Abstract
Selenium, an essential trace element for mammals, is incorporated into a selected class of selenoproteins as selenocysteine. All known isoenzymes of mammalian thioredoxin (Trx) reductases (TrxRs) employ selenium in the C-terminal redox center -Gly-Cys-Sec-Gly-COOH for reduction of Trx and other substrates, whereas the corresponding sequence in Drosophila melanogaster TrxR is -Ser-Cys-Cys-Ser-COOH. Surprisingly, the catalytic competence of these orthologous enzymes is similar, whereas direct Sec-to-Cys substitution of mammalian TrxR, or other selenoenzymes, yields almost inactive enzyme. TrxRs are therefore ideal for studying the biology of selenocysteine by comparative enzymology. Here we show that the serine residues flanking the C-terminal Cys residues of Drosophila TrxRs are responsible for activating the cysteines to match the catalytic efficiency of a selenocysteine-cysteine pair as in mammalian TrxR, obviating the need for selenium. This finding suggests that the occurrence of selenoenzymes, which implies that the organism is selenium-dependent, is not necessarily associated with improved enzyme efficiency. Our data suggest that the selective advantage of selenoenzymes is a broader range of substrates and a broader range of microenvironmental conditions in which enzyme activity is possible.
Selenium is an essential trace element for mammals due to vital roles played by one or several selenoproteins, which is illustrated by the lethal phenotype of mice lacking the tRNA needed for selenocysteine (Sec; U in one-letter code) incorporation (1). Sec functions as an extraordinarily reactive cysteine homologue; the increased reactivity of oxidoreductases having Sec in place of cysteine is usually regarded as the raison d'etre for selenoproteins, despite their costly and inefficient synthesis machinery (2).
Mammalian thioredoxin (Trx) reductase (TrxR) enzymes are important selenoproteins that, together with Trx and additional Trx-dependent enzymes, carry out several antioxidant and redox regulatory roles in cells. These roles include synthesis of deoxyribonucleotides with ribonucleotide reductase, reduction of peroxides or oxidized methionine residues with peroxiredoxins or methionine sulfoxide reductases, respectively, regulation of several transcription factor or protein kinase activities, as well as regeneration of many low molecular weight antioxidant compounds (see ref. 3 and references therein). The catalytic activities of mammalian TrxR isoenzymes depend on a redox-active Cys-Sec couple within the C-terminal tetrapeptide motif, -Gly-Cys-Sec-Gly-COOH (4–6). Compared with sulfur, selenium is not only generally more reactive but also exhibits an ≈15% longer bond length, which facilitates formation of a selenenyl-sulfide bridge between the adjacent Cys-Sec residues, which is a necessary intermediate in the catalytic cycle (7–11). Mutational studies of the mammalian enzyme in which Sec was replaced by Cys showed a marked decrease in the catalytic rate (kcat) for Trx reduction (10–12). This result was expected, because disulfide bridges between two sequentially adjacent cysteines are generally not favored (13). Indeed, the disulfide bond between the two Cys residues of the eight-member cyclo-Cys-Cys ring is not compatible with a standard peptide-bond geometry (14).
In Drosophila melanogaster, TrxR is particularly important because its product, Trx(SH)2, in place of glutathione reductase, serves as the principal reductant of glutathione (15, 16). TrxR of the fruit fly is closely related to mammalian TrxR but carries a redox-active C-terminal -Ser-Cys-Cys-Ser-COOH motif that presumably involves a strained disulfide as a catalytic intermediate within the C-terminal Cys-Cys-sequence. Thus, it is surprising that the kcat for oxidized Trx of the Drosophila enzyme is ≈50% that of the human enzyme for the same substrate (15), which is in strong contrast to the Sec498 → Cys mutants of the mammalian enzyme, which have very low catalytic activity (10–12).
In this study, we have demonstrated that very slight changes in the active site of the insect TrxR are sufficient to yield a high catalytic activity without involvement of selenium. The results lead to general questions regarding the role and necessity of Sec in mammalian TrxRs and, possibly, in other selenoproteins as well.
Materials and Methods
Cloning, Expression, and Purification of D. melanogaster TrxR-2 (DmTrxR-2) C-Terminal Mutants. Mutants were designed to yield enzymes with the following C-terminal tetrapeptide sequences: GCCG, SCCG, GCCS, GCCD, DCCG, DCCD, SCSS, GCUG, SCUG, GCUS, and SCUS. These C-terminal mutants of (N-terminally His-tagged) wild-type DmTrxR-1 (National Center for Biotechnology Information accession no. AF301144) were cloned, expressed, and purified by using standard techniques and the recently developed methodology for heterologous expression of mammalian selenoproteins in Escherichia coli (17). Technical details are given in Supporting Methods, which is published as supporting information on the PNAS web site, www.pnas.org. For an efficient selenoprotein synthesis, the pSUABC-plasmid was used as described in ref. 17, and metabolic labeling with 75Se showed that only Sec-containing mutants incorporated selenium (see Table 1 and Fig. 5, which are published as supporting information on the PNAS web site). Enzyme purity was confirmed by silver-stained SDS/PAGE.
Determination of Enzyme Concentration. Subunit concentrations of the Cys-Cys mutants were determined by measuring absorbance at 462 nm by using an assumed ε462 nm of 11.3 mM–1·cm–1 for the flavoprotein subunits (7). By using the Bio-Rad protein assay procedure with BSA as standard, it was confirmed that all variants of DmTrxR contained stoichiometric amounts of FAD (1 ± 0.2) per 53.9-kDa subunit. Concentrations of Cys-Sec mutants were determined from the selenium concentration as measured by atomic absorption spectroscopy (see Table 1). The concentration of oxidized DmTrx-2, purified as described earlier (18), was measured enzymatically by end-point determination by using excess NADPH and 1 unit/ml of wild-type DmTrxR.
Enzyme Kinetics. All standard assays were carried out at 25°C. Recombinant N-terminally His-tagged DmTrx-2 was prepared as described (18). NADPH was from Biomol (Hamburg, Germany). Buffer T, which was used in most assays consisted of 100 mM potassium phosphate, 2 mM EDTA, 100 μM NADPH, pH 7.4. In all assays, the oxidation of NADPH was monitored as the decrease in absorbance at 340 nm (εNADPH, 340 nm = 6.22 mM–1·cm–1). One unit is defined as the consumption of 1 μmol of NADPH per min. Unless otherwise stated ≈10–20 milliunits/ml of enzyme were used in each assay.
Trx assay. The enzyme samples were added to a cuvette containing buffer T. The reaction was started by adding DmTrx-2 (50 μM final concentration).
GHOST (glutathione as substrate of Trx) assay.¶ Enzyme samples were added to a cuvette containing 1 mM glutathione disulfide in buffer T. The reaction was started by adding oxidized DmTrx-2 (30 μM final concentration).
To minimize the effects of ionic strength due to pH differences, a modified buffer was used for the pH profiles: 400 mM NaCl, 50 mM sodium phosphate, 1 mM EDTA and 100 μM NADPH. The pH was adjusted from 9.0 to 6.0 in ΔpH steps of 0.2 or 0.5 by using HCl.
Methylseleninate reduction assay. Methylseleninic acid (CH3SeO2H) was obtained from PharmaSe (Lubbock, TX). One nanomole (127 mg) was weighed out and dissolved in 500 μl 2 M KOH. Buffer T (500 μl) was added to yield a stock solution with a final concentration of 1 M CH3SeO2K. Enzyme samples were added to a cuvette containing buffer T. The reaction was started by adding methylseleninate (100 μM final concentration).
Hg2+reduction assay. EDTA-free enzyme samples (50–100 milliunits/ml as determined in the GHOST assay) were assayed as described (19).
Titration and Stopped-Flow Experiments. All titrations and stopped-flow experiments were conducted in 100 mM potassium phosphate buffer, pH 7.0, anaerobically, as described (7). The methods of data analysis are outlined in ref. 20.
Reduction of enzymes with NaBH4. DmTrxR mutants were reduced by using a sodium borohydride solution (100 mM in 0.02 M NaOH) in ≈100-fold excess to the enzyme solution under anaerobic conditions. Excess NaBH4 is completely hydrolyzed at neutral pH within a few minutes. The reduced enzyme could be used directly for titrations and stopped-flow experiments.
Stopped-flow-state kinetics. Rapid reaction kinetics were all conducted in a Hi-Tech SF-61DX2 stopped-flow photometer (Hi-Tech, Salisbury, Wiltshire, U.K.) under anaerobic conditions at 10°C. The DmTrxR concentration used was 15–20 μM after mixing. Data from kinetic traces were analyzed by fitting to multiple exponential functions with the program a, which was written by R. Chang, C.Y. Chiu, J. Dinverno, and D. P. B. (University of Michigan).
The reaction of NADPH with oxidized enzyme was carried out in the stopped-flow spectrophotometer. Time-dependent spectra were followed by diode array detection. Single-wavelength kinetic traces were recorded by using a photomultiplier tube. For the oxidative half-reaction, NaBH4-reduced enzyme was prepared and subsequently reacted with oxidized DmTrx-2.
Auranofin Inhibition. Enzyme samples were assayed for their Trx-reduction activity in the absence or presence of 1 μM auranofin (ICN Biochemicals, Aurora, OH) prepared from a 1-mM stock solution in dimethyl sulfoxide.
Results
The two C-terminal redox active cysteines in DmTrxR are flanked by polar serines, rather than by the Gly residues in the corresponding active site of mammalian TrxR. We reasoned that this slight difference in the microenvironment of the C-terminal cysteines might compensate for the tension in the presumably unfavorable disulfide bond of adjacent cysteines (14). Therefore, we expressed mutants of DmTrxR in which the flanking Ser residues were replaced by Gly residues in all possible permutations. We also replaced these residues by Asp residues to introduce groups with negative charges capable of forming hydrogen bonds. Finally, to probe the possible advantage of a Sec residue in the redox-active center, we expressed Cys-to-Sec mutant forms of DmTrxR, by using a rather intriguing methodology, because Sec is incorporated by specific UGA codons in a species-specific manner (2, 17). We achieved good yield of pure enzyme with all mutants (see Table 1 and Fig. 5), and this approach circumvents the potential pitfalls that may arise when comparing enzymes from different species such as humans and Drosophila.∥
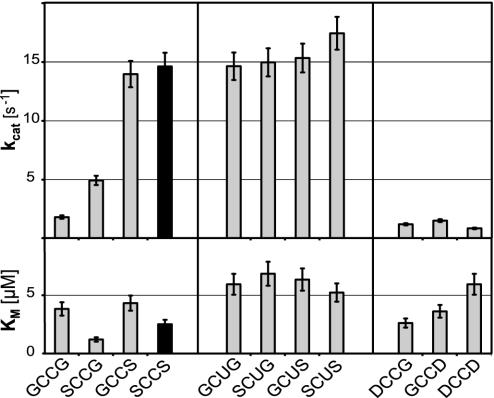
Determination of Kinetic Constants with Different Substrates. All of the variants of DmTrxR were catalytically active (see Fig. 6, which is published as supporting information on the PNAS web site). To the extent that Km is related to Kd, the affinity for Trx appeared to be similar for all mutants, because all Km values were within the range of 2–6 μM (Fig. 1). This finding shows that the changes in the C-terminal tetrapeptide do not significantly affect substrate binding and that all mutants are likely to have an intact quaternary structure capable of binding Trx with similar affinity as the wild-type enzyme. The kcat was, however, highly affected by the sequence of the C-terminal tetrapeptide. By analyzing the GCCG, SCCG, and GCCS mutants, it became apparent that each of the two serine residues present in the wild-type sequence improved the catalytic activity. The second serine (Ser491), however, was far more important in this respect than the first (Ser488). In marked contrast, the kcat values for Trxs of Sec-containing mutants were consistently in the same range as the kcat for the wild-type SCCS enzyme, and were thus largely unaffected by the nature of the flanking residues (Fig. 1). The aspartic acid-containing mutants (DCCG, GCCD, and DCCD) all had a significantly lower kcat values than the wild-type enzyme, some even below the already poorly active GCCG mutant (Fig. 1).
Fig. 1.
Comparison of kinetic parameters of wild-type and mutant DmTrxR. The kcat (Upper) and Km (Lower) values shown here were determined by using DmTrx-2 as a substrate (GHOST assay). The C-terminal tetrapeptide sequences of the respective enzyme are indicated on the x axis. The results for the wild-type enzyme represented by SCCS are highlighted in black. The kcat values shown for the selenoenzymes were normalized for the presence of truncated, inactive enzyme. Note that the wild-type enzyme's (SCCS) activity does not differ significantly from the kcat values of the Sec mutants.
By using methylseleninate as substrate, the absolute differences between the Cys-Cys and the Cys-Sec variants were more pronounced, because the latter showed a 2- to 5-fold higher turnover per subunit than did the Cys-Cys-homologues. For example, the Sec mutant, SCUS, showed a 5.2-fold higher turnover rate for methylseleninate than did the wild-type enzyme (5.2 s–1 vs. 1 s–1).
Because the C-terminal sequence of mercuric ion reductase is -SCCAG, and thereby exhibits some resemblance with the C terminus of DmTrxR-SCCG, we also analyzed Hg2+-reductase activity of all mutants. Under the conditions used, none of the enzymes however showed any mercuric ion reductase activity above background (data not shown).
pH Profile. The Cys-Sec mutants were found to show little variation in activity within a broad physiological pH range (pH 6.0–9.0), whereas the selenium-free mutants lost significant activity at pH values <7.0. (see Fig. 7, which is published as supporting information on the PNAS web site) The pH profile obtained with the different mutants should in part reflect the pKa values of the C-terminal redox-active residues.
Reductive Half-Reaction. The C-terminal redox-active motif in one subunit of the dimeric high Mr TrxR enzymes receives electrons from two N-terminal redox-active cysteines proximal to an FAD in the other subunit, subsequently transferring these electrons to Trx or other substrates of the enzyme (7, 8, 11, 21). By using stopped-flow kinetic methods, the rates of these separate events can be determined.
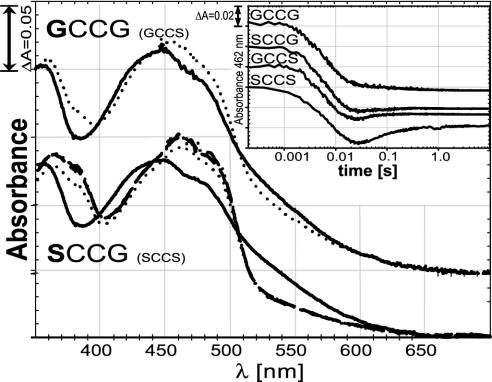
Stopped-flow analysis of both wild-type enzyme (SCCS) and the mutants GCCG, SCCG, and GCCS showed nearly identical first phases during reduction of the flavin with four equivalents of NADPH per subunit (Fig. 2 Inset), with an apparent rate constant, k1 = 150 ± 9 s–1, describing the first step for all four enzymes. The second phase, in which the N-terminal disulfide is reduced by the flavin, was characterized by a rate constant, k2 = 47 ± 14 s–1. The wild-type enzyme exhibited a distinct third phase, with an apparent rate constant, k3, of ≈21 s–1. This phase is most likely attributable to interchange between the N-terminal thiols and the C-terminal disulfide of the enzyme. As the interchange reaction progresses, the charge–transfer interaction between Cys-57 and the FAD decreases, and the absorbance at 462 nm increases slightly. The relative forward and reverse rates of the interchange reaction determine the amount of absorbance increase observed in this step. Thus, the GCCS and SCCG mutant forms showed third phases, but they were smaller than that of the wild-type and were difficult to quantify. Consistent with the interpretation that the third phase is due to interchange between C- and N-terminal cysteine pairs, mutants such as DmTrxR-SSCS and DmTrxR-SCSS, which lack a catalytically functional C-terminal redox-active site, clearly show no third phase, as expected when disulfide–dithiol interchange with the C-terminal group is not possible (22). The kinetic traces at 540 nm, reflecting the state of the charge–transfer complex, (data not shown) were similar in all mutants with rate constants, k1 = 135 s–1 and k2 = 42 s–1.
Fig. 2.
Half-reactions of wild-type enzyme and mutant forms. Each protein was reduced with sodium borohydride. Reduced protein was mixed in the stopped-flow spectrophotometer with various concentrations of oxidized DmTrx. Solid lines represent the reduced enzyme species with no Trx added. Dotted spectra depict the enzyme species after addition of two equivalents of oxidized Trx. In the SCCG case, the dashed line represents three equivalents of Trx, and the dash-dotted line represents five equivalents. For clarity, the spectra of GCCG are shifted by 0.05 units. The absorbance of the GCCS mutant was essentially identical to that of GCCG, whereas the SCCS mutant had essentially the same absorbance as SCCG (data not shown). (Inset) The reductive half-reaction of wild-type enzyme (SCCS) compared with three mutant forms (GCCG, SCCG, and GCCS). The protein was mixed in a stopped-flow spectrophotometer with four equivalents of NADPH per subunit, and the change in absorbance at 462 nm, representing the redox state of the flavin, was recorded over time. All enzymes were used at similar concentrations (≈18 μM subunits). The absorption starting point (0.2) was shifted for the GCCS, SCCG, and GCCG mutants for clarity. Note that the time axis is in logarithmic scale. See text for details.
Oxidative Half-Reaction. Reduction with borohydride yields enzyme having both disulfides reduced to dithiols (EH4). Thus, it is equivalent to enzyme reduced with two equivalents of NADPH, but lacking the NADP+. The spectrum is perturbed due to the charge transfer interaction of the thiolate of Cys-62 with the flavin. Furthermore, a small fraction of the borohydride-reduced enzyme may be present in the fully reduced EH6 form. However, EH6 is unlikely to be formed in vivo due to the low redox potential of the enzyme-bound FAD (22).
Reoxidation on reaction with Trx is indicated by disappearance of the charge-transfer band at 540 nm. The borohydride-reduced enzyme species could not be fully reoxidized, even by the addition of up to five equivalents of oxidized Trx per subunit (Fig. 2). Oxidation is indicated by the loss of charge-transfer absorbance of ≈550 nm and an increase in absorbance of ≈460 nm. Indeed, the mutants with a Gly at position 488 showed only a marginal shift from the fully reduced spectrum toward the spectrum of oxidized enzyme, whereas the forms with Ser at this position led to more pronounced, yet incomplete, reoxidation (Fig. 2). The only spectrum shown for GCCG is that for two equivalents of Trx, but the spectra for three and five equivalents, while of poor quality, were similar. The wild-type enzyme, SCCS, resulted in spectra essentially as shown for SCCG (22). GCCS behaved almost identically to GCCG.
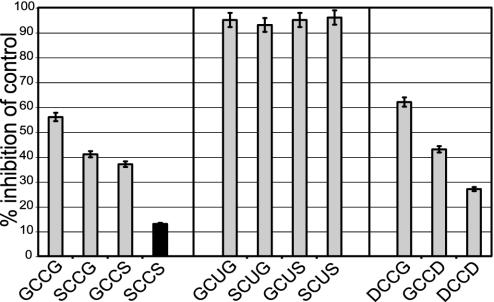
Auranofin Inhibition. Auranofin (1 μM) efficiently inhibited all Cys-Sec mutants, which was similar to the inhibition of mammalian TrxR by this compound (9). The selenium-free mutants were inhibited much less by auranofin (Fig. 3). A considerable difference was observed between the individual mutants, with less inhibition with increasing polarity of the flanking residues in the C-terminal motif.
Fig. 3.
Inhibition by auranofin. Auranofin, an effective inhibitor of human TrxRs, was analyzed for its inhibitory effect on wild-type DmTrxR (highlighted in black) and the DmTrxR mutants. Shown is the percentage of inhibition compared with an auranofin-free control. In all cases, 1 μM auranofin was used and the assays were carried out as in Fig. 1.
Discussion
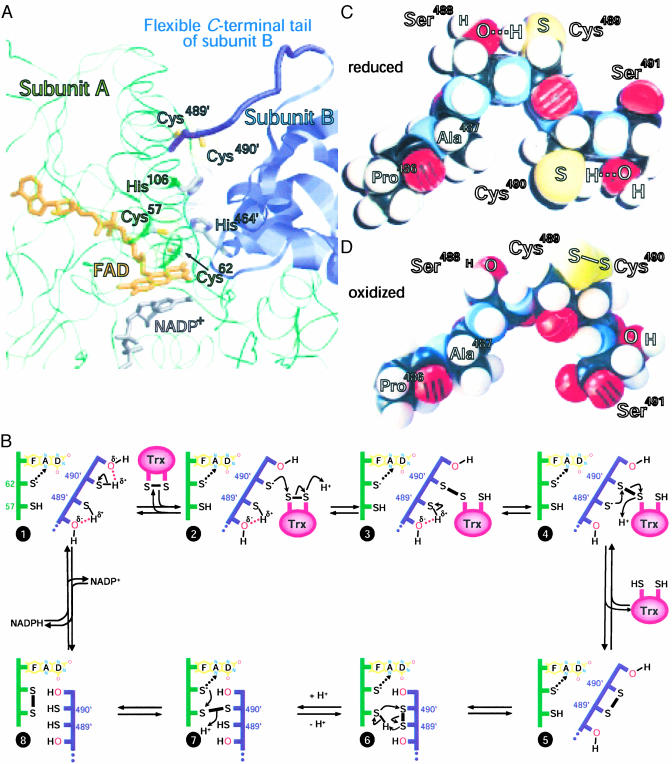
This study shows that the polar serine residues present within the C-terminal tetrapeptide sequence of DmTrxR play a key role in making this selenium-free protein almost as active as its mammalian selenoprotein counterpart. It also shows that with the Sec-containing variant of the C-terminal redox-active motif, the impact of the two flanking residues is diminished. How can these pronounced effects of such subtle changes at the active site be explained? The active species in dithiol-disulfide-exchange reactions is the thiolate (or selenolate for Sec). The pKa value of a protein cysteine thiol is typically ≈8.5, unless affected by neighboring residues (23), whereas the pKa of a selenol group is normally ≈5.3 (24). Because of this occurrence, Sec is normally ionized at physiological pH, and in contrast to cysteine, does not require assistance from the protein environment. The pKa of the interchange thiol of the closely related enzyme glutathione reductase is low, due to interactions with a nearby His residue (25). The analogous residue in DmTrxR is His464′, which seems to be positioned exactly as it is in glutathione reductase (Fig. 4A). Furthermore His106, which is also conserved among large TrxRs, is almost certainly involved (Fig. 4A); its modeled position is relative to the C-terminal thiols, just as His464′ is relative to the N-terminal thiols. The pH profiles of the mutants analyzed herein are consistent with the requirement for ionizing the cysteine(s), because increased pH leads to an increase in turnover rate of the Cys-Cys-containing enzyme species up to a plateau above pH 8.0. By contrast, the activity of the Cys-Sec enzymes have very little dependence on pH in the physiological range. The flanking Ser residues in DmTrxR are crucial for dithiol–disulfide-interchange reactions, possibly by kinetically facilitating transient thiolate formation at the C-terminal redox-active cysteines.
Fig. 4.
(A) Computer model of DmTrxR. The protein backbones of the two subunits in the dimeric enzyme are shown as thin green strands (subunit A) and blue ribbons (subunit B). The flexible C-terminal tail of subunit B is shown as a thick blue strand. The FAD (orange) and nicotinamide (gray) of NADP+, bound to subunit A, are represented as stick models. The side chains of the N-terminal and C-terminal redox-active site (Cys57 and Cys62 and Cys489′ and Cys490′, respectively), as well as the putative base catalysts, His106 and His464′, are also shown as stick models, with the sulfurs in yellow and the nitrogens of His in blue. The van der Waals radii of these amino acids are dotted. This model is based on the crystal structure of rat TrxR U498C (21) (Protein Data Bank ID code 1H6V) by using raswin v2.7.2 for visualization (www.bernstein-plus-sons.com/software/RasMol_2.7.2/doc/rasmol.html). (B) A proposed mechanism for DmTrxR. The dotted arrows between the thiolate of Cys-62 and the flavin indicate charge–transfer interactions. The change in conformation is indicated by the juxtaposition of the two polypeptide chains. See text for other details. (C) Corey–Pauling–Koltun space-filling model of the C terminus of wild-type-DmTrxR in the dithiol state. (D) Corey-Pauling-Koltun space-filling model of the C terminus in the disulfide state. All peptide bonds shown in C and D are trans-configured.
In principle, either cysteine Cys489 or Cys490 could be the thiolate that attacks and reduces the substrate. It has been shown by site-directed mutagenesis and chemical modification studies that Cys490, the penultimate residue of the enzyme, is the thiolate that reacts with both the N-terminal interchange thiol, Cys57, and with Trx (22). The significant differences in kcat values between the SCCG mutant, with ≈30% of wild-type DmTrxR activity and the GCCS-mutant with ≈90%, makes it highly unlikely that the two cysteines in the selenium-free mutants are functionally interchangeable. Consistent with these findings, all mutant forms containing Cys-Sec have essentially the full activity of the wild-type Cys-Cys-enzyme (Fig. 1).
It was surprising to find in the stopped-flow experiments (Fig. 2), that in the enzymes with a serine at position 488, SCCG, and SCCS (wild-type), the flavin was more oxidized than in GCCS, the mutant showing the higher catalytic activity (Fig. 1). The equilibria in Fig. 4B describe the dithiol–disulfide-interchange reactions in the oxidative half-reaction of catalysis. All species, except for species 8, have thiolate-flavin charge-transfer absorbance. Therefore, the equilibria for GCCS result in less species 8 being formed than in SCCG. It is species 8 that is reduced by NADPH. However, this unfavorable equilibrium does not appreciably affect kcat, because the reductive half reaction is not rate limiting in the overall catalysis.
As stated above, both the reduction of Trx by Cys490′ to form a mixed disulfide and the cleavage of the mixed disulfide by Cys489′ require that the thiols be deprotonated; the data indicate that this reaction is promoted by the adjacent Ser residues. The actual acid base catalyst is most likely His464′ in the case of the N-terminal thiols, and His106 for the C-terminal thiols (see above and Fig. 4A). The serine probably assists the deprotonation of either Cys489′ or Cys490′ in various steps of catalysis, as shown in Fig. 4B.
The catalytic mechanism that we propose involves two uncommon configurations as intermediates; that of hydrogen bonding between the Cys residue thiol groups and hydroxyl groups of flanking Ser residues, and a disulfide between two contiguous Cys residues. Although the conformations required for the essential intermediates may seem unfavorable, Corey–Pauling–Koltun space-filling models (Fig. 4 C and D) of the C terminus of wild-type DmTrxR show that interactions, such as those proposed in Fig. 4B, are possible. All peptide bonds are shown in trans-configuration. We have only shown two of the many conformations the peptide permits; many others could be consistent with the proposed mechanism.
The scheme in Fig. 4B shows how the serine residues might potentiate deprotonation (species 1–3). Each Ser-Cys couple serves a specific purpose. We propose that the hydroxyl of the C-terminal Ser forms a hydrogen bond with the neighboring cysteine thiol (Cys490′) to facilitate formation of a reactive thiolate (species 2). This thiolate attacks the disulfide bridge of the substrate, Trx, forming a mixed disulfide and a free thiolate on the substrate, with the thiolate of Trx taking up a proton, probably from His106 (species 3). The thiol of Cys489′ is similarly activated by its adjacent Ser488′ hydroxyl (species 4), and the resulting thiolate of Cys489′ cleaves the intermolecular disulfide between TrxR and Trx, leading to the reduced product, Trx(SH)2, and the C-terminal cysteine residues as a disulfide (species 5). As shown, (22) Cys490′ is also responsible for the dithiol–disulfide interchange with the N-terminal redox-active site, and is promoted by His464′ (species 6 and 7). The x-ray structure of the rat enzyme indicates that the C-terminal tail is quite flexible; the distance between the redox-active Cys497′ and Cys59 could, in that case, be modeled to vary between 3 and 12 Å (20). We suggest that DmTrxR uses this flexibility to optimize interactions of the C-terminal Cys pair alternately with Trx, and with the N-terminal Cys pair as indicated in Fig. 4B (20). The Sec-containing redox-active motif appears to act independent of the nature of the flanking residues, presumably due to the larger atomic radius, a lower pKa, and higher reactivity of selenium (24) compared with sulfur (compare Fig. 4 with Fig. 8, which is published as supporting information on the PNAS web site).
It is tempting to speculate why Diptera, like D. melanogaster and Anopheles gambiae, as well as some other organisms (e.g., Plasmodium falciparum), use selenium-free TrxRs. Alternatively, why do the mammalian TrxR enzymes contain selenium, if a -Ser-Cys-Cys-Ser-COOH motif (as in DmTrxR) is essentially as efficient in Trx reduction as a -Gly-Cys-Sec-Gly-COOH motif? A slightly lowered catalytic activity could easily be compensated for by slightly increased expression of enzyme. In view of their energy-demanding synthesis machineries and dependence on selenium supply, the occurrence of Sec must be explained by some other requirements of the cell. One possibility is that the activity of the Cys-Sec-containing enzymes is less affected by the pH of the surrounding milieu, which would imply that the mammalian Trx system is more resistant to acidic conditions than the corresponding insect system. The importance of pH resistance may have been previously underestimated. Intracellular pH values significantly below neutral as reported for mammalian cells (see e.g., ref. 26) may have imposed a selective genetic pressure that favors Sec over Cys.
Alternatively, the answer might be found in reduction of substrates other than Trx, as impressively indicated by the efficient turnover of non-disulfide compounds, such as the important metabolites, dehydroascorbate, cytosolic ubiquinone, and methylseleninate. These tasks are apparently almost exclusively performed by mammalian (Sec-) TrxRs under physiological conditions (see ref. 3 for a review). The recycling of these intermediates cannot be performed as well with the Cys-Cys-TrxR variants. Thus, it is clear that facilitating the formation of a thiolate cannot fully compensate for the chemical properties of selenium. This finding is revealed, for example, in the reduction of methylseleninate, where the Cys-Sec variants were 3- to 5-fold more efficient than the corresponding Cys-Cys variants. These findings are consistent with the notion that the mammalian Sec-TrxRs have important roles in reduction of low molecular weight substrates and that this might not be as important in insects.
To conclude, we have shown here how a sulfur homolog can approach the catalytic efficiency of a selenium-dependent enzyme. Note that direct sulfur-to-selenium substitution in a selenium-dependent enzyme typically reduces the catalytic efficiency to 5% or less; such dramatic loss of activity has been demonstrated for mammalian TrxR (10–12), as well as for all other natural selenoproteins, where this aspect has been studied (27). The lower activity of cysteine, compared with Sec, has therefore been considered a main reason for the evolution and existence of selenoproteins, despite intricate and resource-demanding translation machineries. Our findings, surprisingly, show that Sec is not necessarily required for efficient catalysis of a high Mr TrxR, and that small changes in the active site microenvironment are sufficient to yield high, selenium-independent activity. We therefore propose that Sec may not be essential for a particular enzyme reaction per se, but that this amino acid expands the metabolic capacities in terms of activity toward a wider variety of substrates and over a broader range of pH. This fact alone could explain why organisms that can rely on a continuous and adequate supply of nutritional selenium have either retained or developed selenium-dependent enzymes.
Supplementary Material
Acknowledgments
We thank Prof. F. Jakob for helpful comments. This work was supported by Deutsche Forschungsgemeinschaft Grant GR 2028/1-1 (to S.G.), the Karolinska Institute, the Swedish Society of Medicine, and Swedish Cancer Society Project Grants 3775 and 4056 (to E.S.J.A.) and by National Institute of General Medical Sciences Grants GM11106 (to D.P.B.) and GM21444 (to C.H.W.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Trx, thioredoxin; TrxR, Trx reductase; DmTrxR, Drosophila melanogaster TrxR; Sec, selenocysteine; U, one-letter code for Sec.
Footnotes
This assay is essentially a Trx assay, yet, due to the recycling of oxidized Trx by means of the nonenzymatic reduction of glutathione disulfide, a constant level of Trx is maintained.
Note that mammalian TrxR and DmTrxR share only ≈55% identity. Thus, >200 residues (45%) are different. Attributing the difference in enzymatic activity, as done earlier, solely to the exchange of one amino acid (Sec ⇌ Cys) is therefore an obvious oversimplification. Our results clearly support this point.
References
- 1.Bösl, M. R., Takaku, K., Oshima, M., Nishimura, S. & Taketo, M. M. (1997) Proc. Natl. Acad. Sci. USA 94 5531–5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Böck, A., Forchhammer, K., Heider, J. & Baron, C. (1991) Trends Biochem. Sci. 16 463–467. [DOI] [PubMed] [Google Scholar]
- 3.Gromer, S., Urig, S. & Becker, K. (2003) Med. Res. Rev. 24 in press. [DOI] [PubMed]
- 4.Gladyshev, V. N., Jeang, K. T. & Stadtman, T. C. (1996) Proc. Natl. Acad. Sci. USA 93 6146–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tamura, T. & Stadtman, T. C. (1996) Proc. Natl. Acad. Sci. USA 93 1006–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhong, L., Arnér, E. S. J., Ljung, J., Åslund, F. & Holmgren, A. (1998) J. Biol. Chem. 273 8581–8591. [DOI] [PubMed] [Google Scholar]
- 7.Arscott, L. D., Gromer, S., Schirmer, R. H., Becker, K. & Williams, C. H., Jr. (1997) Proc. Natl. Acad. Sci. USA 94 3621–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gromer, S., Wissing, J., Behne, D., Ashman, K., Schirmer, R. H., Flohé, L. & Becker, K. (1998) Biochem. J. 332 591–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gromer, S., Arscott, L. D., Williams, C. H., Jr., Schirmer, R. H. & Becker, K. (1998) J. Biol. Chem. 273 20096–20101. [DOI] [PubMed] [Google Scholar]
- 10.Lee, S. R., Bar-Noy, S., Kwon, J., Levine, R. L., Stadtman, T. C. & Rhee, S. G. (2000) Proc. Natl. Acad. Sci. USA 97 2521–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhong, L., Arnér, E. S. J. & Holmgren, A. (2000) Proc. Natl. Acad. Sci. USA 97 5854–5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhong, L. & Holmgren, A. (2000) J. Biol. Chem. 275 18121–18128. [DOI] [PubMed] [Google Scholar]
- 13.Schulz, G. E. & Schirmer, R. H. (1978) Principles of Protein Structure (Springer, New York).
- 14.Wang, X., Connor, M., Smith, R., Maciejewski, M. W., Howden, M. E., Nicholson, G. M., Christie, M. J. & King, G. F. (2000) Nat. Struct. Biol. 7 505–513. [DOI] [PubMed] [Google Scholar]
- 15.Kanzok, S. M., Fechner, A., Bauer, H., Ulschmid, J. K., Müller, H. M., Botella-Munoz, J., Schneuwly, S., Schirmer, R. & Becker, K. (2001) Science 291 643–646. [DOI] [PubMed] [Google Scholar]
- 16.Missirlis, F., Ulschmid, J. K., Hirosawa-Takamori, M., Grönke, S., Schäfer, U., Becker, K., Phillips, J. P. & Jäckle, H. (2002) J. Biol. Chem. 277 11521–11526. [DOI] [PubMed] [Google Scholar]
- 17.Arnér, E. S. J., Sarioglu, H., Lottspeich, F., Holmgren, A. & Böck, A. (1999) J. Mol. Biol. 292 1003–1016. [DOI] [PubMed] [Google Scholar]
- 18.Bauer, H., Kanzok, S. M. & Schirmer, R. H. (2002) J. Biol. Chem. 277 17457–17463. [DOI] [PubMed] [Google Scholar]
- 19.Moore, M. J., Distefano, M. D., Walsh, C. T., Schiering, N. & Pai, E. F. (1989) J. Biol. Chem. 264 14386–14388. [PubMed] [Google Scholar]
- 20.Rietveld, P., Arscott, L. D., Berry, A., Scrutton, N. S., Deonarain, M. P., Perham, R. N. & Williams, C. H., Jr. (1994) Biochemistry 33 13888–13895. [DOI] [PubMed] [Google Scholar]
- 21.Sandalova, T., Zhong, L., Lindqvist, Y., Holmgren, A. & Schneider, G. (2001) Proc. Natl. Acad. Sci. USA 98 9533–9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bauer, H., Massey, V., Arscott, L. D., Schirmer, R. H., Ballou, D. & Williams, C. H., Jr. (2003) J. Biol. Chem. 278 33020–33028. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert, H. F. (1990) Adv. Enzymol. Relat. Areas Mol. Biol. 63 69–172. [DOI] [PubMed] [Google Scholar]
- 24.Huber, R. E. & Criddle, R. S. (1967) Arch. Biochem. Biophys. 122 164–173. [DOI] [PubMed] [Google Scholar]
- 25.Arscott, L. D., Thorpe, C. & Williams, C. H., Jr. (1981) Biochemistry 20 1513–1520. [DOI] [PubMed] [Google Scholar]
- 26.Bevensee, M. O., Cummins, T. R., Haddad, G. G., Boron, W. F. & Boyarsky, G. (1996) J. Physiol. 494 315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stadtman, T. C. (1996) Annu. Rev. Biochem. 65 83–100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.