Abstract
Optical nanostructures have enabled the creation of subdiffraction detection volumes for single-molecule fluorescence microscopy. Their applicability is extended by the ability to place molecules in the confined observation volume without interfering with their biological function. Here, we demonstrate that processive DNA synthesis thousands of bases in length was carried out by individual DNA polymerase molecules immobilized in the observation volumes of zero-mode waveguides (ZMWs) in high-density arrays. Selective immobilization of polymerase to the fused silica floor of the ZMW was achieved by passivation of the metal cladding surface using polyphosphonate chemistry, producing enzyme density contrasts of glass over aluminum in excess of 400:1. Yields of single-molecule occupancies of ≈30% were obtained for a range of ZMW diameters (70–100 nm). Results presented here support the application of immobilized single DNA polymerases in ZMW arrays for long-read-length DNA sequencing.
Keywords: fluorescence, metal passivation, microscopy, polyvinyl phosphonic acid, single molecule
Nanofabrication techniques have enabled new approaches to interrogate individual biomolecules by fluorescence techniques (reviewed in refs. 1 and 2). The extremely small size scale of the associated devices results in a drastic illumination volume reduction, allowing single-molecule investigations to take place at fluorophore concentrations increased to biologically relevant levels. In addition to higher temporal resolution and higher signal-to-noise ratios, they also provide spatial resolution beyond diffraction-limited optics.
The zero-mode waveguide (ZMW) is one such nanophotonic confinement structure often consisting of a circular hole in a metal cladding film on a solid transparent substrate (3). In conjunction with laser-excited fluorescence, they provide observation volumes on the order of zeptoliters (10−21 l), three to four orders of magnitude smaller than far-field excitation volumes. Applications of circular ZMWs have included the detection of single-molecule DNA polymerase activity by using labeled nucleotides at micromolar concentrations (3), the study of λ-repressor oligomerization dynamics (4), two-color cross-correlation to rapidly screen for DNA restriction enzyme activity (5), and diffusion analysis of labeled membrane proteins in lipid bilayers of model membranes and living cells (6–11). C-shaped apertures have been described to study DNA hybridization interactions (12).
ZMW technology applications have been limited by the unavailability of selective immobilization methods to position molecules exclusively in the observation volume, immediately above the transparent ZMW floor. One approach to selective immobilization exploits a feature inherent in the ZMW architecture. The transparent substrate and metal cladding are made of different materials, opening the possibility for a selective derivatization that will direct protein adsorption to the floor and not to the nearby metal walls. The nanometer size scale and three-dimensional nature of ZMWs present challenges to generate such selective attachment sites on otherwise nonfouling surfaces. The derivatization chemistry contrast must be high, because the size of the target is small relative to the cladding surface area. The derivatized surfaces must be compatible with protein activities that take place only a few nanometers away. Additionally, the surface coatings should (i) exhibit high stability in the aqueous solution conditions required for the enzyme assays, (ii) exhibit low-fluorescence background, and (iii) provide low nonspecific adsorption to fluorescently labeled substrates.
An extensive literature exists on suppressing nonspecific protein adsorption to either glass or metal oxide surfaces (13, 14). Selective, localized passivation on mixed material nanoscale surfaces with the above-mentioned characteristics constitutes an area of intensive research, most commonly using glass in conjunction with gold (15–18). Aluminum is preferred for ZMWs because of its short optical skin depth and high reflectivity, which provide the best optical confinement; however, it is more difficult to functionalize differentially and can be susceptible to corrosion (19). Organophosphorus acids and their derivatives are known to react with a variety of metal oxides, such as titanium or aluminum oxide, while not binding to SiO2 surfaces in an aqueous medium (20–22), thereby representing a potential approach for targeted molecular placement of proteins in ZMW nanostructures.
Here, we demonstrate the suitability of polyvinylphosphonic acid (PVPA) chemistry to selectively protect the aluminum surfaces of ZMW arrays against protein adsorption, using neutravidin as an example. We show both the bias and the biocompatibility of this nanostructure derivatization by observing extended polymerization of long DNA by single φ29 DNA polymerase molecules immobilized on the floors of ZMWs in high-density arrays. Preservation of enzyme activity was demonstrated by detecting DNA synthesis products by fluorescence microscopy after incorporation of base-linked, fluorescently labeled nucleotides. DNA product strands observed to emanate into the solution above ZMW holes were associated with polymerases found within the ZMW observation volume, proving a strong polymerase adsorption bias. The length of the generated DNA was quantitated by fluorescence brightness analysis. Under the experimental conditions used, typical lengths of DNA synthesis were several thousand bases, consistent with the bulk performance of the enzyme.
Results
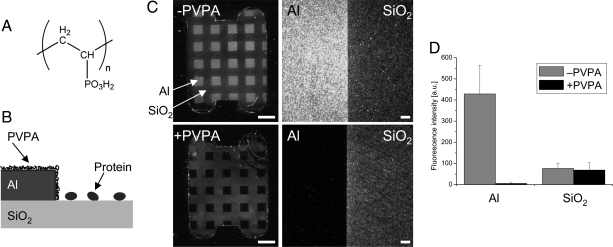
The molecular structure of PVPA is shown in Fig. 1A. Each molecule contains ≈200 phosphonate groups, imparting high water solubility. The derivatization is very fast (2 min) and proceeds at the native pH of the phosphonic acid (pH ≈2) at high temperatures (90°C), indicating rapid formation of a protective layer to prevent corrosion. It was followed by a dry annealing step intended to support formation of covalent aluminophosphonate bonds (23, 24). Ellipsometric measurements of PVPA-treated aluminum showed the formation of a very thin layer (≈0.5 nm) and no change in the native aluminum oxide layer thickness (data not shown).
Fig. 1.
Passivation of aluminum surfaces from protein adsorption by PVPA derivatization. (A) Molecular structure of PVPA. (B) Scheme of protein passivation by selective PVPA coating of aluminum on a mixed-material surface. (C) Patterned chips containing 0.5-mm aluminum squares (Al) on fused silica (SiO2) were untreated (Upper) or treated (Lower) with PVPA as described in Materials and Methods. Passivation was assayed by neutravidin adsorption and visualized using biotinylated fluorescent latex beads. (Left) The entire chip. (Scale bars, 1 mm.) (Right) Wide-field epifluorescence microscopy images of the boundary regions of the two surface materials. (Scale bars, 10 μm.) (D) Quantitative fluorescence intensity comparison of the two surface materials as a function of PVPA treatment (n = 6 chips for each condition; background-corrected; error bars indicate standard deviation).
Initial studies investigating PVPA-mediated aluminum passivation from protein adsorption used macroscopic patterned surfaces and neutravidin as a test protein (Fig. 1B). Fused silica chips containing a regular pattern of aluminum squares were manufactured by using the same thermal evaporation process that is used for ZMW array fabrication [see supporting information (SI) Text]. The chips were treated with PVPA as described in Materials and Methods. Phosphonate deposition on aluminum was confirmed by x-ray photoelectron spectroscopy, whereas it was undetectable on fused silica (data not shown). Upon incubating PVPA-treated or control chips with neutravidin solution and subsequent washing, physisorbed neutravidin was assayed by using biotinylated 40-nm fluorescent latex beads (Fig. 1C). Without deposition of the phosphonate polymer, neutravidin bound to both the fused silica and the aluminum surfaces with high densities. Fluorescence from the beads was enhanced by the proximity of the metal (25, 26), resulting in higher signal levels from the aluminum surface regions. In contrast, excellent bias of neutravidin adsorption toward the fused silica surface was observed for PVPA-treated samples, translating to very few biotinylated beads detectable by fluorescence microscopy on the aluminum surface. Average fluorescence intensities across the aluminum portions of the entire PVPA-treated chip were close to background levels, whereas signal levels on the fused silica surface regions, within the error of the measurement, were unaffected (Fig. 1D). Control experiments omitting neutravidin from the protein immobilization step confirmed the specificity of the observed signal to neutravidin adsorption (data not shown). Immobilization bias was confirmed by measurements of the ellipsometric thickness of the adsorbed protein layers on the two different surfaces (SI Fig. 6).
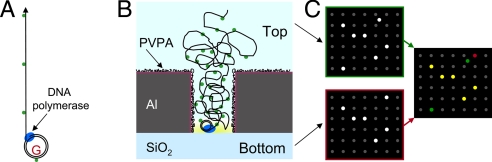
Similar protein physisorption bias was observed for φ29 DNA polymerase (SI Fig. 7), enabling the application of PVPA passivation to ZMW nanostructure arrays designed for DNA sequencing applications. To test the immobilization and activity of DNA polymerase in ZMWs, we developed the assay system shown schematically in Fig. 2. A small, circular DNA template and a base-linked fluorescent nucleotide, Alexa Fluor A488-dCTP, alongside dATP, dGTP, and dTTP, were used as substrates (Fig. 2A). The minicircle template was designed to contain only one guanine site, resulting in the generation of a long, fluorescently labeled single strand of DNA by polymerase-mediated, processive rolling-circle DNA strand displacement synthesis, with an Alexa Fluor label at regularly spaced intervals along the length of the synthesized DNA strand (72 bases). This strategy enabled correlation of fluorescence brightness to DNA product length (see below). Unextended φ29 DNA polymerase/template complexes were immobilized into high-density ZMW arrays by physisorption (Fig. 2B). The PVPA treatment minimized polymerase binding to the aluminum top and ZMW side wall surfaces, biasing polymerase localization toward the ZMW bottom. Upon subsequent DNA extension, the polymerase generated long single strands of repetitive DNA, eventually emanating into the solution above the ZMW. The fluorescently labeled DNA was imaged from both the glass and the solution side of ZMW arrays, henceforth referred to as bottom and top sides, respectively (Fig. 2C). This strategy leverages the optical confinement by ZMWs: bottom side fluorescence identifies DNA polymerase/DNA complexes located on the ZMW floor, and observation of fluorescence from the top surface identifies long DNA products that exit the ZMW. Colocalization analysis of the two superimposed images was used to detect the presence of active polymerases in ZMWs and to determine the bias of polymerase immobilization toward glass over aluminum surfaces.
Fig. 2.
Principle of observing DNA synthesis inside ZMWs. (A) Template design. The minicircle DNA template contained a single guanine site, allowing incorporation of a base-linked fluorescent nucleotide, Alexa Fluor 488-dCTP. Rolling circle, DNA strand displacement synthesis by φ29 DNA polymerase produced DNA with fluorescent labels at regular DNA length intervals (72 bases). (B) ZMW nanostructures were treated with PVPA, enabling selective immobilization of DNA polymerase at the bottom of ZMWs, followed by DNA extension reactions. The ZMW observation volume is highlighted in yellow. (C) Fluorescent DNA products were imaged from both sides of ZMW arrays. Image superposition and colocalization analysis was used to determine the bias of immobilization toward the glass floor (SiO2, yellow and red dots) over the side wall and top surfaces (Al, green dots) and to demonstrate single molecule occupancy. The DNA length was determined by fluorescence-brightness analysis.
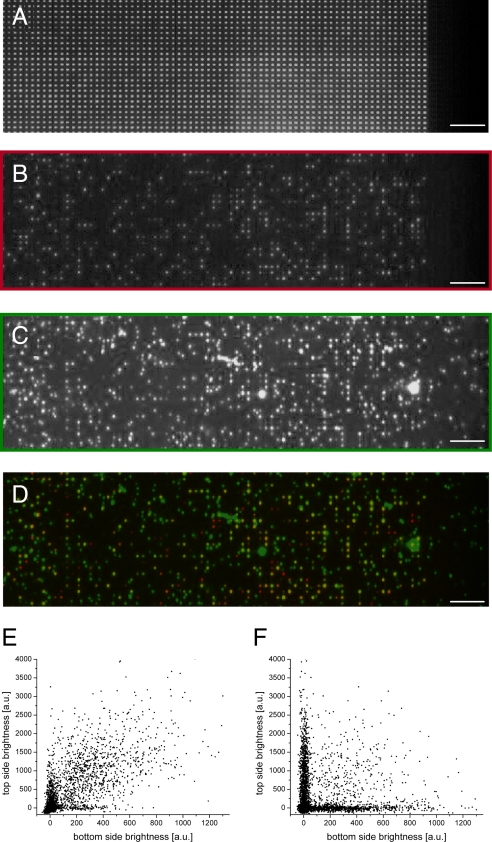
The colocalization approach successfully identified polymerase molecules that directed efficient DNA synthesis in a ZMW (Fig. 3). A section of a high-density array, containing 2,000 ZMWs (1.6-μm ZMW spacing) is shown by transmitted light microscopy to illustrate nanostructure uniformity (Fig. 3A). Under the experimental conditions used in this study, a DNA polymerase/template complex concentration of 35 nM yielded an average of approximately one polymerase per ZMW (see Materials and Methods). Following DNA synthesis, epifluorescence microscopy of the bottom side (Fig. 3B) showed Alexa Fluor 488-labeled DNA products as bright spots in some ZMWs, whereas ZMWs not containing DNA remained dark. The corresponding top side (Fig. 3C) exhibited signal from ZMW locations as well as from DNA not localized to ZMW positions. The density of non-ZMW-localized DNA, indicating polymerases immobilized on aluminum, was consistent with the density observed on blank aluminum surfaces treated with PVPA (0.12/μm2, Table 1 and SI Fig. 7).
Fig. 3.
DNA synthesis in PVPA-passivated ZMW arrays. (A) Section of a ZMW array (2,000 ZMWs shown) viewed by transmitted light microscopy to show ZMW locations. (B and C) Fluorescence microscopy image of the ZMW array viewed from the bottom (B) and the corresponding top side (C). (D) Colocalization image of superimposed bottom (false colored red) and top sides (green). (Scale bars, 10 μm.) (E) Scatter plot of bottom- and top-side fluorescence brightness intensities. Points at the origin correspond to empty ZMWs, and points away from both axes are colocalized DNA signals. (F) The strong colocalization disappears by intentionally randomizing bottom- and top-side data point pairs.
Table 1.
PVPA-mediated bias of DNA polymerase adsorption to the fused silica ZMW floor
| Variable | Value |
|---|---|
| Total no. of ZMWs | 34,835 |
| No. of polymerases in ZMWs (SiO2)* | 9,341 |
| ZMW bottom (SiO2) area, μm2 | 175 |
| Polymerase density on SiO2, μm−2 | 53 |
| No. of polymerases on Al† | 10,763 |
| Al area, μm2 | 88,344 |
| Polymerase density on Al, μm−2 | 0.12 |
| Immobilization bias (SiO2/Al) | 438 |
ZMW arrays were analyzed for localizations of polymerase-mediated DNA synthesis products, as shown in Fig. 3. DNA signals were corrected for the relative SiO2 and aluminum areas on the ZMW array to determine polymerase densities on the two different surface materials. Immobilization bias was defined as the ratio between polymerase densities on SiO2 over aluminum. The standard deviation on the obtained average bias was 150 (n = 8 chips).
*DNA signal from bottom-side images.
†DNA not located on ZMW locations, plus top-side ZMW-localized DNA without bottom-side signal.
False-colored colocalization of the bottom (red) and top (green) images showed a high degree of colocalization (yellow spots), consistent with polymerase attachment to the ZMW floor and production of long DNA emanating through the confined ZMW volume into the top-side solution (Fig. 3D). Brownian motion of the fluorescently labeled DNA, tethered in this way to the ZMW floor by the polymerase, was observed from the top side (SI Movie 1). Of bottom-side detected DNA, 66 ± 7% showed a corresponding fluorescence signal on the top side (n = 8 chips). DNA molecules detected from the bottom but lacking top-side signal (red spots), were either due to polymerase stalling during the extension reaction or to possible release of the DNA strand after the first bottom-side imaging step (27). ZMW-localized molecules without a bottom-side signal (green spots at ZMW locations) were attributed to aluminum surface, top-side attachments within the optical resolution of the ZMW location or ZMW side wall attachments. They were present at a similar density observed for PVPA-treated blank aluminum control surfaces (0.09 ± 0.03/μm2), ruling out additional top side signal contributions from the presence of ZMWs. Control experiments omitting essential components of the reaction (dNTPs, polymerase, or DNA template) exhibited no fluorescence signals on either side of the arrays (data not shown).
The level of colocalization was analyzed further by plotting all ZMW-localized DNA molecules in a scatter plot of bottom- vs. top-side fluorescence intensities (Fig. 3E). The population near the origin corresponds to empty ZMWs. Active polymerases attached to the ZMW floor contribute to the population located off of either axis around the 45° diagonal, representing a colocalized DNA signal. As a control, the colocalization signal disappeared upon intentionally randomizing the assignments of top- and bottom-side fluorescence-intensity data pairs (Fig. 3F). The observed level of colocalization was 10 standard deviations above values expected from random top- and bottom-side distributions (SI Table 2).
From the colocalization images, the chemical bias for DNA synthesis toward the fused-silica ZMW floor over the aluminum ZMW side wall and top surfaces was derived (Table 1). DNA detected from the two sides of the array was corrected for the different surface areas of the two materials on the ZMW array. An average bias of DNA synthesis density of >400:1 on SiO2 over aluminum was obtained. Without the phosphonate treatment, a dense, highly fluorescent DNA layer was formed on the top surface, ruling out any colocalization analysis. Thus, PVPA provides sufficient protection against nonspecific adsorption of DNA polymerase to aluminum to impart a strong bias of protein binding to the ZMW floor.
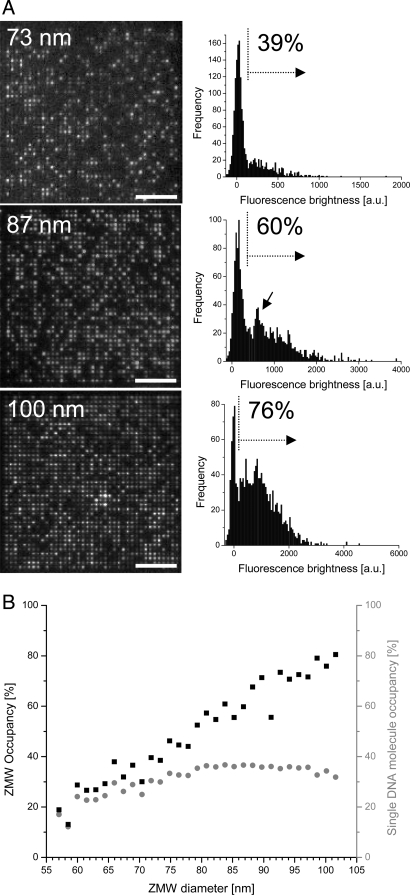
DNA polymerase loading into ZMWs was investigated as a function of ZMW diameter by analyzing fluorescence images of the bottom side of the arrays, with three representative examples shown in Fig. 4A. Histogram analysis of the integrated fluorescence brightness of each ZMW gave rise to a narrow peak around zero, corresponding to unoccupied ZMWs (Fig. 4A Right). The variable fraction of ZMWs with fluorescence intensities beyond this background contained one or more DNA molecules. Consistent with the appearance of discrete levels of fluorescence brightness in the images, an additional peak could sometimes be discerned, particularly in cases of ≈60% occupancy, indicating occupancy levels of one DNA molecule per ZMW (e.g., Fig. 4A Middle Right, arrow). The magnitudes of these populations agreed well with expectations based on Poisson-distributed deposition statistics (see SI Text). It is worth noting that the existence of these modulations is further proof that the polymerases responsible for DNA synthesis are immobilized exclusively on the ZMW floor. Increased DNA polymerase loading was observed with larger ZMW diameters (Fig. 4B, black squares). The fraction of single DNA polymerase molecular occupancies was derived from these data by using Poisson statistics (Fig. 4B, gray circles). Single active polymerase loading to ≈30% yield was achieved over a wide range of ZMW diameters (70–100 nm).
Fig. 4.
DNA polymerase loading into ZMW arrays. (A) (Left) DNA synthesis (30-min extension time) in ZMW arrays of different diameters imaged from the bottom (SiO2) side (1,521 ZMWs; 1.1-μm spacing; average diameters are indicated). (Scale bars, 10 μm.) (Right) Corresponding histograms of integrated fluorescence brightness from each ZMW. The peak around zero corresponds to empty ZMWs; brightness values beyond the zero peak (dotted vertical lines) are DNA-containing ZMWs from which ZMW occupancy fractions were derived. (Middle Right) The arrow marks the population with single molecular occupancy. (B) Polymerase occupancy of ZMW arrays as a function of ZMW diameter (black squares). The gray circles indicate single molecular occupancy fractions assuming Poisson-distributed depositions (see SI Text).
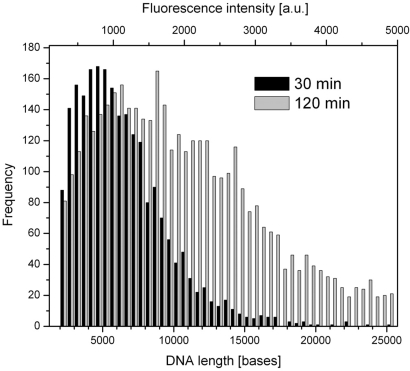
The length of DNA synthesized in the ZMW was determined by comparison with the fluorescence brightness of DNA-length standards (Fig. 5). The specific sequence design of the circular template used in this study (Fig. 2A), with a singular guanine site in the circular DNA template, allowed 100% replacement of dCTP by Alexa Fluor 488-dCTP, generating a product strand containing one fluorophore at regular length intervals of 72 bases. Conversion of fluorescence brightness to DNA length is described in detail in SI Text. Briefly, DNA extension reactions were carried out in free solution. The samples were split and analyzed (i) for length of DNA product by agarose gel electrophoresis analysis by using DNA length markers as standards and (ii) for fluorescence brightness by wide-field microscopy on PVPA-treated aluminum surfaces by using the same instrument settings applied to ZMW chips (SI Figs. 8 and 9). The resulting standard curve was applied to ZMW colocalized DNA signals in top-side images, as shown by example in Fig. 3 (yellow spots), with an associated average error of DNA-length assignment from fluorescence intensities of ±500 bases (95% confidence interval). The polymerase concentration was reduced 3-fold to yield lower total occupancies (average of ≈25%) to minimize contributions from multiply occupied ZMWs (SI Table 2 and SI Fig. 10). The histogram of DNA lengths synthesized in ZMWs (100-nm average diameter) shows that each polymerase synthesized several kilobases of DNA (Fig. 5). DNA synthesis rates were consistent with bulk measurements (average size of ≈5 kb in 30 min, SI Fig. 8), implying that the enzyme remained active for the duration of the DNA extension period. Average DNA lengths were increased beyond 10 kb by lengthening the reaction time to 120 min (Fig. 5). The measured DNA lengths represent a lower estimate, because this method detects fluorescence of only the DNA portion emanating into the top solution and does not account for DNA inside the ZMW volume.
Fig. 5.
Length of DNA synthesis in ZMWs. The top x axis shows integrated fluorescence intensities from top-side images of ZMW colocalized products after 30 and 120 min of DNA extension (100-nm average ZMW diameter, ≈25% total ZMW occupancy). Intensities were converted to DNA size (bottom x axis) by generating a standard curve using known DNA length samples with the same template design described in Fig. 2A (SI Figs. 8 and 9).
Discussion
Prior studies using ZMWs have relied on random, nonselective modes of enzyme immobilization across the entire nanostructure surface. Here, we have demonstrated selective immobilization of proteins at the bottom of high-density ZMW arrays. Polyphosphonate chemistry was found to provide excellent polymerase physisorption bias toward the fused silica ZMW floor over the metal oxide surface, in excess of 400:1. Such high levels of surface selectivity ensure that the proteins are located exclusively inside the ZMW observation volume, providing the highest signal-to-noise ratios for fluorescence events at the active site and eliminating signal variability due to polymerase position in the ZMW. The phosphonate coatings also provided low nonspecific adsorption of fluorescently labeled nucleotides to aluminum surfaces, to levels similar to fused silica or polyacrylic acid terminated polyelectrolyte multilayers (data not shown and ref. 28).
This surface-coating procedure is brief in comparison with self-assembled monolayer (SAM) phosphonate protocols. Similar to nonpolymeric phosphonates, initial electrostatic interactions in solution are likely stabilized in the subsequent dehydration step by the formation of covalent aluminophosphonate bonds (Al-O-P), which are hydrolytically more stable than Si-O-P bonds (23, 24, 29). To date, phosphonate derivatizations of metal oxide surfaces have predominantly used monophosphonate-terminated alkanes to form SAMs (24, 30–33) or complex polyethylenglycol–polyalkylphosphonate graft copolymers (34, 35). To our knowledge, simple aliphatic polymers containing many phosphonate side groups have not previously been described for protein antifouling applications on metal oxide surfaces. As both electrostatic and hydrophobic interactions mediate protein adsorption, PVPA passivation efficacies, and thus the resulting immobilization bias, will likely depend on the particular protein under investigation. The presented passivation results for neutravidin, in addition to the specific example of φ29 DNA polymerase, point toward possible generalizations to many enzymes immobilized via avidin-biotin coupling. It would also provide a specific protein-immobilization scheme to circumvent potential artifacts in biochemical functions that could arise from nonspecific, physisorbed immobilization strategies such as used here.
For applications of ZMW arrays in conjunction with DNA polymerase for real-time, single-molecule DNA sequencing, the polymerase must remain active and synthesize long DNA in the confined space of the ZMW. Thousands of successive base incorporations by individual φ29 DNA polymerase molecules were obtained by using this selective protein immobilization method, demonstrating that the enzymatic activity was not inhibited substantially by the phosphonate surface coating or the ZMW geometrical confinement. The observed DNA sizes translate directly to projected DNA sequence read lengths (3). DNA polymerase from bacteriophage φ29 was chosen because of its intrinsically high processivity, with a single polymerase/DNA template complex capable of synthesizing DNA tens of kilobases in length (36). In this study, the observed DNA lengths were largely restricted by the applied reaction times. Therefore, longer potential read lengths to the limit of enzymatic processivity levels are anticipated.
Materials and Methods
Surface Passivation.
Patterned mixed material and ZMW array chip fabrication is described in SI Text. Surfaces were derivatized by thermal deposition of PVPA, adapted from printing plate manufacture processes (Diversitec). Chips were cleaned by successive acetone and isopropanol rinses, dried with a nitrogen stream, and subjected to an oxygen plasma (Harrick Plasma). The chips were immersed in preheated 2% (vol/vol) aqueous solution of PVPA (molecular weight = 24,000) (Polysciences) for 2 min at 90°C. They were rinsed briefly with HPLC-grade water, dried with nitrogen, and annealed in a dry oven at 80°C for 10 min.
In some cases, an electrochemical deposition was used with equivalent results. In this technique, a standard 250-ml, three-electrode electrochemical cell was used, consisting of a saturated calomel reference electrode, a graphite counter electrode, and a ZMW working electrode immersed in 2% aqueous PVPA solution. The potential was controlled by a Gamry Instruments series G300 Potentiostat/Galvanostat/ZRA. PVPA was deposited either by cyclic polarization or a 200-s potentiostatic pulse to a final anodic voltage of 2 V. A copolymer of PVPA and polyacrylic acid (Rhodia-Novecare) gave similar results (data not shown).
Neutravidin Derivatization and Detection.
The top chip surface was incubated in a humid chamber for 15 min with 8 μl of 50 nM neutravidin (Pierce) in a buffer containing 50 mM MOPS, pH 6.5, 75 mM potassium acetate, and 5 mM DTT (buffer A) and rinsed with HPLC-grade water. Neutravidin binding was detected by 15 min incubation with 0.01% 40-nm biotinylated latex beads (Invitrogen) in the same buffer, rinsing with water and drying with nitrogen. Fluorescent beads were used instead of bare dyes to maximize fluorescence signals and to avoid effects of fluorescence quenching by close proximity to the aluminum surface. Detection was carried out by using a Typhoon scanner (GE Healthcare) and a wide-field fluorescence microscope (see below; Olympus).
DNA Synthesis.
φ29 DNA polymerase and circular, primed DNA template were prepared and purified as described in SI Text. φ29 DNA polymerase (35 nM) was bound to 100 nM DNA template on ice for 10 min in buffer A. PVPA-passivated chips were incubated on ice for 15 min with the prebound polymerase/DNA template complex and rinsed 3× with ice-cold buffer A to remove unbound polymerase, followed by incubation with a solution containing 10 μM each of dATP, ChromaTide Alexa Fluor 488–7-OBEA-dCTP (Invitrogen), dGTP, and dTTP, 0.7 mM MnCl2, and 20 mM ammonium acetate in buffer A. DNA synthesis reactions with incorporation of the fluorescently labeled dCTP were carried out in the dark at room temperature (23°C) for 30 min (unless indicated otherwise), followed by 5× rinsing in buffer A. For indirect DNA staining (SI Fig. 7), Alexa Fluor 488-dCTP was replaced by dCTP, with an additional incubation step using SybrGold DNA stain at 1:102 dilution in buffer A for 10 min at room temperature, followed by 5× washing in buffer A (Invitrogen).
Image Acquisition and Analysis.
Chips were imaged from both sides by using a wide-field fluorescence microscope equipped with a mercury arc lamp, standard filters for Alexa Fluor 488 (488/10x, Q505LP, and two HQ510LP; Chroma), a 60 × 1.2 N.A. water immersion objective for bottom side imaging (60 × 0.9 N.A., water-immersion dip objective for top side; both from Olympus), and a Hamamatsu EM-CCD camera for detection (C9100; Hamamatsu). Images of the bottom and top sides were background subtracted and superimposed by using transmitted light images to assign ZMW positions. A mask was applied to filter any defects present on the array. Integrated fluorescence intensities were extracted from each spot by using a Gaussian fitting algorithm. The colocalization threshold for top and bottom side DNA signals was 1.5 pixels (380 nm).
Supplementary Material
ACKNOWLEDGMENTS.
We thank the entire staff at Pacific Biosciences. We also thank Prof. J. Puglisi (Stanford University, Palo Alto, CA) for many insightful comments and helpful discussions. This work was supported in part by National Human Genome Research Institute Grant R01 HG003710.
Footnotes
Conflict of interest statement: The authors are employed by Pacific Biosciences, Inc., which has carried out the research described as part of a for-profit commercial development program.
This article contains supporting information online at www.pnas.org/cgi/content/full/0710982105/DC1.
References
- 1.Craighead H. Future lab-on-a-chip technologies for interrogating individual molecules. Nature. 2006;442:387–393. doi: 10.1038/nature05061. [DOI] [PubMed] [Google Scholar]
- 2.Mannion JT, Craighead HG. Nanofluidic structures for single biomolecule fluorescent detection. Biopolymers. 2007;85:131–143. doi: 10.1002/bip.20629. [DOI] [PubMed] [Google Scholar]
- 3.Levene MJ, et al. Zero-mode waveguides for single-molecule analysis at high concentrations. Science. 2003;299:682–686. doi: 10.1126/science.1079700. [DOI] [PubMed] [Google Scholar]
- 4.Samiee KT, Foquet M, Guo L, Cox EC, Craighead HG. Lambda-repressor oligomerization kinetics at high concentrations using fluorescence correlation spectroscopy in zero-mode waveguides. Biophys J. 2005;88:2145–2153. doi: 10.1529/biophysj.104.052795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wenger J, et al. Dual-color fluorescence cross-correlation spectroscopy in a single nanoaperture: Towards rapid multicomponent screening at high concentrations. Optics Express. 2006;14:12206–12216. doi: 10.1364/oe.14.012206. [DOI] [PubMed] [Google Scholar]
- 6.Edel JB, Wu M, Baird B, Craighead HG. High spatial resolution observation of single-molecule dynamics in living cell membranes. Biophys J. 2005;88:L43–L45. doi: 10.1529/biophysj.105.061937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samiee KT, Moran-Mirabal JM, Cheung YK, Craighead HG. Zero-mode waveguides for single-molecule spectroscopy on lipid membranes. Biophys J. 2006;90:3288–3299. doi: 10.1529/biophysj.105.072819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wenger J, et al. Diffusion analysis within single nanometric apertures reveals the ultrafine cell membrane organization. Biophys J. 2007;92:913–919. doi: 10.1529/biophysj.106.096586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moran-Mirabal JM, Torres AJ, Samiee KT, Baird BA, Craighead HG. Cell investigation of nanostructures: Zero-mode waveguides for plasma membrane studies with single molecule resolution. Nanotechnology. 2007;18:195101–195607. [Google Scholar]
- 10.Wenger J, Rigneault H, Dintinger J, Marguet D, Lenne PF. Single-fluorophore diffusion in a lipid membrane over a subwavelength aperture. J Biol Phys. 2006;32:SN1–SN4. doi: 10.1007/s10867-006-2909-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danelon C, Perez JB, Santschi C, Brugger J, Vogel H. Cell membranes suspended across nanoaperture arrays. Langmuir. 2006;22:22–25. doi: 10.1021/la052387v. [DOI] [PubMed] [Google Scholar]
- 12.Fore S, Yuen Y, Hesselink L, Huser T. Pulsed-interleaved excitation FRET measurements on single duplex DNA molecules inside C-shaped nanoapertures. Nano Lett. 2007;7:1749–1756. doi: 10.1021/nl070822v. [DOI] [PubMed] [Google Scholar]
- 13.Love JC, Estroff LA, Kriebel JK, Nuzzo RG, Whitesides GM. Self-assembled monolayers of thiolates on metals as a form of nanotechnology. Chem Rev. 2005;105:1103–1169. doi: 10.1021/cr0300789. [DOI] [PubMed] [Google Scholar]
- 14.Herrwerth S, Eck W, Reinhardt S, Grunze M. Factors that determine the protein resistance of oligoether self-assembled monolayers–internal hydrophilicity, terminal hydrophilicity, and lateral packing density. J Am Chem Soc. 2003;125:9359–9366. doi: 10.1021/ja034820y. [DOI] [PubMed] [Google Scholar]
- 15.Bruckbauer A, et al. An addressable antibody nanoarray produced on a nanostructured surface. J Am Chem Soc. 2004;126:6508–6509. doi: 10.1021/ja0317426. [DOI] [PubMed] [Google Scholar]
- 16.Gardner TJ, Frisbie CD, Wrighton MS. Systems for orthogonal self-assembly of electroactive monolayers on Au and Ito–an approach to molecular electronics. J Am Chem Soc. 1995;117:6927–6933. [Google Scholar]
- 17.Liu Y, Bishop J, Williams L, Blair S, Herron J. Biosensing based upon molecular confinement in metallic nanocavity arrays. Nanotechnology. 2004;15:1368–1374. [Google Scholar]
- 18.Xia YN, Whitesides GM. Shadowed sputtering of gold on V-shaped microtrenches etched in silicon and applications in microfabrication. Adv Mat. 1996;8:765–768. [Google Scholar]
- 19.Vargel C. Corrosion of Aluminium. Amsterdam: Elsevier; 2004. [Google Scholar]
- 20.Michel R, et al. A novel approach to produce biologically relevant chemical patterns at the nanometer scale: Selective molecular assembly patterning combined with colloidal lithography. Langmuir. 2002;18:8580–8586. [Google Scholar]
- 21.Michel R, et al. Selective molecular assembly patterning: A new approach to micro- and nanochemical patterning of surfaces for biological applications. Langmuir. 2002;18:3281–3287. [Google Scholar]
- 22.Mutin PH, et al. Selective surface modification of SiO2-TiO2 supports with phosphonic acids. Chem Mater. 2004;16:5670–5675. [Google Scholar]
- 23.Ramsier RD, Henriksen PN, Gent AN. Adsorption of phosphorus-acids on alumina. Surf Sci. 1988;203:72–88. [Google Scholar]
- 24.Goetting LB, Deng T, Whitesides GM. Microcontact printing of alkanephosphonic acids on aluminum: Pattern transfer by wet chemical etching. Langmuir. 1999;15:1182–1191. [Google Scholar]
- 25.Aslan K, et al. Metal-enhanced fluorescence: An emerging tool in biotechnology. Curr Opin Biotechnol. 2005;16:55–62. doi: 10.1016/j.copbio.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hellen EH, Axelrod D. Fluorescence emission at dielectric and metal-film interfaces. J Opt Soc Am B. 1987;4:337–350. [Google Scholar]
- 27.Akerman B, Tuite E. Single- and double-strand photocleavage of DNA by YO, YOYO and TOTO. Nucleic Acids Res. 1996;24:1080–1090. doi: 10.1093/nar/24.6.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krieg A, Ruckstuhl T, Seeger S. Towards single-molecule DNA sequencing: Assays with low nonspecific adsorption. Anal Biochem. 2006;349:181–185. doi: 10.1016/j.ab.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 29.Mutin PH, Guerrero G, Vioux A. Hybrid materials from organophosphorus coupling molecules. J Mat Chem. 2005;15:3761–3768. [Google Scholar]
- 30.Gao W, Dickinson L, Grozinger C, Morin FG, Reven L. Self-assembled monolayers of alkylphosphonic acids on metal oxides. Langmuir. 1996;12:6429–6435. [Google Scholar]
- 31.Gawalt ES, Avaltroni MJ, Koch N, Schwartz J. Self-assembly and bonding of alkanephosphonic acids on the native oxide surface of titanium. Langmuir. 2001;17:5736–5738. [Google Scholar]
- 32.Allara DL, Nuzzo RG. Spontaneously organized molecular assemblies. 1. Formation, dynamics, and physical-properties of normal-alkanoic acids adsorbed from solution on an oxidized aluminum surface. Langmuir. 1985;1:45–52. [Google Scholar]
- 33.Spori DM, et al. Influence of alkyl chain length on phosphate self-assembled monolayers. Langmuir. 2007;23:8053–8060. doi: 10.1021/la700474v. [DOI] [PubMed] [Google Scholar]
- 34.Gnauck M, et al. Carboxy-terminated oligo(ethylene glycol)-alkane phosphate: Synthesis and self-assembly on titanium oxide surfaces. Langmuir. 2007;23:377–381. doi: 10.1021/la0606648. [DOI] [PubMed] [Google Scholar]
- 35.Zoulalian V, et al. Functionalization of titanium oxide surfaces by means of poly(alkyl-phosphonates). J Phys Chem B. 2006;110:25603–25605. doi: 10.1021/jp066811s. [DOI] [PubMed] [Google Scholar]
- 36.Blanco L, et al. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J Biol Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.