Abstract
Since the discovery of γ-tubulin, attention has focused on its involvement as a microtubule nucleator at the centrosome. However, mislocalization of γ-tubulin away from the centrosome does not inhibit mitotic spindle formation in Drosophila melanogaster, suggesting that a critical function for γ-tubulin might reside elsewhere. A previous RNA interference (RNAi) screen identified five genes (Dgt2–6) required for localizing γ-tubulin to spindle microtubules. We show that the Dgt proteins interact, forming a stable complex. We find that spindle microtubule generation is substantially reduced after knockdown of each Dgt protein by RNAi. Thus, the Dgt complex that we name “augmin” functions to increase microtubule number. Reduced spindle microtubule generation after augmin RNAi, particularly in the absence of functional centrosomes, has dramatic consequences on mitotic spindle formation and function, leading to reduced kinetochore fiber formation, chromosome misalignment, and spindle bipolarity defects. We also identify a functional human homologue of Dgt6. Our results suggest that an important mitotic function for γ-tubulin may lie within the spindle, where augmin and γ-tubulin function cooperatively to amplify the number of microtubules.
Introduction
Microtubule (MT) nucleation from centrosomes requires γ-tubulin and its several associated proteins (γ-tubulin ring complex [γ-TuRC]), which are docked onto the pericentriolar material (Wiese and Zheng, 2006). Influential work by Kirschner and Mitchison (1986) suggested that MT growth and shrinkage from the centrosome allows chromosomes to capture and stabilize MTs and thus build a mitotic spindle. However, centrosome-mediated MT nucleation is dispensable for functional mitotic spindle formation (Khodjakov et al., 2000; Megraw et al., 2001) and centrosome-based “search and capture” may be insufficient to capture all the kinetochores in the rapid time frame observed in living cells (Wollman et al., 2005).
Recent observations have revealed that spindle MTs can also be generated by noncentrosomal pathways. The best studied is the chromatin-mediated pathway, in which high concentration of Ran-GTP on chromatin stimulates γ-tubulin–dependent MT nucleation proximal to chromatin in meiotic and somatic cells (Karsenti and Vernos, 2001; Gadde and Heald, 2004; Groen et al., 2004). Another recently suggested mechanism for MT generation involves nucleation from existing MTs in the spindle by docking γ-tubulin onto those MTs. This idea stemmed from observations that growing MT plus ends visualized by EB1-GFP emerge throughout the body of the spindle, not solely at centrosomes and chromosomes (Mahoney et al., 2006), and that γ-tubulin is localized throughout the mitotic spindle (Lajoie-Mazenc et al., 1994; Luders et al., 2006). Imaging of EB1-GFP also revealed that MT growth within the spindle is preferentially directed toward the chromosomes (Mahoney et al., 2006). Interestingly, directional growth of MTs from existing MTs has been observed in interphase Schizosaccharomyces pombe (Janson et al., 2005) and plant cells (Murata et al., 2005), although it is unclear whether this mechanism is the same as the one that operates within the mitotic spindle.
Using a genome-wide RNAi screen for mitotic spindle morphology in the Drosophila melanogaster S2 cell line, we identified several genes involved in γ-tubulin localization in metaphase (Goshima et al., 2007). Among them are the outer γ-TuRC subunits (Dgrip71, 75, 128, and 163) and five uncharacterized proteins (called Dgt2–6, with Dgt an acronym for dim γ-tubulin) that are necessary for localizing γ-tubulin to spindle MTs but not to the centrosomes. Dgt proteins do not have any known motifs from which their molecular activity can be predicted. In this paper, we demonstrate that the Dgt proteins form an MT binding complex involved in MT generation within the spindle and that this process plays a vital role in mitotic spindle assembly. We also identify human Dgt6, indicating that the Dgt-dependent MT generation pathway is conserved in human mitosis.
Results and discussion
The five Dgt proteins form a stable complex that interacts with MTs
We constructed S2 cell lines stably expressing HA-tagged Dgt proteins (Dgt2–6) and Dgrip71 and also generated antibodies that detect endogenous Dgt3–6 (Fig. S1, A and B, available at http://www.jcb.org/cgi/content/full/jcb.200711053/DC1). Immunoprecipitation of HA-tagged proteins followed by immunoblotting revealed that Dgt2–6 efficiently coprecipitated with each other (Fig. 1 A). The only exception was that Dgt3-HA brought down Dgt4–6 much less efficiently. However, we suspect that tagging of Dgt3 interferes with its function because endogenous Dgt3 was coprecipitated with the other HA-tagged Dgts. Immunoprecipitations, however, failed to detect interactions between the Dgts and γ-tubulin, Dgrip91 (a component of the γ-tubulin small complex [γ-TuSC] together with Dgrip84 and γ-tubulin), or Dgrip71 (Fig. 1 A), suggesting that Dgt proteins do not interact tightly with γ-TuRC.
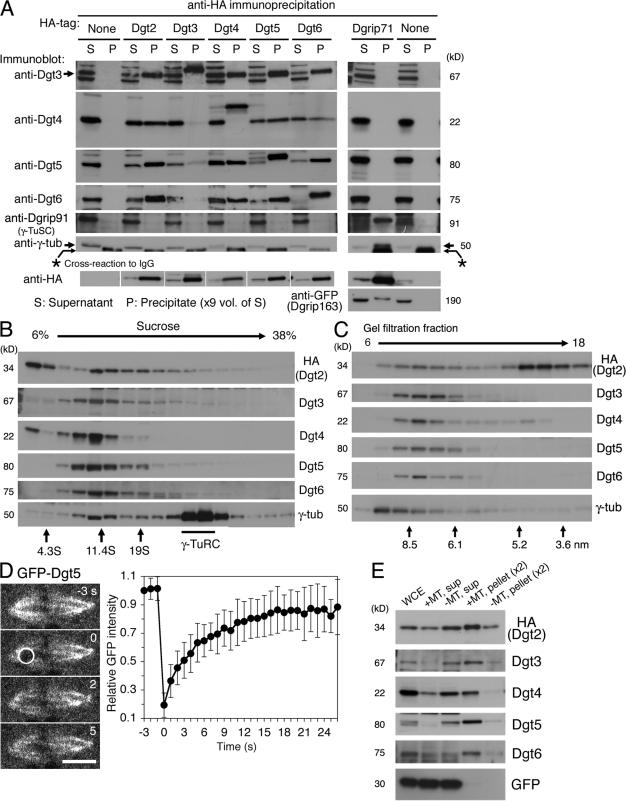
Figure 1.
Dgt2–6 proteins form a complex and associate with MTs. (A) Dgt2–6 proteins coimmunoprecipitate with one another but not with γ-TuRC subunits. Supernatant (S) and precipitated (P) fractions after extract incubation with anti-HA beads were immunoblotted for specific subunits. White lines indicate that intervening lanes have been spliced out. (B and C) Sucrose gradient sedimentation (B) and gel filtration chromatography (C) of Dgt2–6 proteins and γ-tubulin. Dgt proteins had a common peak at 11S and 7.5 nm, whereas the majority of γ-tubulin was in the larger γ-TuRC complex. The positions of standard markers are indicated. (D) FRAP analysis of GFP-Dgt5 on the spindle (n = 17). Bleached area is indicated by a white circle. Bar, 5 μm. Error bars show SD. See also Video 1 (available at http://www.jcb.org/cgi/content/full/jcb.200711053/DC1). (E) MT cosedimentation assay. Significant portions of Dgt2–6 proteins, but not control GFP, cosedimentated with MTs (pellet). WCE; whole cell extract.
Dgt2–6 proteins in S2 extracts comigrated during sucrose gradient centrifugation (11S) and gel filtration chromatography (Stokes radius = 7.5 nm; Fig. 1, B and C). γ-TuRC was detected as a much bigger complex in both assays, as previously reported (Oegema et al., 1999). We also found that reduction of one Dgt reduced the levels of other Dgts (an exception was Dgt2-HA; Fig. S1 C). Multisubunit complexes are often unstable when one component is absent (De Wulf et al., 2003), thus likely explaining this result. Knockdown of γ-TuRC components, in contrast, did not significantly affect the levels of Dgt proteins. Collectively, these experiments suggest that Dgt2–6 proteins coassemble into a stable complex. The size of the complex was estimated from the size-based fractionation (Siegel and Monty, 1966) as 340 kD, which is a little larger than that predicted from the sum of the individual molecular masses of the five proteins (265 kD).
Our previous study showed that GFP-tagged Dgt2 and 4–6 localize to spindle MTs. We confirmed spindle localization of endogenous Dgt4 by immunofluorescence using a specific polyclonal antibody (Fig. S1 D). GFP-Dgt5 signal was clearly detected on kinetochore MTs (kMTs) but was greatly reduced at the spindle equator where only non-kMTs are present (Fig. S1, E and F), suggesting that the Dgt complex may preferentially localize to stable MTs within the spindle. FRAP showed a fast turnover of GFP-Dgt5 on the metaphase spindle (t1/2 = 4 s; n = 17), suggesting a rapid binding and dissociation of the Dgt complex with MTs in vivo (Fig. 1 D; Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200711053/DC1; and Fig. S1 G for early anaphase spindles). We then tested whether the Dgt complex interacts with exogenous MTs added to S2 cell extracts and found that all of the Dgts cosedimented with taxol-stabilized MTs, whereas expressed GFP (as a control) remained in the supernatant (Fig. 1 E). However, additional work will be required to establish whether the Dgt–MT interaction is direct or mediated by other proteins.
Loss of the Dgt complex impairs MT generation within the spindle, bipolar spindle formation, and chromosome alignment
In our previous study, we reported live imaging of Dgt5-depleted cells and showed an increased number of monopolar spindles, reduced density of MTs within bipolar spindles, and chromosome misalignment (Goshima et al., 2007). In this paper, we extend our studies of Dgt-depleted cells to better understand the mechanisms that underlie these phenotypes. In the experiments described in the subsequent sections, a single Dgt was depleted. However, we also find that knockdown of any of the five Dgts generates identical phenotypes (Fig. S2, A and E; and Video 2, available at http://www.jcb.org/cgi/content/full/jcb.200711053/DC1).
MT generation in the spindle is impaired after Dgt knockdown.
MT staining intensity within the spindle was significantly reduced after Dgt RNAi (Fig. 2 A), but the spindle phenotype was not as severe as that seen after knockdown of γ-tubulin. This phenotypic difference may be caused by γ-tubulin continuing to nucleate MTs from centrosomes and chromosomes after Dgt RNAi. Indeed, Dgt knockdown did not diminish MT nucleation at centrosomes, and centrosome-nucleated astral MTs were, on average, much longer in Dgt5-depleted cells (7.8 ± 2.8 μm; n = 200) than in control cells (3.8 ± 1.3 μm; n = 122), which might be because of an increase in the pool of soluble tubulin resulting from impaired nucleation and decreased MT polymer mass within the spindle (Fig. S2, B and C). We therefore reasoned that the Dgt phenotype might become more severe if γ-tubulin–mediated nucleation at the centrosome was simultaneously inhibited.
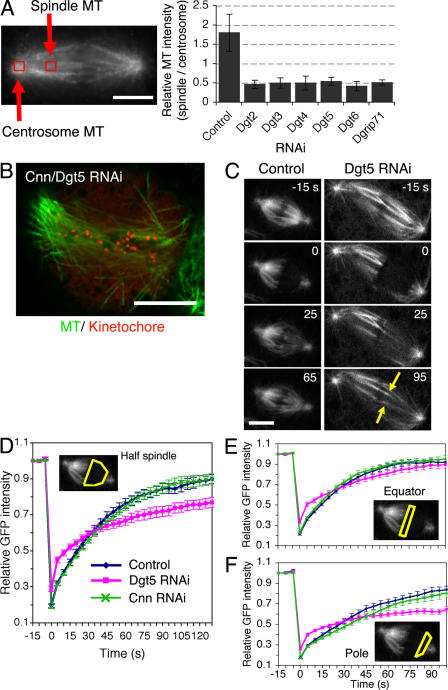
Figure 2.
Dgt2–6 proteins are important for MT generation within the spindle and bipolar spindle formation. (A) MT density specifically inside the spindle decreased in the absence of Dgt2–6 or Dgrip71. Signal intensities at the centrosome and within the spindle (red boxes) were measured. Ratio of spindle and centrosome signal intensity after Dgt2–6 and Dgrip71 knockdowns (0.43–0.55; n = 5 each) were significantly (P < 0.0001) lower than in control (1.81 ± 0.15 SEM; n = 10). (B) A high-resolution still image of MT (green) and kinetochore marker CENP-ACid (red) after double Cnn/Dgt5 RNAi. Spindle morphology and chromosome alignment were severely impaired. (C–F) FRAP analysis of GFP-tubulin for control (n = 13), Dgt5 (n = 14), and Cnn (n = 17) RNAi spindles. Cells were arrested in metaphase by Cdc16 RNAi, entire half spindles (excluding the centrosome) were photobleached (time 0), and relative GFP intensities were plotted (with SEM) for the half spindles (D, yellow), spindle equators (E, yellow), and pole regions (F, yellow). Only partial fluorescence recovery, mostly from the kinetochores (C, arrows), was observed in the absence of Dgt5, whereas recovery was seen in the whole bleached area in control or Cnn spindles (t1/2 = 30 s). The initial steep recovery (0–5 s) is likely because of diffusion of GFP-tubulin in the cytoplasm because it was also detected for the cells in which MTs were depolymerized by colcemid (not depicted). See also Video 4 (available at http://www.jcb.org/cgi/content/full/jcb.200711053/DC1). Bars, 5 μm.
To test this idea, we performed double RNAi of Dgt and Cnn (centrosomin), the protein responsible for docking γ-tubulin to the centrosomes (Megraw et al., 2001). Cnn depletion alone has little effect on spindle morphology and function. However, after depletion of both Cnn and Dgt, fixed cell analysis and time-lapse imaging (1–4 h) revealed that spindle morphology was dramatically impaired (Fig. 2 B, Fig. S2 D, and Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200711053/DC1). MTs were longer, sparse, and wavy, spindle poles were frayed, and the spindle displayed an overall disorganized morphology. This is one of the most severe mitotic defects reported in S2 cells, virtually phenocopying RNAi of γ-tubulin itself. These results suggest that Dgt and Cnn, as γ-tubulin–localizing factors, both contribute to making the MTs that comprise the spindle but that the Dgt–γ-tubulin activity in the spindle plays a dominant role.
To better understand the function of the Dgt complex in the metaphase spindle, we performed FRAP analysis of GFP-tubulin in the spindle. In control cells, the fluorescence recovered after photobleaching of a half spindle with a t1/2 = 30 s (n = 13; Fig. 2, C and D). Further analyses revealed that the recovery took place everywhere within the half spindle, including the spindle pole regions, although the recovery was faster near the spindle equator than spindle poles, likely reflecting polymerization at the plus ends of kMTs (Fig. 2, E and F; and Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200711053/DC1). MT turnover throughout the spindle also has been reported in mammalian cells and sea urchin embryos (Salmon et al., 1984a; Wadsworth and Salmon, 1986; Zhai et al., 1995). Interestingly, we found very similar recovery curves for Cnn-depleted cells, indicating that centrosomes play a relatively minor role in generating new MTs during metaphase (Fig. 2, D–F). In contrast, fluorescence recovery of the half spindle was slower in the absence of Dgt5, particularly near the spindle pole (Fig. 2, D–F). Relatively fast recovery took place near the spindle equator, suggesting that kMT polymerization is not impaired in cells lacking Dgt5 (Fig. 2 C, arrows). However, the recovery was slower than that of control cells also at this region, implicating that Dgt5 is also involved in chromatin-proximal MT generation. FRAP analysis in Cnn/Dgt5-codepleted cells showed slow recovery of GFP-labeled spindle MTs, which was similar to single Dgt5 RNAi (unpublished data). Collectively, these results indicate that the Dgt complex, more prominently than the centrosome, is required for the MT generation taking place within the metaphase spindle.
Dgts are critical for making spindles from chromosome-nucleated MTs.
To better understand the monopolar phenotype (Fig. S2 E), we performed long time-lapse imaging of monopolar spindles and found that all (n = 6) remained in a monopolar state for >150 min and did not convert to monastral bipolar spindles (Video 2). This is quite different from wild-type S2 cells, which undergo monopolar to monastral bipolar conversion within 30 min, a process that involves MT generation around the chromatin and then focusing of these MTs to create an acentrosomal pole (Goshima and Vale, 2003; Maiato et al., 2004). Klp61F/Kinesin-5 is critical for this conversion process. However, because this kinesin localized normally to spindle MTs and the centrosome after Dgt5 RNAi (Fig. S2 F), the lowered bipolar conversion cannot be explained by mislocalization of this kinesin in the absence of Dgt.
Other potential explanations for the severe defect in monopolar to monastral bipolar conversion are that Dgt depletion interferes either with chromosome-mediated MT nucleation or the subsequent stabilization/amplification of chromatin-nucleated MTs. To test these possibilities, we established an assay for observing the nucleation of MTs from chromatin. First, Cnn-depleted cells (to eliminate centrosome-based nucleation) were arrested in prometaphase by colcemid-induced MT depolymerization. After colcemid washout, cells were fixed and stained for MT and γ-tubulin every 15 min (Fig. 3 A). In the single Cnn RNAi cells, MTs appeared around chromatin after 45 min, followed by formation of robust bipolar spindles by 75 min. γ-Tubulin was localized to the newly formed MTs. In cells depleted of both Dgt5 and Cnn, MTs appeared around chromatin with the same timing and in a manner indistinguishable from the single Cnn RNAi cells. However, γ-tubulin did not colocalize with these MTs, and cells did not form robust bipolar spindles. Identical results were obtained after RNAi of other Dgt proteins (unpublished data). These data strongly suggest that the Dgt complex is dispensable for the initial nucleation of MTs around chromatin but is important in the subsequent amplification and/or stabilization of MTs, thereby producing sufficient numbers of MTs for robust chromatin-mediated bipolar spindle formation.
Figure 3.
Dgt is not involved in chromatin-mediated MT nucleation but is needed for chromosome alignment and K-fiber formation. (A) MT regrowth assay after control Cnn (top) or double Cnn/Dgt5 (bottom) RNAi. MTs (green) initially appeared around chromatin (blue) in both samples, whereas γ-tubulin (red) failed to localize to the MTs in the absence of Dgt5. The double RNAi cells failed to form robust bipolar spindles. (B) Delayed chromosome alignment and anaphase onset in the bipolar spindle in the absence of Dgt6 (mCherry-tubulin [red] and Mis12-GFP [green]). See also Video 5 (available at http://www.jcb.org/cgi/content/full/jcb.200711053/DC1). Mean duration (and SEM) from NEBD to metaphase and from metaphase to anaphase onset is plotted (control; RNAi against pBluescript sequences). (C) kMTs visualized after 10 min of colcemid treatment. Some kinetochores (Mis12-mCherry) do not associate with K-fibers (GFP-tubulin) in the absence of Cnn/Dgt5. See text for quantitation. Boxes on the left indicate the enlarged areas shown on the right. Bars, 5 μm.
Dgt-depleted cells are defective in chromosome alignment and kMT formation.
We applied high-throughput long-term live-cell microscopy on a cell line stably expressing both mCherry-tubulin and Mis12-GFP (a kinetochore marker) to follow the time course of chromosome positioning immediately after nuclear envelope breakdown (NEBD; Fig. 3 B and Video 5, available at http://www.jcb.org/cgi/content/full/jcb.200711053/DC1). In control cells, Mis12-GFP congressed to the metaphase plate at 18 ± 2 min (SEM; n = 14) after NEBD. Sister chromatid separation took place at 11 ± 2 min after congression. In contrast, Dgt6 knockdown cells took 35 ± 4 min for congression (n = 17; P < 0.0005) and a further 19 ± 2 min (P < 0.017) for separation. However, despite this significant delay, the majority of the Dgt6-depleted cells (18/20) eventually achieved metaphase congression and successful sister chromatid separation, suggesting that kinetochore–MT interactions were still present. The moderate misalignment phenotype was also found in other Dgt RNAi (Fig. S3 A, available at http://www.jcb.org/cgi/content/full/jcb.200711053/DC1). However, in the absence of both centrosomes and Dgt, chromosomes were much more severely misaligned and rarely congressed to form a stable metaphase plate (six of seven Cnn/Dgt6 double RNAi cells observed never formed a clear metaphase plate within a 4-h span of observation; Video 6)
Defects in chromosome congression after Dgt depletion may be the consequence of decreased numbers of kinetochore-MT associations. To explore this idea, we applied an MT depolymerization assay (Salmon et al., 1984b), in which colcemid treatment rapidly and preferentially destabilizes dynamic non-kMTs while stable kMT bundles (K-fibers) remain intact for several minutes. In control Cnn-depleted cells, K-fibers (visualized by GFP-tubulin) were detected for all the kinetochores (Mis12-mCherry) in 17/20 cells, whereas only 3 cells had a kinetochore that was not associated with MTs (Fig. 3 C). However, in 14/20 cells after Cnn/Dgt5 depletion, a subset of kinetochores (2 ± 0.9) did not associate with K-fibers. Even when K-fibers were still present, they were disorganized and the kinetochores were not aligned in the middle of the spindle (for live imaging see Fig. S3 B and Video 7, available at http://www.jcb.org/cgi/content/full/jcb.200711053/DC1). This assay revealed that Dgt5-depleted spindles lack robust K-fibers, which may account for the chromosome alignment defects found in these cells.
Identification of the Dgt6 orthologue in humans
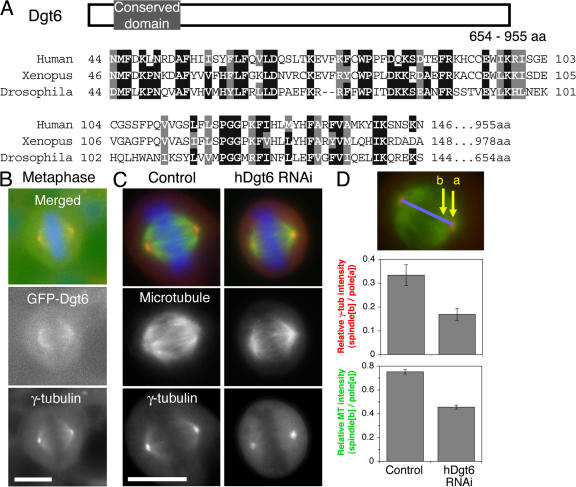
The basic local alignment search tool (BLAST) identified human FAM29A (family with sequence similarity, member 29A), an uncharacterized gene that has an 80-aa domain with 41% identity to D. melanogaster Dgt6 (Fig. 4 A). Similar to D. melanogaster Dgt6, we found that GFP-tagged FAM29A localized throughout the metaphase spindle of HeLa cells (Fig. 4 B). We also performed siRNA treatment of FAM29A and found that γ-tubulin and MT signals in the spindle, but not on the centrosomes, were significantly diminished (Fig. 4 C, D). RNAi of FAM29A also caused a mitotic delay (two- to threefold higher mitotic index than control cells). Collectively, we conclude that FAM29A is the human orthologue of Dgt6 (referring to it as hDgt6) and suggest that Dgt-dependent MT generation also plays an important role in human mitosis.
Figure 4.
Characterization of human Dgt6 in HeLa cells. (A) Alignment of N-terminal regions of Dgt6 proteins from D. melanogaster, Xenopus laevis, and Homo sapiens. Identical amino acids are boxed and the similar ones are hatched. (B) Uniform spindle localization of GFP-tagged human Dgt6 in metaphase (green). (C) Dim γ-tubulin and sparse MT phenotypes after RNAi knockdown of hDgt6 in HeLa cells (72 h). (D) Quantitation of γ-tubulin or MT level at centrosomes and spindles. Ratio of spindle and centrosome signal intensity in hDgt6 RNAi cells (γ-tubulin, 0.17 ± 0.02 SEM [n = 14]; MT, 0.46 ± 0.02 SEM [n = 19]) was significantly (P < 0.0005) lower than in control (γ-tubulin, 0.33 ± 0.04 SEM [n = 7]; MT, 0.75 ± 0.02 SEM [n = 15]). γ-tubulin (red) and DNA (blue) were counterstained. Bars, 10 μm.
The role of the Dgt complex (augmin) in mitotic spindle function
We show here that Dgt proteins associate to form a stable complex. Based upon its role in increasing MT numbers in the spindle, we propose that this complex be called “augmin” from the Latin verb augmentare, which means to increase. The role of augmin in increasing spindle MT density appears to be very important for building K-fibers and enabling chromatin-mediated MT nucleation to proceed toward the formation of a bipolar-shaped spindle, particularly when centrosome function is attenuated.
Our results suggest a model for how centrosome-, chromosome-, and spindle-based MT nucleation processes may cooperate to build mitotic spindles (Fig. 5). After NEBD in somatic cells, the most obvious generation of new MTs occurs at centrosomes, producing astral MT arrays. MTs are also nucleated in the vicinity of chromosomes immediately after NEBD. These processes are critical for generating the first set of mitotic MTs, enabling the process of spindle assembly to begin (Fig. 5 A). However, after the first sets of MTs are formed, we propose that augmin–γ-TuRC nucleates MT growth from existing MTs, thus providing a powerful mechanism for rapidly amplifying the number of MTs within the spindle to facilitate chromosome capture and K-fiber formation (Fig. 5 B).
Figure 5.
Model for augmin-dependent MT amplification in the spindle. (A) MTs are initially nucleated from centrosomes (brown) and near chromosomes/kinetochores (blue) in early prometaphase. γ-TuRC (red) is the major nucleator of these MTs. (B) The augmin/γ-TuRC machinery (yellow/red) then binds to those MTs, preferably to stable ones such as kMTs, and nucleates new MTs. The new MTs, which are oriented parallel or slightly angled to the template MTs, contribute to K-fiber generation and net kinetochore capture by the search-and-capture mechanism.
We postulate that kMTs, initially generated via the centrosome or chromatin pathway, are used as templates for binding augmin, which in turn recruits and activates γ-TuRC for MT nucleation. The outer γ-TuRC subunits appear to be critical for this specific spindle function of γ-tubulin because knockdown of these subunits produces a very similar phenotype to augmin knockdown (e.g., Fig. S2 A; Luders et al., 2006), although we have yet to obtain evidence for a direct interaction between augmin and γ-TuRC. An intriguing possibility is that augmin may dock onto an MT in a manner that positions γ-TuRC to preferentially nucleate new MT growth with the same polarity as the parent MT.
As an alternative to this model, augmin might increase the number of spindle MTs by activating an MT-severing enzyme (McNally et al., 2006; Srayko et al., 2006), although we feel that this is unlikely because RNAi of katanin and other AAA ATPases (e.g., spastin) does not give rise to the same mitotic phenotype as Dgt or γ-TuRC RNAi (Goshima et al., 2007; Zhang et al., 2007). Another class of models is that augmin increases spindle MT density by either stabilizing, elongating, or transporting MTs that are nucleated at the kinetochore/chromatin and that augmin–γ-tubulin are not involved in MT nucleation within the spindle. Although we cannot rule out this possibility, our FRAP results showing the concomitant recovery of fluorescence at the center of the spindle and the spindle poles suggest that MTs are being formed throughout the spindle and not just near the chromatin. The notion of an MT amplification process is also supported by in vitro studies of the assembly of MT asters in X. laevis extract and accompanying computational simulations (Clausen and Ribbeck, 2007). The exact mechanism by which augmin increases MT density awaits future work, most decisively by in vitro reconstitution of the process with purified proteins.
In addition to the mitotic spindle, there are other cellular MT networks in animals that are unlikely to be exclusively built by centrosome-based nucleation and assembly processes, such as axons or the meiotic spindle in oocytes. An MT-templated MT nucleation reaction could constitute an excellent means for generating new MTs while preserving the polarity of an existing MT array. Augmin is a candidate to play a role in this and numerous other cases where polarized noncentrosomal MTs are generated.
Materials and methods
Cell culture, RNAi, and cell staining
D. melanogaster Schneider cell line (S2) was cultured and RNAi was performed as previously described (Goshima and Vale, 2005; Goshima et al., 2007). Double-stranded RNA sequences used in this study are available at http://rnai.ucsf.edu. At the end of RNAi treatment (day 4), cells were resuspended and transferred to glass-bottom concanavalin A–coated plates and allowed to adhere for 2.5 h before fixation. S2 cells adopt a flat fibroblast-like morphology on concanavalin A–coated surfaces, increasing the possibility to image two spindle poles in a single focal plane. Colcemid (10 μg/ml solution; Sigma-Aldrich) was added to the cell at a final concentration of 3.3–5 μg/ml and washed out by replacing the medium three times (nocodazole, which is conventionally used in mammalian cells to depolymerize MTs, does not affect MTs in D. melanogaster cells). Cells were fixed in 6.4% PFA for 15 min, permeabilized with 0.5% SDS in PBS for 15 min, and incubated overnight at 4°C with anti–α-tubulin (YOL1/34; 1:150; AbD Serotec), anti–γ-tubulin (GTU-88; 1:1,000; Sigma-Aldrich), anti-Cid (Henikoff et al., 2000), and anti-Cnn (Li and Kaufman, 1996) antibodies in PBS containing 0.1% Triton X-100 and 5% goat serum, followed by staining with secondary antibodies and 1 μg/ml DAPI. For Dgt4 or Klp10A staining, methanol fixation was used (Rogers et al., 2004). HeLa cells were cultured in DME supplemented with 10% serum. RNAi was performed using DharmaFECT-1 (Dharmacon). siRNAs of hDgt6 (CAGUUAAGCAGGUACGAAATT) and control luciferase (CGUACGCGGAAUACUUCGATT) were synthesized by JbioS. Fixation and staining were performed in the identical way to that for S2 cells.
Plasmid construction and transfection
HA tagging of Dgt genes was performed using the Gateway system (Invitrogen). The fusion genes were inserted behind the copper-inducible metallothionein promoter (pMT vector; Invitrogen). Stably transformed cell lines were selected for each construct (Goshima and Vale, 2005). 50 μM CuSO4 was added to moderately induce gene expression (3 d). GFP-hDgt6 was constructed by PCR and ligation into pEGFP-C1 plasmid (Clontech Laboratories, Inc.). Transient transfection into HeLa cells was performed with Fugene 6 (Roche), and cells were fixed after 40 h.
Microscopy
Live imaging of the GFP-tubulin/Mis12-mCherry cell line was performed using a cooled charge-coupled device camera (MicroMax; Roper Scientific) attached to an inverted microscope (TE2000; Nikon) with perfect focus system. Image acquisition was controlled by μManager software (http://www.micro-manager.org). Fixed images were obtained by a spinning-disk confocal microscope (anti-Cid staining) or wide-field microscope (others). The automated ImageXpress Micro (MDS Analytical Technologies) was used for multisite per well multiwell time-lapse imaging as described previously (Goshima et al., 2007). Fixed kMT imaging after colcemid treatment was performed with a confocal microscope (FV1000; Olympus). Seven focal planes with 0.3-μm intervals were imaged and z projection was done with ImageJ (National Institutes of Health). FRAP was performed with the FV1000 with a 100×/1.40 NA objective lens. Images were acquired every 1 (GFP-Dgt5) or 5 s (GFP-tubulin; cells were treated with Cdc16 double-stranded RNA to arrest in metaphase). Bleaching was conducted for 200 ms (GFP-Dgt5) or 1 s (GFP-tubulin) after three frames of prebleach imaging. The bleached area was 1.6 ± 0.5 (metaphase) or 1.9 ± 0.6 μm2 (anaphase) for GFP-Dgt5 and entire half spindles for GFP-tubulin (14 ± 3, 19 ± 8, 16 ± 3, and 20 ± 7 μm2 for control, Dgt5, Cnn, and Cnn/Dgt5 RNAi cells, respectively). Fluorescence recovery was also measured at spindle equator (4.4 ± 1.1, 5.4 ± 1.7, and 4.1 ± 0.8 μm2) and spindle pole regions (2.6 ± 0.8, 3.7 ± 1.2, and 3.7 ± 1.0 μm2) for control, Dgt5, and Cnn RNAi cells, respectively. GFP intensity of the bleached area was normalized using the intensity of a nonbleached area of similar size after background subtraction.
Biochemistry
For immunoprecipitation, 4 ml of culture expressing HA-tagged proteins was harvested and washed once with serum-free medium. Cell extracts were prepared by incubating for 10 min with 600 μl of extraction buffer HB (50 mM Hepes-KOH, pH 7.6, 100 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 1% Triton X-100, 1 mM 2-mercaptoethanol, 100 μM GTP, and protease inhibitor cocktails; Oegema et al., 1999), followed by centrifugation at 5,000 rpm for 5 min, followed by centrifugation at 15,000 rpm for 20 min. The supernatant was mixed with 10 μl of anti–HA-conjugated agarose beads (Sigma-Aldrich) for 3 h, followed by washing three times with extraction buffer. Immunoblot was performed with rabbit antisera against Dgt5 (1:500), Dgt6 (1:500), affinity-purified anti-Dgt3 (1:100), Dgt4 (1:300), Dgrip91 (1:1,000; a gift from M. Moritz, University of California, San Francisco, San Francisco, CA), mouse monoclonal anti-HA (12CA5), anti-GFP (1:200; Roche), and anti–γ-tubulin (GTU-88; 1:500) antibodies. Sucrose gradient sedimentation and gel filtration chromatography were performed according to Oegema et al. (1999). Cell extracts prepared by HB buffer were applied to 6–38% sucrose gradient and centrifuged with a rotor (P28S; Hitachi) at 27,000 rpm (131 kG) for 11 h at 4°C. 17 fractions were collected and used for SDS-PAGE and immunoblot. All of the Dgt migrated predominantly as a single species except for Dgt2-HA, which exhibited a trailing large molecular mass component. We suspect that this broadened peak might be caused by protein aggregation as the result of ectopic expression, although it is possible that a subpopulation of Dgt2 might act independently of other Dgts. Gel filtration chromatography was performed with a Superdex 200 10/300 column attached to the AKTA system (GE Healthcare). Molecular size markers are described in Oegema et al. (1999). The MT cosedimentation experiment was conducted from 4 ml of culture expressing Dgt2-HA. Cells were suspended by BRB25 (25 mM K-Pipes, pH 6.8, 1 mM MgCl2, and 1 mM EGTA) and mechanically disrupted by a syringe with a 27G needle, followed by centrifugation at 15,000 rpm for 20 min. The supernatant was further centrifuged using a rotor (TLA100.3; Beckman Coulter) at 70,000 rpm for 20 min, and the supernatant was mixed with taxol-stabilized MTs for 15 min at room temperature. MTs were then sedimented at 50,000 rpm for 10 min.
Online supplemental material
Fig. S1 shows the characterization of anti-Dgt antibodies, codepletion of Dgt proteins, and Dgt localization. Fig. S2 shows identical RNAi phenotypes of Dgt2–6, Dgrip71, and Dgrip75. Fig. S3 shows chromosome misalignment and kMT defects after Dgt RNAi. Video 1 shows FRAP of GFP-Dgt5 on the spindle. Video 2 shows spindle defects after RNAi of Dgt2–6, Dgrip71, 75 (γ-TuRC), or Dgrip84 (γ-TuSC). Video 3 shows severe spindle defects after double RNAi of Cnn and Dgt2–6, Dgrip71, 75 (γ-TuRC), or Dgrip84 (γ-TuSC). Video 4 shows FRAP of GFP-tubulin after Dgt5 and Cnn RNAi. Video 5 shows delay of chromosome alignment and anaphase onset after Dgt6 RNAi. Video 6 shows abnormal spindle morphology and severe chromosome misalignment after double Cnn/Dgt6 RNAi. Video 7 shows defective kMT bundles after RNAi of double Cnn/Dgt5. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200711053/DC1.
Supplemental Material
Acknowledgments
We are grateful to Motoyuki Itoh, Toshifumi Inaba, Seiji Kojima, other members of the Department of Biological Science at Nagoya University, Kazuhiro Yagita, Mitsuhiro Yanagida, and Michelle Moritz for reagents and the use of their devices, to Alexandra Zidovska and Chuanhai Fu for their help with microscopy, to Eric Griffis for comments on the manuscript, and to Karen Dell for suggesting the name augmin.
This work is supported by the Special Coordination Funds for Promoting Science and Technology (MEXT, Japan), the Global COE Program “Advanced Systems-Biology: Designing the Biological Function” (MEXT), and the Uehara Memorial Foundation.
Abbreviations used in this paper: γ-TuRC, γ-tubulin ring complex; γ-TuSC, γ-tubulin small complex; kMT, kinetochore MT; MT, microtubule; NEBD, nuclear envelope breakdown.
References
- Clausen, T., and K. Ribbeck. 2007. Self-organization of anastral spindles by synergy of dynamic instability, autocatalytic microtubule production, and a spatial signaling gradient. PLoS ONE. 2:e244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wulf, P., A.D. McAinsh, and P.K. Sorger. 2003. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 17:2902–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadde, S., and R. Heald. 2004. Mechanisms and molecules of the mitotic spindle. Curr. Biol. 14:R797–R805. [DOI] [PubMed] [Google Scholar]
- Goshima, G., and R.D. Vale. 2003. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 162:1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., and R.D. Vale. 2005. Cell cycle-dependent dynamics and regulation of mitotic kinesins in Drosophila S2 cells. Mol. Biol. Cell. 16:3896–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., R. Wollman, S.S. Goodwin, N. Zhang, J.M. Scholey, R.D. Vale, and N. Stuurman. 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 316:417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen, A.C., L.A. Cameron, M. Coughlin, D.T. Miyamoto, T.J. Mitchison, and R. Ohi. 2004. XRHAMM functions in ran-dependent microtubule nucleation and pole formation during anastral spindle assembly. Curr. Biol. 14:1801–1811. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., K. Ahmad, J.S. Platero, and B. van Steensel. 2000. Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA. 97:716–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson, M.E., T.G. Setty, A. Paoletti, and P.T. Tran. 2005. Efficient formation of bipolar microtubule bundles requires microtubule-bound γ-tubulin complexes. J. Cell Biol. 169:297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenti, E., and I. Vernos. 2001. The mitotic spindle: a self-made machine. Science. 294:543–547. [DOI] [PubMed] [Google Scholar]
- Khodjakov, A., R.W. Cole, B.R. Oakley, and C.L. Rieder. 2000. Centrosome-independent mitotic spindle formation in vertebrates. Curr. Biol. 10:59–67. [DOI] [PubMed] [Google Scholar]
- Kirschner, M., and T. Mitchison. 1986. Beyond self-assembly: from microtubules to morphogenesis. Cell. 45:329–342. [DOI] [PubMed] [Google Scholar]
- Lajoie-Mazenc, I., Y. Tollon, C. Detraves, M. Julian, A. Moisand, C. Gueth-Hallonet, A. Debec, I. Salles-Passador, A. Puget, H. Mazarguil, et al. 1994. Recruitment of antigenic gamma-tubulin during mitosis in animal cells: presence of gamma-tubulin in the mitotic spindle. J. Cell Sci. 107:2825–2837. [DOI] [PubMed] [Google Scholar]
- Li, K., and T.C. Kaufman. 1996. The homeotic target gene centrosomin encodes an essential centrosomal component. Cell. 85:585–596. [DOI] [PubMed] [Google Scholar]
- Luders, J., U.K. Patel, and T. Stearns. 2006. GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 8:137–147. [DOI] [PubMed] [Google Scholar]
- Mahoney, N.M., G. Goshima, A.D. Douglass, and R.D. Vale. 2006. Making microtubules and mitotic spindles in cells without functional centrosomes. Curr. Biol. 16:564–569. [DOI] [PubMed] [Google Scholar]
- Maiato, H., C.L. Rieder, and A. Khodjakov. 2004. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J. Cell Biol. 167:831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, K., A. Audhya, K. Oegema, and F.J. McNally. 2006. Katanin controls mitotic and meiotic spindle length. J. Cell Biol. 175:881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw, T.L., L.R. Kao, and T.C. Kaufman. 2001. Zygotic development without functional mitotic centrosomes. Curr. Biol. 11:116–120. [DOI] [PubMed] [Google Scholar]
- Murata, T., S. Sonobe, T.I. Baskin, S. Hyodo, S. Hasezawa, T. Nagata, T. Horio, and M. Hasebe. 2005. Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat. Cell Biol. 7:961–968. [DOI] [PubMed] [Google Scholar]
- Oegema, K., C. Wiese, O.C. Martin, R.A. Milligan, A. Iwamatsu, T.J. Mitchison, and Y. Zheng. 1999. Characterization of two related Drosophila γ-tubulin complexes that differ in their ability to nucleate microtubules. J. Cell Biol. 144:721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, G.C., S.L. Rogers, T.A. Schwimmer, S.C. Ems-McClung, C.E. Walczak, R.D. Vale, J.M. Scholey, and D.J. Sharp. 2004. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 427:364–370. [DOI] [PubMed] [Google Scholar]
- Salmon, E.D., R.J. Leslie, W.M. Saxton, M.L. Karow, and J.R. McIntosh. 1984. a. Spindle microtubule dynamics in sea urchin embryos: analysis using a fluorescein-labeled tubulin and measurements of fluorescence redistribution after laser photobleaching. J. Cell Biol. 99:2165–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon, E.D., M. McKeel, and T. Hays. 1984. b. Rapid rate of tubulin dissociation from microtubules in the mitotic spindle in vivo measured by blocking polymerization with colchicine. J. Cell Biol. 99:1066–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel, L.M., and K.J. Monty. 1966. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim. Biophys. Acta. 112:346–362. [DOI] [PubMed] [Google Scholar]
- Srayko, M., E.T. O'Toole, A.A. Hyman, and T. Muller-Reichert. 2006. Katanin disrupts the microtubule lattice and increases polymer number in C. elegans meiosis. Curr. Biol. 16:1944–1949. [DOI] [PubMed] [Google Scholar]
- Wadsworth, P., and E.D. Salmon. 1986. Analysis of the treadmilling model during metaphase of mitosis using fluorescence redistribution after photobleaching. J. Cell Biol. 102:1032–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese, C., and Y. Zheng. 2006. Microtubule nucleation: gamma-tubulin and beyond. J. Cell Sci. 119:4143–4153. [DOI] [PubMed] [Google Scholar]
- Wollman, R., E.N. Cytrynbaum, J.T. Jones, T. Meyer, J.M. Scholey, and A. Mogilner. 2005. Efficient chromosome capture requires a bias in the ‘search-and-capture’ process during mitotic-spindle assembly. Curr. Biol. 15:828–832. [DOI] [PubMed] [Google Scholar]
- Zhai, Y., P.J. Kronebusch, and G.G. Borisy. 1995. Kinetochore microtubule dynamics and the metaphase–anaphase transition. J. Cell Biol. 131:721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D., G.C. Rogers, D.W. Buster, and D.J. Sharp. 2007. Three microtubule severing enzymes contribute to the “Pacman-flux” machinery that moves chromosomes. J. Cell Biol. 177:231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.