Abstract
Regulatory pathways for protein glycosylation are poorly understood, but expression of branchpoint enzymes is critical. A key branchpoint enzyme is the T-synthase, which directs synthesis of the common core 1 O-glycan structure (T-antigen), the precursor structure for most mucin-type O-glycans in a wide variety of glycoproteins. Formation of active T-synthase, which resides in the Golgi apparatus, requires a unique molecular chaperone, Cosmc, encoded on Xq24. Cosmc is the only molecular chaperone known to be lost through somatic acquired mutations in cells. We show that Cosmc is an endoplasmic reticulum (ER)–localized adenosine triphosphate binding chaperone that binds directly to human T-synthase. Cosmc prevents the aggregation and ubiquitin-mediated degradation of the T-synthase. These results demonstrate that Cosmc is a molecular chaperone in the ER required for this branchpoint glycosyltransferase function and show that expression of the disease-related Tn antigen can result from deregulation or loss of Cosmc function.
Introduction
Metabolic pathways are commonly regulated at key steps termed branchpoints, in which several key pathways diverge from a single precursor. Such a branchpoint occurs in the biosynthesis of O-glycans of animal cell glycoproteins, in which the common precursor to all mucin-type O-glycans, N-acetylgalactosamine (GalNAc) α1-Ser/Thr (Tn antigen), may be modified by one of several enzymes to generate different core structures known as core 1, core 2, core 3, etc. The core 1 O-glycan is generated by the T-synthase, also known as the core 1 β3 galactosyltransferase (Gal-T), which adds galactose to the Tn antigen to generate Galβ1-3GalNAcα1-Ser/Thr (T-antigen; Ju et al., 2002a,b). This is a key precursor for all core 1 and 2 mucin-type O-glycans in vertebrates and invertebrates.
The overall pathway of mucin glycosylation regulated by the branchpoint T-synthase function is developmentally important because disruption of the T-synthase in mice is embryonic lethal (Xia et al., 2004). Alternatively, another branchpoint enzyme that utilizes the Tn antigen is the C3GnT (core 3 β3-N-acetylglucosaminyltransferase), specifically expressed in the GI tract, which generates the disaccharide N-acetylglucosamine (GlcNAc) β1-3GalNAcα1-Ser/Thr. Disruption of the core 3 O-glycan biosynthesis by eliminating C3GnT in mice is associated with increased susceptibility to colitis and colorectal tumors (An et al., 2007).
We observed that some cells and tissues in patients with Tn syndrome or in some tumor cells lack T-synthase activity, although they have the T-synthase transcript, and express the Tn antigen, indicating a lack of branchpoint enzyme activity. In deciphering the regulation of the T-synthase, we discovered that the T-synthase requires a unique and apparently client-specific chaperone that we termed Cosmc (core 1 β3–Gal-T specific molecular chaperone), which is required for formation of active T-synthase (Ju et al., 2002a,b). Thus, the expression of the Tn antigen in patients with Tn syndrome and in human tumor cells results from mutations in Cosmc (Ju and Cummings, 2005; Ju et al., 2008). These discoveries prompted us to explore the molecular nature of the potential chaperone function for Cosmc and its role in assisting the T-synthase in acquiring its active form.
Somatic mutations in Cosmc result in loss of T-synthase activity, apparently because of degradation of newly synthesized T-synthase via proteasome-dependent pathways (Ju and Cummings, 2002). Cosmc appears to be specific for the T-synthase because expression of many other glycosyltransferase activities is unaffected by loss of functional Cosmc (Piller et al., 1990; Ju and Cummings, 2002), and the only consequence in terms of glycoconjugate biosynthesis in cells accompanying loss of function in Cosmc is an increase in Tn and Sialyl Tn antigen expression. However, the precise location and function of Cosmc as a specific molecular chaperone in T-synthase biosynthesis has not yet been elucidated.
In this paper, we report that Cosmc predominantly localizes in the ER and that the T-synthase is mainly localized in the Golgi apparatus. Importantly, Cosmc has ATP-binding activity that is consistent with its chaperone function for maturation of T-synthase. When the T-synthase is expressed in cells expressing a mutated Cosmc that results in loss of function or in insect cells that constitutively lack Cosmc, the enzyme aggregates and is subsequently degraded in the proteasome. Thus, Cosmc serves a unique function in the ER as the key posttranslational regulator for expression of the T-synthase and may represent a new type of chaperone functioning in regulating protein glycosylation.
Results
Human Cosmc and T-synthase are expressed coordinately in human tissue
To define whether Cosmc and the T-synthase are coordinately expressed, we examined expression using Northern blot analysis of multiple human tissue samples. Cosmc is expressed in all tissues examined (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200711151/DC1), with the highest expression in heart, skeletal muscle, kidney, liver, and placenta, which mirrors the expression of T-synthase (Fig. S1 B; Ju et al., 2002a). The coordinate expression of Cosmc and T-synthase in human tissues is consistent with a close relationship between these two proteins.
Cosmc only exists in vertebrates and is highly conserved across species
Upon BLASTP searching in the National Center for Biotechnology Information database using human Cosmc, we identified Cosmc orthologues in monkey, cow, mouse, rat, dog, bird, frog, and zebrafish (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200711151/DC1). No orthologues were found in Caenorhabditis elegans or Drosophila melanogaster. All Cosmc orthologues are predicted to be type II transmembrane proteins, with a short N-terminal cytoplasmic domain (CD) and transmembrane domain (TMD) and a large C-terminal domain (Fig. S2). Human and primate Cosmc both have 318 aa, and there is only a single amino acid difference at V191I. In contrast, rodent Cosmc contains 316 aa and has >95% identity to human Cosmc but has a 2-aa gap between positions 33 and 34. These 2 aa are located at the beginning of the lumenal domain, which we show to be the functional domain of Cosmc. Frog Cosmc is a 317-aa protein with a gap in the sequence at position 33 compared with human. Zebrafish Cosmc lacks 3 aa at the C terminus compared with human Cosmc, ending at position 315. Except for mouse Cosmc, which can act as a molecular chaperone for both human and mouse T-synthase, the functions of other Cosmc orthologues have not been tested.
Interestingly, T-synthase orthologues were identified in both vertebrates and invertebrates such as C. elegans and D. melanogaster. Whereas T-synthase in some lower vertebrates has nonconserved N-glycosylation sites, mammalian T-synthases lack N-glycosylation sites. In contrast, invertebrate T-synthases, as in C. elegans (Ju et al., 2002a, 2006), contain multiple N-glycosylation sites, some of which appear to be conserved. Expression of invertebrate T-synthase in insect cells does not require Cosmc for active protein formation (Ju et al., 2006), which may suggest that the presence of N-glycosylation on invertebrate T-synthases may allow them to be folded in a Cosmc-independent pathway.
Human Cosmc localizes in the ER and T-synthase localizes to the Golgi apparatus in CHO K1 cells
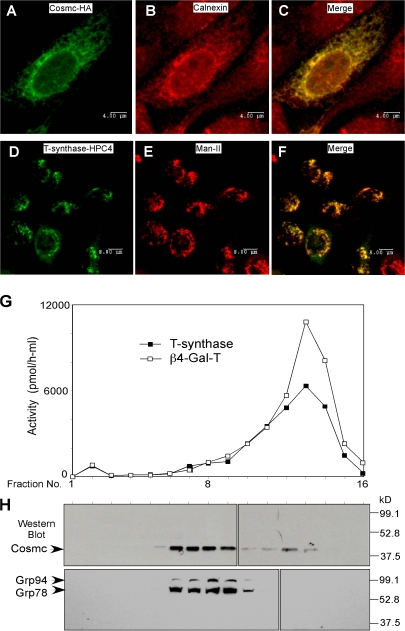
To explore the subcellular localization of Cosmc and T-synthase, we prepared a C-terminal HA-tagged Cosmc (Cosmc-HA) and a C-terminal human protein C (HPC) 4–tagged T-synthase (T-synthase–HPC4). CHO K1 cells were transiently transfected with both constructs and immunostained with anti-HA and anti-HPC4 antibodies. Anti-HA staining revealed a perinuclear pattern of expression for HA-Cosmc, and a merge of this image showed colocalization with calnexin, an ER marker (Fig. 1, A–C). In contrast, anti-HPC4 staining revealed a punctate pattern for T-synthase that was coincident with the localization of α–mannosidase II, a Golgi marker (Fig. 1, D–F). These results demonstrate that human Cosmc localizes in the ER, whereas the T-synthase localizes to the Golgi apparatus.
Figure 1.
Localization of human Cosmc and T-synthase. (A–F) Immunofluorescent staining. CHO K1 cells cultured on chambered slides were transiently transfected with Cosmc-HA or with T-synthase–HPC4 and stained with rat anti-HA IgG1 (green) and rabbit anti-calnexin IgG (red; A–C; bars, 4 μm) or with mouse anti-HPC4 (green) and rabbit anti–α-ManII (red; D–F; bars, 8 μm). (G and H) Sucrose gradient subcellular fractionation. 293T cells transiently transfected with HPC4-Cosmc were harvested and homogenized. The postnuclear supernatant (PNS) was loaded onto a sucrose gradient. After ultracentrifugation, 16 fractions (∼0.6 ml/fraction) were collected and measured for both T-synthase and β4–Gal-T activity (G) and analyzed on Western Blot with anti-HPC4 and anti-KDEL (H).
We also stained with Alexa 488–labeled anti-HA (Fig. S3, green, available at http://www.jcb.org/cgi/content/full/jcb.200711151/DC1) and Alexa 568–labeled Anti-HPC4 (Fig. S3, red) in the same cotransfected cells. Cosmc expression was perinuclear, corresponding to ER localization, and T-synthase stained in a punctate pattern, corresponding to Golgi localization (Fig. S3), which is consistent with the results from Fig. 1. This result further demonstrates that Cosmc and T-synthase localize to different cellular compartments. To confirm the localization of Cosmc in the ER and T-synthase in the Golgi apparatus, we stained human 293T and MDA-MB-231 cells with rabbit polyclonal IgG prepared against peptides from human Cosmc and T-synthase. The staining pattern for anti-Cosmc was similar to that for anti-Calnexin (ER marker), whereas the staining pattern for anti–T-synthase was similar to that for anti–mannosidase II (Golgi marker; unpublished data). These results are consistent with our observations using anti–epitope-tagged forms of Cosmc and T-synthase and further support the conclusion that Cosmc is primarily localized in the ER and T-synthase is in the Golgi apparatus.
Cosmc is in heavy membrane fractions corresponding to the ER and T-synthase is in lighter membrane fractions corresponding to the Golgi apparatus
To further establish the localization of Cosmc in the ER, we performed subcellular fractionation on sucrose gradients of 293T cells expressing full-length HPC4-Cosmc. T-synthase activity was mainly recovered in fractions 12–14, corresponding to the fractions containing the Golgi enzyme marker β4–Gal-T (Fig. 1 G). In contrast, HPC4-Cosmc was recovered primarily in fractions 6–9, corresponding to fractions containing the ER markers GRP78 and GRP94 (Fig. 1 H). These results indicate that Cosmc is primarily localized in the ER, whereas the T-synthase is primarily localized in the Golgi apparatus, which is consistent with the immunofluorescent data (Fig. 1, A–F). Some Cosmc was observed in light membrane fractions and may reflect a small fraction that shuttles between the ER and Golgi during potential retrieval, but this remains to be explored.
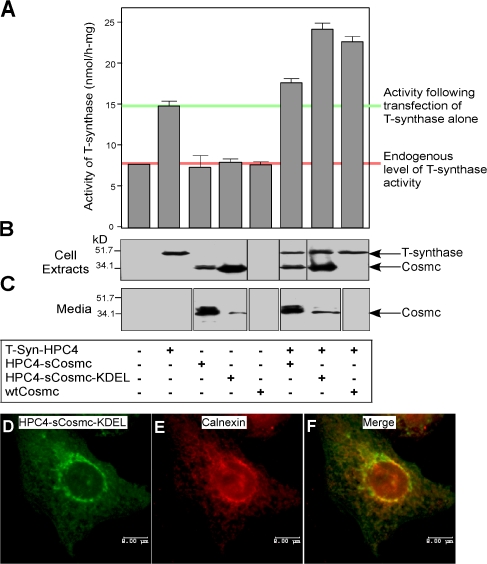
Soluble Cosmc with an engineered ER retention signal promotes T-synthase activity equivalently to wild-type Cosmc
To test whether the TMD of Cosmc was important to localization and function, we made two constructs: one encoding a soluble N-terminal HPC4-tagged Cosmc (HPC4-sCosmc) and the other encoding a soluble Cosmc with a C-terminal KDEL-ER retention sequence (HPC4-sCosmc-KDEL). Cosmc function is generally rate limiting for T-synthase activity (Ju and Cummings, 2002). Therefore, we cotransfected human 293T cells with either HPC4-tagged full-length T-synthase alone or in addition to the Cosmc constructs. Endogenous T-synthase activity was only slightly promoted by HPC4-sCosmc, whereas coexpression with the HPC4-sCosmc-KDEL significantly increased T-synthase activity (Fig. 2 A). The HPC4-sCosmc was mainly secreted into the media, whereas HPC4-sCosmc-KDEL was mainly retained in cells (Fig. 2, B and C). These results show that in the absence of the CD, TMD, and an ER retention signal, Cosmc is secreted from cells. The slight promotion of T-synthase activity by HPC4-sCosmc may result from the small amount of protein that transiently resides in the ER during its biosynthesis. Importantly, both HPC4-sCosmc and HPC4-sCosmc-KDEL were functional in insect cells in rescuing T-synthase activity (unpublished data). As a control, we coexpressed T-synthase with sCosmc lacking its engineered signal sequence. That form of the protein, which would be expected to remain in the cytosol, had no effect on T-synthase activity either in mammalian cells or insect cells (unpublished data). Together, these results show that the CD and TMD of Cosmc are important for ER retention but are not important in chaperone function and that a C-terminal KDEL retention signal is as effective as the full-length protein in its chaperone function toward T-synthase.
Figure 2.
KDEL-tagged soluble form of Cosmc functions as wtCosmc and localizes in ER. (A–C) Function of KDEL-tagged soluble form of Cosmc. 293T cells were transiently transfected with plasmids encoding T-synthase–HPC4, HPC4-sCosmc, HPC4-sCosmc-KDEL, and wtCosmc. Cell extracts were prepared and T-synthase activity was measured. Bar 1 represents a single value, bar 5 represents a mean of duplicate values with error bar shown, and the remaining bars represent a mean of triplicate values with error bars shown. The pink line indicates the endogenous level of T-synthase activity and the green line represents the activity after transfection of T-synthase alone (A). T-synthase and Cosmc in cell extracts and media were also analyzed by Western blot with anti-HPC4 (B and C). Black lines indicate that intervening lanes have been spliced out. (D–F) Localization of sCosmc-KDEL. CHO K1 cells cultured on chambered slides transiently transfected with HPC4-sCosmc-KDEL were immunofluorescently stained with anti-HPC4 (green; D) and rabbit anti-calnexin IgG (red; E) and merged (F). The images were collected by confocal microscopy. Bars, 8 μm.
To confirm the localization of HPC4-sCosmc-KDEL in the ER, CHO K1 cells were transiently transfected and stained with Alexa 488–labeled HPC4 mAb. The HPC4-sCosmc-KDEL was expressed in a perinuclear pattern that generally overlapped that of the ER marker calnexin, a transmembrane protein (Fig. 2, D–F; Wada et al., 1991; Bergeron et al., 1994). Some lack of overlap in HPC4-sCosmc-KDEL expression and calnexin could result from the fact that the HPC4-sCosmc-KDEL is soluble, and some escapes to the Golgi and is secreted to media (Fig. 2 C).
Cosmc contains a single potential site of N-glycosylation in its extreme C terminus at N313. Thus, we examined whether Cosmc was N-glycosylated. About one-third of HPC4-sCosmc and HPC4-sCosmc-KDEL were N-glycosylated (unpublished data), based on sensitivity to Peptidyl N-glycanase F, an enzyme that removes N-glycans. However, the normal transmembrane form of Cosmc was not detectably N-glycosylated because there was no change of electrophoretic mobility upon treatment with Peptidyl N-glycanase F (unpublished data). Inefficient N-glycosylation of Cosmc may result from the fact that the potential N-glycosylation site is very close to the C terminus and is surrounded by acidic amino acids, which can impair the efficiency of N-glycosylation (Whitley et al., 1996; Nilsson and von Heijne, 2000).
To test whether N-glycosylation might be important to Cosmc function, we eliminated the N-glycosylation sequon by substitution of N313 to Q313 (N313Q-Cosmc). The chaperone function of N313Q-Cosmc was similar to that of the wtCosmc both in mammalian cells and insect cells (unpublished data). Thus, N-glycosylation of Cosmc is not required for its function, which is consistent with the fact that membrane form or full-length Cosmc is inefficiently N-glycosylated.
Cosmc itself does not have T-synthase activity
A previous study (Kudo et al., 2002) indicated that the gene we initially reported as Cosmc (Ju and Cummings, 2002) possibly encoded a second T-synthase (termed core 1 β3–Gal-T2), based upon some sequence identity (∼22%) with T-synthase and apparent restoration of T-synthase activity in human Jurkat and LSC cells, which lack T-synthase activity. To clarify this point, we directly tested whether HPC4-sCosmc contained core 1 β3–Gal-T activity. Cosmc lacked any detectable activity (Fig. S4, A and B, available at http://www.jcb.org/cgi/content/full/jcb.200711151/DC1), whereas the soluble form of T-synthase contained high enzymatic activity (Fig. S4, A and B). Thus, Cosmc is not a second T-synthase. This is consistent with our recent analysis of mouse null mutants lacking the T-synthase, in which all T-synthase activity in the homozygous animals is missing (Xia et al., 2004). Although Cosmc may have an evolutionary sequence relationship to T-synthase, Cosmc functions as a chaperone, and not as a second T-synthase.
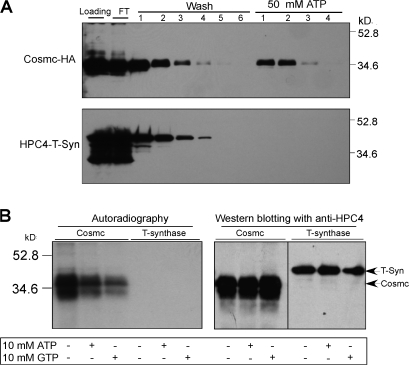
Cosmc binds ATP
Many chaperones have ATP binding or ATPase activity, some with low affinity. Thus, we tested whether Cosmc has ATP binding activity (Itoh et al., 1995). Cosmc binds to ATP-Sepharose and can be eluted with ATP, although the protein does not bind well (Fig. 3 A). In contrast, there was no significant binding of recombinant T-synthase to ATP-Sepharose. To further test whether Cosmc binds ATP directly, we performed photolabeling using 8-azido α-[32P]ATP. Under ultraviolet exposure, the activated azido group directly cross-links to the peptide bond in the ATP binding region of Cosmc. Purified Cosmc, but not T-synthase, could be photo–cross-linked with 8-azido α-[32P]ATP. This binding is specific because it was significantly inhibited by excesses of cold ATP or GTP (Fig. 3 B). Typically, either ATP or GTP can inhibit specific ATP binding to molecular chaperones (Csermely and Kahn, 1991; Saira Mian, 1993; Soti et al., 2002). These results demonstrate that Cosmc can bind ATP, which is similar to other chaperones, whereas T-synthase does not bind ATP.
Figure 3.
Cosmc has ATP-binding activity. (A) Hi-5 insect cells were infected with Baculovirus encoding either Cosmc-HA or HPC4-sT-synthase plus wild-type Cosmc. Cells for Cosmc-HA and media from cell coexpressing HPC4–sT-synthase and wild-type Cosmc were harvested. The cell extracts and media were loaded on ATP-Sepharose column for chromatography and eluted with 50 mM ATP, respectively. The washes and eluates were analyzed by Western blot with anti-HA for Cosmc and anti-HPC4 for T-synthase, as indicated. (B) HPC4-sCosmc and HPC4–sT-synthase (coexpressed with wtCosmc) were expressed in Hi-5 cells and purified from the media. 3 μg of Cosmc and T-synthase were photolabeled with 8-Azido α-[32P]ATP, analyzed on SDS-PAGE, and transferred to nitrocellulose membrane for radioautogram and Western blot with anti-HPC4. Black line indicates that intervening lanes have been spliced out.
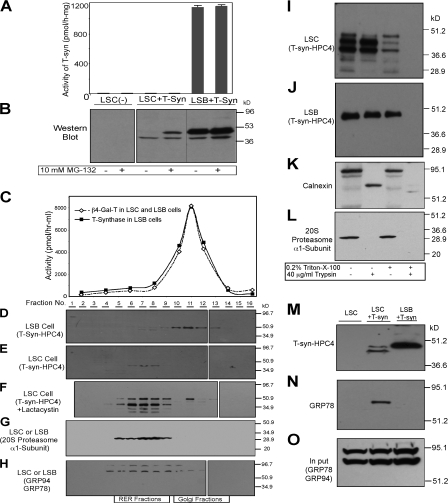
Proteasome inhibitors cause accumulation of full-length T-synthase expression but fail to restore activity
Our previous study showed that T-synthase in Jurkat cells, which have a mutated Cosmc, accumulates upon treatment of the cells with lactacystin, a proteasome inhibitor (Ju and Cummings, 2002), suggesting that the degradation of misfolded T-synthase is targeted to the proteasome. Human carcinoma LSC cells express the Tn antigen because of a lack of T-synthase activity (Brockhausen et al., 1998), whereas LSB cells contain T-synthase activity, which implies that LSC cells lack Cosmc function. In other studies, we have found that LSC cells have dysfunctional Cosmc, caused by an insertional mutation in the Cosmc gene (Ju et al., 2008). To investigate T-synthase in LSC cells, both LSC and LSB cells were stably transfected with C-terminal HPC4-tagged full-length human T-synthase. Human recombinant T-synthase from LSB cell extract was enzymatically active and full length (∼45 kD); however, T-synthase from LSC cell extracts was smaller (∼38 kD), inactive, and there was a reduced protein expression (Fig. 4 A), potentially caused by truncation or cleavage of the peptide from its N terminus by an unknown protease. The low level of protein expression could be caused by either a low rate of protein expression or high rate of degradation. Interestingly, the C. elegans T-synthase, which does not require mammalian Cosmc, is expressed at equal levels recombinantly in both LSB and LSC cells (Ju et al., 2006). Therefore, to explore whether degradation was involved in reduced T-synthase expression in LSC cells, the cells were treated with the proteasome inhibitor MG-132. In the presence of MG-132, more full-length T-synthase was recovered, although it was still inactive (Fig. 4, A and B). In contrast, the steady-state level of T-synthase expression in LSB cells and activity of the protein remained unchanged. The truncated N-terminal peptide in LSC cells could result in a soluble T-synthase, which might be secreted into the media. Thus, the media was Western blotted with HPC4 mAb. There was no T-synthase detected in the media from LSC cells; however, a substantial amount of soluble T-synthase was secreted into the media from LSB cells (Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200711151/DC1). These data indicate that T-synthase synthesized in LSC cells is rapidly degraded by the proteasome because degradation was blocked by MG-132. Interestingly, truncated T-synthase from LSC cells was the same size as the soluble form of T-synthase from LSB cell media, suggesting that the truncation of T-synthase in LSC cells results from loss of the cytoplasmic and TMDs.
Figure 4.
Characterization of T-synthase expressed in LSC cells containing a dysfunctional Cosmc. (A and B) MG-132 causes accumulation of full-length T-synthase protein in LSC cells but failed to restore its activity. LSC and LSB cells stably expressing T-synthase–HPC4 were treated with 10 μM MG-132 for 12 h and harvested. Cell extract was made and one portion was used for measuring T-synthase activity (A), whereas the other portion was analyzed on SDS-PAGE (30 μg protein/lane) by Western blot with anti-HPC4 (B). Error bars represent ±1 SD from the mean. Black lines indicates that intervening lanes have been spliced out. (C–H) T-synthase expressed in LSC cells resides mainly in heavy membrane fractions (RER), whereas T-synthase from LSB cells is in light membrane fractions (Golgi). LSC and LSB cells as in A and B treated with 10 μM lactacystin were homogenized and cell homogenate was fractionated on a sucrose gradient by ultracentrifugation. The fractions were collected and the activities of β4–Gal-T and T-synthase in fractions were determined (C). The fractions were also analyzed by Western blotting with anti-HPC4 (D–F), anti-20S proteasome α1-subunit (G), and anti-KDEL (GRP78 and GRP94; H). In F, lane 2 represents the combination of fractions 1 and 2. (I–O) T-synthase expressed in LSC cells resides mainly in the lumen of the RER and was associated with GRP78. The cell PNS of LSC and LSB cells as in A and B was digested with 40 μg/ml trypsin in the presence or absence of 0.2% Triton X-100, and T-synthase was analyzed under reducing SDS-PAGE by Western blotting with HPC4 mAb (I and J). As controls, calnexin and 20S proteasome α1-subunit were also analyzed by Western blotting with their respective antibody (K and L). T-synthase was purified from LSC and LSB cells and Western blotted with anti-HPC4 (M) and anti-KDEL (GRP78; N). GRP78 in the cell extract corresponding to one-tenth of the material was detected to confirm comparable starting amounts of material (O).
Truncated and full-length T-synthase from LSC cells localizes in rough ER, whereas T-synthase from LSB cells localizes in the Golgi apparatus
As shown in the previous section, T-synthase is a transmembrane protein localized in the Golgi, where it functions to synthesize the core 1 structure (T-antigen). To explore the localization of the inactive T-synthase in LSC cells, we performed subcellular fractionation using sucrose gradient ultracentrifugation. Both LSC and LSB cells were transfected to express human T-synthase-HPC4. Because of the low expression of T-synthase in LSC cells and less clear/organized images of the ER/Golgi in these tumor cells, we used sucrose gradient subcellular fractionation to localize Cosmc and the T-synthase in these cells. Subcellular fractionation studies, by Western blotting against the HPC4 epitope, showed that both the protein and activity of T-synthase from LSB cells were in the light membrane fractions (fractions 10–13) as is the Golgi marker β4–Gal-T (Fig. 4, C and D). Interestingly, truncated T-synthase from LSC cells was recovered in the heavy membrane fractions (fractions 5–8; Fig. 4 E), along with the ER markers 20S proteasome α1-subunit, GRP94, and GRP78 (Fig. 4, G and H). In addition, the full-length T-synthase from LSC cells treated with lactacystin was mainly retained in heavy membrane fractions, with a small amount found in the light membrane fractions corresponding to the Golgi (Fig. 4 F). These data support the conclusion that Cosmc functions as an ER chaperone to assist the folding and/or acquisition of T-synthase activity.
Truncated T-synthase from LSC cells resides in the lumen of the rough ER associated with GRP78
The soluble, truncated, and inactive forms of T-synthase from LSC cells were recovered in heavy membrane fractions; however, they could be present in the lumen of the ER or associated with the cytosolic face of the ER membrane. To address this question, we treated the membranes with trypsin in the absence or presence of Triton X-100. We anticipated that the soluble truncated T-synthase in LSC cells might be associated with the proteasome on the cytosolic side of the ER. Unexpectedly, T-synthase from LSC cells, which was recovered in heavy ER membranes, was not cleaved by trypsin in the absence of Triton X-100 but was susceptible to trypsin action in the presence of Triton X-100 (Fig. 4 I). As expected, T-synthase from LSB cells, which is Golgi localized, was insensitive to trypsin in the absence of Triton X-100 but was quantitatively lost by trypsin treatment in the presence of Triton X-100 (Fig. 4 J). Calnexin, a type-I ER membrane protein containing a cytoplasmic tail of 87 aa, was cleaved by trypsin with a shift of ∼10.5 kD and retained the intact transmembrane and lumenal domains (Fig. 4 K). In contrast, the 20S proteasome α1-subunit, which associates with the cytosolic side of the ER, was sensitive to trypsin (Fig. 4 L). Similarly, calnexin and the 20S proteasome α1-sunbunit from both of the cell types were digested by trypsin in the presence of Triton X-100 (Fig. 4, I–L). T-synthase from LSC cell extracts (in the presence of Triton X-100) was decreased significantly even without trypsin treatment, which might be caused by partial digestion by cellular (perhaps lysosomal) proteases. Collectively, these results show that T-synthase from LSC cells, which is inactive, is mainly retained within the ER lumen.
In spite of the data in the previous paragraph, T-synthase lacks any described ER retention signals (Gomord et al., 1999). Thus, we considered whether retention of the inactive and misfolded T-synthase in the ER of LSC cells might be a result of nonproductive association with known molecular chaperones. Many chaperones, such as calnexin/calreticulin and the UDP-Glc:glycoprotein glucosyltransferase system, retain misfolded N-glycosylated proteins in the ER and help them fold correctly. However, this system cannot function with human T-synthase because it lacks N-glycans. Thus, we tested whether the GRP78 or BiP machinery might associate with the inactive T-synthase in the ER of LSC cells. We conducted coimmunoprecipitation experiments in which the HPC4-tagged T-synthase expressed in LSC cells was purified, and this material was analyzed by Western blot to probe for coprecipitating chaperones. GRP78 was coimmunoprecipitated with T-synthase from LSC cells but not from LSB cells (Fig. 4, M–O). We did not detect HSP40 in this coprecipitation, but it is possible that it might be present because the antibody to HSP40 has low affinity. More comprehensive proteomic approaches will be needed to identify all the proteins that coprecipitate with inactive T-synthase. Nevertheless, these results show that inactive T-synthase in LSC cells is associated with a protein chaperone, but that association clearly cannot productively result in active folded enzyme.
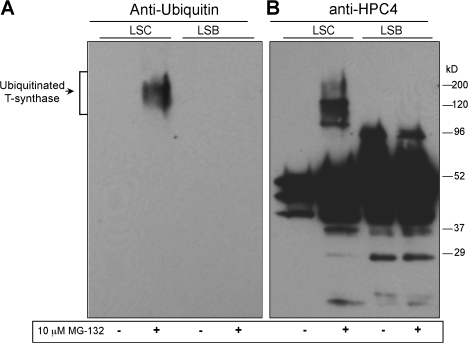
T-synthase purified from LSC cells treated with MG-132 is partially ubiquitinated
The results in the previous section show that misfolded T-synthase is degraded by an ER-associated degradation (ERAD) pathway, which usually requires ubiquitination. To probe whether the misfolded T-synthase is ubiquitinated, we performed a Western blot using anti-ubiquitin antibody against the affinity-purified full-length HPC4-tagged T-synthase from LSC cells treated with MG-132. The gels were intentionally overloaded to enhance detectability of ubiquitinated species. No ubiquitinated protein was detected in T-synthase purified from LSB cell extracts with or without MG-132 treatment (Fig. 5, A and B). In contrast, a portion of the inactive T-synthase from LSC cells treated with MG-132 was observed as high molecular mass species. Importantly, high molecular mass species from LSC cells in the presence of MG-132 were stained with ubiquitin antibody. These results demonstrate that a fraction of the T-synthase that accumulates in LSC cells in the presence of a proteasome inhibitor is ubiquitinated.
Figure 5.
T-synthase expressed in LSC Cells is partially ubiquitinated. LSC and LSB cells stably expressing T-synthase–HPC4 were treated with MG-132 for 12 h and subsequently harvested. T-synthase from 200 μl of cell extract (500 μg of protein total) was purified and analyzed by Western blot with anti-ubiquitin (A) first, and then with anti-HPC4 (B) after stripping of the membrane.
T-synthase is present as disulfide-bonded covalent oligomers in the absence of Cosmc
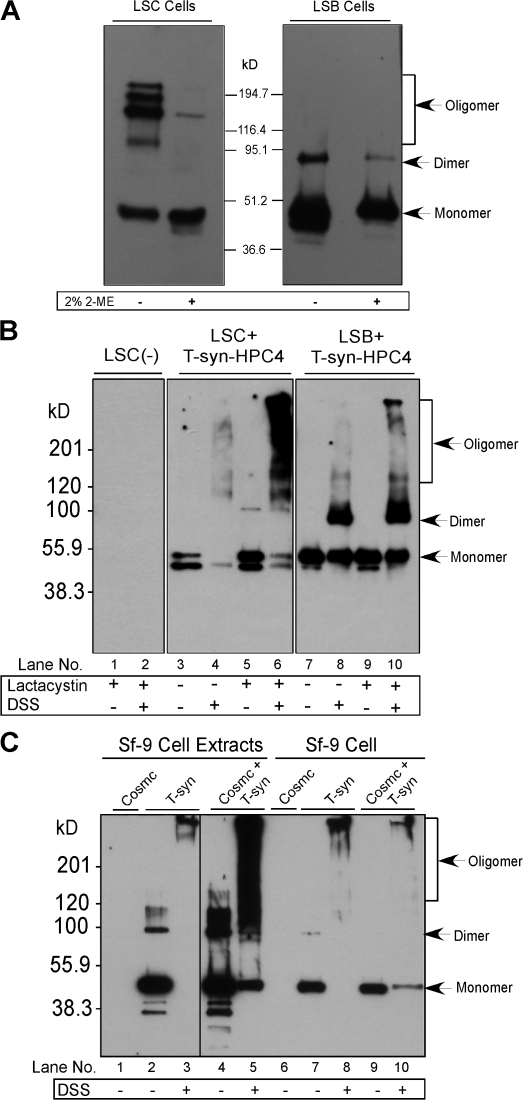
Many molecular chaperones function to prevent protein oligomerization and subsequent degradation. To further investigate the nature of T-synthase synthesized in the absence of Cosmc, purified T-synthase from LSC and LSB cells were compared by Western blot under reducing conditions and after cross-linking with disuccinimidyl suberate (DSS). The T-synthase from LSB cells, which have normal wtCosmc, appears mainly as a monomer under reducing conditions and as both monomers and dimers in nonreducing gels (Fig. 6 A). In contrast, mainly the monomeric form of T-synthase is seen under reducing conditions from LSC cells (Fig. 6 A). More striking is the presence of high molecular mass oligomeric forms of T-synthase in LSC cells observed in nonreducing gels (Fig. 6 A). These results demonstrate that in the absence of Cosmc, T-synthase cannot form dimeric enzyme but appears to form oligomeric complexes.
Figure 6.
T-synthase from cells in the absence of a functional Cosmc is mainly in covalent oligomers. T-synthase–HPC4 expressed in LSC and LSB cells treated with 10 μM lactacystin was affinity purified and analyzed on SDS-PAGE under reducing and nonreducing conditions and Western blotted with anti-HPC4 (A). Cell extracts were made and cross-linking was performed by adding 5 mM DSS. The recombinant T-synthase was purified and analyzed by Western blotting under reducing conditions (B). Sf-9 cells infected with Baculovirus-expressing T-synthase-HPC4 or coinfected with Baculovirus encoding wtCosmc were harvested and one portion was made for cell extracts. The cell extracts and the other portion of intact cells were treated with DSS. T-synthase was purified from the cell extracts and analyzed by Western blot with anti-HPC4 (C). Black line indicates that intervening lanes have been spliced out.
We explored the oligomeric state of T-synthase in the absence of Cosmc using the noncleavable cross-linker DSS. The T-synthase from LSB cells was mainly found in dimeric form in the presence of DSS (Fig. 6 B, lanes 7 and 8). However, most of the T-synthase from LSC cells was recovered as high molecular mass aggregates in the presence of DSS (Fig. 6 B, lanes 3 and 4). To confirm these results, we explored the oligomeric state of T-synthase generated in an insect expression system, which lacks a Cosmc orthologue. We examined T-synthase under reducing conditions in both intact Sf-9 cells and cell extracts treated with permeable DSS. In the absence of DSS or Cosmc, T-synthase is recovered primarily as a monomer in cells and extracts (Fig. 6 C, lanes 2 and 7). When coexpressed with Cosmc, we also recovered T-synthase mainly as a monomer (Fig. 6 C, lanes 4 and 9). In contrast, treatment with DSS of cells expressing T-synthase but lacking Cosmc coexpression resulted in a quantitative shift of T-synthase to high molecular mass complexes (Fig. 6 C, lanes 3 and 8). However, coexpression of Cosmc and T-synthase rescued much of the T-synthase from cross-linking and a significant amount of T-synthase monomer was recovered (Fig. 6 C, lanes 5 and 10). These results indicate that in the absence of Cosmc, T-synthase appears in oligomeric and inactive complexes, whereas in the presence of Cosmc, T-synthase appears as a monomer and dimer, demonstrating that Cosmc helps to prevent nonproductive oligomerization of T-synthase.
We also explored the effects of blocking proteasomal degradation on the oligomeric state of T-synthase in LSC and LSB cells. When LSB cells were treated with lactacystin, there was little change in the amount of monomeric T-synthase recovered (Fig. 6 B, lane 9), and after DSS treatment of cells, there were increased amounts of the dimeric form, as expected because natural T-synthase is a homodimer, and a small amount of oligomers form perhaps because of limited endogenous Cosmc (Fig. 6 B, lane 10). In contrast, treatment of LSC cells with lactacystin caused a larger increase in total T-synthase (Fig. 6 B, lane 5), with enhanced recovery of high molecular mass aggregates of T-synthase in DSS-treated cells (Fig. 6 B, lane 6). These results show that T-synthase in cells expressing functional Cosmc is primarily in active monomeric and dimeric forms, whereas in the absence of Cosmc, the T-synthase is partly degraded and recoverable in high molecular weight complexes, and no dimeric active forms of enzyme are present, indicating that proteolysis occurs in the context of the oligomeric complexes.
Cosmc directly interacts with T-synthase
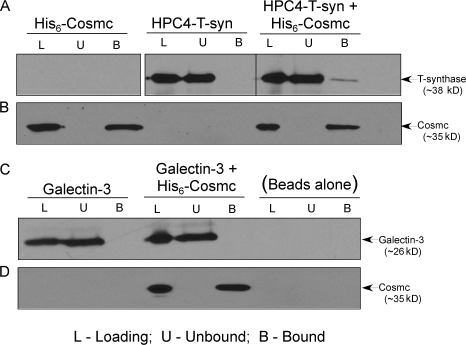
Many molecular chaperones function to prevent protein oligomerization by directly interacting with their client proteins. To investigate the nature of Cosmc and T-synthase interaction, we purified soluble HPC4-tagged T-synthase and His-tagged Cosmc from Hi-5 cells and tested for their direct interaction in vitro. As a control, we used galectin-3, which would not be expected to bind Cosmc. After mixing Cosmc with either of these proteins, Cosmc was quantitatively captured by adsorption on Ni-NTA beads and eluted with imidazole. The input (loading), unbound, and bound materials were analyzed by Western blot with anti-Cosmc, anti-HPC4, or anti–galectin-3 antibodies. Cosmc was quantitatively bound by Ni-NTA beads, whereas T-synthase did not bind to Ni-NTA beads (Fig. 7, A and B). In contrast, from the mixture of Cosmc with T-synthase, a fraction of T-synthase coprecipitated with Cosmc (Fig. 7, A and B). We tested whether this interaction was specific to the client T-synthase using galectin-3 as a potential client. Galectin-3 did not bind to Ni-NTA beads and also did not coprecipitate with Cosmc on Ni-NTA beads (Fig. 7, C and D). These results show that Cosmc directly binds to T-synthase and that this interaction is specific, supporting the conclusion that Cosmc is a specific molecular chaperone for its client T-synthase and that they directly interact.
Figure 7.
Cosmc directly interacts with T-synthase. The N-terminal His-tagged soluble Cosmc were expressed in Hi-5 insect cells. The human N-terminal HPC4 epitope-tagged soluble T-synthase was expressed by cotransfecting WT Cosmc in Hi-5 insect cells. Proteins were purified directly from the media and used in coprecipitation experiments to test physical association. Galectin-3 was used as a negative control for specificity. Cosmc, T-synthase, Cosmc and T-Synthase, Galectin-3, Cosmc and Galactin-3 were incubated with Ni-NTA Superflow. Ni-NTA Superflow beads were washed and proteins were eluted and analyzed by Western blot with mAb to the HPC4, anti-Galectin-3, and anti-Cosmc. Cosmc interactions with T-synthase were determined (A and B). As a control, Cosmc and Galectin-3 interactions were determined (C and D). Black line indicates that intervening lanes have been spliced out. L, loading; U, unbound; B, bound.
Discussion
Our interest in T-synthase and its regulation as a branchpoint enzyme arose from observations that the lack of T-synthase activity, which leads to expression of the truncated Tn antigen, is correlated with several autoimmune diseases including IgA nephropathy (Allen et al., 1997) and Tn-Syndrome (Berger, 1999). In addition, the Tn and sialyl-Tn antigen are recognized as tumor-associated antigens in mucins and other glycoproteins from many human tumors (Springer, 1984; Nakagoe et al., 2001; Schietinger et al., 2006). In examining human Jurkat cells that lacked T-synthase activity, we found that expression of the T-synthase transcript was normal but, surprisingly, active forms of recombinant T-synthase could not be expressed in cells. This led to our finding of Cosmc as a required molecular chaperone for active T-synthase formation and the finding that the X-linked Cosmc gene [Xq24] is mutated in cells lacking T-synthase activity (Ju and Cummings, 2002). Thus, acquired somatic or potentially inherited mutations in Cosmc could account for the defects in core 1 O-glycan biosynthesis and expression of Tn antigen. We recently showed that mutations in Cosmc are associated with Tn antigen expression in patients with Tn syndrome (Ju and Cummings, 2005) and in human tumor cells (Ju et al., 2008). Expression of the Tn and sialyl-Tn antigens in human tumors are associated with poor prognosis, including human breast carcinoma, and colorectal and esophageal cancer (Springer, 1984; Nakagoe et al., 2001; Schietinger et al., 2006). However, until now the nature of Cosmc function in providing active T-synthase was unknown. We originally considered that Cosmc might be a “subunit” of the T-synthase, but we found that purified active T-synthase from rat liver and purified recombinant soluble form of T-synthase from 293T cell media were fully active and devoid of Cosmc (Ju et al., 2002a,b). Thus, Cosmc is not a required cofactor or subunit of active T-synthase enzyme.
Our studies demonstrate that Cosmc is a resident ER protein that binds ATP, whereas the T-synthase is a resident Golgi protein. In the absence of Cosmc, the T-synthase is retained in the ER lumen and accumulates as an enzymatically inactive oligomeric disulfide-bonded complex associated with GRP78. Proteins that are misfolded are eliminated from the ER by an ERAD pathway, in which misfolded proteins are targeted for reverse translocation out of the ER, where they are ubiquitinated, and then disposed of in the cytoplasmic proteasome (Ahner and Brodsky, 2004). The inactive T-synthase is subjected to degradation by the ERAD pathway, as indicated by the presence of ubiquitinated T-synthase, which accumulates in the presence of proteasome inhibitors. The ER retention of Cosmc requires its transmembrane and cytosolic domains because soluble Cosmc is secreted from cells. However, soluble Cosmc engineered to contain a KDEL ER-retrieval signal is retained in the ER and functions as a chaperone for T-synthase. Human Cosmc and T-synthase are coordinately expressed in human tissues, indicating the close relationship between these two proteins. Active T-synthase cannot be expressed without the presence of functional Cosmc (Ju and Cummings, 2002). More importantly, purified recombinant Cosmc directly interacts with purified recombinant T-synthase (Fig. 7). Thus, Cosmc is an ER-localized molecular chaperone that prevents degradation and aggregation of mammalian T-synthase, and the overall proposed pathway is shown in Fig. 8. It is important to note that Cosmc itself does not have any T-synthase activity, which is in contrast to a previous study that erroneously concluded that the Cosmc gene encoded a second core 1 β3–Gal-T (T-synthase) in humans named C1Gal-T2 (Kudo et al., 2002). The T-synthase activity is encoded by a single gene, which is consistent with our studies in this paper showing that loss of Cosmc function is associated with a complete loss of T-synthase activity, that deletion of T-synthase in mice eliminates all T-synthase activity and results in embryonic death (Xia et al., 2004), and that mutations in Cosmc are associated with loss of T-synthase in patients with Tn syndrome (Ju and Cummings, 2005).
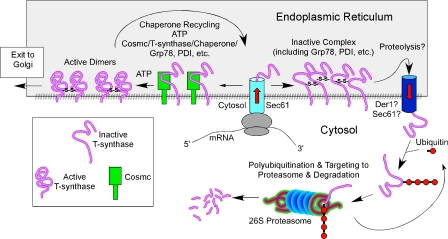
Figure 8.
Working model for Cosmc function. Human Cosmc is localized predominantly in the ER where it interacts with the nascent polypeptide of human T-synthase. Cosmc, along with other ER chaperones such as HSP70 (BiP)/HSP40 and protein disulfide isomerase (PDI), assists its folding properly. Native T-synthase, which occurs mainly as a homodimer, exits to the Golgi apparatus, where it functions in synthesizing core 1 O-glycans (T-antigen). When Cosmc is mutated and dysfunctional, the nascent polypeptides of T-synthase form inactive aggregates or oligomers, which are associated with GRP78 (BiP), and subsequently proceed to the ERAD pathway where they are retrotranslocated from the ER to the cytosol, ubiquitinated, and subsequently degraded by the proteasome.
The T-synthase is not a glycoprotein and has no canonical motifs for N-glycosylation, which is in contrast to most glycosyltransferases. Thus, the T-synthase, unlike other newly synthesized glycoproteins in the ER, cannot interact with the known quality control systems composed of several lectin-based chaperones, such as the calnexin/calreticulin system, and other glycan-recognizing or modifying proteins (Ruddock and Molinari, 2006; Schroder, 2006; Otsu and Sitia, 2007). The absence of such interactions may underscore the molecular function of Cosmc in specifically preventing oligomerization and inactivation of the T-synthase and provides the first specific evidence for the quality control of nonglycosylated protein folding in the ER.
It is interesting that although the ER contains many chaperones that can promote proper protein folding, in the absence of Cosmc these chaperones are not successful in promoting folding of the T-synthase. We found that GRP78 (BiP) could be coimmunoprecipitated with inactive T-synthase generated in the absence of Cosmc. GRP78 is known to associate with unfolded proteins in the ER to aid in their exit (Kleizen and Braakman, 2004; Ni and Lee, 2007). The inactive oligomers of T-synthase accumulate within the ER in the absence of Cosmc and reside within disulfide-bonded complexes. We also found that Cosmc binds directly to ATP, thus indicating that Cosmc is a chaperone. In other experiments, when we purified the T-synthase from rat liver microsomes, which included the ER and Golgi compartments, we found that both Cosmc and HSP40 copurified in early steps with the T-synthase (unpublished data). Although this further supports a role for Cosmc as a T-synthase chaperone, it also indicates that HSP40 could be a cochaperone for Cosmc as it is for HSP70 (Kleizen and Braakman, 2004).
In order for the misfolded T-synthase to be ubiquitinated by ubiquitination system, it must be retrotranslocated from the lumen back to the cytosol. However, little is known about the mechanisms of retrotranslocation for type II membrane proteins and how such unfolded proteins that accumulate in the ER are degraded by ERAD pathway. The degradation of T-synthase, the first identified enzyme with a deficiency in its ER chaperone protein that leads to disease, may provide a novel system to explore these pathways.
Thus, Cosmc represents a protein-specific quality control factor and joins a growing number of such chaperones in the ER (Hendershot and Bulleid, 2000; Ellgaard and Helenius, 2003). Cosmc is not a T-synthase subunit, lacks glycosyltransferase activity, and, among glycosyltransferases, it is only known to associate with the inactive T-synthase. We noted that Cosmc does not associate with fully folded T-synthase (Ju and Cummings, 2002), but it can be coimmunoprecipitated with a subset of T-synthase proteins, likely representing folding intermediates in the pathway to fully active enzyme. Most importantly, we have shown that purified recombinant Cosmc can directly interact with purified T-synthase. As expected, only a small fraction of recombinant T-synthase coprecipitated with purified recombinant His-tagged Cosmc, which could result from the possibility that only a fraction of purified recombinant T-synthase might be in unfolded state upon purification. This fraction therefore transiently interacts with active recombinant His-tagged Cosmc, supporting the fact that Cosmc is an ER molecular chaperone, but this issue will require extensive studies, including exploring the role of ATP in these interactions. Loss-of-function of Cosmc results in the formation of misfolded aggregates of T-synthase polypeptide, which would be predicted to be retrotranslocated back to the cytosol where ubiquitination occurs, and the ubiquitinated protein is then degraded in the proteasome, as shown in the model (Fig. 8). Overall, our study shows that Cosmc is a key regulator of the expression of functional T-synthase, which represents a novel mechanism for controlling a branchpoint glycosyltransferase in O-glycan biosynthesis.
Materials and methods
Materials
GalNAc-α-phenyl, UDP-Gal, GlcNAc-β-S-pNp, and ATP-Sepharose were obtained from Sigma-Aldrich. UDP-6-[3H]Gal (40–60 Ci/mmol) was obtained from American Radiolabeled Chemicals, Inc. Human embryonic kidney cell line (HEK293T), Chinese hamster ovary cells (CHO K1), and insect cells (Hi-5 and Sf-9) were obtained from American Type Culture Collection. Human colon carcinoma cell lines LSC and LSB were provided by S. Itzkowitz (Mount Sinai School of Medicine, New York, NY). Sep-Pak C18 Cartridges were obtained from Waters Corporation. Restriction enzymes were obtained from New England Biolabs, Inc. FuGENE6, Taq DNA polymerase, and rat anti-HA mAb were obtained from Roche. TNM-FH and EX-Cell 405 media, BaculoGold Transfection kit, vector pVL1393, mouse anti–human calnexin mAb (IgG1), mouse anti-–human HSP40, and anti-ubiquitin mAb (IgG1) were purchased from BD Biosciences. Galactin-3 antibody was purchased from Santa Cruz Biotechnology, Inc. Rabbit anti–human calnexin antiserum and mouse anti-KDEL (GRP78 and GRP94) mAb (10C3) were purchased from Assay Designs. Alexa Fluor–labeled secondary antibodies were purchased from Invitrogen. Peroxidase-labeled secondary antibodies were obtained from KPL. Proteasome inhibitors MG-132 and lactacystin and rabbit anti–human proteasome 20S α-type1 subunit (IgG) were purchased from EMD. Vector pcDNA3.1(+), PCR TOPO4 cloning kit, SuperScript One-Step RT-PCR kit and SDS-PAGE gels were obtained from Invitrogen. Ni-NTA Superflow, Plasmid Purification kit, and QIAquick Gel Extraction kits were obtained from QIAGEN. 8-Azido α-[32P]ATP was purchased from Affinity Labeling Technologies. Chemiluminescent Substrate, BCA protein assay kit, DSS, and UltraLink Support Media were purchased from Thermo Fisher Scientific. Rabbit anti–mannosidase II antibody was provided by K. Moremen (University of Georgia, Athens, GA).
Northern blot of human Cosmc and T-synthase
Human 12-lane multiple tissue Northern blot (Clontech Laboratories, Inc.) was incubated with 32P-labeled probes for full length of human Cosmc (970 bp) as previously described (Ju et al., 2002a) at 68°C overnight using PerfectHyb solution (Sigma-Aldrich). After washing, the blot was exposed to BioMax x-ray film (Kodak) for 36 h and developed. β-Actin in the same blot was probed with 32P-labeled human Actin cDNA as previously described (Ju et al., 2002a).
Preparation of expression constructs
A construct encoding C-terminal HA-tagged Cosmc (Cosmc-HA) was made by introducing HA-epitope (YPYDVPDYA) into wild-type Cosmc at its C terminus by PCR. The product was cloned into PCR3.1. The insert was cut with BamHI (partially)–XbaI and cloned into pcDNA3.1(+) or pVL1393. The construct expressing C-terminal HPC4 epitope-tagged T-synthase was made as previously described (Ju and Cummings, 2002). A construct encoding N-terminal HPC4-tagged Cosmc (HPC4-Cosmc) was made using a similar strategy to Cosmc-HA. The HPC4 epitope tag was introduced into the N terminus of Cosmc by PCR. The PCR product was digested with BamHI and ligated with the vector fragment generated by BamHI digestion of pcDNA3.1(+) containing wild type Cosmc (Ju et al., 2002a). A construct encoding a soluble N-terminal HPC4 epitope-tagged Cosmc (HPC4-sCosmc) was made using the similar strategy to making soluble N-terminal HPC4 epitope-tagged T-synthase (HPC4–sT-synthase; Ju et al., 2002a). The construct in pcDNA3.1(+) or pVL1393 with an HPC4-epitope tag was fused inframe with the lumenal domain of Cosmc. HPC4-sCosmc with the KDEL-retrieval signal at its C terminus (HPC4-sCosmc-KDEL) was constructed using PCR and subcloning into pcDNA3.1(+). The PCR primers are listed in Table I.
Table I. PCR primers used for making expression constructs.
| Constructs | Forward primer | Reverse primer |
|---|---|---|
| Cosmc-HA | 5′-GCGGATCCACCATGCTTTCTGAAAGCAGC TCCTTTTTG-3′ |
5′-GCTCTAGACTAAGCGTAGTCTGGGACGTCGTATGGGTAG TCATTGTCAGAACCATTTG-3′ (containing HA-tag sequence) |
| HPC4-Cosmc | 5′-GCGGATCCACCATGCTTGAGGACCAGGTGGA CCCCAGGCTGATCGACGGCAAGATGCTTTCTGA AAGCAGCTCC-3′ (containing HPC4-tag sequence) |
5′-GGTCTCCAGATTTTATAGTGTGGC-3′ |
| HPC4-sCosmc-KDEL | 5′-TCCTTGCACGCCCCACTACG-3′ | 5′-GCTCTAGAGTTCATCTTTGTCATTGTCAGAACCATTTG-3′ (containing KDEL sequence) |
The construct for expression of a soluble N-terminal 6× His-tagged Cosmc was prepared using the following strategy. The N-terminal cytoplasmic and TMDs of Cosmc were replaced with transferrin N-terminal signal sequence followed by 6× His tag. The lumenal domain of Cosmc was prepared by the digestion of full-length Cosmc cDNA in pcDNA3.1(+) with BsmI–XbaI. The Vector with pVL1393 backbone and transferrin N-terminal signal sequence was obtained by digestion of the plasmid encoding human soluble T-synthase with EcoNI– XbaI. The DNA oligos (IDT [Integrated DNA Technologies]) encoding 6× His coding sequence with EcoNI and BsmI at each site were synthesized and would fill in the gap between the EcoNI site from the vector and the BsmI site from the Cosmc fragment. The DNA oligos were the following: sense strain, 5′-CCCATCACCATCACCATCACGACGATGACGATAAGAGGATTGGTCATGGAAATAGAATGCA-3′; and the complementary strain, 5′- CATTCTATTTCCATGACCAATCCTCTTATCGTCATCGTCGTGATGGTGATGGTGATGG-3′. Then the vector fragment, the annealed oligos, and the Cosmc fragment were ligated and the construct was made and confirmed by sequencing.
Cell culture
293T, CHO K1, LSC, and LSB cells were cultured in DME plus 10% FBS at 37°C in 5% CO2. Sf-9 cells were cultured in TNM-FH, whereas Hi-5 cells were cultured in Ex-Cell 405 media at 27°C.
Transfection and glycosyltransferase activity
Mammalian cells were transfected with Fugene6 transfection reagent and insect cells were transfected with a BaculoGold Transfection kit (Ju and Cummings, 2002). T-synthase activity was measured using GalNAc-α-phenyl as the acceptor (Ju et al., 2002b). β4–Gal-T activity was measured using pNP-β-S-GlcNAc as the acceptor (Kawar et al., 2002).
Immunofluorescent staining of CHO K1 cells
CHO K1 cells were cultured on a chambered slide and transiently transfected with the expression constructs using Fugene 6 transfection reagent according to the manufacturer's protocol. At 48 h after transfection, cells were washed with PBS and fixed with 4% PFA on ice for 45 min and permeabilized with 0.1% Triton X-100 for 30 min on ice. After blocking with 1% BSA in PBS for 1 h at RT, the cells were incubated with primary antibodies for 1 h at RT. The cells were washed with PBS three times and incubated with Alexa Fluor–labeled secondary antibodies at RT for 1 h. In some cases, the transfected cells were stained with Alexa Fluor–labeled primary antibodies directly, as indicated in the figures. Cells were then washed four times with PBS and mounted with Prolong Antifade Media (Invitrogen). After drying at RT for 12–16 h, cells were visualized on a confocal microscope (TCS NT; Leica) at RT under 40× Plan Fluotar 1.0 NA oil immersion or 100× Plan APO 1.4 NA oil immersion objective lenses. The images were maximum projection collected with a pinhole of 1 using 0.5-μm step size. Images were analyzed using the TCS and Volocity software (Leica).
Preparation of cell extracts
Cell extracts were prepared as previously described (Ju and Cummings, 2002). Protein concentration was determined by BCA assay according to manufacturer's protocol.
Western blot
Western blotting with anti-HPC4, Calnexin, GRP78 (BiP), HSP40, or 20S proteasome α1-subunit was performed as previously described (Ju and Cummings, 2002) or as otherwise stated.
Subcellular fractionation
About 5 × 106 293T cells transiently transfected with HPC4-Cosmc for 48 h or LSB and LSC cells stably expressing T-synthase–HPC4 were harvested and washed with cold PBS. Cells were homogenized in 25 mM Hepes, pH 7.5, containing 250 mM sucrose. Then PNS was made by centrifugation at 20,000 g for 30 min. Then the concentration of sucrose in PNS was adjusted to 50% (wt/vol) and loaded on 70, 60, 40, and 20% sucrose gradient. After centrifugation at 100,000 g for 20 h, 16 fractions were collected from the bottom of the tube. The fractions were analyzed by Western blotting with antibodies indicated in the results and also measured for both T-synthase and β4–Gal-T activity.
T-synthase activity of soluble N-terminal HPC-tagged Cosmc purified from media
293T cells in a T75 flask were transiently transfected with constructs encoding soluble HPC4-tagged Cosmc with or without KDEL-signal and soluble HPC4-tagged T-synthase. The epitope-tagged proteins were absorbed on HPC4-Ultralink resin and washed. Some of the sample was used for T-synthase activity assay directly and the other part was eluted for Western blotting by HPC4 mAb.
ATP-Sepharose chromatography of Cosmc
Hi-5 insect cells were infected with Baculovirus encoding either Cosmc-HA or soluble N-terminal HPC4 epitope-tagged T-synthase plus wild-type Cosmc. At 96 h after infection, cells expressing Cosmc-HA were harvested and the media from cells expressing soluble T-synthase and wild-type Cosmc was collected. The cell extract and media were chromatographed on a 0.1-ml ATP-Sepharose column preequilibrated with 50 mM Tris-HCl buffer, pH 7.8, containing 100 mM NaCl and 5 mM MgCl2, respectively. The column was washed with 0.4 ml of the buffer six times. Bound proteins were eluted with 50 mM ATP in the buffer (0.1 ml/fraction). The loading, flowthrough, washes, and eluates were analyzed by Western blot with either anti-HA and or anti-HPC4 mAbs, as indicated.
Photolabeling of Cosmc by 8-Azido α-[32P]ATP
HPC4-sCosmc and HPC4–sT-synthase coexpressed with wtCosmc were expressed in Hi-5 cells and affinity purified from the media. 3 μg Cosmc and T-synthase were photolabeled in 20 μl of reaction containing 50 mM Tris-HCl, pH 7.8, 3 mM MgCl2 100 mM NaCl, and 1 mM ATP plus 1 μCi of 8-Azido α-[32P]-ATP in the absence or presence of 10 mM ATP and 10 mM GTP for 30 min on ice. Then the reactions were exposed under UV (wavelength ∼254 nm) for 90 s on ice followed by immediate addition of 250 μl TBS containing 50 mM DTT. The reactions were dialyzed with Centricon Concentrator (3 kD cutoff) with TBS and concentrated down to 50 μl. 20 μl of sample were analyzed on SDS-PAGE (4–20%) and transferred to nitrocellulose membrane for radioautogram and Western blot with anti-HPC4.
Proteasome inhibitors treatment
About 106 LSC and LSB cells were seeded in T75 flasks and grown for 24 h. The cells were treated with 10 μM MG-132 or lactacystin (dissolved in 100% DMSO at 2 mM stock) or 0.5% DMSO in complete media for 12–14 h. The cells were harvested for T-synthase activity assay and Western blot.
Trypsin digestion experiment and coimmunoprecipitation of GRP78
About 2 × 106 of LSC and LSB cells stably expressing T-synthase–HPC4 were harvested and cell after nuclear homogenate was obtained. One portion the homogenate was solubilized with 0.2% Triton X-100 to make the extract. The cell homogenate and extract (containing 50 μg of protein) were treated with 40 μg/ml trypsin at 37°C for 4 h, respectively, and then analyzed by Western blotting with HPC4, mouse anti–human 20S α1-subunit mAb, and mouse anti–human Calnexin mAb. The T-synthase from cell extracts was also purified and Western blotted with anti-KDEL (GRP78) mAb.
Cross-linking experiments
LSC, LSB, and Sf-9 cells expressing C-terminal HPC4-tagged T-synthase with and without Cosmc were harvested and washed twice with PBS. Cell extract was made from one portion of the cells and incubated with 5 mM DSS and 1% DMSO at RT for 30 min. The other portion of Sf-9 cells was treated with 5 mM DSS at RT for 30 min and the cell extracts were made as previously described. The reaction was quenched by adding 50 mM Tris-HCl, pH 7.5. After adding 1 mM CaCl2 to all cell extracts, the T-synthase–HPC4 was purified and analyzed on Western blot with anti-HPC4.
In vitro Cosmc and T-synthase coprecipitation
Approximately 2 μg each of His-tagged soluble Cosmc, HPC4-tagged soluble T-synthase, and soluble Galectin-3 (provided by S. Stowell, Emory University, Atlanta, GA) were used. His-Cosmc and HPC4-T-synthase were mixed in buffer (5 mM Tris-HCl and 30 mM NaCl, pH 7.85). Similarly, His-Cosmc and Galectin-3 were mixed. For controls, His-Cosmc and HPC4–T-Synthase were individually prepared in buffer. All preparations were incubated at RT (∼23°C) for 25 min. One-fifth of each mixture was allocated for input (loading). The remaining part was diluted 25× with Ni-NTA washing buffer (50 mM imidazole, 50 mM NaH2PO4, 300 mM NaCl, and 0.1% Triton X-100, pH 7.8). 20 μl Ni-NTA Superflow beads (QIAGEN) were added to the Ni-NTA washing buffer. Proteins were incubated with beads overnight while rotating at 4°C. The beads were collected using bench centrifugation and the beads were washed five times with 400 μl Ni-NTA washing buffer. The bead-bound material was eluted five times with 20 μl of elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole,0.1% Triton X-100). Input, unbound, and bound fractions (in equal proportions) were electrophoresed on an SDS/PAGE (4–20%) and transferred to nitrocellulose membrane (Bio-Rad Laboratories). After blocking with 1% milk, Western blots were performed, first for HPC4–T-synthase and Galectin-3, using 4 ml of 4 μg/ml mAb to HPC4 and 4 ml of 0.2 μg/ml of antibody to Galectin-3. The membranes were incubated for 1 h at RT. For HPC4 blotting, the membrane was washed five times with 20 mM Tris-HCl, 300 mM NaCl, and 1 mM CaCl2, pH 7.2. For Galectin-3, the membrane was washed five times with 20 mM Tris-HCl, 300 mM NaCl, and 0.05% Tween-20, pH 7.2. After washing, the membranes were incubated with 4 ml of 0.17 ng/ml HRP-conjugated goat anti–mouse IgG at 23°C for 45 min. Membranes were washed five times with 20 mM Tris-HCl, 300 mM NaCl, and 1 mM CaCl2, pH 7.2, and incubated with 3 ml of SuperSignal West Pico Chemiluminescent Substrate at RT for 1 min. The blot was exposed to film (Denville Scientific, Inc.) for 1 min, and developed by autoradiography. The membranes were stripped by washing with 2% SDS and 1% 2-mercaptoethanol in deionized water for 30 min, followed by washing three times with 20 mM Tris-HCl and 300 mM NaCl, pH 7.2. The membranes were blocked with 1% milk and washed three times with 20 mM Tris-HCl and 300 mM NaCl, pH 7.2. Membranes were incubated with polyclonal chicken IgG against Cosmc using 4 ml of 0.2 μg/ml antibody for 1 h. Membranes were washed five times with 20 mM Tris-HCl and 300 mM NaCl, pH 7.2, and incubated with 4 ml of 0.17 ng/ml HRP-conjugated goat anti–chicken IgG at 23°C for 45 min. Membranes were washed with 20 mM Tris-HCl and 300 mM NaCl, pH 7.2, and developed accordingly.
Online supplemental material
Fig. S1 shows that human Cosmc is coordinately transcribed with human T-synthase in Northern blot. Fig. S2 shows that protein sequences of Cosmc are highly conserved across species. Fig. S3 shows that Cosmc and T-synthase localize in different cellular compartments. Fig. S4 shows that Cosmc itself does not have any T-synthase activity. Fig. S5 shows that truncated T-synthase protein in LSC cells is not secreted into the media. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200711151/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Steven Itzkowitz for kindly providing human colon carcinoma cell lines LSC and LSB. We thank Dr. Kelley Moremen for his generosity in providing rabbit anti-Man II antisera and Sean Stowell for recombinant Galectin-3. We also thank Dr. Baoyun Xia, Dr. Jamie Heimburg-Molinaro, Ms. Connie Arthur, and Mr. Anthony Luyai for suggestions and critiques of this manuscript.
This work was supported by a National Institutes of Health RO1 grant (RO1 GM068559-01A2) to R.D. Cummings. The authors declare that there is no conflict of interest.
Abbreviations used in this paper: CD: cytoplasmic domain; DSS, disuccinimidyl suberate; ERAD, ER-associated degradation; GalNAc, N-acetylgalactosamine; Gal-T, galactosyltransferase; GlcNAc, N-acetylglucosamine; HPC, human protein C; PNS, Postnuclear supernatant; TMD, transmembrane domain.
References
- Ahner, A., and J.L. Brodsky. 2004. Checkpoints in ER-associated degradation: excuse me, which way to the proteasome? Trends Cell Biol. 14:474–478. [DOI] [PubMed] [Google Scholar]
- Allen, A.C., P.S. Topham, S.J. Harper, and J. Feehally. 1997. Leucocyte beta 1,3 galactosyltransferase activity in IgA nephropathy. Nephrol. Dial. Transplant. 12:701–706. [DOI] [PubMed] [Google Scholar]
- An, G., B. Wei, B. Xia, J.M. McDaniel, T. Ju, R.D. Cummings, J. Braun, and L. Xia. 2007. Increased susceptibility to colitis and colorectal tumors in mice lacking core 3–derived O-glycans. J. Exp. Med. 204:1417–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, E.G. 1999. Tn-syndrome. Biochim. Biophys. Acta. 1455:255–268. [DOI] [PubMed] [Google Scholar]
- Bergeron, J.J., M.B. Brenner, D.Y. Thomas, and D.B. Williams. 1994. Calnexin: a membrane-bound chaperone of the endoplasmic reticulum. Trends Biochem. Sci. 19:124–128. [DOI] [PubMed] [Google Scholar]
- Brockhausen, I., J. Yang, N. Dickinson, S. Ogata, and S.H. Itzkowitz. 1998. Enzymatic basis for sialyl-Tn expression in human colon cancer cells. Glycoconj. J. 15:595–603. [DOI] [PubMed] [Google Scholar]
- Csermely, P., and C.R. Kahn. 1991. The 90-kDa heat shock protein (hsp-90) possesses an ATP binding site and autophosphorylating activity. J. Biol. Chem. 266:4943–4950. [PubMed] [Google Scholar]
- Ellgaard, L., and A. Helenius. 2003. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4:181–191. [DOI] [PubMed] [Google Scholar]
- Gomord, V., E. Wee, and L. Faye. 1999. Protein retention and localization in the endoplasmic reticulum and the golgi apparatus. Biochimie. 81:607–618. [DOI] [PubMed] [Google Scholar]
- Hendershot, L.M., and N.J. Bulleid. 2000. Protein-specific chaperones: the role of hsp47 begins to gel. Curr. Biol. 10:R912–R915. [DOI] [PubMed] [Google Scholar]
- Itoh, H., R. Kobayashi, H. Wakui, A. Komatsuda, H. Ohtani, A.B. Miura, M. Otaka, O. Masamune, H. Andoh, K. Koyama, et al. 1995. Mammalian 60-kDa stress protein (chaperonin homolog). Identification, biochemical properties, and localization. J. Biol. Chem. 270:13429–13435. [DOI] [PubMed] [Google Scholar]
- Ju, T., and R.D. Cummings. 2002. A unique molecular chaperone Cosmc required for activity of the mammalian core 1 beta 3-galactosyltransferase. Proc. Natl. Acad. Sci. USA. 99:16613–16618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju, T., and R.D. Cummings. 2005. Protein glycosylation: chaperone mutation in Tn syndrome. Nature. 437:1252. [DOI] [PubMed] [Google Scholar]
- Ju, T., K. Brewer, A. D'Souza, R.D. Cummings, and W.M. Canfield. 2002. a. Cloning and expression of human core 1 beta1,3-galactosyltransferase. J. Biol. Chem. 277:178–186. [DOI] [PubMed] [Google Scholar]
- Ju, T., R.D. Cummings, and W.M. Canfield. 2002. b. Purification, characterization, and subunit structure of rat core 1 Beta1,3-galactosyltransferase. J. Biol. Chem. 277:169–177. [DOI] [PubMed] [Google Scholar]
- Ju, T., Q. Zheng, and R.D. Cummings. 2006. Identification of core 1 O-glycan T-synthase from Caenorhabditis elegans. Glycobiology. 16:947–958. [DOI] [PubMed] [Google Scholar]
- Ju, T., G.S. Lanneau, T. Gautam, Y. Wang, B. Xia, S.R. Stowell, M.T. Willard, W. Wang, Y. Xia, R.E. Zuna, et al. 2008. Human Tumor Antigens Tn and Sialyl Tn Arise from Mutations in Cosmc. Cancer Res. 68:1636–1646. [DOI] [PubMed] [Google Scholar]
- Kawar, Z.S., I. Van Die, and R.D. Cummings. 2002. Molecular cloning and enzymatic characterization of a UDP-GalNAc: GlcNAcbeta-R beta1,4-N-acetylgalactosaminyltransferase from Caenorhabditis elegans. J. Biol. Chem. 277:34924–34932. [DOI] [PubMed] [Google Scholar]
- Kleizen, B., and I. Braakman. 2004. Protein folding and quality control in the endoplasmic reticulum. Curr. Opin. Cell Biol. 16:343–349. [DOI] [PubMed] [Google Scholar]
- Kudo, T., T. Iwai, T. Kubota, H. Iwasaki, Y. Takayma, T. Hiruma, N. Inaba, Y. Zhang, M. Gotoh, A. Togayachi, and H. Narimatsu. 2002. Molecular cloning and characterization of a novel UDP-Gal:GalNAc(alpha) peptide beta 1,3-galactosyltransferase (C1Gal-T2), an enzyme synthesizing a core 1 structure of O-glycan. J. Biol. Chem. 277:47724–47731. [DOI] [PubMed] [Google Scholar]
- Nakagoe, T., T. Sawai, T. Tsuji, M. Jibiki, A. Nanashima, H. Yamaguchi, N. Kurosaki, T. Yasutake, and H. Ayabe. 2001. Circulating sialyl Lewis(x), sialyl Lewis(a), and sialyl Tn antigens in colorectal cancer patients: multivariate analysis of predictive factors for serum antigen levels. J. Gastroenterol. 36:166–172. [DOI] [PubMed] [Google Scholar]
- Ni, M., and A.S. Lee. 2007. ER chaperones in mammalian development and human diseases. FEBS Lett. 581:3641–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson, I., and G. von Heijne. 2000. Glycosylation efficiency of Asn-Xaa-Thr sequons depends both on the distance from the C terminus and on the presence of a downstream transmembrane segment. J. Biol. Chem. 275:17338–17343. [DOI] [PubMed] [Google Scholar]
- Otsu, M., and R. Sitia. 2007. Diseases originating from altered protein quality control in the endoplasmic reticulum. Curr. Med. Chem. 14:1639–1652. [DOI] [PubMed] [Google Scholar]
- Piller, V., F. Piller, and M. Fukuda. 1990. Biosynthesis of truncated O-glycans in the T cell line Jurkat. Localization of O-glycan initiation. J. Biol. Chem. 265:9264–9271. [PubMed] [Google Scholar]
- Ruddock, L.W., and M. Molinari. 2006. N-glycan processing in ER quality control. J. Cell Sci. 119:4373–4380. [DOI] [PubMed] [Google Scholar]
- Saira Mian, I. 1993. Sequence similarities between cell regulation factors, heat shock proteins and RNA helicases. Trends Biochem. Sci. 18:124–127. [PubMed] [Google Scholar]
- Schietinger, A., M. Philip, B.A. Yoshida, P. Azadi, H. Liu, S.C. Meredith, and H. Schreiber. 2006. A mutant chaperone converts a wild-type protein into a tumor-specific antigen. Science. 314:304–308. [DOI] [PubMed] [Google Scholar]
- Schroder, M. 2006. The unfolded protein response. Mol. Biotechnol. 34:279–290. [DOI] [PubMed] [Google Scholar]
- Soti, C., A. Racz, and P. Csermely. 2002. A Nucleotide-dependent molecular switch controls ATP binding at the C-terminal domain of Hsp90. N-terminal nucleotide binding unmasks a C-terminal binding pocket. J. Biol. Chem. 277:7066–7075. [DOI] [PubMed] [Google Scholar]
- Springer, G.F. 1984. T and Tn, general carcinoma autoantigens. Science. 224:1198–1206. [DOI] [PubMed] [Google Scholar]
- Wada, I., D. Rindress, P.H. Cameron, W.J. Ou, J.J. Doherty II, D. Louvard, A.W. Bell, D. Dignard, D.Y. Thomas, and J.J. Bergeron. 1991. SSR alpha and associated calnexin are major calcium binding proteins of the endoplasmic reticulum membrane. J. Biol. Chem. 266:19599–19610. [PubMed] [Google Scholar]
- Whitley, P., I.M. Nilsson, and G. von Heijne. 1996. A nascent secretory protein may traverse the ribosome/endoplasmic reticulum translocase complex as an extended chain. J. Biol. Chem. 271:6241–6244. [DOI] [PubMed] [Google Scholar]
- Xia, L., T. Ju, A. Westmuckett, G. An, L. Ivanciu, J.M. McDaniel, F. Lupu, R.D. Cummings, and R.P. McEver. 2004. Defective angiogenesis and fatal embryonic hemorrhage in mice lacking core 1–derived O-glycans. J. Cell Biol. 164:451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.