Abstract
Fluorescence correlation spectroscopy (FCS) can resolve the intrinsic fast-blinking kinetics (FBKs) of fluorescent molecules that occur on the order of microseconds. These FBKs can be heavily influenced by the microenvironments in which the fluorescent molecules are contained. In this work, FCS is used to monitor the dynamics of fluorescence emission from Cy5 labeled on DNA probes. We found that the FBKs of Cy5 can be tuned by having more or less unpaired guanines (upG) and thymines (upT) around the Cy5 dye. The observed FBKs of Cy5 are found to predominantly originate from the isomerization and back-isomerization processes of Cy5, and Cy5-nucleobase interactions are shown to slow down these processes. These findings lead to a more precise quantification of DNA hybridization using FCS analysis, in which the FBKs play a major role rather than the diffusion kinetics. We further show that the alterations of the FBKs of Cy5 on probe hybridization can be used to differentiate DNA targets with single-nucleotide differences. This discrimination relies on the design of a probe-target-probe DNA three-way-junction, whose basepairing configuration can be altered as a consequence of a single-nucleotide substitution on the target. Reconfiguration of the three-way-junction alters the Cy5-upG or Cy5-upT interactions, therefore resulting in a measurable change in Cy5 FBKs. Detection of single-nucleotide variations within a sequence selected from the Kras gene is carried out to validate the concept of this new method.
INTRODUCTION
A variety of confocal fluorescence spectroscopic techniques, for instance, two-color coincidence detection (1–4), single-pair fluorescence resonance energy transfer efficiency analysis (5,6), burst-size distribution analysis (7,8), photon counting histogram analysis (9,10), fluorescence intensity distribution analysis (11,12), burst-integrated fluorescence lifetime analysis (13), and fluorescence correlation spectroscopy (FCS) (14–20) have been developed or made spectacular progress in the past decade. The superior sensitivity of these techniques has facilitated many investigations in life science, such as the observation of molecular dynamics (6,12,13,18), enzymatic kinetics (6,21,22), and molecular interactions (16,17,20), as well as the precise quantification of individual biocomponents (1–5,8,12). These spectroscopic techniques not only monitor the evolution of intrinsic parameters of fluorophores (e.g., lifetime, anisotropy, brightness, and spectral properties), but also analyze fluorescence fluctuations. These fluctuations provide further information about distance between fluorophores, diffusivity and concentration, and thus allow characterization of molecular dynamics, interactions, mobilities, and microenvironments around molecules. Fluorescence intermittency (i.e., fluorescence on-off blinking) that occurs on the order of microseconds to milliseconds is among those properties that can be derived from fluorescence fluctuation analysis. For instance, FCS has been used to characterize the blinking kinetics of many organic fluorophores, including fluorescein derivatives (23), rhodamine derivatives (24), oxazine dyes (16), and the cyanine dye, Cy5 (25,26). However, application-based reports using these blinking kinetics have remained in the physical arena, whereas chemical and biological assays remain uncommon, with limited use in a pH sensing probe (23) and a protein binding assay (16).
It has been shown recently that the cyanine dye Cy5 possesses an intriguing long timescale blinking behavior under deoxygenated aqueous conditions that can be used to study molecular conformation dynamics as a new type of molecular ruler (27), to make a reversible molecular optical switch as data storage element (28), and to develop a novel scheme for subdiffraction-limit imaging (29,30). However, these slow-blinking kinetics of Cy5 have on-off durations of hundreds of milliseconds to tens of seconds, often overlapping with timescales of fluorescence fluctuations resulting from biological events (31). Special detection schemes (31) or buffer adjustments (32) are often needed for direct application of these slow-blinking kinetics in biological studies. On the other hand, blinking kinetics that occur on the order of microseconds to tens of microseconds (hereafter, referred to as fast-blinking kinetics, FBKs) that can be altered by biological events, such as molecular association or dissociation, may provide a much more useful indicator for the development of new biological assays.
In this work we use the FBKs of Cy5 to quantify the hybridization of specific-nucleic acids and detect nucleic acid sequences with single nucleotide differences. Alterations of Cy5 FBKs are monitored using FCS and seen as shifts in autocorrelation curves at the fast-relaxation process region, which can be characterized by the FCS parameter mean relaxation time (MRT, 〈τr〉). Specifically, the FBKs of Cy5 labeled on DNA probes are found to be influenced by GT content in the probe sequences, with a general trend for a longer MRT (i.e., slower blinking rate) with increasing GT content (i.e., more Cy5-unpaired guanines (upG) or Cy5-unpaired thymines (upT) interactions). Importantly, hybridization of these probes to complementary sequences results in disruption of the Cy5-upG and Cy5-upT interactions and immediate restoration of the dye's MRT to a lower value. On an FCS autocorrelogram, this hybridization-induced shift at the fast-relaxation region (due to the shortened MRT) is found to be more significant than the corresponding shift at the translational diffusion region (due to the increased mass). This leads to a more precise quantification of DNA hybridization using FCS analysis, in which the MRT plays a major role rather than the diffusion time. Precise quantification of DNA duplex (bias <2.5%) in a binary (double strand/single strand) mixing system is shown. This result transforms FCS autocorrelation binding assays to a relatively mass-insensitive method, lifting previous limitations to molecular interactions involving large changes in mass or mobility.
Taking advantage of these upG- and upT-affected FBKs of Cy5, we have further developed a hybridization-based, homogeneous detection method that is able to differentiate DNA targets with single-nucleotide differences. This method uses a DNA three-way-junction (hereafter, referred to as 3WJ) formed by two DNA probes (one labeled with Cy5) and the target sequence. These two probes cohybridize to the target in juxtaposition around the nucleotide of interest and form a third, short duplex arm with each other. The sequences of the probes are designed so that the substitution of a single nucleotide on the target will result in a basepairing frame shift in the 3WJ structure. The frame shift changes the number of unpaired guanines or thymines that can interact with the end-labeled Cy5 dye, thus allowing discrimination of targets with single-nucleotide differences based on the MRT measurements of Cy5. Advantages of this method for detection of single-nucleotide differences include its separation-free format and elimination of several restrictions that are characteristic of other hybridization-based assays such as molecular beacons (33), including the need for temperature control and special buffer conditions. Furthermore, the FCS parameter used to characterize the FBKs, the MRT, is more stable than brightness measurements that are sensitive to sample volume, the inner filter effect or photobleaching, and more straightforward than fluorescence lifetime measurements, which require more sophisticated and expensive detection system. To our knowledge, this result represents the first assay capable of detecting single-nucleotide differences based on FBKs of a fluorescence probe alone.
MATERIALS AND METHODS
Oligonucleotide sequences
All the Cy5-labeled DNA probes and the unlabeled DNA strands were synthesized by Integrated DNA Technology, with the Cy5 attached to the 5′ phosphate of the DNA probe. The DNA probes and target used in the hybridization quantification experiments and in the investigation of nucleobase influences on the Cy5's FBKs are listed in Table 1. DNA strands used to create various eight-nucleotide overhangs around the Cy5 labeled on probe P5 are GAG AAT GCC ACA GGG GCC GTG CG (X)8, where X is A, C, T, or G. A reporter probe (Cy5-CTC TCC AGC TCC AAC TAC CAC A), a matching probe (TGC CTA CGC CAA GAG GGG G), and four target strands were used in the single-nucleotide variation detection experiments. The sequence of the target is CTT GTG GTA GTT GGA GCT GXT GGC GTA GGC AAG AGT GCC T, where X is G for the wild-type target (KrasG) and T, A, and C for the mutant-type targets (KrasT, KrasA, KrasC, respectively). The reporter probe, the matching probe and the individual Kras targets were mixed at the final concentrations of 5 nM, 50 nM, and 50 nM, respectively.
TABLE 1.
Oligonucleotides used in the study of FBKs of Cy5 labeled on DNA
| Oligo name | Oligo sequence | Guanine and thymine content (G + T) |
|---|---|---|
| P1 | *CAA CCC CAA ACC ACA ACC ATA A | 1 (G-0, T-1) |
| P2 | *CTC TCC AGC TCC AAC TAC CAC A | 5 (G-1, T-4) |
| P3 | *TAA CCG CAA AAT ACG AAC GCG | 6 (G-4, T-2) |
| P4 | *GAC CCC GAA CCG CGA CCGTAA | 6 (G-5, T-1) |
| P5 | *CGC ACGGCC CCTGTGGCA TTC TC | 11 (G-6, T-5) |
| P6 | *ACTTTC CAGGGA GGC GTGGCC TG | 14 (G-9, T-5) |
| P7 | *GAG AGTGGC GTA GGC AAG AGGT | 14 (G-11, T-3) |
| P8 | *GTG CGTTCGGCGGTTGCGGAG A | 16 (G-11, T-5) |
| P9 | *GTGTGTTTGGTGGTTGTGGAG A | 20 (G-11, T-9) |
| P5C | GAG AAT GCC ACA GGG GCC GTG CG |
Labeled with a Cy5 dye.
All hybridization was carried out in a buffer that contains 20 mM Tris-HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2, and 0.1% NP40, with the reaction temperature slowly cooling down from 95°C to room temperature. The samples were then stored in a −20°C freezer until required for binary mixing experiments or for FCS measurements.
FCS setup
FCS measurements were carried out with a custom-built inverted confocal fluorescence spectroscopic system, as described in previous publications (4,20). A He-Ne (633 nm, 25-LHP-151-249, Melles Griot) laser was used to excite the Cy5 dyes. An avalanche photo diode (APD, SPCM-AQR-13, Perkin-Elmer) was used to detect Cy5 emission and a hardware correlator (ALV-5000/EPP, ALV-GmbH) was used compute autocorrelation functions. Unless otherwise noted, the laser excitation intensity was kept low (100 μW before entering the aperture of objective) to avoid fluorescence saturation, triplet-state formation and photobleaching (26,34). The laser beam was focused 10 μm into the sample and was controlled by a piezo-actuator with submicron resolution (P-517.3CL, Physik Instrumente) for all FCS measurements in this work. All the FCS measurements were carried out at a Cy5 concentration of 5 nM and the autocorrelation analysis was carried out for 100 s during each measurement.
Binary mixing and DNA hybridization analysis
A DNA duplex (dsDNA, probe P5 hybridized with strand P5C, prepared separately) was mixed with its corresponding single-stranded probe (ssDNA, probe P5) at various molar ratios, which simulated different hybridization fractions. The total oligo concentration (i.e., (ssDNA) plus (dsDNA)) was fixed at 5 nM in each mixture. The mixing molar fraction, R, was defined as the molar fraction of dsDNA in every mixture and had a known value based on the mixing procedure. On the other hand, the hybridization fraction, Y, was defined as the fraction of Cy5-labeled oligo forming duplex and was estimated from FCS two-component analysis for each binary mixing sample (see Appendix, Eq. 5). Diffusion times (τd) and other FCS parameters (F, τr, α, β) were first determined using the one-component model for samples containing only ssDNA or dsDNA (see Appendix, Eqs. 1–3). The relative brightness (Q) of Cy5 on dsDNA to that on ssDNA was estimated from fluorescence intensity analysis. These parameters were then used in the two-component model to analyze the autocorrelation curves measured from the binary mixtures and to compute the corresponding value of Y.
RESULTS
Effect of GT content on the FBKs of Cy5 (labeled on DNA probe)
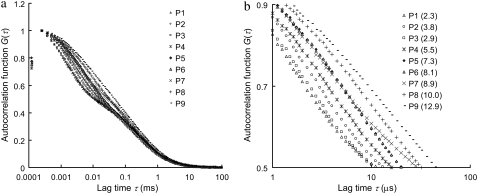
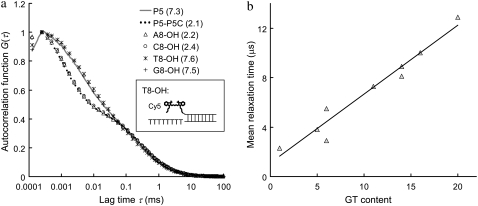
Fig. 1 shows the autocorrelation curves of nine selected single-stranded probes (probe P1–P9, Table 1) measured by FCS. The MRT of each probe is calculated according to Eq. 4 in Appendix and is shown in parentheses in Fig. 1 b. Each of these probes has a unique autocorrelation curve that does not overlap with curves from other probes within the autocorrelogram's fast-relaxation process region (lag time τ from 1 to 10 μs). We view this kind of uniqueness as the “signature” of each probe, and hypothesize that the remarkable variation seen at the fast-relaxation process region may be a result of some altered microsecond-time-scale relaxation processes of Cy5, possibly due to different Cy5-nucleobase interactions within each probe. To investigate this hypothesis, a separate experiment is carried out using complementary DNA sequences to create different eight-nucleotide overhangs around the Cy5 dye labeled on probe P5 through hybridization. These overhangs are eight consecutive adenines (A8), cytosines (C8), thymines (T8), and guanines (G8), respectively. The results are shown in Fig. 2 a, together with the autocorrelation curves from probe P5 alone and the P5-P5C duplex. Fig. 2 a shows nearly identical curves from probe P5, T8-OH (i.e., a P5-P5C duplex DNA with a (T)8 overhang; see Fig. 2 a, inset), and G8-OH, characterized by long MRTs (∼7.5 μs). On the other hand, the curves from P5-P5C duplex, A8-OH, and C8-OH are characterized by shorter MRTs (∼2.3 μs). Based on these results and the fact that free Cy5 dye in the same buffer has a short MRT (∼0.7 μs), we conclude that Cy5 interactions with “unpaired” guanines and thymines in its close proximity (such as unpaired G and T in probe P5 and the (T)8 or (G)8 overhangs in T8-OH and G8-OH) can act to alter the FBKs of Cy5. The Cy5-upG and Cy5-upT interactions tend to slow down the fast-relaxation process of Cy5, leading to an elongated relaxation time measured in FCS. However, Cy5 does not seem to interact with the (A)8 or (C)8 overhangs or the basepaired guanines or thymines within the duplex. A notable observation from this initial experiment is the MRT change from 7.3 μs for the single-stranded P5 probe to 2.1 μs for the double-stranded P5-P5C duplex, which may be due to the disrupted Cy5-upG and Cy5-upT interactions on duplex formation. This is also supported by the observation that on hybridization all the DNA probes show reduced MRTs (∼2 μs), close to the MRT of a free probe with minimum GT content in its sequence (data not shown). Similar disruption in interactions between Cy3, a closely related dye to Cy5, and thymine after duplex formation has been reported previously and was investigated through absorbance measurements (35). Fig. 2 b shows the GT content within each of the nine probes versus the associated MRT. A strong correlation between GT content and MRT is noted. The general trend is for probes with more GT content in their sequences to have a longer MRT. However, as shown in Fig. 1 b, probe P3 (GT content-6) has shorter MRT than that of probe P2 (GT content-5), whereas probes P6 and P7 (GT content-14) have distinct MRTs. The results suggest that the FBKs of Cy5 are not only influenced by the number of GT but also by the spatial arrangement of GT nucleobases within each Cy5-labeled probe. Further investigation, similar to the study done by Nazarenko et al. (36) on fluorescence quenching due to different nucleotide arrangements around fluorescein dye, is necessary to elucidate all the factors that contribute to the changes of FBKs of Cy5 labeled on DNA.
FIGURE 1.
(a) Autocorrelation curves of nine single-stranded DNA probes (probe P1–P9 in Table 1). (b) A zoom-in of fast-relaxation process region on autocorrelogram. The mean relaxation time (MRT, 〈τr〉 in μs) of each probe is shown in parentheses. The autocorrelation curves are normalized at their peaks for comparison purpose.
FIGURE 2.
(a) Autocorrelation curves of six difference Cy5 samples: probe P5, P5-P5C duplex, and P5-P5C hybrids with (A)8, (C)8, (T)8, and (G)8 overhangs, respectively. The MRT of each sample is shown in parentheses (in μs). The inset is a schematic representation of the DNA hybrid with an eight-thymine overhang close to the Cy5 (denoted T8-OH). The concentration of Cy5 is fixed at 5 nM during measurements. (b) GT content within each of the nine DNA probes versus the associated MRT. A strong correlation is seen between GT content and MRT.
Quantification of DNA duplex using FCS two-component analysis
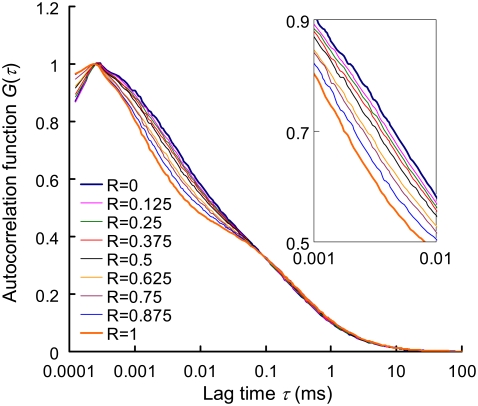
The autocorrelation curves of the dsDNA (P5-P5C duplex), the ssDNA (probe P5) and their binary mixing samples (R ranging from 0 to 1 with an increment of 0.125) are shown in Fig. 3 and the quantification results using FCS two-component model are listed in Table 2. It is noted that on hybridization, a more significant shift in the autocorrelation curve is seen at the fast-relaxation process region compared to the translational diffusion region. From Table 2, it is clear that MRT decreases by nearly 3.5-fold on hybridization whereas characteristic diffusion time only increases by 1.2-fold. This significant change in MRT on hybridization greatly enhances the DNA duplex quantification accuracy, with biases (i.e., Y − R) <2.5% in all binary mixtures. This kind of accuracy is not seen in previous quantification attempts using FCS two-component analysis. The extent of the change in MRT on hybridization is key to quantification accuracy and is influenced by the number and the spatial arrangement of GT nucleobases within the Cy5-labeled probe. In the case where the Cy5-labeled probe contains only one thymine (probe P1), the shift in Cy5 MRT on duplex formation becomes insignificant and quantification accuracy is poor (see Fig. S1 in the Supplementary Material, Data S1).
FIGURE 3.
Autocorrelation curves of nine binary (dsDNA/ssDNA) mixtures. R represents the mixing molar fraction of dsDNA to total DNA. The overall DNA concentration (i.e., (ssDNA) + (dsDNA)) is fixed at 5 nM in each sample. The representative autocorrelation curves are averages from six 100-s measurements. The inset is zoom-in of lag time from 1 to 10 μs.
TABLE 2.
Autocorrelation analysis (see Appendix) of the binary mixing samples
| R | Y† | Bias† (Y-R) |
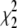 † †
|
〈τr〉* (μs) | 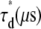 |
β* |
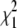 * *
|
|---|---|---|---|---|---|---|---|
| 0 | 0.000 ± 0.020 | 0.000 | 0.46 | 7.3 ± 0.3 | 217.2 ± 5.6 | 0.585 ± 0.009 | 0.47 |
| 0.125 | 0.146 ± 0.012 | 0.021 | 0.47 | 6.6 ± 0.3 | 223.3 ± 9.1 | 0.571 ± 0.016 | 0.44 |
| 0.25 | 0.247 ± 0.014 | −0.003 | 0.49 | 6.3 ± 0.2 | 231.5 ± 7.5 | 0.549 ± 0.012 | 0.47 |
| 0.375 | 0.354 ± 0.011 | −0.021 | 0.53 | 5.9 ± 0.3 | 237.8 ± 9.8 | 0.527 ± 0.021 | 0.53 |
| 0.5 | 0.480 ± 0.008 | −0.020 | 0.85 | 5.2 ± 0.2 | 246.4 ± 9.8 | 0.516 ± 0.013 | 0.74 |
| 0.625 | 0.639 ± 0.006 | 0.014 | 0.88 | 4.2 ± 0.0 | 252.0 ± 2.6 | 0.493 ± 0.012 | 0.85 |
| 0.75 | 0.726 ± 0.014 | −0.024 | 1.31 | 3.6 ± 0.1 | 253.7 ± 5.8 | 0.494 ± 0.014 | 1.14 |
| 0.875 | 0.874 ± 0.006 | −0.001 | 1.41 | 2.8 ± 0.0 | 263.4 ± 2.2 | 0.511 ± 0.011 | 1.41 |
| 1 | 0.999 ± 0.008 | −0.001 | 1.71 | 2.1 ± 0.1 | 265.3 ± 10.0 | 0.548 ± 0.034 | 1.36 |
The values 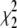 and
and 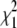 measure the goodness of the fit for the two-component and one-component models, respectively (42).
measure the goodness of the fit for the two-component and one-component models, respectively (42).
From the one-component model.
From the two-component model.
R, mixing molar fraction of dsDNA to total DNA; Y, fraction of Cy5-labeled oligo forming duplex; Bias, Y-R; 〈τr〉, mean relaxation time; τd, diffusion time; β, stretch parameter.
Effects of illumination power and buffer conditions on the FBKs of Cy5 (labeled on DNA probe)
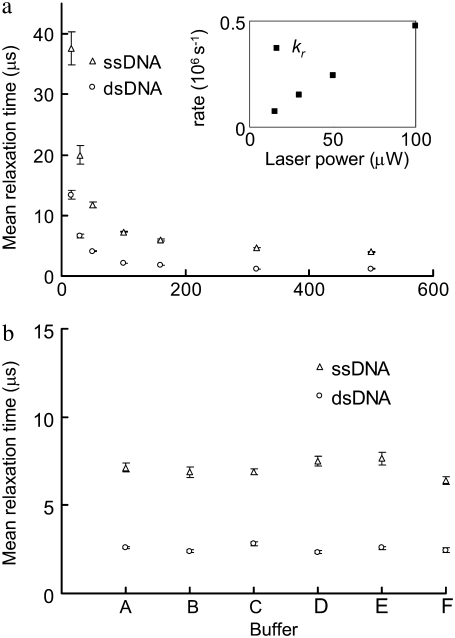
Fig. 4 a shows the MRTs of the dsDNA (P5-P5C duplex) and the ssDNA (probe P5) measured by FCS at various laser illumination powers, ranging from 500 μW to 15 μW. It is noted that the FBKs of Cy5 labeled on DNA are significantly altered by changing the illumination power, with a relaxation rate (kr, defined as 1/MRT) linearly dependent on the illumination power (Fig. 4 a, inset). This suggests that the observed FBKs originate from some photo-induced, fast-relaxation process of Cy5. Fig. 4 b shows the MRTs of the same dsDNA and ssDNA under different buffer conditions. The FBKs of Cy5 labeled on DNA are found insensitive to various buffer conditions. Chemicals such as β-mercaptoethanol (27,28), potassium iodide (25), and Trolox (32) have been reported to alter some photophysical properties of Cy5 in solution. We have added these chemicals to the buffer to evaluate the effect of these chemicals on the MRT measurements. Interestingly, neither apparent shifts in autocorrelation curves (see Fig. S3, Data S1) nor significant changes in MRTs (Fig. 4 b) are seen due to these chemical additives. Addition of sucrose to solution containing Cy5 has been reported to significantly lower the trans-cis isomerization rate of Cy5, leading to an elongated isomerization relaxation time that can be observed within the fast-relaxation process region on autocorrelograms (25). We have also added sucrose to our buffer and a result similar to the one presented by Widengren and Schwille (25) is obtained (see Fig. S4, Data S1). Previous groups have reported a linear dependence of isomerization rate on laser power for Cy5-labeled DNA duplexes (26) and significant reduction in the isomerization rate of Cy5 conjugated to DNA compared to that of free dye (26,34). All of the above evidence suggests that the change in FKBs observed in our FCS measurements originate predominantly from the altered isomerization and back-isomerization processes of Cy5.
FIGURE 4.
(a) MRTs of the ssDNA and the dsDNA estimated by FCS under various laser illumination powers. The inset shows the relaxation rates (kr, 106s−1) of Cy5 on the dsDNA at various laser powers. (b) The MRTs under different buffer conditions. (A) In a buffer containing 100 mM sodium phosphate (pH 7.0) and 50 mM NaCl. (B) In a PCR buffer containing 40 mM Tris-HCl (pH 8.8), 20 mM KCl, 20 mM (MH4)2SO4, 4 mM MgCl2, and 0.2% Triton X-100. (C) In a buffer containing 20 mM Tris-HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2, and 0.1% NP40 and 143 mM β-mercaptoethanol. (D) In a buffer containing 20 mM Tris-HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2, 0.1% NP40, and 2 mM Trolox. (E) In a buffer containing 20 mM Tris-HCl (pH 7.5), 50 mM NaCl, 5 mM MgCl2, 0.1% NP40, and 100 mM KI. (F) In water. The dsDNA is first prepared in a 50 mM Tris-HCl (pH 8.0) buffer at 1 μM. Then it is 200-fold diluted in the above solutions. Error bars represent SD from six FCS measurements.
Detection of single-nucleotide variants
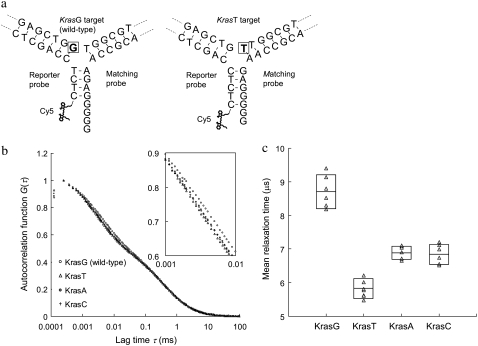
The finding that the FBKs of Cy5 can be “tuned” by having more or less unpaired guanines or thymines in Cy5 proximity has been used to design a simple detection method capable of discriminating targets with single-nucleotide variations. In this method, a pair of probes, which includes a reporter and a matching probe, is used for single-nucleotide variation detection on a target sequence. For proof of concept, synthetic DNA targets that mimic mutational variants of the Kras gene at codon 12 are used in the experiment. Hybridization of these probes to the wild-type (KrasG) versus the mutant-type (KrasT) targets results in an altered conformation of the DNA 3WJs (Fig. 5 a). The 3WJ formed with the wild-type target has a (G)4 DNA overhang in the third arm where Cy5 is labeled. However, the 3WJ based on the mutant-type target has a shorter (G)2 overhang. The change of overhang length is a result of the probe design, which allows a two-nucleotide frame shift when a guanine is replaced with a thymine at the branch point of the target sequence. Thus polymorphisms can be discriminated by measuring the change in MRTs of Cy5, because the frame shift alters the extent of Cy5-upG interactions and thereby the FBKs of Cy5. The autocorrelation curves measured from the wild-type and the mutant-type targets are shown in Fig. 5 b. The MRTs are 8.7 and 5.9 μs for the wild-type (KrasG) and the mutant-type (KrasT) targets, respectively. As expected, a longer MRT is measured in the wild-type experiment where more significant Cy5-upG interactions occur due to a larger number of unpaired guanines in the vicinity of Cy5. In contrast, a MRT of 3.6 μs is measured in a negative control experiment where the targets are absent. It is noteworthy that this MRT is identical to that measured from free Cy5-labeled reporter probe, suggesting that a stable hybrid cannot form between the reporter and the matching probe in the absence of targets. The results of various control experiments are summarized in Table S1, Data S1.
FIGURE 5.
Detection of single-nucleotide variants on Kras targets using the DNA 3WJ method. (a) Schematics of DNA 3WJs expected to form with KrasG (wild-type) and KrasT (mutant type) targets, respectively. In presence of KrasG target in solution, a (G)4 overhang is formed at the end of the third arm. On the other hand, in presence of KrasT target, a shorter (G)2 overhang is formed. The nucleotide under investigation is marked by a square. (b) Autocorrelation curves from samples containing reporter probe, matching probe and any of the four Kras targets. The inset is the zoom-in of lag time from 1 to 10 μs. (c) MRTs measured by FCS for four different Kras targets. Each measured data point is represented by a triangle (Δ). The boxes represent the size of SD and the center lines indicate the averages.
We have also attempted to test the same pair of probes with other mutant-type targets (KrasC and KrasA) that have different nucleotide substitutions at the branch point. Interestingly, we found that the MRTs of the two mutant-type targets, KrasC (6.8 μs) and KrasA (6.9 μs), are also very distinct from that of the wild-type target (Fig. 5 c). This result indicates that this new method enables the use of a single pair of probes to detect multiple mutations. The case of multiple mutations within the same tested sequences is often observed in human cancers. For example, in the Kras gene, 12 different base substitutions resulting in missense mutations could occur within codons 12 and 13 (37). Conventional mutation detection methods require use of multiple probes to analyze a gene sequence that has various polymorphisms. The unique capability of this new method in detecting multiple mutations with only a pair of probes may promise a cost-effective method for diagnosis of genetic diseases using point mutations as markers.
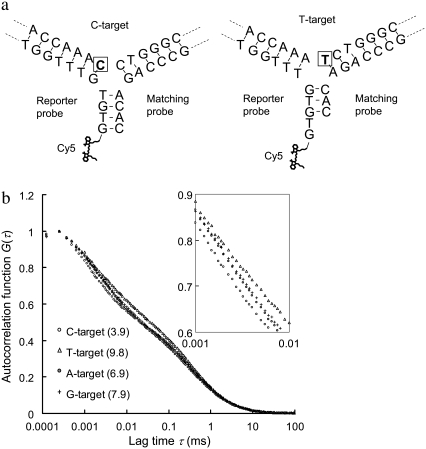
A second verification of using the 3WJ method for discrimination of single-nucleotide variants on a different target sequence is provided in Fig. 6. In this example, a pair of probes is designed mainly to differentiate C to T substitution. The schematics of 3WJs expected to form with the C- and T-targets are shown in Fig. 6 a. The probes are designed to form a blunt-end in the third arm (where Cy5 is labeled) with the C-target, but a sticky-end GT overhang with the T-target. The detection results are presented in Fig. 6 b. The change in Cy5 MRT due to C to T substitution in this case is much more significant than the change in the previous case of G to T substitution. The reason is believed to be that a stable third arm cannot form with only three base pairings in the arm. Because the third arm does not form with the T-target, more upGs and upTs are allowed to interact with the Cy5. More evidence to support this hypothesis and the control experiments of this second case are provided in Fig. S5, Data S1. Similarly, the MRTs measured with the other two single-nucleotide variants (A- and G-target) also show substantial deviations from that with perfectly complementary target (C-target), further showing the unique feature of this method for multiplexed nucleotide variation analysis.
FIGURE 6.
Second verification of using the 3WJ method for discrimination of single-nucleotide variants on a different target sequence. The sequence of the four targets used here is AAT CTC CAC AAC CAC CAA AXC TGG GCA GGC TTC TCT GTA G, where X is A, C, T, or G (denoted A-, C-, T-, and G-target). A reporter probe (Cy5-GTG TGT TTG GTG GTT GTG GAG A) and a matching probe (AGC CTG CCC AGA CAC) are designed to target C to T substitution. (a) Schematics of DNA 3WJs expected to form with the C- and T-targets, respectively. The nucleotide under investigation is marked by a square. (b) Autocorrelation curves from samples containing the reporter probe, the matching probe and any of the four targets. The numbers in parentheses represent the MRTs of each sample in μs.
DISCUSSION
Some organic dyes, such as fluorescein derivatives (23,36), rhodamine derivatives (13,24,38) and oxazine dyes (18,38–40), can be quenched by guanines (basepaired or unpaired) through photo-induced charge transfer (38,41). However, it has been reported that fluorescence quenching by guanine via charge transfer does not happen to Cy5 dye (38,41). In fact, in our experiments Cy5 emission is found to be slightly enhanced through the Cy5-upG and Cy5-upT interactions. It has also been suggested that Cy5 in both trans and cis states can associate with DNA (28,34), and it is the rate of dissociation that determines the observed isomerization rate (34). Given these considerations, we hypothesize that Cy5 in trans conformation (trans is the normal, fluorescent state whereas cis is a weakly fluorescent state) associates better with nearby, unpaired guanines and thymines, resulting in a lower dissociation rate. Not only does this association event slow down the isomerization of Cy5, resulting in the elongated MRT, but it also enhances the Cy5 fluorescence emission because the dye now stays in its trans form longer than typical.
To our knowledge, this is the first report discussing the interactions between Cy5 and specific nucleobases, and the consequential effects on Cy5 FBKs. The significance of this finding is shown herein through evidence that the FBKs of Cy5 labeled on DNA probe can be “tuned” on probe-target hybridization. This newly discovered property of Cy5 is used to improve the FCS quantification accuracy in DNA binary mixing systems, as a precursor for a DNA hybridization assay. Furthermore, the same concept of “tunable blinking kinetics” is used to develop a hybridization-based methodology for the detection of targets with single-nucleotide differences. The key to this assay lies in the observation that only unpaired guanines and thymines can change Cy5 FBKs, not those within the double helix. However, in previous reports using assays that are based on the detection of guanine-induced quenching of fluorescence, both basepaired and unpaired guanines can quench fluorophores through photo-induced charge transfer (36). Our finding allows direct control over the FBKs of Cy5 through design of binding probes for assays, whose sensitivity is only influenced by the number of unpaired guanines or thymines around Cy5 before and after the formation of DNA duplex or 3WJ.
FCS is used commonly for the analysis of molecular binding by measuring the associated change of molecular mobility (16,17,20). However, conventional FCS assays are limited to binding systems involving large mass or mobility changes on molecular association to obtain a precise quantification result (42–44). By taking advantage of the FBKs of Cy5 in FCS analysis, we have achieved the detection and precise quantification of DNA hybridization involving only a twofold mass change on molecular association. These results have opened a new scope in autocorrelation analysis by lifting the previous mass limitation on FCS resolution.
The proposed single-nucleotide discrimination methodology is based on the formation of DNA 3WJ on probe-target hybridization. In practice, the single-stranded target can be produced from asymmetric polymerase chain reaction (45) or from the selective enzymatic hydrolysis of the antisense strand by T7 gene 6 exonuclease (46). Unlike other FCS-based, single-nucleotide variation detection techniques (47), our method does not require the use of additional enzymatic reactions for probe elongation or extension. Other homogeneous, hybridization-based molecular probe techniques for single-nucleotide discrimination, such as locked nucleic acid probe (48) and molecular beacon probe (33,40), rely on the decrease in thermostability caused by a single-pair mismatch in probe-target hybrids. The change in thermostability due to a single-pair mismatch is usually small, therefore leading to ambiguous discrimination results. Enlarging the thermostability difference by carrying out detection at elevated temperature (33) or in a buffer with specific divalent ion concentration is often needed in these methods to obtain the best discrimination results. Another way to achieve better discrimination specificity is through multiparameter fluorescence detection (40); however, this increases assay complexity. In our method, the DNA 3WJ gives very stable and reproducible single-nucleotide discrimination results and there is no need to carry out detection at elevated temperature or to use an optimized buffer system.
CONCLUSION
We report the observation of interactions between Cy5 and unpaired guanines or thymines within Cy5-labeled DNA probes and the resulting effect on Cy5 FBKs. This finding facilitates a new scheme for precise DNA hybridization analysis in a homogeneous format. Such hybridization analysis strategies can be used in the quantification of short amplicons during polymerase chain reaction (PCR). Hybridization analysis and quantification of PCR products using FCS have previously been reported (45,49). However, without taking advantage of FBKs of Cy5, the conventional FCS quantification method is limited to a labeled primer being extended into a relatively large PCR product (for instance, 500 bp) (45). We show that the MRT, an FCS parameter that characterizes the FBKs, can be used as an independent, information-rich parameter in confocal fluorescence detection schemes. Furthermore, we have used this parameter as an indicator in a newly developed assay for detection of single-nucleotide variations. To use the FBKs of Cy5, we design a DNA 3WJ whose basepairing configuration can be altered due to a single-nucleotide substitution on the target. The initial design only gives a two-guanine frame shift on a single-nucleotide replacement. However, this frame shift is sufficient to cause a measurable change in Cy5 MRT. Future work will be focused on various 3WJ designs that can have larger frame shifts due to a single-nucleotide substitution on the target and also on the extension of this technique to detect all single-nucleotide variation combinations. Furthermore, a new range of analytical tools may be developed based on this new finding of hybridization dependant alterations in the FBKs of Cy5. In theory, any binding of ligand to DNA can be analyzed by the FBKs of Cy5, if such a ligand binding disrupts Cy5-upG or Cy5-upT interactions.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation and the Defense Advanced Research Projects Agency.
APPENDIX: DETAILED DESCRIPTION OF THE FSC ANALYSIS
1. The one-component model.
The following one-component FCS model is used to characterize the dynamics or photophysical processes in single fluorescent species systems (20,25,42):
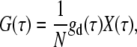 |
(1) |
where G(τ) is the autocorrelation function and τ is the lag time. N is the average number of light-emitting particles residing in the detection volume. gd(τ) is the autocorrelation function arising from fluorescence fluctuations due to translational diffusion (42), for which we use a simplified 2D diffusion model with an anomalous factor to represent (20):
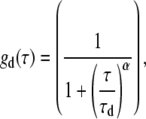 |
(2) |
where α is the anomalous factor and τd is the characteristic diffusion time. X(τ) is the fluctuation due to the fast-blinking kinetics (FBKs), for which we use the following stretched-exponential form to represent (18,34,50):
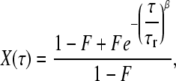 |
(3) |
where F is the effective fraction of molecules in non-fluorescent states and τr is the relaxation characteristic time in dark states. β is the stretch parameter. The mean relaxation time (MRT, 〈τr〉) is calculated as follows (18,34):
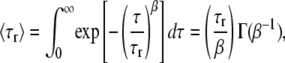 |
(4) |
where Γ(β−1) is the gamma function.
Autocorrelation curves are normalized to common peak amplitudes of one for comparison; thus, the particle number N is not used for further analysis. A similar normalization scheme is used by Haupts et al. (51) for illustrating fluorescence fluctuation dynamics in green fluorescence protein. Although normalization clearly illustrates the change in FBKs among a group of FCS curves, it hides information such as the suppression or enhancement of the autocorrelation amplitude due to FBKs. The unnormalized autocorrelation curves from Fig. 3 are included in the Supplementary Material (Fig. S7, Data S1) with typical amplitudes ranging from 0.3 to 0.6. Further treatment of autocorrelation curves to correct for afterpulsing has been shown to improve accuracy in timescales related to the FBK measurements herein (52); however, the FCS data presented here is not corrected for these detector artifacts.
2. The two-component model.
In the presence of two types of fluorescent species, for instance, the single-stranded probe and the DNA duplex, the molar fraction of the second species (i.e., the DNA duplex), Y, can be determined using the following two-component analytical model (20,42):
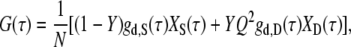 |
(5) |
where Q is the relative brightness of Cy5 on the DNA duplex compared to that on the free, unhybridized single-stranded probe. The subscripts S and D denote the single-stranded and double-stranded species, respectively. The least-squares fit used in this work is based on the Levenberg-Marquardt algorithm within Origin 7.0 (OriginLab).
Editor: Kathleen B. Hall.
References
- 1.Castro, A., and J. G. K. Williams. 1997. Single-molecule detection of specific nucleic acid sequences in unamplified genomic DNA. Anal. Chem. 69:3915–3920. [DOI] [PubMed] [Google Scholar]
- 2.Neely, L. A., S. Patel, J. Garver, M. Gallo, M. Hackett, S. McLaughlin, M. Nadel, J. Harris, S. Gullans, and J. Rooke. 2006. A single-molecule method for the quantitation of microRNA gene expression. Nat. Methods. 3:41–46. [DOI] [PubMed] [Google Scholar]
- 3.Orte, A., R. Clarke, S. Balasubramanian, and D. Klenerman. 2006. Determination of the fraction and stoichiometry of femtomolar levels of biomolecular complexes in an excess of monomer using single-molecule, two-color coincidence detection. Anal. Chem. 78:7707–7715. [DOI] [PubMed] [Google Scholar]
- 4.Yeh, H. C., Y. P. Ho, I. M. Shih, and T. H. Wang. 2006. Homogeneous point mutation detection by quantum dot-mediated two-color fluorescence coincidence analysis. Nucleic Acids Res. 34:e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deniz, A. A., M. Dahan, J. R. Grunwell, T. J. Ha, A. E. Faulhaber, D. S. Chemla, S. Weiss, and P. G. Schultz. 1999. Single-pair fluorescence resonance energy transfer on freely diffusing molecules: observation of Forster distance dependence and subpopulations. Proc. Natl. Acad. Sci. USA. 96:3670–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ha, T. J., A. Y. Ting, J. Liang, W. B. Caldwell, A. A. Deniz, D. S. Chemla, P. G. Schultz, and S. Weiss. 1999. Single-molecule fluorescence spectroscopy of enzyme conformational dynamics and cleavage mechanism. Proc. Natl. Acad. Sci. USA. 96:893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enderlein, J., D. L. Robbins, W. P. Ambrose, and R. A. Keller. 1998. Molecular shot noise, burst size distribution, and single-molecule detection in fluid flow: effects of multiple occupancy. J. Phys. Chem. A. 102:6089–6094. [Google Scholar]
- 8.Van Orden, A., N. P. Machara, P. M. Goodwin, and R. A. Keller. 1998. Single molecule identification in flowing sample streams by fluorescence burst size and intraburst fluorescence decay rate. Anal. Chem. 70:1444–1451. [DOI] [PubMed] [Google Scholar]
- 9.Chen, Y., J. D. Muller, P. T. C. So, and E. Gratton. 1999. The photon counting histogram in fluorescence fluctuation spectroscopy. Biophys. J. 77:553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, Y., M. Tekmen, L. Hillesheim, J. Skinner, B. Wu, and J. D. Muller. 2005. Dual-color photon-counting histogram. Biophys. J. 88:2177–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian, H., and E. L. Elson. 1990. Distribution of molecular aggregation by analysis of fluctuation moments. Proc. Natl. Acad. Sci. USA. 87:5479–5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kask, P., K. Palo, D. Ullmann, and K. Gall. 1999. Fluorescence-intensity distribution analysis and its application in biomolecular detection technology. Proc. Natl. Acad. Sci. USA. 96:13756–13761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eggeling, C., J. R. Fries, L. Brand, R. Gunther, and C. A. M. Seidel. 1998. Monitoring conformational dynamics of a single molecule by selective fluorescence spectroscopy. Proc. Natl. Acad. Sci. USA. 95:1556–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magde, D., E. L. Elson, and W. W. Webb. 1972. Thermodynamics fluctuations in a reacting system—measurement by fluorescence correlation spectroscopy. Phys. Rev. Lett. 29:704–708. [Google Scholar]
- 15.Eigen, M., and R. Rigler. 1994. Sorting single molecules—application to diagnostics and evolutionary biotechnology. Proc. Natl. Acad. Sci. USA. 91:5740–5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheffler, S., M. Sauer, and H. Neuweiler. 2005. Monitoring antibody binding events in homogeneous solution by single-molecule fluorescence spectroscopy. Z. Phys. Chem. 219:665–678. [Google Scholar]
- 17.Stoevesandt, O., K. Kohler, R. Fischer, I. C. D. Johnston, and R. Brock. 2005. One-step analysis of protein complexes in microliters of cell lysate. Nat. Methods. 2:833–835. [DOI] [PubMed] [Google Scholar]
- 18.Kim, J., S. Doose, H. Neuweiler, and M. Sauer. 2006. The initial step of DNA hairpin folding: a kinetic analysis using fluorescence correlation spectroscopy. Nucleic Acids Res. 34:2516–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bacia, K., S. A. Kim, and P. Schwille. 2006. Fluorescence cross-correlation spectroscopy in living cells. Nat. Methods. 3:83–89. [DOI] [PubMed] [Google Scholar]
- 20.Yeh, H. C., C. M. Puleo, T. C. Lim, Y. P. Ho, P. E. Giza, R. C. C. Huang, and T. H. Wang. 2006. A microfluidic-FCS platform for investigation on the dissociation of Sp1-DNA complex by doxorubicin. Nucleic Acids Res. 34:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.English, B. P., W. Min, A. M. van Oijen, K. T. Lee, G. B. Luo, H. Y. Sun, B. J. Cherayil, S. C. Kou, and X. S. Xie. 2006. Ever-fluctuating single enzyme molecules: Michaelis-Menten equation revisited. Nat. Chem. Biol. 2:87–94. [DOI] [PubMed] [Google Scholar]
- 22.Rarbach, M., U. Kettling, A. Koltermann, and M. Eigen. 2001. Dual-color fluorescence cross-correlation spectroscopy for monitoring the kinetics of enzyme-catalyzed reactions. Methods. 24:104–116. [DOI] [PubMed] [Google Scholar]
- 23.Widengren, J., B. Terry, and R. Rigler. 1999. Protonation kinetics of GFP and FITC investigated by FCS—aspects of the use of fluorescent indicators for measuring pH. Chem. Phys. 249:259–271. [Google Scholar]
- 24.Widengren, J., J. Dapprich, and R. Rigler. 1997. Fast interactions between Rh6G and dGTP in water studied by fluorescence correlation spectroscopy. Chem. Phys. 216:417–426. [Google Scholar]
- 25.Widengren, J., and P. Schwille. 2000. Characterization of photoinduced isomerization and back-isomerization of the cyanine dye Cy5 by fluorescence correlation spectroscopy. J. Phys. Chem. A. 104:6416–6428. [Google Scholar]
- 26.Widengren, J., E. Schweinberger, S. Berger, and C. A. M. Seidel. 2001. Two new concepts to measure fluorescence resonance energy transfer via fluorescence correlation spectroscopy: theory and experimental realizations. J. Phys. Chem. A. 105:6851–6866. [Google Scholar]
- 27.Bates, M., T. R. Blosser, and X. W. Zhuang. 2005. Short-range spectroscopic ruler based on a single-molecule optical switch. Phys. Rev. Lett. 94:108101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heilemann, M., E. Margeat, R. Kasper, M. Sauer, and P. Tinnefeld. 2005. Carbocyanine dyes as efficient reversible single-molecule optical switch. J. Am. Chem. Soc. 127:3801–3806. [DOI] [PubMed] [Google Scholar]
- 29.Rust, M. J., M. Bates, and X. W. Zhuang. 2006. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods. 3:793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bates, M., B. Huang, G. T. Dempsey, and X. W. Zhuang. 2007. Multicolor super-resolution imaging with photo-swtichable fluorescent probes. Science. 317:1749–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabanayagam, C. R., J. S. Eid, and A. Meller. 2005. Long time scale blinking kinetics of cyanine fluorophores conjugated to DNA and its effect on Forster resonance energy transfer. J. Chem. Phys. 123:224708. [DOI] [PubMed] [Google Scholar]
- 32.Rasnik, I., S. A. McKinney, and T. Ha. 2006. Nonblinking and longlasting single-molecule fluorescence imaging. Nat. Methods. 3:891–893. [DOI] [PubMed] [Google Scholar]
- 33.Mhlanga, M. M., and L. Malmberg. 2001. Using molecular beacons to detect single-nucleotide polymorphisms with real-time PCR. Methods. 25:463–471. [DOI] [PubMed] [Google Scholar]
- 34.White, S. S., H. T. Li, R. J. Marsh, J. D. Piper, N. D. Leonczek, N. Nicolaou, A. J. Bain, L. M. Ying, and D. Klenerman. 2006. Characterization of a single molecule DNA switch in free solution. J. Am. Chem. Soc. 128:11423–11432. [DOI] [PubMed] [Google Scholar]
- 35.Randolph, J. B., and A. S. Waggoner. 1997. Stability, specificity and fluorescence brightness of multiply-labeled fluorescent DNA probes. Nucleic Acids Res. 25:2923–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nazarenko, I., R. Pires, B. Lowe, M. Obaidy, and A. Rashtchian. 2002. Effect of primary and secondary structure of oligodeoxyribonucleotides on the fluorescent properties of conjugated dyes. Nucleic Acids Res. 30:2089–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogelstein, B., and K. W. Kinzler. 1999. Digital PCR. Proc. Natl. Acad. Sci. USA. 96:9236–9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heinlein, T., J. P. Knemeyer, O. Piestert, and M. Sauer. 2003. Photoinduced electron transfer between fluorescent dyes and guanosine residues in DNA-hairpins. J. Phys. Chem. B. 107:7957–7964. [Google Scholar]
- 39.Knemeyer, J. P., N. Marmé, and M. Sauer. 2000. Probes for detection of specific DNA sequences at the single-molecule level. Anal. Chem. 72:3717–3724. [DOI] [PubMed] [Google Scholar]
- 40.Marme, N., A. Friedrich, M. Muller, O. Nolte, J. Wolfrum, J. D. Hoheisel, M. Sauer, and J. P. Knemeyer. 2006. Identification of single-point mutations in mycobacterial 16S rRNA sequences by confocal single-molecule fluorescence spectroscopy. Nucleic Acids Res. 34:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torimura, M., S. Kurata, K. Yamada, T. Yokomaku, Y. Kamagata, T. Kanagawa, and R. Kurane. 2001. Fluorescence-quenching phenomenon by photoinduced electron transfer between a fluorescent dye and a nucleotide base. Anal. Sci. 17:155–160. [DOI] [PubMed] [Google Scholar]
- 42.Meseth, U., T. Wohland, R. Rigler, and H. Vogel. 1999. Resolution of fluorescence correlation measurements. Biophys. J. 76:1619–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayboroda, O. A., A. van Remoortere, H. J. Tanke, C. H. Hokke, and A. M. Deelder. 2004. A new approach for fluorescence correlation spectroscopy (FCS) based immunoassays. J. Biotechnol. 107:185–192. [DOI] [PubMed] [Google Scholar]
- 44.Pope, A. J., U. M. Haupts, and K. J. Moore. 1999. Homogeneous fluorescence readouts for miniaturized high-throughput screening: theory and practice. Drug Discov. Today. 4:350–362. [DOI] [PubMed] [Google Scholar]
- 45.Kinjo, M. 1998. Detection of asymmetric PCR products in homogeneous solution by fluorescence correlation spectroscopy. Biotechniques. 25:706–712, 714–715. [DOI] [PubMed] [Google Scholar]
- 46.Nikiforov, T. T., R. B. Rendle, M. L. Kotewicz, and Y. H. Rogers. 1994. The use of phosphorothioate primers and exonuclease hydrolysis for the preparation of single-stranded PCR products and their detection by solid-phase hybridization. PCR Methods Appl. 3:285–291. [DOI] [PubMed] [Google Scholar]
- 47.Twist, C. R., M. K. Winson, J. J. Rowland, and D. B. Kell. 2004. Single-nucleotide polymorphism detection using nanomolar nucleotides and single-molecule fluorescence. Anal. Biochem. 327:35–44. [DOI] [PubMed] [Google Scholar]
- 48.Simeonov, A., and T. T. Nikiforov. 2002. Single nucleotide polymorphism genotyping using short, fluorescently labeled locked nucleic acid (LNA) probes and fluorescence polarization detection. Nucleic Acids Res. 30:e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kinjo, M., and R. Rigler. 1995. Ultrasensitive hybridization analysis using fluorescence correlation spectroscopy. Nucleic Acids Res. 23:1795–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, H., G. B. Luo, P. Karnchanaphanurach, T. M. Louie, I. Rech, S. Cova, L. Y. Xun, and X. S. Xie. 2003. Protein conformational dynamics probed by single-molecule electron transfer. Science. 302:262–266. [DOI] [PubMed] [Google Scholar]
- 51.Haupts, U., S. Maiti, P. Schwille, and W. W. Webb. 1998. Dynamics of fluorescence fluctuations in green fluorescent protein observed by fluorescence correlation spectroscopy. Proc. Natl. Acad. Sci. USA. 95:13573–13578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao, M., L. Jin, B. Chen, Y. Ding, H. Ma, and D. Y. Chen. 2003. Afterpulsing and its correction in fluorescence correlation spectroscopy experiments. Appl. Opt. 42:4031–4036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.