable spindles do not assemble in embryos depleted of CeCENP-A or CeCENP-C. (A) Time-aligned stills summarizing spinning disk confocal videos of GFP–α-tubulin expressing wild-type (left) and CeCENP-C–depleted (right) embryos. The time, in seconds after NEBD, appears in the top right hand corner of each panel. In the absence of CeCENP-C, MTs invade the nuclear space after NEBD, but a stable spindle fails to form and the spindle poles move apart abruptly. Identical results are obtained in CeCENP-A–depleted embryos (not shown here). (B) Pole separation in wild-type (diamonds) and CeCENP-C–depleted (squares) embryos. Three sequences are plotted for each condition. The grey arrowhead indicates the average time after NEBD when the spindle initiates posterior movement in wild-type. The black arrowhead indicates the average time of anaphase onset in wild-type. In CeCENP-C–depleted embryos, the poles separate abruptly during the interval marked by the arrowheads. (C) The maximum rate of pole separation was determined for each experimental condition. The mean ± SD from analysis of five videos per condition are plotted. The maximum pole separation rate is significantly different between wild-type and CeCENP-C/CeCENP-A depletions (P = <0.001 and P = <0.05, respectively, in a Student's t test), but is not significantly different between CeCENP-A and CeCENP-C depletions. Online supplemental videos are available at http://www.jcb.org/cgi/content/full/153/6/1209/DC1. Bar, 5 μm.
Figure 9.
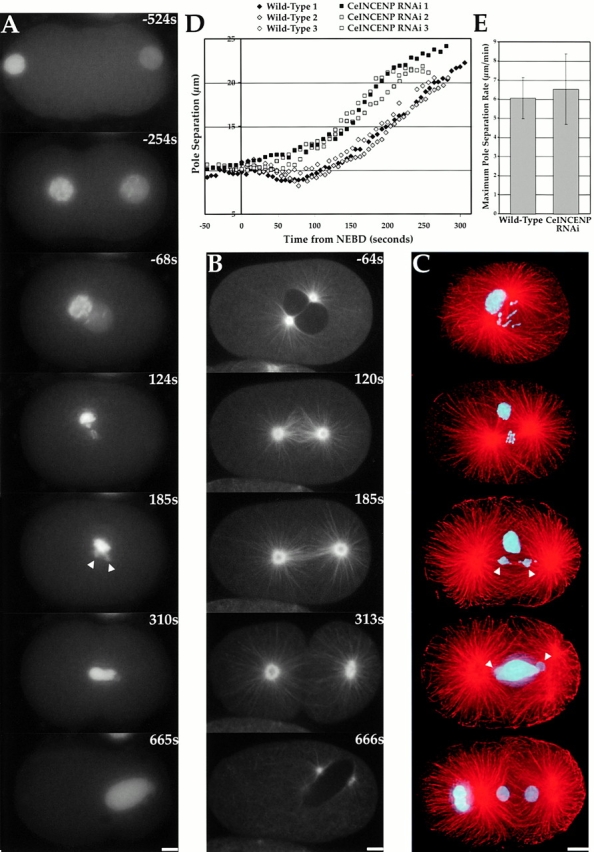
Depletion of CeINCENP by RNAi results in defects distinct from those seen in CeCENP-A/C depletions. Time in A and B is seconds after NEBD. (A) Stills from a video of a CeINCENP-depleted embryo expressing GFP-histone. Meiotic divisions of the oocyte nucleus fail, generating the defective maternal pronucleus on the left. Chromosomes derived from the meiotically normal paternal nucleus on the right condense, but fail to align properly. Leading and lagging chromatin is seen during anaphase. However, separation of these chromosomes into two masses (arrowheads) is always observed. (B) Stills from a video of a CeINCENP-depleted embryo expressing GFP–α-tubulin. A more robust spindle is formed relative to CeCENP-A/C depletions (compare 120s and 185s here to the 120s and 176s CeCENP-C panels in Fig. 7 A). In late anaphase, a spindle midzone fails to form and, unlike in CeCENP-A/C depletions, cytokinesis fails (C) CeINCENP-depleted embryos were fixed and stained for DNA (cyan) and MTs (red). The chromosomes of the maternal pronucleus form a large mass near the spindle. Occasionally, this mass is not in the spindle vicinity (e.g., in the bottom telophase panel). As with the live analysis in A, the paternal chromosomes fail to align to a tight plate and exhibit leading and lagging chromatin during anaphase, but always separate into two equivalently sized masses (arrowheads). All images are projections of 3D stacks. (D) Pole separation in wild-type (diamonds) and CeINCENP-depleted (squares) embryos. Three sequences are plotted for each. (E) The maximum rate of pole separation is not significantly different between wild-type and CeINCENP RNAi embryos. The mean ± SD from analysis of five videos is plotted next to the wild-type values from Fig. 7. Online supplemental videos are available at http://www.jcb.org/cgi/content/full/153/6/1209/DC1. Bars, 5 μm.
Results
CeCENP-A and CeCENP-C Colocalize at Kinetochores During Mitosis
To initiate a molecular analysis of kinetochore assembly and function during the first mitotic division of the C. elegans embryo, we examined the localization of the C. elegans homologues of the conserved centromere–kinetochore components CENP-A and CENP-C. A C. elegans gene homologous to CENP-A (hcp-3) has been localized to diffuse mitotic kinetochores (Buchwitz et al. 1999). We identified a C. elegans gene (T03F1.9) with homology to CENP-C by searching the genome with the sequence of the relatively conserved region I (Dawe et al. 1999). In concert with earlier nomenclature, we propose hcp-4 as the systematic name for the T03F1.9 gene. In this paper, we will refer to the protein products of hcp-3 and hcp-4 as CeCENP-A and CeCENP-C, respectively.
The distribution of CeCENP-A and CeCENP-C during the first mitotic division of C. elegans is shown in Fig. 1. CeCENP-A is associated with chromosomes throughout the cell cycle. In contrast, CeCENP-C is first detected in nuclei and on condensing chromosomes during prophase (Fig. 1 A). During most of prophase, CeCENP-A and CeCENP-C colocalize to a patchy stripe that runs along the chromosome (Fig. 1A and Fig. B). By late prophase/pro-metaphase both proteins colocalize to two stripes on opposite faces of each chromosome (Fig. 1 B). At metaphase, CeCENP-A/C localize to opposite sides of the metaphase plate (Fig. 1 A). Both proteins remain associated with chromosomes from prophase through telophase. These staining patterns are specific, with the exception of the weak pole staining of the anti–CeCENP-A antibody, because they disappear in embryos depleted of the corresponding proteins by RNAi (see below). These results indicate that CeCENP-A and CeCENP-C localize to the diffuse kinetochores of holocentric C. elegans chromosomes and confirm that the CENP-C–like gene we identified by weak sequence homology is indeed a kinetochore component in C. elegans.
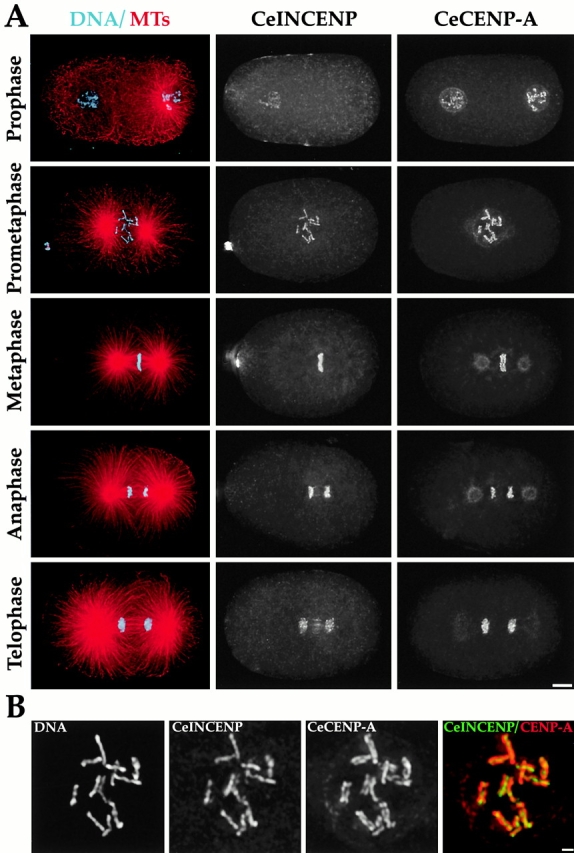
Figure 1.

CeCENP-A and CeCENP-C colocalize to the diffuse kinetochores of one-cell stage C. elegans embryos. (A) Wild-type embryos were stained to visualize DNA (cyan), MTs (red), Ce-CENP-A, and CeCENP-C. Different stages during the first division of the C. elegans embryo are shown. The sperm-derived pronucleus with associated centrosomal asters is to the right and the oocyte-derived pronucleus to the left in the early prophase and prophase panels. The weak spindle pole staining in the Ce-CENP-A metaphase and anaphase panels is nonspecific. (B) Higher magnification panels of nuclei stained for DNA and CeCENP-C. The panels on the left show a prophase pronucleus just before pronuclear meeting. CeCENP-C localizes to a patchy stripe that runs along the partially condensed chromosomes. The panels on the right show a pronucleus after the pronuclei have met and the nuclear envelope has broken down. By this time, CeCENP-C localizes to two plates on opposite faces of the fully condensed chromosomes. All images are projections of three-dimensional stacks. Bars: (A) 5 μm; (B) 1 μm.
Depletion of CeCENP-A or CeCENP-C Results in a Similar Severe Mitotic Chromosome Segregation Defect
To assay the effects of depleting CeCENP-A or CeCENP-C on chromosome segregation, we generated a worm strain expressing GFP-histone H2B. In wild-type embryos after fertilization, the maternal pronucleus completes meiosis and both pronuclei replicate their DNA. Subsequently, the pronuclei migrate towards each other while in pro-phase. After pronuclear meeting (Fig. 2; wild-type, −28s) and nuclear envelope breakdown (NEBD; t = 0 for sequences in Fig. 2), the condensed chromosomes congress to and distribute over the metaphase plate (Fig. 2; wild-type, 49 and 119s). The sister chromatids then separate (Fig. 2; wild-type, 210s), decondense, and cytokinesis partitions the two segregated DNA masses into daughter cells (Fig. 2; wild-type, 603s).
Next, we filmed embryos depleted of either CeCENP-A or CeCENP-C by RNAi. In C. elegans, dsRNA injected into adult hermaphrodites specifically ablates transcripts with high sequence homology, preventing further production of the protein coded by the targeted gene (Montgomery and Fire 1998). Continued embryo production and protein turnover deplete the maternal cytoplasm of the targeted protein within 20–30 h. We confirmed depletions of CeCENP-A and CeCENP-C to levels undetectable by immunofluorescence (see Fig. 3). Interestingly, depletion of either protein resulted in an essentially identical phenotype (Fig. 2, middle and right). In embryos depleted of either CeCENP-A (n = 15 one-cell embryos) or CeCENP-C (n = 8 one-cell embryos), chromosomes derived from the two pronuclei compact into separate discrete masses and fail to distribute over the spindle equator (Fig. 2/119s). The failure of chromosome distribution is particularly clear in the insets which show end-on views of metaphase spindles. These views are derived from the second division, during which the spindle in the anterior cell often orients perpendicular to the focal plane. At the time when wild-type embryos undergo anaphase, chromosomes in the depleted embryos remain in separate compact masses and no chromosome segregation occurs (Fig. 2, 208 and 210s). Nevertheless, cytokinesis initiates at the same time after NEBD as in wild-type. As a consequence of the cytokinetic furrow and cytoplasmic flows, the two unsegregated DNA masses distribute randomly between the two cells. In most cases, some DNA is trapped in the cleavage furrow (Fig. 2, 600 and 602s). CeCENP-A/C–depleted embryos progress through the cell cycle with wild-type kinetics, consistent with previous work suggesting the absence of a mitotic checkpoint during early divisions of the C. elegans embryo (Gonczy et al. 1999).
Figure 3.
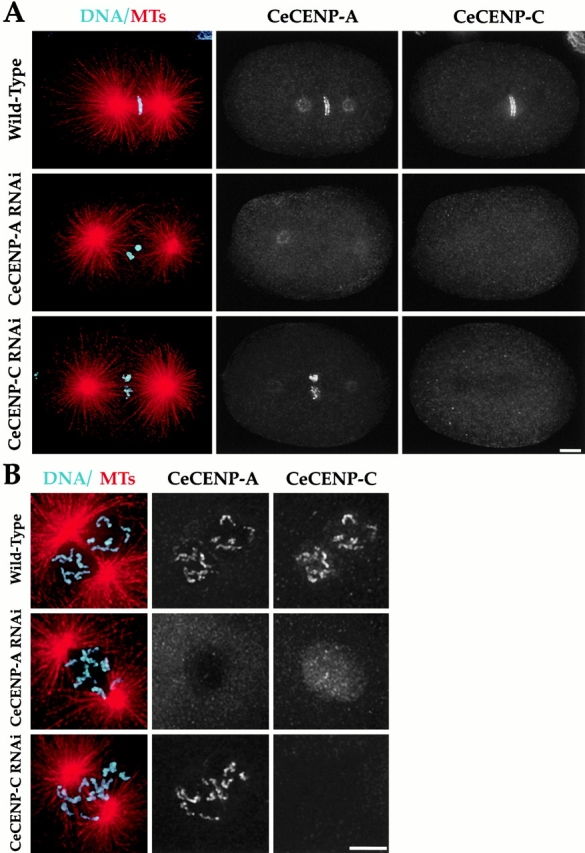
CeCENP-C localization to chromosomes requires CeCENP-A but not vice versa. Wild-type, CeCENP-A–, or CeCENP-C–depleted embryos were fixed and stained to visualize DNA (cyan), MTs (red), CeCENP-A, and CeCENP-C. Metaphase embryos (A) and higher magnification views of prophase/prometaphase nuclei (B) are shown. RNAi of either component reduces it to levels undetectable by immunofluorescence. Persistence of the spindle pole staining in CeCENP-A–depleted embryos indicates that this staining is nonspecific (see also Fig. 10 C). In the absence of CeCENP-A (A and B, middle rows), CeCENP-C is visible in nuclei during prophase, but is not observed associated with chromosomes at any point during mitosis. In contrast, in the absence of CeCENP-C, CeCENP-A associates with chromosomes throughout mitosis (A and B, bottom rows). All images are projections of three-dimensional stacks. Bars, 5 μm.
In CeCENP-A/C–depleted embryos we observed polar body defects consistent with problems in the two meiotic divisions of the oocyte nucleus that occur after fertilization. Surprisingly, however, a maternal pronucleus containing an approximately wild-type amount of DNA was always formed. Chromosomes from the paternal pronucleus are free of any inherited defects because the meiotic divisions leading to sperm formation occur before the dsRNA is injected. We did not observe any significant differences between chromosomes derived from the maternal and paternal pronuclei in CeCENP-A/C–depleted embryos, leading us to conclude that the segregation defect during the first mitotic division is not a consequence of prior meiotic defects.
CeCENP-A Is Required to Recruit CeCENP-C but Not Vice Versa
Although CeCENP-C targets to chromosomes later than CeCENP-A (Fig. 1), depletion of either results in similar segregation defects (Fig. 2). Since CeCENP-A is a histone, these results suggest that recruitment of CeCENP-C by CeCENP-A chromatin is a key intermediate step in kinetochore assembly. To test this hypothesis, we analyzed dependency relationships between these two components during kinetochore assembly. Because antibodies to both CeCENP-A and CeCENP-C were raised in rabbits, we directly labeled them and performed four-color immunofluorescence to visualize chromosomes, MTs, the component depleted by RNAi, and the component being assayed. Three-dimensional widefield images were collected and computationally deconvolved; projections of these three-dimensional stacks are shown.
This type of analysis revealed that depletion of Ce-CENP-A completely blocks the assembly of CeCENP-C onto chromosomes (Fig. 3A and Fig. B; n = 19 one-cell and 22 two-cell embryos). Although CeCENP-C did not associate with chromosomes, it still accumulated in nuclei during prophase (Fig. 3 B), suggesting that CeCENP-A is not required for stability of CeCENP-C. In contrast, the converse experiment showed that CeCENP-A remains associated with chromosomes throughout mitosis in the absence of CeCENP-C (Fig. 3A and Fig. B; n = 22 one-cell embryos and 16 two-cell embryos). Thus, we conclude that chromosomal targeting of CeCENP-C requires CeCENP-A, but not vice versa.
CeMCAK and CeBub1 Require CeCENP-A and CeCENP-C to Target to Chromosomes
To extend our analysis of dependency relationships during kinetochore assembly, we wanted to test whether targeting of other mitotic kinetochore components depended on CeCENP-A or CeCENP-C. To do this, we analyzed two putative mitotic kinetochore components: the C. elegans homologues of Bub1, a protein kinase involved in the mitotic checkpoint, and MCAK, a MT-depolymerizing kinesin. We raised, affinity-purified, and directly labeled antibodies to CeBub1 (R06C7.8) and CeMCAK (K11D9.1). Although both CeBub1 and CeMCAK are essential for embryonic viability, a CeCENP-A/C–like chromosome segregation defect was not observed in embryos depleted of either, consistent with the expectation that they do not perturb CeCENP-A/C targeting and function (Grill et al. 2001; Oegema, K., unpublished data).
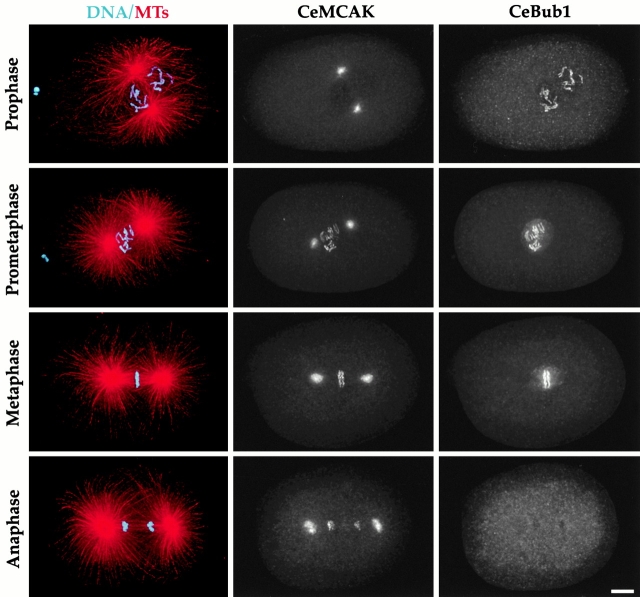
We first characterized localization of CeBub1 and CeMCAK in wild-type embryos (Fig. 4). Like its vertebrate homologues, CeMCAK localizes to centrosomes and kinetochores during mitosis. CeMCAK first localizes to kinetochores after NEBD in prometaphase and remains associated with the chromosomes until telophase (Fig. 4). CeBub1 localizes to kinetochores in prophase before NEBD and remains associated with kinetochores through metaphase (Fig. 4). During metaphase, CeBub1 also localizes to a matrix-like structure around the spindle that does not coalign with spindle MTs. CeBub1 is undetectable on chromosomes by middle to late anaphase and the matrix staining also disappears by this stage. These staining patterns disappear in embryos depleted of the corresponding proteins by RNAi, confirming their specificity (data not shown).
Figure 4.
CeMCAK and CeBub1 are mitotic kinetochore components in C. elegans. Wild-type embryos were stained to visualize DNA (cyan), MTs (red), CeMCAK, and CeBub1. Different stages of the first embryonic division are shown. All images are projections of three-dimensional stacks. Bar, 5 μm.
In embryos depleted of either CeCENP-C (Fig. 5) or CeCENP-A (not shown), neither CeBub1 (n = 11 one-cell and 16 two-cell embryos) or CeMCAK (n = 18 one-cell and 17 two-cell embryos) localized to chromosomes. However, CeMCAK localized normally to centrosomes (Fig. 5 B) indicating that the protein was still present but unable to target to chromosomes. Similarly, CeBub1 accumulated in prophase nuclei (Fig. 5 A) and prominently stained the spindle matrix-type structure (not shown), but did not target to chromosomes. These results suggest that CeCENP-A and CeCENP-C are essential for establishment of kinetochore structure and in their absence kinetochores fail to assemble. Thus, these conserved proteins appear to be at the top of a hierarchy of interactions driving kinetochore assembly, suggesting that the defects observed in their absence represent a “kinetochore null” phenotype.
Figure 5.
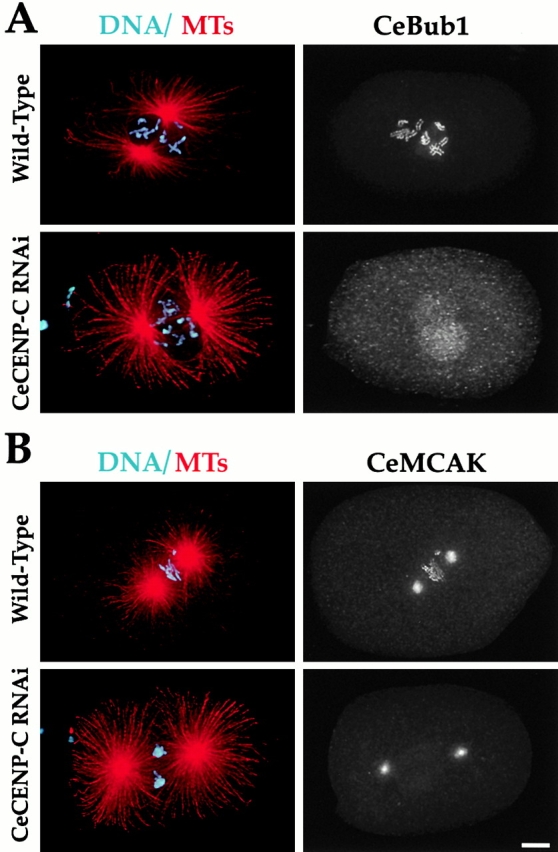
Both CeMCAK and CeBub1 require CeCENP-C to target to chromosomes. Wild-type or CeCENP-C–depleted embryos were fixed and stained to simultaneously visualize DNA (cyan), MTs (red), CeCENP-C, and either CeBub1 (A) or CeMCAK (B). CeCENP-C was depleted to undetectable levels (not shown). Centrosomal localization of CeMCAK and nuclear accumulation of CeBub1 occur in CeCENP-C–depleted embryos. However, neither protein localizes to chromosomes during any mitotic stage. Identical results were obtained in analysis of CeBub1 and CeMCAK localization in CeCENP-A–depleted embryos (not shown). All images are projections of three-dimensional stacks. Bar, 5 μm.
Polarity-cued Forces Provide an Assay for the Contribution of Kinetochores to Spindle Stability
We wanted to use CeCENP-A/C depletions to test if kinetochore–MT interactions contribute to spindle mechanical integrity. One way to assay spindle integrity would be to pull on the spindle poles and assess the consequences. In the asymmetrically dividing C. elegans embryo, polarity-cued forces differentially pull on the two spindle poles to move the spindle towards the posterior during the first mitotic division (Grill et al. 2001). These forces could be used to assay the contribution of kinetochores to spindle stability if they act after kinetochore–MT interactions are established but before anaphase onset.
To determine when polarity-cued forces act, we filmed embryos expressing both GFP-histone and GFP–γ-tubulin (Fig. 6 A). This allowed us to track spindle movement, a readout for the action of polarity-cued forces, and anaphase onset in the same embryo. Analysis of eight embryos revealed that persistent movement of the spindle towards the posterior was initiated between 60 and 100 s before anaphase onset (Fig. 6A and Fig. B). Chromosome congression is almost complete by this time (Fig. 6 A, −64s), suggesting that significant bipolar kinetochore–MT interactions have already been established when polarity-cued forces begin to act. Posterior spindle movement continues for ∼2 min, ending ∼35 s after anaphase onset. These results reveal that polarity-cued forces pull on spindle poles during a time window that is ideal for assessing the contribution of kinetochore–MT interactions to spindle stability.
Interestingly, this analysis also revealed that chromosomes do not move significantly polewards during anaphase in C. elegans (Fig. 6 C). The chromosome-to-pole distance after chromosome segregation (5.32 ± 0.49 μm) was essentially the same as the chromosome-to-pole distance before anaphase onset (5.50 ± 0.35 μm). Thus, chromosome segregation in the one cell C. elegans embryo is primarily due to anaphase B spindle elongation.
Kinetochore Function Is Necessary for Formation of a Mechanically Stable Spindle
To test whether kinetochore–MT interactions are required to assemble a stable spindle, we compared spindle structure and dynamics in wild-type and CeCENP-A/C–depleted embryos expressing GFP–α-tubulin. Spinning disk confocal microscopy was used to image the MT cytoskeleton in the ∼20-μm thick embryos. Images obtained using this technique in wild-type and CeCENP-C–depleted embryos are shown in Fig. 7 A. This analysis was timed relative to NEBD because anaphase onset is difficult to score precisely in GFP–α-tubulin videos. To facilitate interpretation, the interval between the onset of polarity-cued forces (on average 75 s after NEBD) and the initiation of anaphase (on average 155 s after NEBD) is marked on the kinetic traces in Fig. 7 B.
Similar to the GFP-histone analysis, depletion of Ce-CENP-C (n = 8 one-cell embryos) or CeCENP-A (n = 14 one-cell embryos) resulted in an identical phenotype (a CeCENP-C–depleted embryo is shown in Fig. 7 A). During the first minute after NEBD, depleted embryos appeared relatively normal (Fig. 7 A, compare 48s panels). However, during the next minute the spindle poles began to separate rapidly and a robust spindle failed to form (Fig. 7 A, 120s panels). Kinetic traces revealed a slight increase in spindle pole separation in wild-type embryos in the interval between onset of polarity-cued forces and initiation of anaphase (Fig. 6 C and 7 B). In contrast, spindle poles in depleted embryos separated rapidly during this same period (Fig. 7 B). Pole separation in depleted embryos was nearly complete by the time wild-type embryos initiated anaphase (Fig. 7 B). Analysis of the complete kinetic traces revealed that spindle poles in depleted embryos separate at approximately twice the maximum velocity of wild-type (Fig. 7 C). The abrupt separation of spindle poles in CeCENP-A/C–depleted embryos in response to polarity-cued pulling forces indicates that functional kinetochores are required to assemble a stable mitotic spindle.
We also compared MT dynamics in wild-type and depleted embryos during late anaphase and telophase. In wild-type, a tightly packed array of interzonal MTs forms between the separating chromosomes during anaphase (Fig. 7 A, 252s). As the furrow begins to ingress, this interzonal array appears to loosen and individual MT bundles become visible between the separated chromosome masses. MT bundles peripheral to the spindle that presumably form from MTs emanating from the two opposing asters are also observed. As the cleavage furrow ingresses, the MT bundles compact to form the midbody that connects the two daughter cells (Fig. 7 A, 499s). In embryos depleted of CeCENP-A or CeCENP-C, the chromosomes fail to segregate and the interzonal MT array does not form. Instead, the two asters appear to move independently of one another (Fig. 7 A, 176 and 256s). Nevertheless, as the furrow begins to ingress, MT bundles form between the two opposing asters. These bundles compact and a stable midbody results, allowing cytokinesis to complete normally (Fig. 7 A, 504s; online supplemental videos are available at http://www.jcb.org/cgi/content/full/153/6/1209/DC1).
CeINCENP Localizes to Mitotic Chromosomes and Interzonal MTs
Recent work in C. elegans suggested that depletion of a homologue of the chromosomal passenger INCENP (ICP-1; referred to here as CeINCENP) leads to a complete failure of chromosome segregation during the first mitotic division (Kaitna et al. 2000). Therefore, we wanted to determine if there was any connection between CeINCENP and kinetochore assembly and function. We first characterized CeINCENP localization in wild-type embryos. CeINCENP is visible on condensing chromosomes in prophase nuclei (Fig. 8 A). At prometaphase, CeINCENP is in a single stripe coincident with the chromosome between the two kinetochore plates (Fig. 8A and Fig. B). During anaphase and telophase, CeINCENP remains associated with the chromosomes, but is also present on the interzonal MTs (Fig. 8 A). Chromosomal CeINCENP localization is specific and disappears in embryos depleted of CeINCENP by RNAi (see Fig. 10). This localization pattern confirms that CeINCENP is a chromosomal passenger protein in C. elegans.
Figure 8.
CeINCENP localizes to the chromosomal region between the two kinetochore plates. (A) Wild-type embryos were stained to simultaneously visualize DNA (cyan), MTs (red), CeINCENP, and CeCENP-A. Different stages of the first mitotic division are shown. In the first row, the maternal pronucleus is to the left and the paternal pronucleus to the right. CeINCENP localization early in prophase was more prominent in the maternal pronucleus than in the paternal pronucleus. (B) Higher magnification panels of the prometaphase nucleus in A, showing localization of CeINCENP to the chromosomal region between the two kinetochore plates. All images are projections of three-dimensional stacks. Bars: (A) 5 μm; (B) 1 μm.
Figure 10.
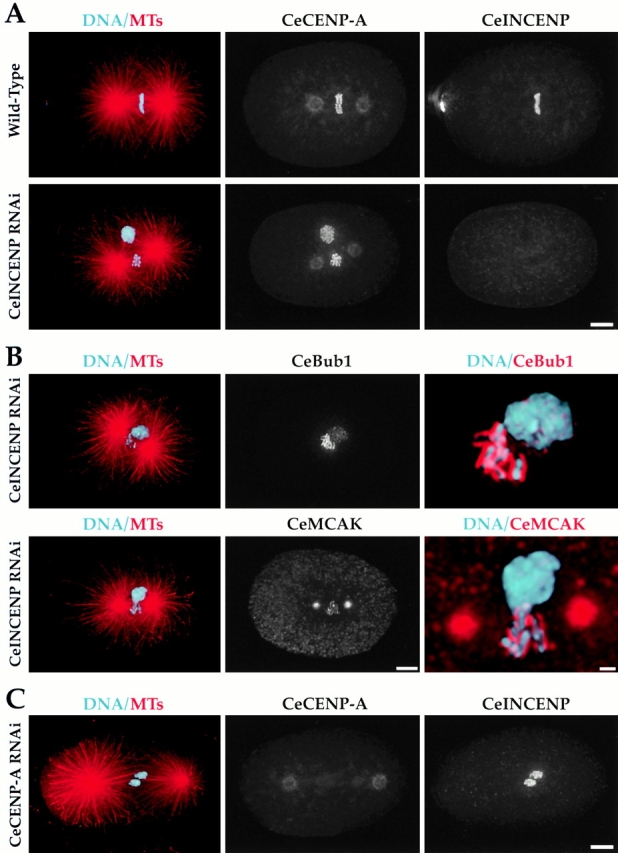
Chromosomal association of CeINCENP and kinetochore assembly are independent processes. (A) Wild-type or CeINCENP–depleted embryos were fixed and stained to visualize DNA (cyan), MTs (red), CeCENP-A, and CeINCENP. CeINCENP is depleted to levels undetectable by immunofluorescence. Nevertheless, CeCENP-A localizes to the condensed mitotic chromosomes. (B) CeINCENP-depleted embryos were fixed and stained to visualize DNA, MTs, CeINCENP, and CeBub1 (top) or CeMCAK (bottom). CeINCENP was depleted to undetectable levels as in A (not shown). The right panels in both rows show higher magnification views. Both CeBub1 and CeMCAK localize to the condensed paternal chromosomes in the absence of CeINCENP and form normal-looking kinetochore plates. Localization to the meiotically defective maternal mass is weak or absent for both markers. (C) A CeCENP-A–depleted embryo stained for DNA (cyan), MTs (red), CeCENP-A, and CeINCENP. CeCENP-A was depleted to undetectable levels, but CeINCENP still localized to chromosomes. Bars: (A) 5 μm; (B, left and middle) 5 μm; (B, right) 1 μm; (C) 5 μm.
CeINCENP Depletion Leads to Mitotic Defects Distinct from CeCENP-A/C Depletions
To test whether CeINCENP influences kinetochore function, we first analyzed mitotic chromosome segregation in depleted embryos (n = 28 one-cell embryos). In CeINCENP-depleted embryos, meiotic divisions of the oocyte nucleus completely fail, resulting in a maternal pronucleus with four times the wild-type DNA content (Fig. 9 A, −524s). To avoid interference from prior meiotic failure, we focused our analysis on the paternal chromosomes derived from the sperm pronucleus. These chromosomes are free of meiotic defects because they complete meiosis before injection of the dsRNA targeting CeINCENP. In CeINCENP-depleted embryos, the paternal chromosomes condense normally and align near the center of the spindle at metaphase (Fig. 9 A, 124s). However, they have difficulty congressing and fail to form a tight metaphase plate. Nevertheless, at anaphase the paternal chromosomes are always pulled apart into two equivalently sized masses (Fig. 9 A, 185s, arrowheads). This segregation is not normal and leading and lagging chromatin is always observed. At telophase, all of the DNA decondenses to form a structure with a “Mickey Mouse”–like appearance due to the presence of small separated paternal DNA–derived “ears” on the large unsegregated maternal DNA mass. The example shown in Fig. 9 A is typical. Infrequently, lateral movement of the maternal DNA mass allows an unobstructed view of paternal chromosome segregation (Online supplemental video 9 available at http://www.jcb.org/cgi/content/full/153/6/1209/DC1). Cytokinesis initiates normally in CeINCENP-depleted embryos, but it does not complete and the furrow regresses. The phenotype of CeINCENP depletion in this assay was indistinguishable from depletion of the aurora B kinase AIR-2, or the survivin BIR-1 (Online supplemental video 9 available at http://www.jcb.org/cgi/content/full/153/6/1209/DC1).
To confirm the conclusions of our live analysis, we obtained three-dimensional deconvolved images of fixed CeINCENP-depleted embryos (Fig. 9 C). This analysis confirmed both the depletion of CeINCENP (not shown here; see Fig. 10) and the phenotype seen in the live assays. In prophase, the maternal chromosomes appear condensed, but well-resolved, normal-looking chromosomes are not formed. In contrast, the paternal chromosomes are clearly visible (Fig. 9 C, top two panels) and appear normally condensed. In metaphase, the paternal chromosomes appear to make MT attachments and align near the center of the spindle, but do not form a tight metaphase plate (Fig. 9 C, second panel). In anaphase and telophase, two paternal chromosome masses of equal size are found associated with the two spindle poles (Fig. 9 C, middle and bottom panels). However, in anaphase, leading and lagging chromatin is always observed (Fig. 9 C, middle panel). Spindle midzone MTs that form between the separating chromosomes during anaphase and telophase are absent (compare Fig. 9 C, middle and bottom panels, with anaphase and telophase panels of Fig. 1 A and 8 A). The combination of live and fixed analysis leads us to conclude that depletion of CeINCENP results in mitotic chromosome segregation defects that are distinct from those observed in the absence of CeCENP-A or CeCENP-C.
CeINCENP-depleted Embryos Form Mechanically Stable Spindles
Phenotypic analysis suggested that bipolar kinetochore-MT attachments are formed by the paternal chromosomes in CeINCENP-depleted embryos. To test if this was the case, we analyzed spindle structure and dynamics in the depleted embryos. Qualitatively, spindle assembly and structure appeared normal until midanaphase (Fig. 9 B, compare −64, 120, and 185s panels with wild-type in Fig. 3 A), except for interference from the defective maternal chromosome mass. Quantitative analysis of CeINCENP-depleted embryos (Fig. 9D and Fig. E) revealed that poles separate at approximately the same rate and with the same timing as in wild-type. However, the spindle poles do not get as close together during spindle assembly as in wild-type, perhaps because only paternal chromosomes interact with MTs. These results suggest that bipolar kinetochore-MT attachments capable of resisting extra-spindle forces are formed in the absence of CeINCENP.
Spindle defects become apparent during anaphase and telophase in CeINCENP-depleted embryos. Although the paternal chromosomes are pulled apart into two discrete masses during anaphase, no interzonal MT array forms between the separating chromosomes. Nevertheless, a cleavage furrow begins to ingress (Fig. 9 B, 313s). In contrast to CeCENP-A/C–depleted embryos, MT bundles also fail to form between the asters during telophase and no midbody is formed. Instead, the MT array between the two asters dissipates in mid- to late telophase and the cleavage furrow regresses (Fig. 9 B, 666s).
CeINCENP Localization and Kinetochore Assembly Are Independent Processes
The paternal chromosomes make bipolar MT attachments and segregate into two masses in CeINCENP-depleted embryos, suggesting that CeCENP-A and CeCENP-C target to chromosomes in the absence of CeINCENP. Consistent with this prediction, we found that depletion of CeINCENP did not affect chromosomal targeting of CeCENP-A (Fig. 10 A; 15 one-cell and 8 two-cell embryos) or CeCENP-C (not shown). Furthermore, CeCENP-A and CeCENP-C localized to two plates on opposing faces of the condensed paternal chromosomes, indicating the absence of any dramatic effects of CeINCENP depletion on higher order chromosome structure.
The segregation defects of paternal chromosomes in CeINCENP-depleted embryos could result from a failure in targeting kinetochore components downstream of Ce-CENP-A and CeCENP-C. To test this, we determined whether CeBub1 and CeMCAK localized normally to the paternal chromosomes in CeINCENP-depleted embryos. We found that both CeBub1 (Fig. 10 B, top; n = 11 one-cell and 9 two-cell embryos) and CeMCAK (Fig. 10 B, bottom; n = 14 one-cell and 10 two-cell embryos) localized normally to the paternal chromosomes. Most likely as a consequence of prior meiotic defects, staining of the maternal chromosome mass was always very weak or absent for both CeBub1 and CeMCAK (Fig. 10 B). Aurora B localization depends on INCENP in C. elegans and vertebrates (Adams et al. 2000; Kaitna et al. 2000), suggesting that chromosomal targeting of neither INCENP nor aurora B is required for kinetochore assembly.
To test if chromosomal targeting of CeINCENP and kinetochore assembly are independent processes, we assayed targeting of CeINCENP to chromosomes in embryos depleted of CeCENP-A, which fail to recruit CeCENP-C, CeBub1, and CeMCAK. We found that CeINCENP localized to chromosomes in CeCENP-A–depleted embryos (Fig. 10 C; n = 11 one-cell and 21 two-cell embryos). Thus, we conclude that kinetochore assembly and chromosomal targeting of CeINCENP are independent processes.
Discussion
Depletion of either CeCENP-A or CeCENP-C Leads to an Identical “Kinetochore Null” Phenotype
CENP-A and CENP-C are conserved proteins that localize to the kinetochore and are required for chromosome segregation in diverse species. However, how these conserved components contribute to kinetochore assembly remains an open and important question. Here, we use RNAi to obtain single cell loss of function phenotypes for the C. elegans homologues of CENP-A and CENP-C. Depletion of either protein leads to an identical phenotype characterized by a complete failure of mitotic chromosome segregation as well as failure to target mitotic kinetochore components and to assemble a mechanically stable spindle. These results lead us to conclude that loss of CeCENP-A or CeCENP-C function results in a “kinetochore null” cell.
An Important Function of CENP-A–containing Chromatin Is to Recruit CENP-C
Using fixed assays to examine dependency relationships during kinetochore assembly, we found that recruitment of CeCENP-C required CeCENP-A, but not vice versa. A similar dependency has also been noted in interphase nuclei of cells derived from CENP-A knockout mice (Howman et al. 2000). This result, combined with the identical phenotypes of embryos depleted of either CeCENP-A or CeCENP-C, suggests that a key role of CENP-A–containing chromatin is the recruitment of CENP-C. A tempting hypothesis arising from these findings is that CENP-A–containing chromatin directly recruits CENP-C. The conservation of these two proteins without clear evidence for an equivalently conserved putative linking intermediate is consistent with this speculation. Mammalian CENP-C directly binds to DNA in vitro (Sugimoto et al. 1994; Yang et al. 1996), but whether CeCENP-C and other CENP-C homologues directly bind DNA and whether DNA binding by CENP-C is important for its recruitment and function at kinetochores is not clear. The recent biochemical reconstitution of nucleosomes containing CENP-A instead of histone H3 (Yoda et al. 2000) should make it possible to address these issues in vitro.
CeCENP-A and CeCENP-C Are at the Top of a Hierarchy of Pathways Driving Kinetochore Assembly
Embryos depleted of either CeCENP-A or CeCENP-C fail to recruit both CeBub1 and CeMCAK to kinetochores. Combined with the severity of the chromosome segregation and spindle defects in their absence, this result suggests that CeCENP-A and CeCENP-C are at the top of a hierarchy of pathways driving kinetochore assembly (Fig. 11). In vertebrates, Bub1 and MCAK localize to distinct structural subdomains of the kinetochore. In C. elegans, we have also observed spatial distinctions in the localization of these two proteins; furthermore, their targeting to kinetochores is independent of one another (Oegema, K., unpublished results). Therefore, we depict CeMCAK and CeBub1 on separate pathways downstream of CeCENP-C (Fig. 11). Previous evidence for multiple parallel pathways participating in later stages of kinetochore assembly comes from the independence of Mad/Bub checkpoint protein localization from ZW10/Rod function (Basu et al. 1999; Chan et al. 2000). Our results also show that chromosomal recruitment of CeINCENP and kinetochore assembly are independent events (Fig. 11). This result is consistent with a report of normal kinetochore formation, as assayed by EM, in tissue culture cells expressing a dominant negative form of INCENP (Mackay et al. 1998).
Figure 11.
A summary of the dependency relationships during kinetochore assembly in C. elegans. The phenotypic similarity and dependency results reveal that a key intermediate in kinetochore assembly is the recruitment, either direct or indirect, of Ce-CENP-C by chromosome-associated CeCENP-A. CeBub1 and CeMCAK are shown on separate pathways downstream of CeCENP-C (see text). The arrows indicate dependency relationships during kinetochore assembly and are not intended to suggest direct interactions between components. Our results also reveal that chromosomal association of CeINCENP and kinetochore assembly are independent processes. The depicted dependency of AIR-2, the C. elegans aurora B kinase, on CeINCENP and BIR-1 is derived from work of other groups (Adams et al. 2000; Kaitna et al. 2000; Speliotes et al. 2000).
Kinetochore Function Is Required for Spindle Integrity
In vitro assays in meiotic extracts have shown that self-organization of MT motor complexes, coupled with chromatin-stimulated MT nucleation, can drive formation of a bipolar spindle in the absence of kinetochores and centrosomes (Heald et al. 1996). However, whether the mechanical integrity of spindles formed without kinetochores is compromised has not been determined. In embryos depleted of either CeCENP-A or CeCENP-C, the onset of polarity-cued extra-spindle forces leads to an abrupt separation of the spindle poles and a stable metaphase spindle does not form. This result demonstrates that kinetochore–MT interactions contribute to the formation of a mechanically stable spindle. A similar effect on spindle structure was noted in a fission yeast centromere protein mutant (Goshima et al. 1999), suggesting that an important function of bipolar kinetochore–MT attachments is to resist forces that tend to elongate spindles. Interestingly, this type of analysis showed that spindles in CeINCENP-depleted embryos are able to resist extra-spindle pulling forces, suggesting that the kinetochores formed in the absence of CeINCENP are functional.
The Kinetochore and the Chromosomal Passengers Make Distinct Contributions to Mitotic Chromosome Segregation
The phenotypes arising from depletions of CeCENP-A/CeCENP-C and passenger proteins are remarkably distinct. In the absence of any of the three chromosomal passengers, meiotic chromosome segregation completely fails. In contrast, although embryos depleted of CeCENP-A/C form abnormal-looking polar bodies, a maternal pronucleus containing an apparently normal amount of DNA is formed. Conversely, during the first mitotic division, embryos depleted of CeCENP-A/C completely fail to segregate their chromosomes. In contrast, the paternal chromosomes in embryos depleted of the chromosomal passengers always segregate into two equivalently sized masses. The difficulty of following the paternal chromosomes in the presence of the large mass of maternal chromatin may explain why their separation was not noted in a previous study (Kaitna et al. 2000). An additional difference is that stable mitotic spindles assemble in CeINCENP, but not in CeCENP-A/C–depleted embryos. These results suggest an independence between the functions of the kinetochore and the chromosomal passengers. They also suggest that meiotic and mitotic chromosome segregation use fundamentally different mechanisms in C. elegans. The latter suggestion is supported by ultrastructural work showing that meiotic C. elegans chromosomes lack recognizable diffuse kinetochores; instead, MTs terminate within the chromatin near the ends of the chromosomes (Albertson and Thomson 1993).
Although the paternal chromosomes segregate into two apparently equivalent masses in CeINCENP-depleted embryos, they fail to congress properly and during anaphase leading and lagging chromatin is always seen. These defects are remarkably similar to those observed when a dominant negative INCENP fragment was expressed in tissue culture cells (Mackay et al. 1998) and are consistent with chromosome missegregation, but not a total lack of segregation, in mutants of chromosomal passenger homologues in budding yeast (Biggins et al. 1999; Kim et al. 1999).
Possible Contributions of Chromosomal Passengers to Mitotic Chromosome Segregation
Our results suggest that kinetochores assemble and make bipolar attachments to spindle MTs in the absence of CeINCENP. What is then the basis for the defects in chromosome congression and segregation observed in the absence of the chromosomal passengers? A popular hypothesis, based on the involvement of aurora B in histone H3 phosphorylation (Hsu et al. 2000; Giet and Glover 2001), is a role for these proteins in chromosome condensation (Cheung et al. 2000; Adams et al. 2001). In our analysis of CeINCENP-depleted embryos, the paternal chromosomes condensed and assembled well-defined kinetochore plates. In addition, depletion of C. elegans condensin subunits leads to more severe mitotic segregation defects than those resulting from depletion of the chromosomal passengers (Oegema, K., unpublished observations). These results lead us to conclude that a majority of the large scale restructuring of interphase chromatin into mitotic chromosomes can occur in the absence of chromosomal passenger function. This conclusion is consistent with the results of Severson et al. 2000, who showed that aurora B function in C. elegans is necessary only during the 2 min before anaphase onset.
The formation of condensed chromosomes in CeINCENP-depleted embryos does not rule out a role for the chromosomal passengers in higher order chromosome structure. An influence on the mechanical properties of chromosomes could perturb their response to MT-mediated forces or cause an increase in chromosome malorientations, leading to missegregation. Perturbation of higher order chromosome structure could also lead to defects in the establishment or dissolution of cohesion. Our analysis of spindle dynamics in CeINCENP-depleted embryos suggests that paternal sister chromatids are paired and make bipolar attachments to spindle MTs. Subsequent separation of the paternal chromosomes into two equivalently sized masses suggests dissolution of chromatid cohesion at the metaphase–anaphase transition. These results are consistent with normal cohesin dissociation in mutants of budding yeast aurora B (Biggins et al. 1999). Establishing that sister chromatids segregate to opposite spindle poles in CeINCENP-depleted embryos will require direct analysis using either FISH or a GFP-marked chromosome.
A final possibility is that chromosomal passengers regulate chromosome–MT interactions. CeINCENP is not required for assembly of a kinetochore–MT interface, but the passengers could regulate the dynamics of this interface, as suggested by analysis of the aurora B homologue in budding yeast (Biggins et al. 1999). The only argument against this possibility is that the chromosomal passengers and the kinetochore–MT interface are spatially distinct on the chromosome surface. Alternatively, the chromosomal passengers could contribute to a kinetochore-independent mechanism for chromosome–MT interactions, such as the one described recently for a Xenopus chromokinesin (Funabiki and Murray 2000).
Supplemental Material
Acknowledgments
We are very grateful to Yuji Kohara (National Institute of Genetics, Mishima, Japan) for cDNAs used for RNA and antibody production, G. Seydoux (Johns Hopkins University, Baltimore, MD) for vectors and advice on germline expression, Theresa Stiernagle at the Caenorhabditis Genetics Center (University of Minnesota, St. Paul, MN) for strains, the Wellcome Trust C. elegans Course for much-needed instruction, Paul Maddox and Ted Salmon for use of their confocal during early stages of this study, Eva Hannak for the GFP–α-tubulin construct, and Martin Srayko for comments on the manuscript.
K. Oegema is supported by a Helen Hay Whitney Fellowship. A. Desai was supported by fellowships from the European Molecular Biology Organization and the American Cancer Society.
Footnotes
The online version of this article contains supplementary material.
K. Oegema and A. Desai contributed equally to this work.
Abbreviations used in this paper: DIC, differential interference contrast; GFP, green fluorescent protein; MT, microtubule; NEBD, nuclear envelope breakdown; RNAi, RNA-mediated interference.
References
- Adams R.R., Wheatley S.P., Gouldsworthy A.M., Kandels-Lewis S.E., Carmena M., Smythe C., Gerloff D.L., Earnshaw W.C. INCENP binds the aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol. 2000;10:1075–1078. doi: 10.1016/s0960-9822(00)00673-4. [DOI] [PubMed] [Google Scholar]
- Adams R.R., Carmena M., Earnshaw W.C. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 2001;11:49–54. doi: 10.1016/s0962-8924(00)01880-8. [DOI] [PubMed] [Google Scholar]
- Albertson D.G., Thomson J.N. The kinetochores of Caenorhabditis elegans . Chromosoma. 1982;86:409–428. doi: 10.1007/BF00292267. [DOI] [PubMed] [Google Scholar]
- Albertson D.G., Thomson J.N. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans . Chromosome Res. 1993;1:15–26. doi: 10.1007/BF00710603. [DOI] [PubMed] [Google Scholar]
- Basu J., Bousbaa H., Logarinho E., Li Z., Williams B.C., Lopes C., Sunkel C.E., Goldberg M.L. Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila . J. Cell Biol. 1999;146:13–28. doi: 10.1083/jcb.146.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S., Severin F.F., Bhalla N., Sassoon I., Hyman A.A., Murray A.W. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwitz B.J., Ahmad K., Moore L.L., Roth M.B., Henikoff S. A histone-H3-like protein in C. elegans . Nature. 1999;401:547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- Chan G.K., Jablonski S.A., Starr D.A., Goldberg M.L., Yen T.J. Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores. Nat. Cell Biol. 2000;2:944–947. doi: 10.1038/35046598. [DOI] [PubMed] [Google Scholar]
- Cheung P., Allis C.D., Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- Comings D.E., Okada T.A. Holocentric chromosomes in Oncopeltuskinetochore plates are present in mitosis but absent in meiosis. Chromosoma. 1972;37:177–192. doi: 10.1007/BF00284937. [DOI] [PubMed] [Google Scholar]
- Cutts S.M., Fowler K.J., Kile B.T., Hii L.L., O'Dowd R.A., Hudson D.F., Saffery R., Kalitsis P., Earle E., Choo K.H. Defective chromosome segregation, microtubule bundling and nuclear bridging in inner centromere protein gene (Incenp)-disrupted mice. Hum. Mol. Genet. 1999;8:1145–1155. doi: 10.1093/hmg/8.7.1145. [DOI] [PubMed] [Google Scholar]
- Dawe R.K., Reed L.M., Yu H.G., Muszynski M.G., Hiatt E.N. A maize homolog of mammalian CENPC is a constitutive component of the inner kinetochore. Plant Cell. 1999;11:1227–1238. doi: 10.1105/tpc.11.7.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis-Lang H., Minden J., Sullivan W., Oegema K. Live confocal analysis with fluorescently labeled proteins. Methods Mol. Biol. 1999;122:223–239. doi: 10.1385/1-59259-722-x:223. [DOI] [PubMed] [Google Scholar]
- Funabiki H., Murray A.W. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell. 2000;102:411–424. doi: 10.1016/s0092-8674(00)00047-7. [DOI] [PubMed] [Google Scholar]
- Giet R., Glover D.M. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 2001;152:669–682. doi: 10.1083/jcb.152.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P., Schnabel H., Kaletta T., Amores A.D., Hyman T., Schnabel R. Dissection of cell division processes in the one cell stage Caenorhabditis elegans embryo by mutational analysis. J. Cell Biol. 1999;144:927–946. doi: 10.1083/jcb.144.5.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Saitoh S., Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 1999;13:1664–1677. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill S.W., Gonczy P., Stelzer E.H., Hyman A.A. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 2001;409:630–633. doi: 10.1038/35054572. [DOI] [PubMed] [Google Scholar]
- Harlow E., Lane D. AntibodiesA Laboratory Manual. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1988. [Google Scholar]
- Heald R., Tournebize R., Blank T., Sandaltzopoulos R., Becker P., Hyman A., Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Howman E.V., Fowler K.J., Newson A.J., Redward S., MacDonald A.C., Kalitsis P., Choo K.H. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. USA. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J.Y., Sun Z.W., Li X., Reuben M., Tatchell K., Bishop D.K., Grushcow J.M., Brame C.J., Caldwell J.A., Hunt D.F. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000;102:279–291. doi: 10.1016/s0092-8674(00)00034-9. [DOI] [PubMed] [Google Scholar]
- Kaitna S., Mendoza M., Jantsch-Plunger V., Glotzer M. Incenp and an aurora-like kinase form a complex essential for chromosome segregation and efficient completion of cytokinesis. Curr. Biol. 2000;10:1172–1181. doi: 10.1016/s0960-9822(00)00721-1. [DOI] [PubMed] [Google Scholar]
- Kalitsis P., Fowler K.J., Earle E., Hill J., Choo K.H. Targeted disruption of mouse centromere protein C gene leads to mitotic disarray and early embryo death. Proc. Natl. Acad. Sci. USA. 1998;95:1136–1141. doi: 10.1073/pnas.95.3.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen G.H., Allshire R.C. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- Kelly W.G., Xu S., Montgomery M.K., Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caenorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Kang J.S., Chan C.S. Sli15 associates with the ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae . J. Cell Biol. 1999;145:1381–1394. doi: 10.1083/jcb.145.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D., Strunnikov A. Mitotic chromosome condensation. Annu. Rev. Cell Dev. Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- Mackay A.M., Ainsztein A.M., Eckley D.M., Earnshaw W.C. A dominant mutant of inner centromere protein (INCENP), a chromosomal protein, disrupts prometaphase congression and cytokinesis. J. Cell Biol. 1998;140:991–1002. doi: 10.1083/jcb.140.5.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney T., Ginkel L.M., Hunter A.W., Wordeman L. The kinetochore of higher eucaryotesa molecular view. Int. Rev. Cytol. 2000;194:67–131. doi: 10.1016/s0074-7696(08)62395-5. [DOI] [PubMed] [Google Scholar]
- Mello C.C., Kramer J.M., Stinchcomb D., Ambros V. Efficient gene transfer in C. elegansextrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P.B., Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P.B., Yang P., Glowczewski L., Koshland D., Smith M.M. Cse4p is a component of the core centromere of Saccharomyces cerevisiae . Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- Montgomery M.K., Fire A. Double-stranded RNA as a mediator in sequence-specific genetic silencing and co-suppression. Trends Genet. 1998;14:255–258. doi: 10.1016/s0168-9525(98)01510-8. [DOI] [PubMed] [Google Scholar]
- Moore L.L., Morrison M., Roth M.B. HCP-1, a protein involved in chromosome segregation, is localized to the centromere of mitotic chromosomes in Caenorhabditis elegans . J. Cell Biol. 1999;147:471–480. doi: 10.1083/jcb.147.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K., Peters J.M., Uhlmann F. Splitting the chromosomecutting the ties that bind sister chromatids. Science. 2000;288:1379–1385. doi: 10.1126/science.288.5470.1379. [DOI] [PubMed] [Google Scholar]
- Rieder C.L., Salmon E.D. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 1998;8:310–318. doi: 10.1016/s0962-8924(98)01299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh H., Tomkiel J., Cooke C.A., Ratrie H., Maurer M., Rothfield N.F., Earnshaw W.C. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- Severson A.F., Hamill D.R., Carter J.C., Schumacher J., Bowerman B. The aurora-related kinase AIR-2 recruits ZEN-4/CeMKLP1 to the mitotic spindle at metaphase and is required for cytokinesis. Curr. Biol. 2000;10:1162–1171. doi: 10.1016/s0960-9822(00)00715-6. [DOI] [PubMed] [Google Scholar]
- Skibbens R.V., Hieter P. Kinetochores and the checkpoint mechanism that monitors for defects in the chromosome segregation machinery. Annu. Rev. Genet. 1998;32:307–337. doi: 10.1146/annurev.genet.32.1.307. [DOI] [PubMed] [Google Scholar]
- Skoufias D.A., Mollinari C., Lacroix F.B., Margolis R.L. Human survivin is a kinetochore-associated passenger protein. J. Cell Biol. 2000;151:1575–1582. doi: 10.1083/jcb.151.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes E.K., Uren A., Vaux D., Horvitz H.R. The survivin-like C. elegans BIR-1 protein acts with the Aurora-like kinase AIR-2 to affect chromosomes and the spindle midzone. Mol. Cell. 2000;6:211–223. doi: 10.1016/s1097-2765(00)00023-x. [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Yata H., Muro Y., Himeno M. Human centromere protein C (CENP-C) is a DNA-binding protein which possesses a novel DNA-binding motif. J. Biochem. (Tokyo) 1994;116:877–881. doi: 10.1093/oxfordjournals.jbchem.a124610. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Chen E.S., Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–2219. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- Tomkiel J., Cooke C.A., Saitoh H., Bernat R.L., Earnshaw W.C. CENP-C is required for maintaining proper kinetochore size and for a timely transition to anaphase. J. Cell Biol. 1994;125:531–545. doi: 10.1083/jcb.125.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler-Smith C., Floridia G. Many paths to the top of the mountaindiverse evolutionary solutions to centromere structure. Cell. 2000;102:5–8. doi: 10.1016/s0092-8674(00)00004-0. [DOI] [PubMed] [Google Scholar]
- Uren A.G., Wong L., Pakusch M., Fowler K.J., Burrows F.J., Vaux D.L., Choo K.H. Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr. Biol. 2000;10:1319–1328. doi: 10.1016/s0960-9822(00)00769-7. [DOI] [PubMed] [Google Scholar]
- Vafa O., Sullivan K.F. Chromatin containing CENP-A and alpha-satellite DNA is a major component of the inner kinetochore plate. Curr. Biol. 1997;7:897–900. doi: 10.1016/s0960-9822(06)00381-2. [DOI] [PubMed] [Google Scholar]
- Warburton P.E., Cooke C.A., Bourassa S., Vafa O., Sullivan B.A., Stetten G., Gimelli G., Warburton D., Tyler-Smith C., Sullivan K.F., Poirier G.G., Earnshaw W.C. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- Wordeman L., Mitchison T.J. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J. Cell Biol. 1995;128:95–104. doi: 10.1083/jcb.128.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.H., Tomkiel J., Saitoh H., Johnson D.H., Earnshaw W.C. Identification of overlapping DNA-binding and centromere-targeting domains in the human kinetochore protein CENP-C. Mol. Cell. Biol. 1996;16:3576–3586. doi: 10.1128/mcb.16.7.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda K., Ando S., Morishita S., Houmura K., Hashimoto K., Takeyasu K., Okazaki T. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc. Natl. Acad. Sci. USA. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.