Abstract
Nuclear-coded valyl-tRNA synthetase (ValRS) of eukaryotes is regarded of mitochondrial origin. Complete ValRS sequences obtained by us from two amitochondriate protists, the diplomonad, Giardia lamblia and the parabasalid, Trichomonas vaginalis were of the eukaryotic type, strongly suggesting an identical history of ValRS in all eukaryotes studied so far. The findings indicate that diplomonads are secondarily amitochondriate and give further evidence for such conclusion reached recently concerning parabasalids. Together with similar findings on other amitochondriate groups (microsporidia and entamoebids), this work provides critical support for the emerging notion that no representatives of the premitochondrial stage of eukaryotic phylogenesis exist among the species living today.
Keywords: amitochondriate protists, diplomonads, parabasalia
According to the generally accepted serial endosymbiotic theory, the complex eukaryotic cell is the result of the merger of an eubacterium, probably an α-proteobacterium, into a poorly understood “host,” assumed to be related to the archaebacteria. According to this theory the eubacterium became the mitochondrion and the “host” provided the nucleocytoplasmic compartment of the eukaryotic cell (1–3). This integration was accomplished by endosymbiotic transfer of a considerable portion of the genes of the eubacterium to the genome of the “host” (1, 3, 4). However, certain unicellular organisms belonging to diverse taxonomic groups, while they display a number of characteristics of typical eukaryotic cells, lack mitochondria, as demonstrated by biochemical and morphological findings (5–7). Whenever studied, no evidence of a second, organellar genome was found in such organisms (8, 9).
Amitochondrial eukaryotes can be assigned to at least two major categories, based on the subcellular organization of their metabolism (5, 6, 10, 11). Both groups comprise of several independent taxa. Type I organisms lack any compartmentation of their core (energy) metabolism. Diplomonads (including Giardia lamblia) and entamoebids belong to this type. Although the core metabolism of microsporidia remains unknown, their morphology was taken as evidence suggesting that they also are of type I (7, 11). Type II organisms, such as parabasalids (including Trichomonas vaginalis), certain heterolobosea, some ciliates and chytrid fungi, contain a distinct, double-membrane bounded organelle, the hydrogenosome (12, 13). Although hydrogenosomal metabolism markedly differs from that of mitochondria, the two organelles share a number of functional properties (12, 13).
The existence of amitochondriate organisms challenged the notion that the mitochondrion is a sine qua non attribute of eukaryotic cells (7, 14). It also raised the question of the evolutionary origins of the amitochondriate condition. Before any molecular phylogenetic data became available, cytological considerations led to the proposal by Cavalier-Smith that diplomonads, parabasalids, and microsporidia could be primitively amitochondriate (15, 16), and to the erection of the taxon Archezoa for these organisms. These three amitochondriate lineages represented the first branches on phylogenetic trees based on rRNA (17, 18) and some protein sequences (19). This observation was regarded as compelling evidence for an early separation of these groups from the main eukaryotic lineage, preceding the acquisition of mitochondria (20). Other amitochondriate organisms, notably the type II ciliates and chytrid fungi, however, belong to lineages that consist dominantly of mitochondriate species, strongly suggesting their secondary amitochondriate nature (21). One of the goals of current research on such organisms is to obtain information, which could settle the question whether the amitochondriate condition, at least in some organisms, represents an ancestral, “premitochondrial” feature or that all these organisms descended from ancestors that once harbored mitochondria and lost secondarily the characteristic bioenergetic functions of the mitochondrion and also the organelle itself. A widely exploited approach is the search in amitochondriate species for genes that could have been part of the genome of the ancestral mitochondrion but were subsequently transferred to the nuclear genome.
Such genes have been recently detected in several amitochondriate organisms (discussed in refs. 22–25). The most compelling data concern heat shock proteins, molecular chaperones, known to play a key role in mitochondrial biogenesis. Genes for such proteins are present in the nuclear genomes of amitochondriate organisms belonging to microsporidia (26, 27), parabasalids (28–31), and entamoebids (32). These results argued strongly against an ancestral amitochondriate nature of these three groups. Of the better studied “early-branching” organisms only G. lamblia and other diplomonads remained as possible candidates for being primitively amitochondrial, seemingly in accord with widespread views that these organisms represent one of the most ancestral eukaryotic group (17, 20, 33).
Aminoacyl-tRNA synthetases are essential enzymes for protein synthesis, and are useful for tracing the early evolution of life (34). To explore the status of G. lamblia, we selected valyl-tRNA synthetase (ValRS) as a potential marker for a mitochondrial endosymbiotic event in the past. Only a single gene for ValRS is present in fungi, and the product of this gene has been shown to be functional not only in cytosol but also in mitochondria (35, 36). A single ValRS gene has been detected also in Homo sapiens with its product similarly assumed to have cytosolic and mitochondrial functions (37). A ValRS phylogeny rooted by isoleucyl-tRNA synthetases (IleRS) revealed that eukaryotic ValRS sequences form a monophyletic clade that shows the closest affinity with Gram-negative bacteria (38). On the basis of these findings Brown and Doolittle (38) concluded that the gene coding for this protein in mitochondrion-containing eukaryotes has been transferred to the nucleus from the ancestral mitochondrion (38, 39). Thus the presence or absence of typical eukaryotic ValRS in amitochondriate protists could provide an important clue to determine whether any given organism did harbor mitochondria in its evolutionary past.
We sequenced the complete gene encoding ValRS from G. lamblia and also from T. vaginalis and aligned the deduced amino acid sequences with all ValRS and IleRS sequences available in the databases.The evidence obtained shows that G. lamblia represents no exception and also carries the relics of a past mitochondrial endosymbiotic event. This conclusion is in accord with earlier data pointing in the same direction (40–42). The very recent demonstration that this species contains and expresses a chaperonin 60 gene closely related to the mitochondrial clade is in good agreement with our results (43). Our data also confirm earlier conclusions based on a partial ValRS sequence from T. vaginalis (38) and provide additional evidence for the postmitochondrial nature of this species.
MATERIALS AND METHODS
Preparation of Genomic DNA and Genomic Gene Libraries.
G. lamblia, strain WB, clone 6 (ATCC 300957) and T. vaginalis, strain C-1:NIH (ATCC 30001) were used. Genomic DNA was extracted from G. lamblia with the use of a Blood & Culture DNA kit (Qiagen, Chatsworth, CA) according to the manufacturer’s protocol and from T. vaginalis according to a protocol for nuclease-rich protozoa (44). A ZAPII library of G. lamblia was kindly provided by F. D. Gillin and S. A. Aley (University of California, San Diego). A ZAPII and a ZAP Express libraries of T. vaginalis were kindly provided by P. J. Johnson (University of California, Los Angeles, Medical School) and J. M. Logsdon (Dalhousie University, Halifax, NS, Canada), respectively. These libraries were prepared from the same strains used to prepare genomic DNA.
Cloning and Sequencing of ValRS Genes.
Two sets of degenerate primers were synthesized based on highly conserved regions of the ValRS sequences: PPPNVTG (sense) and CGTDALRF (anti-sense); and CISRQLWWG (sense) and FCNK(L/I)W(N/Q)A (anti-sense). Corresponding fragments were amplified by PCR with the two sets of primers and genomic DNA of G. lamblia or T. vaginalis. The fragments were purified by using a Gel Extraction kit (Qiagen) and cloned into pT7 vector (Novagen). The excised inserts were labeled with the use of a Random Primers DNA Labeling System (GIBCO/BRL) and were used to probe genomic DNA libraries and Southern blots. The sequences of the isolated clones were determined for both strands with the use of an automated DNA sequencer (Applied Biosystems model 377, Perkin–Elmer/Cetus) by primer walking.
Sequence Alignment.
For a preliminary sequence alignment of the ValRS/IleRS proteins, we used the sam program that applies a linear hidden Markov model to facilitate recognition of conserved subdomains within a protein family (45). The alignment was further improved by visual editing. Sequences included in the alignment are listed below with their database accession numbers except for those obtained from genome project databases. [ValRS]: Homo sapiens (M98326), Fugu rubripes (X91856), Saccharomyces cerevisiae (J02719), Neurospora crassa (M64703), Arabidopsis thaliana (U89986), T. vaginalis (this work), G. lamblia (this work), Escherichia coli (X05891), Thermus thermophilus (Y10900), Bacillus subtilis (X77239), Bacillus stearothermophilus (M16318), Lactobacillus casei (L08854); [IleRS]: H. sapiens (D28473), Caenorhabditis elegans (Z81038), S. cerevisiae (X07886), Schizosaccharomyces pombe (AB004538), Tetrahymena thermophila (M30942), Nosema locustae (L37097), Mycobacterium tuberculosis (Z74020), Staphylococcus aureus (plasmid) (X75439), Sulfolobus acidocaldarius (L37106), Methanobacterium thermoautotrophicum (M59245), Pyrococcus furiosus (L37105), E. coli (X00776), Campylobacter jejuni (U15295), Pseudomonas fluorescens (X80132), Aquifex pyrophilus (L37096), S. aureus (X74259), Thermotoga maritima (L37104),and S. cerevisiae (mitochondrion) (L38957).
For the following bacterial species, the ValRS and/or IleRS sequences were obtained from the Institute for Genomic Research Microbial Databases (http://www.tigr.org/tdb/): Haemophilus influenzae, Helicobacter pylori, Synechocystis sp., Chlamydia trachomatis, Borrelia burgdorferi, B. subtilis (IleRS), Mycoplasma genitalium, Mycoplasma pneumoniae, Archaeoglobus fulgidus, Methanobacterium thermoautotrophicum (ValRS), and Methanococcus jannaschii.
Phylogenetic Analysis.
The maximum likelihood (ML) method of protein phylogeny (46) was used to infer phylogenetic relationships among related sequences. The analysis was performed with the protml 2.3 program (47). The Jones-Taylor-Thornton (JTT) model was assumed for amino acid substitutions (48). The actual amino acid frequencies (“F” option) of the protein under analysis were used as equilibrium frequencies (JTT-F model). Because the number of operational taxonomic units to be analyzed was very large, we first constructed a neighbor-joining tree (49) using a distance matrix estimated by the ML method. The neighbor-joining tree thus obtained was further analyzed by the local rearrangement method of the ML analysis. The quartet puzzling (QP) method in the puzzle3.1 program (50) was also used to infer a tree topology, in which multifurcations are used for ambiguous branchings. The JTT-F + γ model of amino acid substitutions (48, 50) was assumed in the analysis. Rate heterogeneity among sites was approximated by the discrete gamma distribution (with four categories), and the ML estimate of its shape parameter (α) was calculated by using the puzzle program. On the basis of the alignment constructed, we selected 555 and 320 positions, respectively, for independently analyzing the ValRS and IleRS data sets, and 270 positions for the ValRS/IleRS composite data set. Preliminary evaluation with the puzzle program demonstrated that amino acid composition of the ValRS from Th. thermophilus, M. genitalium, and M. pneumoniae are highly and significantly different from the frequency distribution of the ValRS data set (P < 0.001, χ2 test), and that IleRS sequences from M. genitalium and M. pneumoniae also show significantly different amino acid compositions (P < 0.01, χ2 test). Because the inclusion of these extremely biased data was very likely to violate the result of phylogenetic placings especially among eubacteria, we excluded these five sequences from subsequent analysis.
RESULTS
Cloning and Sequencing of ValRS Genes.
With the use of two pairs of degenerate primers we amplified ≈1.9- and 0.7-kb fragments with G. lamblia and T. vaginalis genomic DNAs as template. The fragments were purified, cloned, sequenced, and used to screen genomic DNA libraries. Using additional probes based on information obtained by sequence analysis, we finally isolated four positive clones for G. lamblia and five clones for T. vaginalis. Nucleotide sequences of both the G. lamblia and T. vaginalis ValRS genes determined from overlapping clones contained putative ORFs not interrupted by intron-like sequences, which coded for proteins 1,218 and 1,003 amino acids in length, respectively. Three different but highly similar sequences were found in T. vaginalis, showing <2% nucleotide differences within the ORFs. Genomic Southern hybridization revealed that the gene is present in a single copy in G. lamblia and that T. vaginalis contains at least three copies of the gene (data not shown).
Comparison of ValRS Sequences.
The alignment (available on request from T.H.) revealed a high overall identity (32–89% for common positions) among all available ValRS sequences. The lengths of these proteins varied from 837 to 1,263 residues with the differences located primarily around the N-terminal portion of the eukaryotic sequences. The G. lamblia sequence contained several insertions compared with all other sequences. Two remarkable insertions consisting of ≈60 amino acids were found around the positions 270–330 and 802–865 on the G. lamblia sequence. The G. lamblia and T. vaginalis sequences contained the conserved KMSKS and HIGH peptide motives characteristic of all class I aminoacyl-tRNA synthetases, regardless of their amino acid specificity (51). A major sequence signature differentiated the eukaryotic and γ-proteobacterial ValRS sequences from all others. In the region downstream of the KMSKS motif ValRS sequences from eukaryotes (cytosolic/mitochondrial) and γ-proteobacteria contained a 37-residue insertion (Fig. 1). The insertion in these shows a 22–73% amino acid identity and is clearly homologous. No similarly long insertion has been seen in any IleRS sequence, although the S. cerevisiae mitochondrial sequence has an eight residue insertion in the same place.
Figure 1.
A 37-residue insertion present only in eukaryotic (cytosolic, mitochondrial) and γ-proteobacterial ValRS. The KMSKS motif is boxed by a dashed line. The 37-residue sequences of the amitochondriate protists are shadowed. Numbers on the top of the alignment indicate sequence positions of H. sapiens ValRS. Abbreviations of the species: Hs, H. sapiens; At, A. thaliana; Tv, T. vaginalis; Gl, G. lamblia; Sc, S. cerevisiae; Nc, N. crassa; Ec, E. coli; Hi, Ha. influenzae; Hp, He. pylori; Ss, Synechococcus sp.; Tht, The. thermophilus; Ct, C. trachomatis; Bb, Bo. burgdorferi; Bs, B. subtilis; Mg, M. genitalium; Af, Ar. fulgidus; Mj, Me. jannaschii; Tet, Te. thermophila; Sa, St. aureus. Asterisks (★) denote amitochondriate eukaryotes. mit, Mitochondrion; cyt, cytosol; plasmid, gene carried on a plasmid.
Phylogenetic Relationships Among All the ValRS/IleRS Sequences.
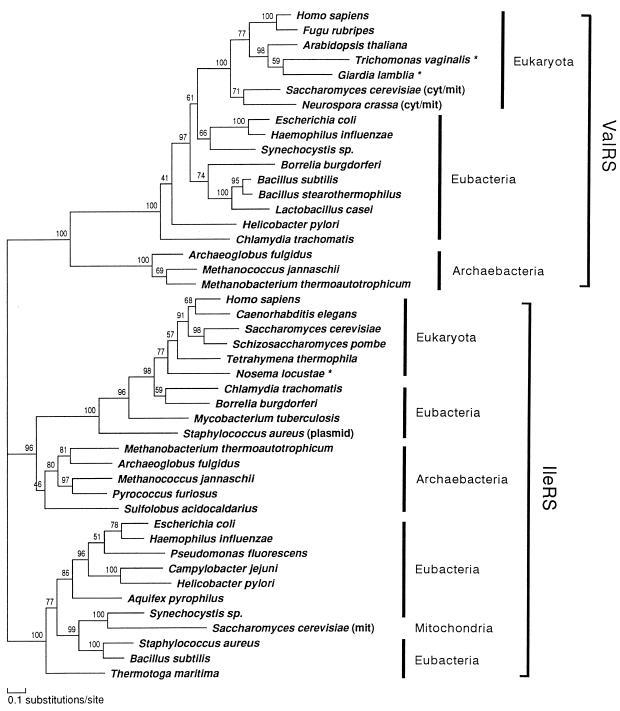
We first inferred a composite tree of ValRS/IleRS by the ML method of protein phylogeny using protml program. The final tree (ProtML tree) generated by the local rearrangement method of the ML analysis (Fig. 2) placed the gene families for ValRS and IleRS into separate and statistically supported monophyletic clades. Each of the two clades could be used for mutually rooting the other one, placing the root of the tree between the ValRS and IleRS branches (38). The ML estimate of the gamma shape parameter (α) for the ValRS/IleRS data set was 1.14, suggesting a weak rate heterogeniety in this data set. The QP topology obtained by the puzzle program did not resolve clearly the relationships among groups within the eubacterial and eukaryotic domains in either the ValRS or the IleRS portions (data not shown).
Figure 2.
Composite tree of ValRS and IleRS using the ML method of protein phylogeny (JTT-F model). A ProtML tree finally selected by the local rearrangement method of the protml program was shown. The horizontal length of each branch is proportional to the estimated number of substitutions. Local bootstrap probabilities calculated by the protml program are attached to the internal branches. Asterisks (★) denote amitochondriate eukaryotes. cyt, Cytosol; mit, mitochondrion; plasmid, gene carried on a plasmid. Unambiguously aligned 270 positions were selected and used. These correspond to the following positions in the ValRS sequence of G. lamblia: 142–143, 145–153, 171–253, 409–438, 469–487, 491–493, 497–525, 554–581, 624–642, 657–666, 669–683, and 686–708.
In the IleRS portion of the tree, the archaebacterial sequences clearly clustered with the subtree that contains all of the eukaryotic and four of the peculiar eubacterial ones with most of the eubacterial sequences constituting the outgroup. Within the subtree the common ancestor of C. trachomatis and Bo. burgdorferi IleRS appeared as a sister group of eukaryotic IleRS with a high local bootstrap support, while the other two eubacterial sequences (My. tuberculosis IleRS and St. aureus plasmid-coded IleRS) also branched deeply from the line leading to eukaryotes, and were rather divergent from most eubacterial and mitochondrial sequences. The existence of eukaryotic type IleRS genes in some eubacteria suggests the possibility of lateral gene transfer events from eukaryotes to eubacteria (52).
Rather than being the deepest emerging eubacterial branch as in rRNA trees (53, 54), IleRS of the hyperthermophile, Aq. pyrophilus, appeared to be specifically allied with the proteobacterial sequences. IleRS of another hyperthermophile, Th. maritima, represented the outgroup to all eubacterial sequences, being consistent with the rRNA phylogeny, but this placement was not highly supported by the local bootstrap value. Inclusion of the Mycoplasma IleRS data, which showed significant biases of amino acid composition (see Materials and Methods), placed Mycoplasma as a sister group to the common ancestor of Gram-positive bacteria, and slightly changed the branchings among proteobacteria, but the positions of the two hyperthermophiles were robust (data not shown).
In contrast to the relationships of the IleRS sequences, ValRS of archaebactera formed a monophyletic clade and its common ancestor deeply branched among all the other ValRS sequences, followed by the branching of the eubacterial sequences. The tree topology demonstrated with a high local bootstrap value that all eukaryotic ValRS share a most recent common ancestor, and that this monophyletic group is located within the eubacterial ValRS clade. Relationships among the groups in the eubacterial ValRS were poorly resolved, but the γ-proteobacteria and the cyanobacterium Synechocystis sp. appeared to show the closest affinity to the eukaryotic homologs. This phylogenetic position coupled with the mitochondrial function of the fungal homologs (35, 36) suggests that this gene most likely entered eukaryotes with the proteobacterial endosymbiont that gave rise to mitochondria.
The robust placement of ValRS of the two amitochondriate protists studied, G. lamblia and T. vaginalis, within the eukaryotic clade indicates that these genes shared their past history with ValRS genes of other eukaryotes, a conclusion in good agreement with the shared insertion mentioned above. The data are in agreement with conclusions based on a partial ValRS sequence from T. vaginalis (38).
Phylogenetic Relationships Within the ValRS Portion.
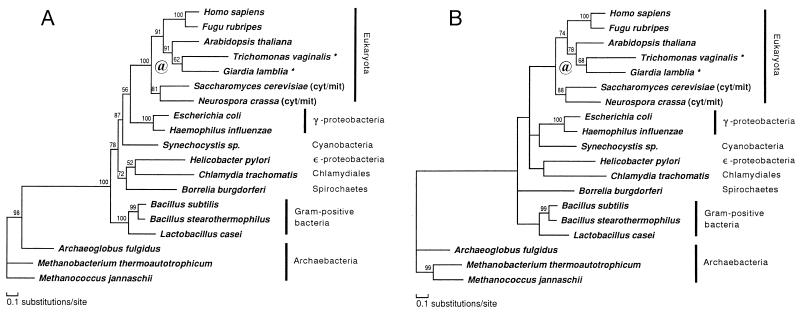
Relationships among the ValRS genes were further analyzed after the exclusion of the IleRS sequences. The ProtML tree based on a much larger number of common positions (Fig. 3A) changed the placements of Synechocystis sp., Bo. burgdorferi, He. pylori, and C. trachomatis, compared with the previous composite tree. In contrast to the composite tree in Fig. 2, only γ-proteobacteria represented the closest relative of eukaryotes excluding Synechocystis sp., further suggesting the proteobacterial origin of eukaryotic ValRS, but with low local bootstrap support. The placement of He. pylori (ɛ-proteobacteria) as a sister group of C. trachomatis (Chlamidiales) was quite anomalous, although it was poorly supported by the local bootstrap value.
Figure 3.
ValRS trees obtained with the ML method of protein phylogeny (JTT-F model). See also the legend of Fig. 2. Asterisks (★) denote amitochondriate eukaryotes. @, Putative gene duplication (see Results). Unambiguously aligned 555 positions were selected and used. These correspond to the following positions in the ValRS sequence of G. lamblia: 61–82, 97–143, 145–153, 166–253, 358–393, 402–406, 409–452, 469–531, 554–581, 624–645, 657–683, 686–720, 760–801, 892–940, 968–990, 1002–1009, and 1067–1073. (A) A ProtML tree finally selected by the local rearrangement method of the protml program. (B) A ProtML tree estimated on the basis of the QP topology.
The gamma shape parameter (α) for the ValRS data set was 0.66, suggesting that rate variation among sites is extreme. On the basis of the QP topology obtained branch lengths were estimated by the ML method and the tree was shown in Fig. 3B. The relationship among eukaryotes was the same as that in the ProtML tree shown in Fig. 3A, but the placements of Synechocystis and Borrelia were slightly changed. The QP topology indicates that the relationships are still uncertain among groups within eubacteria.
Two aspects of the eukaryotic ValRS part of both trees in Figs. 2 and 3 are discordant with accepted taxonomic relationships. The sistergroup relationship of the two amitochondriate species and their sharing a common ancestor with a plant are probably due to insufficient species sampling. The data place fungi as an outgroup of all other eukaryotes, a result in disagreement with the accepted phylogeny based on extensive molecular information (55–58). It is possible that the fungal genes are paralogs deriving from gene duplication, probably after the emergence of the eukaryotes (Fig. 3, @). By fixing the relationship among eubacteria and archaebacteria shown in Fig. 3B and by using this relationship as an outgroup, branching order among five eukaryotic lineages (G. lamblia, T. vaginalis, A. thaliana, fungi, and animals) was further examined by the ML method, using an exhaustive tree search for possible 105 topologies. The same tree was obtained as the ML tree, but 25 alternative trees could not be excluded by the criterion of 2SE of log-likelihood difference. In most of these trees fungi represent the deepest branching among eukaryotes, and the total of bootstrap probabilities by the resampling estimated log-likelihood method (59) for the trees in which fungi are the outgroup to other eukaryotes amounts to 76%. However, we cannot exclude the possibility for the accepted phylogeny that fungi are the closest relative of animals, because the sum of bootstrap probabilities for these trees was 23%.
DISCUSSION
Sequence comparisons and phylogenetic reconstructions of ValRSs revealed that this enzyme in two amitochondriate protists, G. lamblia and T. vaginalis belongs to a clade that encompasses all ValRSs of eukaryotic cells. The closest outgroup of this clade consists of γ-proteobacteria (38). This is in contrast to several other aminoacyl-tRNA synthetases of the eukaryotic nucleocytoplasmic compartment, which show a closer relationship to their archaebacterial homologs than to their eubacterial ones (39, 51). The ValRS tree is regarded as evidence for an endosymbiotic, mitochondrial origin of this enzyme in all eukaryotes (38). The robust placement of ValRS of the two amitochondriate species studied by us among their eukaryotic homologs and the signature insertion shared by these genes reveals a shared history of ValRS in all eukaryotes, to wit that both G. lamblia and T.vaginalis experienced a mitochondrial endosymbiotic event in their past. This finding supports the notion that G. lamblia, thus the diplomonads will have to be removed from the ever dwindling group of putatively ancestral amitochondrial lineages, as also shown by recent data on chaperonin 60 (43). The case of T. vaginalis already has been taken as convincingly adjucated in favor of its secondary amitochondriate status (28–31) with the present data further supporting this judgment. However, we only have an evidence that the γ-proteobacterial ValRS is the closest relative of eukaryotic counterparts, thus we cannot entirely exclude a possibility of lateral transfer from a γ-proteobacterium rather than transfer from mitochondria as the source of this gene. A solving of the relationships between α-proteobacteria and eukaryotes in the ValRS phylogeny will be necessary to support or dispose the mitochondrial origin of eukaryotic ValRS.
The subsequent fate of nuclear genes acquired from a mitochondrial endosymbiosis and their products will depend on the nature of the gene and also on the organizational type of the amitochondriate lineage in question. In the case of chaperones involved in organellar biogenesis there is a marked difference between type I and type II amitochondriates, because in the former, e.g., G. lamblia, no major organelle can be recognized as the remnant of the ancestral endosymbiont, while in type II organisms, e.g., T. vaginalis, the hydrogenosome is regarded as a direct descendant of the endosymbiont (22, 24). This means that chaperones could retain their original function in organisms containing hydrogenosomes, and indeed they are localized in the organelle (28, 60). In contrast, in type I organisms no such organellar targets have been recognized yet and the role of the chaperones remains unknown (32, 43). The case of the tRNA synthetases is clearly different because in both groups of amitochondriate protists they have an exclusively cytosolic function. Other cytosolic enzymes in G. lamblia, triosephosphate isomerase (41) and glyceraldehyde-3-phosphate dehydrogenase (42), have been assumed to have an endosymbiotic, mitochondrial origin. These inferences are further supported by the present ValRS results. The proteobacterial endosymbiont may have contributed more genes functioning in the cytosol of extant eukaryotes than previously thought.
The possible endosymbiotic origin of ValRS in T. vaginalis has been inferred from a partial sequence (38), a suggestion supported by this study. The enzyme in this species is also clearly cytoplasmic, because there is no need for ValRS in hydrogenosome. It demonstrates that evidence for the ancestral endosymbiotic event can be detected among both hydrogenosomal and cytoplasmic components.
The results presented here strongly support the notion that the critical endosymbiotic event occurred before the divergence of the extant lineages of eukaryotes and make it unlikely that we will discover living descendants of a putative premitochondrial state in eukaryotic evolution. The aminoacyl-tRNA synthetase databases are still too meager to make a contribution to our understanding of the interrelationships of various protist lineages. The fact that some amitochondriate groups emerge deep in phylogenetic trees based on rRNA (18, 33) and on some proteins (19) was discussed as strong evidence for the premitochondrial nature of these lineages. With this inference dramatically weakened, the emerging view is of a eukaryotic world encompassing a number of diverging lineages, the emergence of which cannot be ordered on the basis of increasing cellular complexity. The indications are that all extant lineages separated after eukaryotic evolution attained a relatively advanced stage and that the reconstruction of early eukaryotic evolution will be a daunting task.
Acknowledgments
We thank Ms. J. A. Lee for preparing high quality gDNA from T. vaginalis; Drs. B. ter Kuile and G. Wu for technical support and discussion; and Ms. N. Arisue and Ms. A. Deguchi for technical assistance. We also thank Drs. F. D. Gillin and S. A. Aley (University of California, San Diego), P. J. Johnson (University of California, Los Angeles, Medical School) and J. M. Logsdon (Dalhousie University, Halifax, NS, Canada) for provision of the gDNA libraries. This work was carried out under the Institute of Statistical Mathematics Cooperative Research Program (96ISM CRP-A93 and 97ISM CRP-A71). T.H. and M.H. were supported by grants from the Ministry of Education, Science, Sports, and Culture of Japan. Work in the New York laboratory was supported by National Institutes of Health Grant AI11942 to M.M. This study was made possible in part by funds granted by the Norman and Rosita Winston Foundation. The statements made and views expressed, however, are solely the responsibility of the authors.
ABBREVIATIONS
- IleRS
isoleucyl-tRNA synthetase
- JTT
Jones-Taylor-Thornton
- ML
maximum likelihood
- QP
quartet puzzling
- ValRS
valyl-tRNA synthetase
Footnotes
References
- 1.Gray M W, Doolittle W F. Microbiol Rev. 1982;46:1–42. doi: 10.1128/mr.46.1.1-42.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Margulis L. Symbiosis in Cell Evolution. 2nd Ed. New York: Freeman; 1993. [Google Scholar]
- 3.Gray M W, Spencer D F. Symp Soc Gen Microbiol. 1996;54:109–126. [Google Scholar]
- 4.Martin W, Schnarrenberger C. Curr Gen. 1997;32:1–18. doi: 10.1007/s002940050241. [DOI] [PubMed] [Google Scholar]
- 5.Müller M. Annu Rev Microbiol. 1988;42:465–488. doi: 10.1146/annurev.mi.42.100188.002341. [DOI] [PubMed] [Google Scholar]
- 6.Müller M. In: Christian Gottfried Ehrenberg-Festschrift. Schlegel M, Hausmann K, editors. Leipzig: Leipziger Universit; 1996. pp. 63–76. [Google Scholar]
- 7.Cavalier-Smith T. Nature (London) 1987;326:332–333. doi: 10.1038/326332a0. [DOI] [PubMed] [Google Scholar]
- 8.Turner G, Müller M. J Parasitol. 1983;69:234–236. [PubMed] [Google Scholar]
- 9.van der Giezen M, Sjollema K A, Artz R R E, Alkema W, Prins R A. FEBS Lett. 1997;402:147–150. doi: 10.1016/s0014-5793(97)00409-2. [DOI] [PubMed] [Google Scholar]
- 10.Fenchel T, Finlay B J. Ecology and Evolution in Anoxic Worlds. Oxford: Oxford Univ. Press; 1995. [Google Scholar]
- 11.Martin W, Müller M. Nature (London) 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- 12.Müller M. J Gen Microbiol. 1993;139:2879–2889. doi: 10.1099/00221287-139-12-2879. [DOI] [PubMed] [Google Scholar]
- 13.Biagini G A, Finlay B J, Lloyd D. FEMS Microbiol Lett. 1997;155:133–140. doi: 10.1016/s0378-1097(97)00333-9. [DOI] [PubMed] [Google Scholar]
- 14.Cavalier-Smith T. Microbiol Rev. 1993;57:953–994. doi: 10.1128/mr.57.4.953-994.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cavalier-Smith T. In: Endocytobiology II. Schenk H E A, Schwemmler W, editors. Berlin: de Gruyter; 1983. pp. 265–279. [Google Scholar]
- 16.Cavalier-Smith T. In: Endocytobiology II. Schenk H E A, Schwemmler W, editors. Berlin: de Gruyter; 1983. pp. 1027–1034. [Google Scholar]
- 17.Sogin M L. Curr Opin Gen Dev. 1991;1:457–463. doi: 10.1016/s0959-437x(05)80192-3. [DOI] [PubMed] [Google Scholar]
- 18.Sogin M L, Silberman J D, Hinkle G, Morrison H G. Symp Soc Gen Microbiol. 1996;54:167–184. [Google Scholar]
- 19.Hashimoto T, Hasegawa M. Adv Biophys. 1996;32:73–120. doi: 10.1016/0065-227x(96)84742-3. [DOI] [PubMed] [Google Scholar]
- 20.Cavalier-Smith T, Chao E E. J Mol Evol. 1996;43:551–562. doi: 10.1007/BF02202103. [DOI] [PubMed] [Google Scholar]
- 21.Embley T M, Finlay B J, Dyal P L, Hirt R P, Wilkinson M, Williams A G. Proc R Soc London B. 1995;262:87–93. doi: 10.1098/rspb.1995.0180. [DOI] [PubMed] [Google Scholar]
- 22.Müller M. Parasitol Today. 1997;13:166–167. doi: 10.1016/s0169-4758(97)01036-3. [DOI] [PubMed] [Google Scholar]
- 23.Müller M. Parasitol Today. 1997;13:455–456. doi: 10.1016/s0169-4758(97)01123-x. [DOI] [PubMed] [Google Scholar]
- 24.Palmer J D. Science. 1997;275:790–791. doi: 10.1126/science.275.5301.790. [DOI] [PubMed] [Google Scholar]
- 25.Sogin M L. Curr Biol. 1997;7:R315–R317. doi: 10.1016/s0960-9822(06)00147-3. [DOI] [PubMed] [Google Scholar]
- 26.Germot A, Philippe H, Le Guyader H. Mol Biochem Parasitol. 1997;87:159–168. doi: 10.1016/s0166-6851(97)00064-9. [DOI] [PubMed] [Google Scholar]
- 27.Hirt R P, Healy B, Vossbrinck C R, Canning E U, Embley T M. Curr Biol. 1997;7:995–998. doi: 10.1016/s0960-9822(06)00420-9. [DOI] [PubMed] [Google Scholar]
- 28.Bui E T N, Bradley P J, Johnson P J. Proc Natl Acad Sci USA. 1996;93:9651–9656. doi: 10.1073/pnas.93.18.9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Germot A, Philippe H, Le Guyader H. Proc Natl Acad Sci USA. 1996;93:14614–14617. doi: 10.1073/pnas.93.25.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horner D S, Hirt R P, Klivington D, Lloyd D, Embley T M. Proc R Soc London B. 1996;263:1053–1059. doi: 10.1098/rspb.1996.0155. [DOI] [PubMed] [Google Scholar]
- 31.Roger A J, Clark C G, Doolittle W F. Proc Natl Acad Sci USA. 1996;93:14618–14622. doi: 10.1073/pnas.93.25.14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark C G, Roger A J. Proc Natl Acad Sci USA. 1995;92:6518–6521. doi: 10.1073/pnas.92.14.6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sogin M L, Gunderson J H, Elwood H J, Alonso R A, Peattie D A. Science. 1989;243:75–77. doi: 10.1126/science.2911720. [DOI] [PubMed] [Google Scholar]
- 34.Brown J R. In: Thermophiles: The Keys to Molecular Evolution and the Origin of Life? Wieger J, Adams M, editors. London: Taylor & Francis; 1998. , in press. [Google Scholar]
- 35.Chatton B, Walter P, Ebel J-P, Lacroute F, Fasiolo F. J Biol Chem. 1988;263:52–57. [PubMed] [Google Scholar]
- 36.Kubelik A R, Turcq B, Lambowitz A M. Mol Cell Biol. 1991;11:4022–4035. doi: 10.1128/mcb.11.8.4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsieh S-L, Campbell R D. Biochem J. 1991;278:809–816. doi: 10.1042/bj2780809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown J R, Doolittle W F. Proc Natl Acad Sci USA. 1995;92:2441–2445. doi: 10.1073/pnas.92.7.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown J R, Doolittle W F. Microbiol Mol Biol Rev. 1997;61:456–502. doi: 10.1128/mmbr.61.4.456-502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soltys B J, Gupta R S. J Parasitol. 1994;80:580–590. [PubMed] [Google Scholar]
- 41.Keeling P J, Doolittle R F. Proc Natl Acad Sci USA. 1997;94:1270–1275. doi: 10.1073/pnas.94.4.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henze K, Badr A, Wettern M, Cerff R, Martin W. Proc Natl Acad Sci USA. 1995;92:9122–9126. doi: 10.1073/pnas.92.20.9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roger A J, Svärd S G, Tovar J, Clark C G, Smith M W, Gillin F D, Sogin M L. Proc Natl Acad Sci USA. 1998;95:229–234. doi: 10.1073/pnas.95.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Riley D E, Krieger J N. Mol Biochem Parasitol. 1992;51:161–164. doi: 10.1016/0166-6851(92)90212-3. [DOI] [PubMed] [Google Scholar]
- 45.Hughey R, Krogh A. Comput Appl Biosci. 1996;12:95–107. doi: 10.1093/bioinformatics/12.2.95. [DOI] [PubMed] [Google Scholar]
- 46.Kishino H, Miyata T, Hasegawa M. J Mol Evol. 1990;30:151–160. doi: 10.1007/BF02109497. [DOI] [PubMed] [Google Scholar]
- 47.Adachi J, Hasegawa M. molphy Version 2.3: Programs for Molecular Phylogenetics Based on Maximum Likelihood, Computer Science Monographs, No. 28. Tokyo: Inst. Statistical Mathematics; 1996. [Google Scholar]
- 48.Jones D T, Taylor W R, Thornton J M. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 49.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 50.Strimmer K, von Haesler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 51.Moras D. Trends Biochem Sci. 1992;17:159–164. doi: 10.1016/0968-0004(92)90326-5. [DOI] [PubMed] [Google Scholar]
- 52.Shiba K, Motegi H, Schimmel P. Trends Biochem Sci. 1997;22:453–457. doi: 10.1016/s0968-0004(97)01135-3. [DOI] [PubMed] [Google Scholar]
- 53.Burggraf S, Olsen G J, Stetter K O. Syst Appl Microbiol. 1992;15:352–356. doi: 10.1016/S0723-2020(11)80207-9. [DOI] [PubMed] [Google Scholar]
- 54.Olsen G J, Woese C R, Overbeek R. J Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hasegawa M, Hashimoto T, Adachi J, Iwabe N, Miyata T. J Mol Evol. 1993;36:380–388. doi: 10.1007/BF00182185. [DOI] [PubMed] [Google Scholar]
- 56.Baldauf S L, Palmer J D. Proc Natl Acad Sci USA. 1993;90:11558–11562. doi: 10.1073/pnas.90.24.11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wainright P O, Hinkle G, Sogin M L, Stickel S K. Science. 1993;260:340–342. doi: 10.1126/science.8469985. [DOI] [PubMed] [Google Scholar]
- 58.Nikoh N, Hayase N, Iwabe N, Kuma K, Miyata T. Mol Biol Evol. 1994;11:762–768. doi: 10.1093/oxfordjournals.molbev.a040156. [DOI] [PubMed] [Google Scholar]
- 59.Hasegawa M, Kishino H. Mol Biol Evol. 1994;11:142–145. [Google Scholar]
- 60.Bozner P. J Parasitol. 1997;83:224–229. [PubMed] [Google Scholar]