Abstract
Tetraspanins regulate cell migration, sperm–egg fusion, and viral infection. Through interactions with one another and other cell surface proteins, tetraspanins form a network of molecular interactions called the tetraspanin web. In this study, we use single-molecule fluorescence microscopy to dissect dynamics and partitioning of the tetraspanin CD9. We show that lateral mobility of CD9 in the plasma membrane is regulated by at least two modes of interaction that each exhibit specific dynamics. The majority of CD9 molecules display Brownian behavior but can be transiently confined to an interaction platform that is in permanent exchange with the rest of the membrane. These platforms, which are enriched in CD9 and its binding partners, are constant in shape and localization. Two CD9 molecules undergoing Brownian trajectories can also codiffuse, revealing extra platform interactions. CD9 mobility and partitioning are both dependent on its palmitoylation and plasma membrane cholesterol. Our data show the high dynamic of interactions in the tetraspanin web and further indicate that the tetraspanin web is distinct from raft microdomains.
Introduction
Lateral segregation of constituents of the plasma membrane of eukaryotic cells is now widely accepted as a requirement for the function of biological membranes. However, the mechanisms underlying this membrane organization are still a matter of debate, and several theories have been proposed to explain the lateral segregation of lipids and proteins. The most popular remains the lipid raft microdomains model (Simons and van Meer, 1988). These microdomains are currently defined as small, heterogeneous, highly dynamic sterol- and sphingolipid-enriched domains that compartmentalize cellular processes (Jacobson et al., 2007). According to this model, plasma membrane may alternate liquid-ordered (lo; rafts) and fluid (nonraft) phases, the former forming functional platforms (for review see London, 2005). Lipids, especially cholesterol (Chl), are considered key elements in the sorting of proteins into rafts. In the fence picket model, the underlying membrane skeleton is proposed to create or stabilize membrane domains that participate in the confinement of proteins (for review see Kusumi and Suzuki, 2005). Recent results using single-molecule approaches have confirmed that protein–protein interactions also play an important role in the organization of macromolecular structures in the plasma membrane of eukaryotic cells (Douglass and Vale, 2005). Indeed, protein clustering of the transmembrane molecules CD2 and linker for activation of T cells (LAT) was shown to form discrete subdomains during T cell activation (Douglass and Vale, 2005), and these domains are distinct from lipid rafts. They appear after activation of T cell receptors and result from the formation of an interaction network recruiting cytoplasmic signaling molecules without involvement of the cytoskeleton. Furthermore, a recent study has also described the dynamics of supramolecular protein clusters formed by syntaxin 1 (Sieber et al., 2007). It was suggested that proteins could spontaneously self-associate based on simple physical principles, namely a balance between weak homophilic interactions and crowding-induced steric repulsion (Sieber et al., 2007). The existence of small, stable clusters of membrane proteins is also supported by numerical simulations (Destainville, 2008).
Tetraspanins compose a family of proteins with four transmembrane domains delineating two extracellular domains of unequal size. All mammalian cells tested so far express several of these proteins; thus, not surprisingly, these molecules have been implicated in numerous physiological processes. In many examples, tetraspanins have been shown to share similar functions. For example, antibodies to several tetraspanins modulate cell migration, induce a costimulatory signal, or trigger a transduction signal (for reviews see Boucheix and Rubinstein, 2001; Hemler, 2003; Levy and Shoham, 2005). In addition, it has been shown that the tetraspanins CD9 and CD81 act in concert for an effective sperm–egg fusion (Rubinstein et al., 2006). Tetraspanins are also implicated in different diseases. Several of them, including CD9, can modulate metastasis formation (for review see Boucheix and Rubinstein, 2001). Antibodies to tetraspanins have been shown to interfere with virus-induced cell–cell fusion, including HIV (Gordon-Alonso et al., 2006), and HIV assembly takes place in tetraspanin-enriched plasma membrane domains that may serve as gateways for the virus (Nydegger et al., 2006). CD81 is also required for infection of hepatocytes by the hepatitis C virus and malaria sporozoites (Silvie et al., 2003; Cocquerel et al., 2006).
The function of tetraspanins is thought to be related to their ability to interact with one another and with various other surface proteins, forming a network of molecular interactions referred to as the tetraspanin web. Inside the tetraspanin web, small primary complexes composed of particular tetraspanins associated with partner nontetraspanin proteins have been identified (for review see Boucheix and Rubinstein, 2001). The fact that primary complexes are observed only under conditions disrupting tetraspanin–tetraspanin interactions using digitonin and, in some cases, Triton X-100 (Serru et al., 1999) suggests that most interactions in the tetraspanin web are secondary to tetraspanin–tetraspanin interactions. The demonstration that CD151 contributed to the interaction of the integrin α3β1 with other tetraspanins, as did CD9 for one of its molecular partners, CD9P-1, provided strong support for this hypothesis (Berditchevski et al., 2002; Charrin et al., 2003b). Tetraspanin–tetraspanin interactions have been shown to be dependent at least in part on lipids, including gangliosides and Chl (Charrin et al., 2003c; Odintsova et al., 2006). The interaction of tetraspanins with Chl is functionally important because modulation of cellular Chl levels was shown to modify the extent of tyrosine phosphorylation observed upon antibody ligation of tetraspanins (Charrin et al., 2003c; Delaguillaumie et al., 2004) and the CD81-dependent entry of malaria sporozoites into hepatocytic cells (Silvie et al., 2006). Additionally, palmitoylation of tetraspanins participates in the interaction with one another, although other mechanisms of interaction are likely (Berditchevski et al., 2002; Charrin et al., 2002; Yang et al., 2002). Based on these data and their ability to associate with detergent-resistant membranes, tetraspanins have been suggested to laterally segregate the proteins to which they associate, possibly by forming microdomains physically and functionally distinct from lipid rafts (for reviews see Boucheix and Rubinstein, 2001; Hemler, 2003; Levy and Shoham, 2005).
So far, tetraspanins have been mainly studied using biochemical techniques such as coimmunoprecipitation or conventional fluorescence microscopy on fixed cells, which only provide a snapshot of tetraspanin-mediated interactions. To understand how tetraspanins functionally regulate the molecules to which they associate, it is important to determine how they laterally segregate in the plasma membrane. Single-molecule fluorescence microscopy on living cells, a technique pioneered by Sako et al. (2000) and Schutz et al. (2000), can provide insights into spatial and temporal behavior of membrane components of eukaryotic cells. Diffusion coefficients of membrane proteins as well as the different modes of motion yield information on their local environment and allow the study of factors that limit diffusion (Lommerse et al., 2004).
In this paper, we focused on the membrane behavior of CD9, a prototype tetraspanin. Thanks to a dual-view–based detection, we compared the behavior of CD9 in single molecules to its ensemble membrane distribution. We demonstrate that tetraspanin assemblies form dynamic interaction platforms in permanent exchange with the rest of the membrane and also highlight different modes of interaction in the tetraspanin web.
Results
Characterization of tetraspanin complexes in PC3 cells
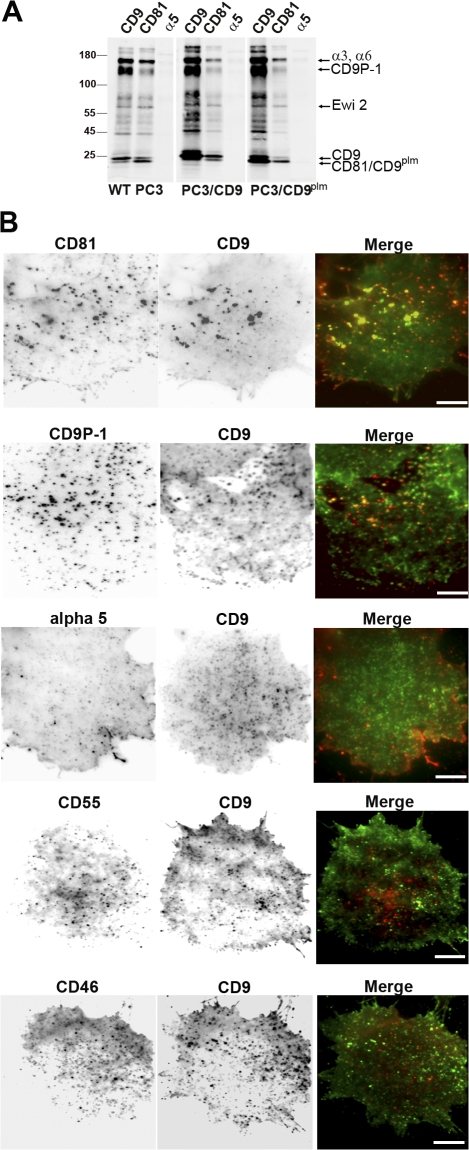
PC3 is a well-characterized human prostatic carcinoma cell line that expresses several tetraspanin molecules, including CD81 and CD9. Loss of CD9 expression in prostate cancer cells has been associated with prostate cancer progression (Wang et al., 2007). Before carrying out single-molecule experiments, tetraspanin assemblies in PC3 cells were first characterized using immunoprecipitation and immunofluorescence experiments. Immunoprecipitation experiments were performed with anti-CD9 and anti-CD81 antibodies on biotin-labeled cells lysed with the detergent Brij97 (Fig. 1 A). Because Brij97 preserves tetraspanin–tetraspanin interactions, any tetraspanin antibody can precipitate the entire set of proteins present in the tetraspanin web (Fig. 1 A, compare CD81 and CD9 profiles). Among these proteins are most likely CD9P-1, EWI-2, and the integrin α3β1, which associate with tetraspanins in all cells studied so far (Fig. 1 A; Charrin et al., 2001, 2003a; Stipp et al., 2001). As a negative control, the integrin α5β1 did not immunoprecipitate CD9 or CD81 (no band comigrating with CD9 or CD81 was detected even at higher exposure). As one of the aims of this study was to determine the role of CD9 palmitoylation, we expressed a nonpalmitoylatable CD9 mutant (CD9plm) in PC3 cells (Charrin et al., 2002) so that CD9plm molecules represented at least 75% of the total surface CD9 fraction (unpublished data). We also overexpressed wild-type (WT) CD9 as a control (PC3/CD9). As expected (Rubinstein et al., 1996), quantitatively more proteins were coimmunoprecipitated with CD9 (or CD9plm) in transfected cells as compared with WT cells. We note that, as already observed (Charrin et al., 2002; Kovalenko et al., 2004), the apparent molecular weight of CD9plm was lower than that of WT CD9.
Figure 1.
Analysis of tetraspanin assemblies in PC3 cells. (A) Immunoprecipitation experiments in WT PC3 cells or in cells overexpressing CD9 (PC3/CD9) or a nonpalmitoylated form of CD9 (PC3/CD9plm). Biotin-labeled cells were lysed in Brij97 and incubated with anti-CD9, anti-CD81, or anti-α5 antibodies (the latter is used as a negative control). Immunoprecipitated proteins were detected using peroxidase-coupled streptavidin. (B) Immunofluorescence images of PC3/CD9 living cell basal membrane by TIRF microscopy at 37°C. Cells were incubated with the anti-CD9 Cy3B-conjugated antibody SYB-1 (middle; green in the merge image) and with various antibodies labeled with Atto647N (left; red in the merge images) and raised against (top to bottom) CD81, CD9P-1, the α5 chain of integrin, CD55, or CD46. Bars, 10 μm.
The distribution of tetraspanins at the basal cell surface of PC3 cells plated on fibronectin was also analyzed by immunofluorescence on living cells using multicolor total internal reflection fluorescence (TIRF) microscopy (Fig. 1 B). CD9 molecules were distributed along the whole cell surface, with a higher concentration in dotlike areas. These enriched zones also contain CD81 as well as its molecular partner CD9P-1. These structures are unlikely to be adhesion structures because they do not contain the fibronectin receptor, the integrin α5 subunit. The CD9-enriched areas did not contain CD55 or CD46, two closely related members of the regulator of complement activation family. CD55 is a glycosyl-phosphatidylinositol (GPI)–anchored protein that is a raft resident protein and does not interact with tetraspanins (Charrin et al., 2002). CD46 is a type I membrane protein excluded from rafts (Manie et al., 2000) and sometimes associated with tetraspanins, albeit indirectly through the β1 integrin (Lozahic et al., 2000). Collectively, all of these results indicate that the behavior of CD9 in PC3 cells is comparable with its behavior in other cell lines.
Dynamics of CD9 molecules in PC3 plasma membrane
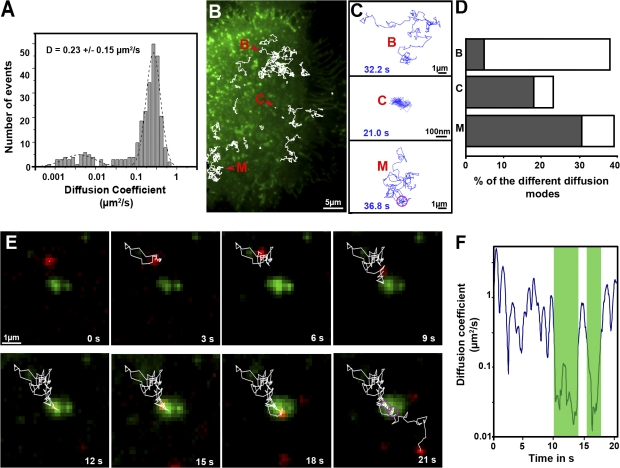
The general strategy for the tracking of single fluorescent CD9 molecules was as follows. PC3 cells were plated on fibronectin-coated glass coverslips to allow attachment and cell spreading. To label single CD9 molecules, Fab fragments of the mAb SYB-1 were prepared and labeled with a single Atto647N fluorescent probe. Fig. 2 A represents the distribution of the apparent diffusion coefficient (ADC) of 261 individual CD9 molecules (10–20 trajectories per cell) calculated using a linear fit to the mean squared displacement (MSD) versus time plots (ADC mean value of 0.23 ± 0.15 μm2/s). The type of diffusion was evaluated by the analysis of MSD versus time plots of the trajectories of individual molecules (for details see the Data analysis section in Materials and methods). Three different diffusion modes were observed for CD9 trajectories (Fig. 2, C and D; and Table I): (1) pure Brownian diffusion (38% of the total trajectories) characterized by an ADC mean value of 0.33 μm2/s; (2) pure confined or restricted diffusion (23% of the total trajectories) with a very low ADC mean value of 0.03 μm2/s and a confinement diameter of 254 nm on average; and (3) diffusion with different combinations of Brownian and confined modes referred as “mixed trajectories” (39% of total trajectories; Fig. 2 C). The confinement diameter mean value for mixed trajectories was generally larger than that of pure confined trajectories (462 nm). Under our experimental conditions, no directed diffusion of CD9 molecules was clearly identified.
Figure 2.
Single-molecule analysis of CD9 in PC3/CD9 cells. (A) ADC distribution and mean value (±SD) for CD9 molecules at 100-ms acquisition time. Note that all determinations were made by linear fitting in the MSD-τ plot using points two to four of the total curve, and 261 trajectories were analyzed. As delineated by the dashed line, two populations can be identified. Trajectories with the lower diffusion coefficient correspond to pure confined molecules, whereas the other trajectories correspond to molecules with Brownian or mixed (Brownian with a transient confinement in the same trajectory) behavior. D is the mean value of the ADC. (B) Superposition of ensemble CD9 labeling achieved with anti-CD9 Cy3B-labeled antibodies (green) and several fluorescent CD9 single-molecule trajectories obtained after tracking Atto647N-conjugated Fab fragments of SYB-1 (white thin lines). The letters in red correspond to the different diffusion modes (shown in C). (C, top to bottom) Trajectories of various single CD9 molecules exhibiting different diffusion modes: Brownian (B), confined (C), and mixed (M), a combination of Brownian and confined diffusion. The purple ring in trajectory M corresponds to transient confinement, and the blue numbers correspond to the duration of the trajectories. Video 1 corresponds to this experiment (available at http://www.jcb.org/cgi/content/full/jcb.200803010/DC1). Note that the scale bar is different for trajectory C. (D) Histograms (open boxes) representing the percentage of each CD9 diffusion mode relative to the total number of trajectories. The gray part corresponds to the proportion of trajectories associated with TEAs (identified with the ensemble membrane labeling) for each diffusion mode. (E) Time lapse of a mixed trajectory. The red spots correspond to a single CD9 molecule entering and exiting from a TEA (green). The purple rings in the last micrograph correspond to the confined areas in which CD9 ADC was decreased (see F). The duration of Video 1 is 21 s. (F) Plot of the instantaneous diffusion coefficient of the particle analyzed in the time lapse of E as a function of time (total duration of the time lapse). The green shaded part corresponds to the time period when the position of the single CD9 molecule overlaps with the ensemble CD9 labeling. A clear decrease in the instantaneous ADC of the particle is detected during this period.
Table I. ADC of single molecules and percentage of the different diffusion modes.
| Proteins and treatments |
D ± SD | DBrownian ± SD | Brownian | Confined | Mixed |
|---|---|---|---|---|---|
| μm2/s | μm2/s | μm2/s | % | % | |
| CD9 | 0.23 ± 0.15 | 0.33 ± 0.12 | 38 (5) | 23 (18) | 39 (30) |
| CD9plm | 0.28 ± 0.18a | 0.43 ± 0.18b | 38 (4) | 21 (14) | 41 (22) |
| CD9 + MβCD | 0.08 ± 0.09b | 0.19 ± 0.12b | 32 (3) | 31 (19) | 37 (18) |
| CD9 + MβCD–Chl | 0.41 ± 0.33b | 0.64 ± 0.23b | 40 (8) | 28 (23) | 32 (28) |
| CD55 | 0.24 ± 0.26 | 0.37 ± 0.31 | 40 (4) | 18 (4) | 42 (9) |
| CD55 + MβCD | 0.06 ± 0.05b | 0.08 ± 0.06 | 47 (ND) | 19 (ND) | 34 (ND) |
| CD46 | 0.13 ± 0.08b | 0.18 ± 0.07 | 52 (ND) | 13 (ND) | 35 (ND) |
| CD46 + MβCD | 0.05 ± 0.05b | 0.09 ± 0.04 | 39 (ND) | 27 (ND) | 34 (ND) |
D is the ADC mean value of all trajectories (corresponding to the three modes of diffusion), and DBrownian is the ADC mean value of pure Brownian trajectories. Numbers in parentheses correspond to the percentage of total trajectories associated with TEAs. Error estimates represent the SD for the different cells analyzed.
P < 10−3; a different distribution as compared with control cells (CD9) using the Mann-Whitney U test.
P < 10−4; a different distribution as compared with control cells (CD9) using the Mann-Whitney U test.
Individual trajectories were also compared with the ensemble distribution of CD9 thanks to dual-view microscopy. Under these conditions, as shown in Fig. 1 B (green), CD9 molecules appeared distributed along the whole cell surface, with a higher concentration in dotlike areas. These tetraspanin-enriched areas (TEAs) appeared very stable in position and shape, even for observation times >10 min using intermittent laser excitation to prevent photobleaching. We note that this CD9 ensemble membrane labeling did not interfere with single-molecule behavior because the distribution and the mean values of ADC as well as the percentage of the different diffusion modes for single Atto647N-labeled CD9 molecules were not modified by this labeling (unpublished data). It is also important to notice that a similar ensemble labeling was obtained with fluorescent Fab fragments. The ability to simultaneously observe single CD9 molecules and their ensemble distribution allowed us to evaluate their behavior when they cross the TEAs. We clearly observed that the confinement zones of single CD9 molecules were mainly restricted to these TEAs (Fig. 2 D, Table I, and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200803010/DC1). In contrast, only a minority of the Brownian trajectories crossed these areas (Fig. 2 D). Within the mixed trajectories, restricted diffusion (confined) in CD9-enriched dotlike structures alternated with more mobile diffusion (Brownian), with CD9 molecules exploring several times the same TEAs or switching between different areas (Video 1). The ADC of CD9 molecules when crossing a TEA was frequently decreased (see time-lapse and instantaneous ADC analysis in Fig. 2, E and F). The dwell time of CD9 molecules in these areas was highly variable (the mean value was 3.7 ± 4.8 s with a maximum of 20 s). Thus, although the TEA are relatively stable in time and place on the minute time scale, their majority appear to be highly dynamic structural domains in that numerous exchanges in or out of these structures take place.
The dynamic behaviors of CD9 and CD55, a raft resident protein, are different
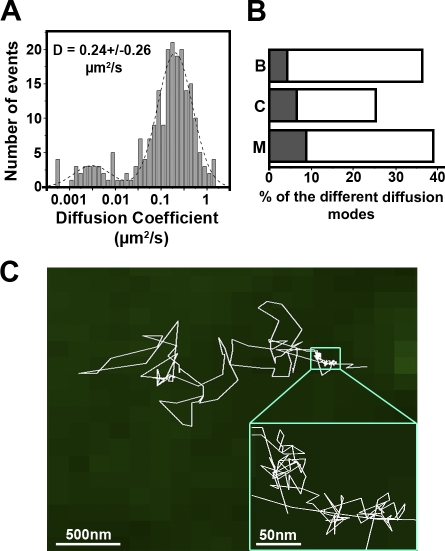
To compare CD9 behavior with that of proteins in raft microdomains, single-molecule experiments were performed with the aforementioned GPI-anchored protein CD55 (labeled with the Atto647N-conjugated mAb 12A2) and CD9 ensemble labeling. The mean value of CD55 ADC was in the same range as that of CD9 (0.24 vs. 0.23 μm2/s, respectively; Figs. 2 A and 3 A, reproduced in Fig. 4 A for comparison). The percentages of Brownian, confined, and mixed trajectories were also comparable for both membrane proteins (compare Fig. 2 D with Fig. 3 B; and Table I). However, major differences were observed: (1) ∼10% of directed trajectories was clearly identified with CD55, whereas such trajectories were never detected with CD9; (2) the distribution of CD55 ADC for Brownian trajectories was much more heterogeneous than that of CD9 (Fig. 4 A and Table I); and (3) CD55 trajectories were notably excluded from TEAs. In particular, only 9% of the mixed trajectories was associated with these areas (compare Fig. 2 D with Fig. 3 B; and Table I). CD55 sometimes crossed these areas but was never confined to them. Similarly, the trajectories with pure confined modes seldom overlapped with CD9-enriched areas (Fig. 3 B and Table I), and, finally, just a few CD55 Brownian trajectories were associated with these areas. Altogether, these observations show that at this time scale, the dynamics of CD9 molecules and their partitioning in TEAs is clearly different from that of a GPI-anchored protein, a classical raft marker.
Figure 3.
Single-molecule analysis of CD55 in PC3/CD9 cells. (A) ADC distribution and mean value (±SD) of CD55 molecules labeled with Atto647N-conjugated mAb 12A12. D is the mean value of the ADC calculated from a linear fit of the MSD-τ plot, and the dashed line delineates two different populations corresponding to pure confined trajectories (lower ADC) or mixed and Brownian trajectories. (B) Histograms (open boxes) representing the percentage of each CD55 diffusion mode as compared with the total number of trajectories. The gray part corresponds to the proportion of trajectories associated with TEAs (identified with the ensemble membrane labeling) for each diffusion mode (B, Brownian; C, confined; M, mixed). Compare with Fig. 2 D. (C) Trajectories of a single CD55 molecule. The inset is a magnification of the transient confinement area delineated by the boxed area.
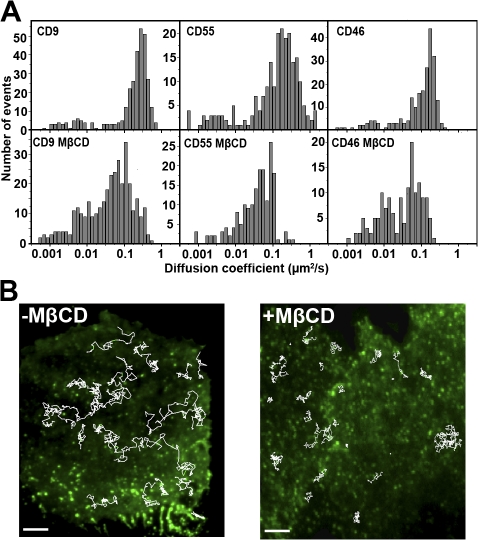
Figure 4.
Influence of MβCD on membrane dynamics. (A) Distribution of the ADC of CD9, CD55, and CD46 treated or not treated with MβCD (∼50% of the membrane Chl was removed). CD55 is a raft marker, and CD46 is excluded from rafts and TEAs. Mean values of ADC of all the molecules are available in Table I. (B) Comparison of trajectories (thin white lines) in living PC3 cells before (left) or after (right) MβCD treatment. Bars, 7.5 μm.
Chl influences CD9 membrane dynamics and partitioning
To better understand the molecular mechanisms underlying CD9 dynamics and partitioning into plasma membrane, Chl concentration was modified using methyl-β-cyclodextrin (MβCD), a cyclic oligosaccharide that removes this lipid from the cell plasma membrane (Yancey et al., 1996).
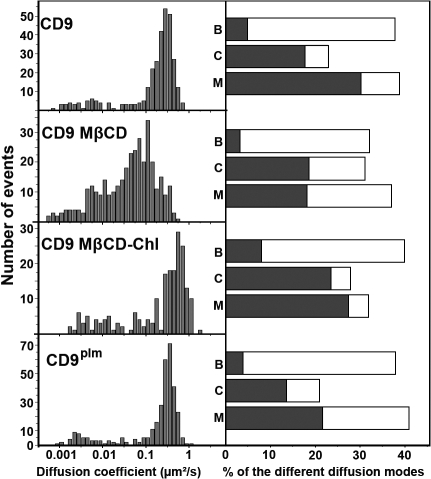
Treatment with 20 mM MβCD removed ∼50% of total Chl (see Treatment of cells with drugs section in Materials and methods) and did not dramatically alter the cells as shown by the preservation of the actin network and the conservation of dotlike structures enriched in CD9 (Fig. S2 B, available at http://www.jcb.org/cgi/content/full/jcb.200803010/DC1). Decrease of the membrane Chl content led to a threefold reduction of the mean value of CD9 ADC, which can be easily observed by eye (from 0.23 to 0.08 μm2/s; Fig. 4 A, Table I, and Video 2). The effect on CD9 ADC was partly reversed by a treatment of cells with preformed MβCD–Chl complexes (from 0.08 μm2/s after MβCD treatment to 0.16 μm2/s after Chl repletion). Reduction of the ADC mean value observed after Chl depletion partly resulted from an increase in the number of confined trajectories (where the ADC is lower) from 23 to 31% of the total trajectories (Table I). Moreover, the ADC mean value of Brownian trajectories was decreased from 0.33 to 0.19 μm2/s. Importantly, the proportion of CD9 confined trajectories associated with CD9-enriched areas was not changed upon MβCD treatment (Fig. 5). This indicates that (1) the maintenance of CD9 confinement in TEAs does not require MβCD-accessible Chl, which is consistent with the conservation of the dotlike structures as determined with the ensemble labeling (Fig. S2), and (2) the additional confined molecules are located outside TEAs. In contrast to the confined trajectories, the proportion of mixed trajectories was not changed upon Chl depletion, but the proportion associated with TEAs was clearly reduced from 30 to 18% of the total trajectories (Fig. 5 and Table I). No significant modification of the dwell time of CD9 molecules in TEAs was observed (3.7 ± 4.8 s vs. 3.3 ± 2.9 s for untreated vs. treated cells, respectively).
Figure 5.
Distribution of the ADC and diffusion modes of CD9 and CD9plm and their partitioning in tetraspanin-enriched compartments. (left) ADC distribution of CD9 in control cells (CD9), cells treated with MβCD (CD9 MβCD), cells treated with MβCD loaded with Chl (CD9 MβCD–Chl), or cells transfected with nonpalmitoylated CD9 (CD9plm). 50% of the membrane Chl was removed by MβCD treatment, and MβCD–Chl treatment increased the Chl content to 130% as compared with control cells. All of the palmitoylation sites have been mutated in CD9plm cells. (right) Histograms (open boxes) representing the percentage of each diffusion mode of the molecules as compared with the total number of trajectories (B, Brownian; C, confined; M, mixed). The gray part corresponds to the proportion of trajectories associated with TEAs (identified with the ensemble membrane labeling) for each diffusion mode.
To further investigate the role of Chl, cells were treated with preformed MβCD–Chl complexes that raised by 30% the cellular Chl concentration. In contrast to Chl depletion, this treatment increased the mean value of CD9 ADC from 0.23 to 0.41 μm2/s as well as the mean value of ADC for Brownian trajectories from 0.33 to 0.64 μm2/s (Table I). This treatment also increased the number of confined trajectories at the expense of mixed trajectories. Contrary to MβCD treatment, the large majority of confined and mixed trajectories remained associated with TEAs (Fig. 5 B).
To complete our view of the effect of Chl on membrane dynamics and as a control, we sought to investigate the effect of Chl depletion on the behavior of proteins associated with rafts (CD55) or excluded from these microdomains (CD46). Recent single-molecule studies have demonstrated that diffusion of several membrane proteins could be reduced by Chl depletion, especially in the case of raft (Orr et al., 2005; Lenne et al., 2006; Nishimura et al., 2006) but also nonraft proteins (Kwik et al., 2003; Nishimura et al., 2006). In our hands, as observed with CD9, MβCD treatment induced a significant reduction in the ADC mean value of CD55 from 0.24 to 0.06 μm2/s (Fig. 4). Similarly, a decrease of the ADC mean value from 0.13 to 0.05 μm2/s (Fig. 4) was also observed with CD46, a type I transmembrane protein that is not present in TEAs in PC3 cells (Fig. 1). These results strongly suggest that Chl depletion affects the organization of a large part of the plasma membrane, including nonraft membrane areas, and therefore extend the aforementioned studies. These results are consistent with the hypothesis that membrane Chl provides a dynamic environment that facilitates the motion of transmembrane proteins by increasing membrane fluidity through its preferential interaction with lipids, presenting a high order parameter.
Collectively, our results demonstrate that Chl greatly influences CD9 membrane dynamics and partitioning into TEAs. Decrease of CD9 dynamics by Chl depletion is likely to be the result of a general effect on the plasma membrane, probably by modifying its fluidity, whereas modification of CD9 partitioning appears to be linked directly to the organization of the tetraspanin web.
Palmitoylation modifies CD9 membrane behavior
Palmitoylation plays a key role in the association of tetraspanins with each other, and indirect evidence suggests that it may contribute to the interaction with Chl (Berditchevski et al., 2002; Yang et al., 2002; Charrin et al., 2003b). Taking into account the aforementioned influence of Chl in CD9 dynamics, we evaluated the role of this posttranslational modification in CD9 behavior using PC3 cells expressing a nonpalmitoylatable CD9 mutant (PC3/CD9plm; Charrin et al., 2002).
Single-molecule analysis reveals a slight but significant difference in the ADC mean value between WT CD9 and its nonpalmitoylatable form CD9plm (0.23 μm2/s and 0.28 μm2/s, respectively). This difference mainly corresponds to an increase of the ADC of Brownian trajectories (from 0.33 to 0.43 μm2/s), suggesting that such lipid modification contributes to restricting the free diffusion of CD9. Palmitoylation is also involved in CD9 partitioning. Indeed, the absence of CD9 palmitoylation largely decreased the percentage of total trajectories with a mixed (from 30 to 22%) and a confined (from 18 to 14%) diffusion mode that are localized in or associated with TEAs (Fig. 5 and Table I). As already observed after Chl depletion, modification of CD9 partitioning was not associated with a modification of the dwell time of CD9plm molecules in confined areas of mixed trajectories (3.7 ± 4.8 s vs. 3.7 ± 2.7 s for WT CD9 vs. CD9plm cells, respectively). Because the distribution of the different diffusion modes for CD9plm was similar to that of the WT protein (Fig. 5 and Table I), these results strongly suggest that palmitoylation promotes CD9 confinement within TEAs.
Dynamic colocalization of two CD9 molecules
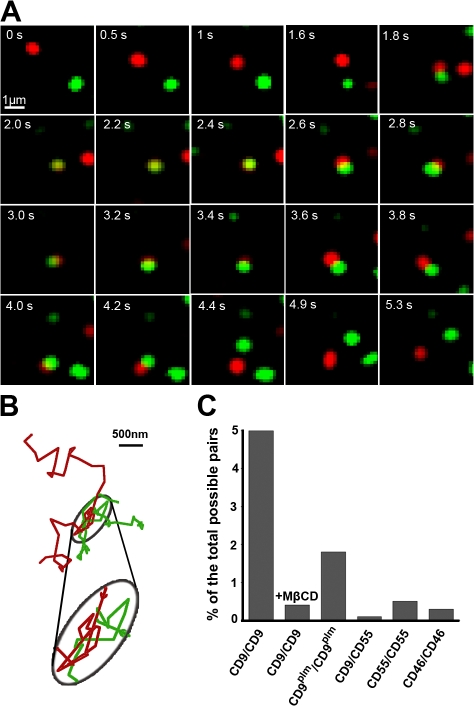
In addition to the confinement of CD9 into tetraspanin-enriched stable membrane structures described above (Dynamics of CD9 molecules in PC3 plasma membrane section), we also were able to clearly detect the overlap of trajectories of two diffusing CD9 molecules labeled with spectrally distinct fluorophores. For quantification, two molecules were arbitrarily considered as codiffusing when at least one pixel of their fluorescence signals were overlapped during at least seven frames (700 ms). An example of dynamic interaction is shown in the time lapse of Fig. 6 A, where two molecules were diffusing close together (Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200803010/DC1). All of the molecules identified as codiffusing exhibited a Brownian diffusion mode, even when the two molecules were in close proximity, and their ADC was not significantly modified during their codiffusion (unpublished data).
Figure 6.
Real-time dynamic observation of CD9 colocalization. (A) Time lapse showing a simultaneous single-molecule tracking of two differentially labeled CD9 molecules with a Fab fragment conjugated with Atto647N (red) or with Cy3B (green); see Video 2 (available at http://www.jcb.org/cgi/content/full/jcb.200803010/DC1). (B) Representative trajectory of CD9 dynamic colocalization. The parts of trajectories where the fluorescence signal of two particles overlap at least for one pixel (160 nm) are encircled in gray and magnified in the ellipse underneath (colored arrows indicate the trajectory direction). (C) Quantitative analysis of single-molecule colocalization. Two particles were considered spatially colocalized when at least one pixel of their fluorescence signals was overlapped during at least seven frames corresponding to 700 ms (the two molecules were colocalized during 24 frames in the time lapse shown in A). Different combinations of proteins were tested: CD9/CD9 on cells treated or not treated with MβCD, CD9plm/CD9plm, and irrelevant pairs such as CD9/CD55, CD55/CD55, and CD46/CD46.
In PC3 cells, codiffusing CD9 molecules were observed in ∼15% of the Brownian and mixed trajectories. However, for comparison with other molecules and to prevent any drawbacks that could be caused by a difference in the density of molecules expressed in different analyzed cells (the higher the density, the higher the probability of two molecules to diffuse together), we express our results as the percentage of colocalized pairs of trajectories as compared with the number of total possible pairs of single-labeled molecules per cell (see Dynamic colocalization section in Materials and methods). Under these conditions, dynamic CD9 colocalization was observed in 5% of the total possible pairs (number of pairs, n = 14,535; Fig. 6 C, histogram). The specificity of CD9 behavior was assessed by measuring the percentage of trajectories displaying dynamic colocalization for different pairs of membrane proteins. As expected, the percentage with the pair CD9/CD55 was very low (Fig. 6 C), which is consistent with the fact that CD9 exhibits specific features that are not shared by GPI proteins in raft microdomains. No dynamic colocalization was observed either with the CD55/CD55 and CD46/CD46 pairs (n = 17,766 and n = 16,290 possible pairs, respectively), strongly suggesting that this phenomenon does not correspond to a random dynamic colocalization of diffusing membrane proteins of similar ADC. Interestingly, when a similar analysis was performed on MβCD-treated PC3 cells, the percentage of colocalization was decreased to 0.4% (n = 11,628; vs. 5% for untreated cells; Fig. 6 C), highlighting the importance of Chl in CD9–CD9 interactions, whether lipid mediated or not. Similarly, although to a lesser degree, the dynamic colocalization of CD9 was decreased from 5 to 1.8% of the possible pairs (n = 14,028) when the molecule was not palmitoylated (CD9plm). However, it is worth noting that this percentage is certainly overestimated because WT CD9 molecules are still expressed on the surface of PC3-CD9plm cells and can contribute to the observed colocalization. In addition to its influence in CD9 dynamics and partitioning, palmitoylation also seems to be involved in the physical interaction between CD9 molecules. All of these results show CD9–CD9 interactions outside the TEAs and point to the role of Chl and palmitoylation in this interaction.
Discussion
Most recent data suggest that the tetraspanin web is organized into several levels of interactions, with tetraspanins directly associated with specific partner molecules and mediating the interaction of these partners with other tetraspanins through tetraspanin–tetraspanin interactions. Here, we have studied in live cells the CD9–CD9 interaction as a model using advanced fluorescence microscopy techniques, namely TIRF microscopy, single-molecule detection, tracking, and diffusion analysis coupled to ensemble labeling.
Dynamic interaction in the tetraspanin web
Ensemble labeling of living PC3 cells clearly revealed the presence of areas highly enriched in the two tetraspanins CD9 and CD81 as well as one of their primary partner CD9P-1, similar to what was published in several previous studies (for reviews see Boucheix and Rubinstein, 2001; Hemler, 2003; Levy and Shoham, 2005). These structures are stable over a range of minutes and are preserved after cytoskeleton disruption by latrunculin B (LTB) treatment (Fig. S2 C) and after Chl depletion, suggesting that their cohesion is probably driven by a network of protein–protein interactions. These TEAs were identified as confinement or restricted diffusion areas of CD9 molecules and can be classified in two categories that differ by their dynamics and their size. The first type corresponds to pure confined trajectories of CD9 molecules with no apparent exchanges with the rest of the membrane and relates to a minority of the trajectories. The second type of TEA is larger and very dynamic in the sense that frequent escape or trapping of CD9 molecules occurs in these regions (Video 1). It therefore represents dynamic interaction platforms in which molecules can be transiently confined. Inside these structures, the intimate interaction of two CD9 molecules or, more generally, of two tetraspanins is supported by experiments using intact antibodies instead of Fab fragments (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200803010/DC1). Under these conditions, CD9 molecules could be tracked, but once associated with TEAs, they were never able to exit. This behavior is probably caused by the stabilization of two CD9 molecules through bivalent antibody-induced cross-linking. mAb to various tetraspanins has been shown to trigger several functional effects that, to some extent, may be the consequence of stabilization of the target tetraspanin in TEAs (for review see Boucheix and Rubinstein, 2001).
Outside of the TEAs, tracking CD9 molecules reveals that 77% of the analyzed molecules displayed a Brownian diffusion mode, at least during part of their trajectory, underlining the very dynamic behavior of CD9. Our results also indicate that two CD9 molecules can transiently interact outside TEAs. This conclusion is supported by the observation that two CD9 molecules often codiffuse for short periods of time. Several controls indicate that this interaction is not the consequence of the random association of membrane proteins (see the negative results with different pairs of proteins in Fig. 6). Although under our labeling conditions no more than 15% of the analyzed diffusing molecules interacted with another CD9 molecule, it is noteworthy that only a minor fraction of the molecules is analyzed in single-molecule experiments (as a result of extremely low labeling levels), and thus, this CD9–CD9 interaction is probably a frequent event. Importantly, no abrupt modification of CD9 velocity was observed when the molecules started to codiffuse. Because the diffusion of transmembrane proteins in liquid membranes should decrease with the size of the diffusant (Gambin et al., 2006; Guigas and Weiss, 2006), this observation seems to exclude the interaction of two isolated CD9 molecules. Therefore, we suggest that CD9 diffuses in the membrane embedded in a small membrane cluster that could accommodate diverse proteins (in particular, tetraspanins and their partners) and lipids. A similar membrane organization in small entities has also been proposed for GPI-anchored proteins in raft microdomains (Mayor and Rao, 2004) and in transient confinement zones (Dietrich et al., 2002). The codiffusion of two CD9 molecules could correspond to the transient association of two diffusing clusters or the transfer of a labeled CD9 molecule from one diffusing cluster to another. This latter hypothesis is supported by the permanent exchange of CD9 molecules in and out of TEAs. However, further studies are clearly needed to discriminate between these two hypotheses.
Influence of Chl and palmitoylation on tetraspanin–tetraspanin interactions
The interaction between tetraspanins has been shown to be modulated by lipids, including gangliosides, Chl, and palmitoylation of tetraspanins (Berditchevski et al., 2002; Charrin et al., 2002, 2003c; Yang et al., 2002; Odintsova et al., 2006; Silvie et al., 2006). Our results bring new insights into the mechanisms whereby Chl and palmitoylation contribute to tetraspanin–tetraspanin interactions.
We have demonstrated that the number of transient confinements of CD9 in the TEAs (observed in mixed trajectories) was reduced after Chl depletion and mutation of palmitoylation sites. Importantly Chl depletion did not disrupt the dotlike CD9-enriched structures (Fig. S2) and did not change CD9 dwell time in TEAs. Similarly, the dwell time of CD9plm residency was not different from that of WT CD9. Altogether, these data suggest that neither Chl nor palmitoylation contribute significantly to the maintenance of CD9 in these structures and rather further support a role of protein–protein interactions. Chl and palmitoylation may therefore facilitate the initial interaction of CD9 with TEAs. This hypothesis is supported by the fact that increasing Chl concentration increased the number of confined trajectories and the confinement of CD9 in TEAs in the mixed trajectories.
The number of CD9 molecules dynamically interacting with one another outside the TEAs was also strongly diminished after Chl depletion or palmitoylation removal (Fig. 6). The effect of palmitoylation removal can be explained in the context of the model whereby tetraspanins diffuse in the membrane embedded in small clusters. A possibility is that palmitoylation may be involved in the interactions of CD9 in such clusters or in the assembly of these clusters. This hypothesis is consistent with the fact that diffusion of the nonpalmitoylated CD9 is faster than that of WT CD9. A possible explanation for this faster diffusion is that this mutant diffuses in smaller platforms than those containing WT CD9, or possibly as an isolated molecule (a similar increase in the ADC for a nonpalmitoylatable mutant of LAT has also been reported; Douglass and Vale, 2005). One can argue that this faster diffusion could also be explained by the preferential partition of palmitoylated proteins in the lo phase (Melkonian et al., 1999), in which lipids diffuse slower than in a fluid phase. However, even if we cannot completely exclude this possibility, the palmitoylation of tetraspanins does not appear to drive their partition into a lo phase because their solubilization by detergents is not influenced by this modification as determined by sucrose gradient fractionation (Charrin et al., 2002; Yang et al., 2002).
The decrease of the number of dynamically interacting CD9 molecules after Chl depletion cannot be explained by a 30% decrease of CD9 ADC of the Brownian trajectories observed after MβCD treatment. Indeed, simulation of randomly chosen Brownian particles under experimental conditions close to those described with CD9 (in terms of the number of particles in a field and the diffusion coefficients; see Brownian dynamic simulation section in Materials and methods) indicates that the number of associations of particles is not dramatically modified by a twofold decrease of the ADC of these particles (unpublished data). It is therefore tempting to speculate that Chl could mediate direct interaction between two diffusing CD9 molecules in agreement with the observation that tetraspanins associate with this lipid (Charrin et al., 2003c).
We propose that palmitoylation and Chl are especially important for the dynamics of tetraspanin interactions rather than being involved in the maintenance of these interactions, whether they take place inside or outside TEAs. This may explain why mutation of palmitoylation sites of tetraspanins has only a partial effect on tetraspanin–tetraspanin interactions (Berditchevski et al., 2002; Charrin et al., 2002; Yang et al., 2002) and why Chl depletion in living cells did not produce any apparent modification of tetraspanin–tetraspanin interactions (Charrin et al., 2003c). In addition, treatment of living cells with MβCD was shown to inhibit the CD81-dependent infection of liver cells by malaria sporozoites (Silvie et al., 2003), tyrosine phosphorylation induced upon tetraspanin ligation in lymphoid B cells (Charrin et al., 2003c), and functional effects induced by CD82 engagement in Jurkat T cells (Delaguillaumie et al., 2004). These effects may reflect the importance of the dynamic of interactions within the tetraspanin web.
Comparison of CD9 dynamic behavior with other membrane proteins
The dynamic parameters that we have measured for CD9 compare well with other membrane proteins. The mean diffusion coefficient of CD9 is of the same order of magnitude as other membrane proteins such GPI-anchored proteins (CD55 in this study or CD59 in recent publications using single dye tracking; Suzuki et al., 2007; Wieser et al., 2007) or transmembrane proteins (the single-span transmembrane protein CD46 in this study, LAT [Douglass and Vale, 2005], or the G protein–coupled μ-opioid receptor [Suzuki et al., 2005]). Similarly, the different types of diffusion modes that we observed with CD9, namely confined, Brownian, and mixed (a combination of Brownian and confined or restricted diffusion), correspond with a widespread behavior of plasma membrane proteins because similar results were obtained for two other proteins in this study and are described in several publications (for reviews see Kusumi and Suzuki, 2005; Sako, 2006). This behavior certainly reflects the general dynamics of plasma membrane and the ubiquitous membrane compartmentalization in regions or domains that can be enriched in specialized lipids as described for rafts (Jacobson et al., 2007), in specialized proteins as described for T signaling clusters (Douglass and Vale, 2005), or a mixture of both, as suggested here for CD9.
Despite these similarities with other membrane proteins, several major and unique characteristics of CD9 can be noted. The actin cytoskeleton does not appear to be involved in maintenance of the TEAs identified as confinement areas. This feature clearly differentiates TEAs from other transient confinement areas such as transient confinement zones enriched in GPI-anchored protein Thy-1 (Chen et al., 2006) or from regions of temporary arrest of lateral diffusion observed with CD59 (Suzuki et al., 2007). Importantly, the confinement areas observed with the GPI-anchored protein CD55 (pure or transient confinement in mixed trajectories) were different from those of CD9 because they were not enriched in tetraspanins, suggesting that different mechanisms drive the transient confinement of different types of membrane proteins. Moreover, no codiffusion between CD9 and CD55 molecules could be observed. Altogether, these data indicate that the tetraspanin web is physically distinct from the raft microdomains enriched in GPI-anchored proteins.
A model for the tetraspanin web
The tetraspanin web has been defined as a network of molecular interactions organized by tetraspanins. Our data indicate that inside the web, tetraspanin–tetraspanin interactions (and therefore secondary interactions) are transient and highly dynamic. Two modes of interactions have been identified. The first mode is based on tetraspanin assemblies that can form membrane platforms stable in shape and localization. The maintenance of these platforms depends neither on membrane Chl nor on the underlying cytoskeleton, strengthening the involvement of direct or indirect protein–protein interaction. Some of these platforms appear to be unconnected to the rest of the membrane, but the majority of these platforms are in permanent exchange with it. Chl and palmitoylation likely contribute to the initial interaction of diffusing tetraspanins with these platforms. A second mode of interaction is suggested by the codiffusion of two CD9 molecules outside TEAs. We suggest that tetraspanins diffuse in the plasma membrane embedded in small clusters that could contain other tetraspanins, some protein partners, and lipids. These clusters interact with each other and possibly exchange CD9 molecules. This dynamic colocalization is dependent on Chl and palmitoylation, although the precise role of these lipids remains unclear. These results exclude a possible self-organization of tetraspanins based uniquely on protein–protein interactions. Collectively, our characterization of the membrane dynamics and partitioning of the CD9 tetraspanin on the single-molecule level reveals a dynamic web of membrane protein–protein–lipid interactions with an organization distinct from that of raft microdomains.
Materials and methods
Materials
Cell culture reagents, Amplex red Chl assay kit, and TransFluoSpheres were purchased from Invitrogen. Atto647N succinimidyl ester was obtained from Atto-tec, and Cy3B succinimidyl ester and PD10 columns were purchased from GE Healthcare. Glass coverslips were obtained from Dutcher, and fibronectin, MβCD, MβCD–Chl, LTB, and BSA were purchased from Sigma-Aldrich.
Cell culture
Human metastatic prostate PC3 cell lines were grown in DME-F12 medium supplemented with antibiotics and 10% FCS. Cells were plated on 25-mm Ø glass coverslips (precoated with 10 μg/ml fibronectin) 24–48 h before the experiment and used at ∼60% confluence. Before coating, coverslips were successively washed with acetone, methanol, and water, sonicated for 30 min in 1 M KOH, and extensively rinsed with water.
Antibody labeling
The mAbs SYB-1 (CD9), TS81 (CD81), 1F11 (CD9P1), 12A12 (CD55), 11C5 (CD46), and v5-vjf (integrin α5) were previously described (Lozahic et al., 2000; Charrin et al., 2003a). Fab fragments were produced using papain digestion according to the protocol provided by Thermo Fisher Scientific. Antibodies or Fab fragments were labeled with Cy3B or Atto647N. In brief, covalent amine labeling of antibodies was performed by adding the fluorophore (succinimidyl ester) to antibody solution in a 3:2 molar ratio in PBS buffer, pH 7.4. The labeling reaction was performed for 2 h at room temperature, and the nonreacted dye was removed with a PD10 column. The dye/protein ratio after labeling was always inferior to 1.
Treatment of cells with drugs
For Chl depletion, cells were incubated in the DME-F12 medium containing 20 mM MβCD and 2% BSA at 37°C for 30 min. Treatment with MβCD removed ∼50% of the Chl content of PC3 cells, as determined using the Amplex red Chl kit. Increase in membrane Chl was achieved by incubating the cells with 1 mM of a 1:10 complex of Chl and MβCD in serum-free medium at 37°C for 15 min. Treatment with MβCD–Chl led to ∼30% Chl increase in the PC3 membrane.
Single-molecule experimental setup
Cells were incubated in DME/F12 at 37°C for 10 min with 4 ng/ml Cy3B-labeled antibodies for ensemble labeling or/and 4 pg/ml Atto647N-labeled Fab fragments for single-molecule labeling. The dynamics of single tetraspanin molecules was investigated at 37°C using TIRF microscopy, which reduces background fluorescence caused by the cytoplasmic autofluorescence (Sako et al., 2000). A homemade objective-type TIRF setup allowing multicolor single-molecule imaging was used. Excitation was achieved by focusing the 532-nm light from a diode-pumped double Nd–yttrium aluminium garnet laser (Crystalaser) and/or the 632.8-nm light from a HeNe laser (Melles Griot) into the back focal plane of an αPlan Fluor 100×/1.45 NA objective (Carl Zeiss, Inc.). The emitted photons were collected through the same objective, and the beam was split into two regions of the CCD detector (Cascade 512B; Roper Scientific) using two dichroic mirrors (630DRLP; Omega Optical), allowing simultaneous observation of the two fluorescent dyes (dual-view format according to Kinosita et al., 1991). Further selection of the Cy3B and Atto647N emissions was achieved with a 580/40BP (Semrock) and 660LP filter (Omega Optical), respectively. All of the experiments were performed with a 100-ms integration time. For some experiments, to achieve a better specificity in the detection of the two fluorescent signals, alternating laser excitation was performed using an acousto-optical tunable filter and controller (AA Optoelectronics; Margeat et al., 2006). The alternation period was defined by the integration time of the camera (i.e., typically 100 ms.)
Data analysis
All of the videos were analyzed using a homemade software (named PaTrack) implemented in visual C++. Trajectories were constructed using the individual diffraction limited signal of each molecule. The center of each fluorescence peak was determined with subpixel resolution by fitting a two-dimensional elliptical Gaussian function. The accuracy of the position measurement in living cells was estimated to be 50 nm by fitting a 2D Gaussian to the emission intensity distribution of an immobile single molecule conjugated with Atto647N. The 2D trajectories of single molecules were constructed frame per frame. Only trajectories containing at least 40 points and including a one-step photobleaching event were retained (mean duration of trajectories is 15 s, ranging from 4 to 55 s). Diffusion coefficient values were determined from a linear fit to the MSD-τ plots between the second and the fourth points (D2–4) according to the equation MSD(t) = 4Dt (Kusumi et al., 1993). More than 200 trajectories were analyzed for each condition. Instantaneous diffusion coefficient as shown in Fig. 2 F was derived from MSD curves calculated over contiguous trajectory stretches of 10 frames (1 s).
Determination of the motional modes (Brownian, confined, or directed) and parameters was performed according to Kusumi et al. (1993). For each trajectory, we first linearly fitted the MSD on the 10% first points to use sufficiently populated curves. If the MSD-τ plot shows positive or negative deviation from a straight line with a slope of 4D (Brownian diffusion), the MSD is adjusted with a quadratic curve (4Dt + ν2t2) (directed diffusion) or with an exponential curve,
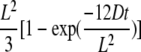 |
(confined diffusion where L is the side of a square domain, the confinement diameter being related to L by dconf = (2/√π)L). For the mixed trajectory exhibiting a combination of Brownian and apparent confined motion mode, the trajectory was split, and the MSD of each segment was adjusted with a linear or an exponential curve.
The software is also implemented to superimpose the two regions of the CCD detector when working in dual-view mode. To achieve the superimposition of the two split images, a few NeutrAvidin-labeled microspheres (TransFluoSpheres) were imaged before each experiment. Pairs of the different peaks were selected and used to calculate a transformation matrix that is applied to our experiment.
Dynamic colocalization
To determine real-time colocalization of particles, PC3 cells were doubly labeled with Cy3B and Atto647N SYB-1 Fab fragments at single-molecule concentrations. Thanks to the dual-view setup and our single-molecule tracking software, the respective trajectories for two different molecules labeled with spectrally distinct fluorophores could be determined with a lateral resolution of ∼50 nm. We chose a scheme in which two particles were considered spatially colocalized when at least one pixel of two fluorescence signals was overlapped during at least seven frames (700 ms). The lateral precision of the superimposition of the particle tracking was ∼50 nm.
Because we cannot exactly control the proportion of labeled particles at the surface of our cells, our results are expressed as a percentage of superimposed particles as compared with the number of possible pairs according to the equation % = [O/(i × j)] × 100, where O is the total number of superimposed red and green particles at the cell surface, i is the number of green-labeled particles, and j is the number of red-labeled particles.
Brownian dynamic simulation
The lateral diffusion of particles was described by Brownian diffusion simulation. At t = 0, particles were placed either randomly with nonoverlapping positions (their number was based on experiments) or using coordinates of CD9 molecules in one frame of videos acquired using a 100-ms time scale. The particle was represented by a 2D Gaussian. Its diffusion between t and t + 1 was defined as a 2D Gaussian with an SD of √2Dt with D = 0.1 or 0.2 μm2/s and t = 100 ms. Two particles were considered spatially colocalized as mentioned in the previous section.
Online supplemental material
Fig. S1 shows ADC distribution and partitioning of CD9 obtained with the intact mAb SYB-1 or with Fab fragments. Fig. S2 shows MβCD and LTB treatment of PC3 cells. Video 1 shows dynamic behavior of CD9 in the context of the tetraspanin-enriched compartment. Video 2 shows that MβCD treatment of PC3 cells decreased membrane dynamics, and Video 3 shows real-time dynamic colocalization of CD9. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200803010/DC1.
Supplementary Material
Acknowledgments
C. Espenel and C. Arduise are recipients of grants from the French Ministry for Research and Technology and from the Association pour la Recherche sur le Cancer. This study was supported by grants from the French Ministry for Research (Action Concertée Incitative [ACI] Biologie Cellulaire, Moléculaire, et Structurale and ACI Dynamique et Réactivité des Assemblages Biologiques), the Association Nationale pour la Recherche Blanc, Institut de Cancérologie et Immunogénétique, l'Association pour la Recherche sur le Cancer, and Nouvelles Recherches Biomédicales Vaincre Le Cancer.
Abbreviations used in this paper: ADC, apparent diffusion coefficient; Chl, cholesterol; GPI, glycosyl-phosphatidylinositol; LAT, linker for activation of T cells; LTB, latrunculin B; MβCD, methyl-β-cyclodextrin; MSD, mean squared displacement; TEA, tetraspanin-enriched area; TIRF, total internal reflection fluorescence; WT, wild type.
References
- Berditchevski, F., E. Odintsova, S. Sawada, and E. Gilbert. 2002. Expression of the palmitoylation-deficient CD151 weakens the association of alpha 3 beta 1 integrin with the tetraspanin-enriched microdomains and affects integrin-dependent signaling. J. Biol. Chem. 277:36991–37000. [DOI] [PubMed] [Google Scholar]
- Boucheix, C., and E. Rubinstein. 2001. Tetraspanins. Cell. Mol. Life Sci. 58:1189–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrin, S., F. Le Naour, M. Oualid, M. Billard, G. Faure, S.M. Hanash, C. Boucheix, and E. Rubinstein. 2001. The major CD9 and CD81 molecular partner. Identification and characterization of the complexes. J. Biol. Chem. 276:14329–14337. [DOI] [PubMed] [Google Scholar]
- Charrin, S., S. Manie, M. Oualid, M. Billard, C. Boucheix, and E. Rubinstein. 2002. Differential stability of tetraspanin/tetraspanin interactions: role of palmitoylation. FEBS Lett. 516:139–144. [DOI] [PubMed] [Google Scholar]
- Charrin, S., F. Le Naour, V. Labas, M. Billard, J.P. Le Caer, J.F. Emile, M.A. Petit, C. Boucheix, and E. Rubinstein. 2003. a. EWI-2 is a new component of the tetraspanin web in hepatocytes and lymphoid cells. Biochem. J. 373:409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrin, S., S. Manie, M. Billard, L. Ashman, D. Gerlier, C. Boucheix, and E. Rubinstein. 2003. b. Multiple levels of interactions within the tetraspanin web. Biochem. Biophys. Res. Commun. 304:107–112. [DOI] [PubMed] [Google Scholar]
- Charrin, S., S. Manie, C. Thiele, M. Billard, D. Gerlier, C. Boucheix, and E. Rubinstein. 2003. c. A physical and functional link between cholesterol and tetraspanins. Eur. J. Immunol. 33:2479–2489. [DOI] [PubMed] [Google Scholar]
- Chen, Y., W.R. Thelin, B. Yang, S.L. Milgram, and K. Jacobson. 2006. Transient anchorage of cross-linked glycosyl-phosphatidylinositol-anchored proteins depends on cholesterol, Src family kinases, caveolin, and phosphoinositides. J. Cell Biol. 175:169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquerel, L., C. Voisset, and J. Dubuisson. 2006. Hepatitis C virus entry: potential receptors and their biological functions. J. Gen. Virol. 87:1075–1084. [DOI] [PubMed] [Google Scholar]
- Delaguillaumie, A., J. Harriague, S. Kohanna, G. Bismuth, E. Rubinstein, M. Seigneuret, and H. Conjeaud. 2004. Tetraspanin CD82 controls the association of cholesterol-dependent microdomains with the actin cytoskeleton in T lymphocytes: relevance to co-stimulation. J. Cell Sci. 117:5269–5282. [DOI] [PubMed] [Google Scholar]
- Destainville, N. 2008. Cluster phases of membrane proteins. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 77:011905. [DOI] [PubMed] [Google Scholar]
- Dietrich, C., B. Yang, T. Fujiwara, A. Kusumi, and K. Jacobson. 2002. Relationship of lipid rafts to transient confinement zones detected by single particle tracking. Biophys. J. 82:274–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass, A.D., and R.D. Vale. 2005. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell. 121:937–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambin, Y., R. Lopez-Esparza, M. Reffay, E. Sierecki, N.S. Gov, M. Genest, R.S. Hodges, and W. Urbach. 2006. Lateral mobility of proteins in liquid membranes revisited. Proc. Natl. Acad. Sci. USA. 103:2098–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon-Alonso, M., M. Yanez-Mo, O. Barreiro, S. Alvarez, M.A. Munoz-Fernandez, A. Valenzuela-Fernandez, and F. Sanchez-Madrid. 2006. Tetraspanins CD9 and CD81 modulate HIV-1-induced membrane fusion. J. Immunol. 177:5129–5137. [DOI] [PubMed] [Google Scholar]
- Guigas, G., and M. Weiss. 2006. Size-dependent diffusion of membrane inclusions. Biophys. J. 91:2393–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemler, M.E. 2003. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 19:397–422. [DOI] [PubMed] [Google Scholar]
- Jacobson, K., O.G. Mouritsen, and R.G. Anderson. 2007. Lipid rafts: at a crossroad between cell biology and physics. Nat. Cell Biol. 9:7–14. [DOI] [PubMed] [Google Scholar]
- Kinosita, K. Jr., H. Itoh, S. Ishiwata, K. Hirano, T. Nishizaka, and T. Hayakawa. 1991. Dual-view microscopy with a single camera: real-time imaging of molecular orientations and calcium. J. Cell Biol. 115:67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko, O.V., X. Yang, T.V. Kolesnikova, and M.E. Hemler. 2004. Evidence for specific tetraspanin homodimers: inhibition of palmitoylation makes cysteine residues available for cross-linking. Biochem. J. 377:407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi, A., and K. Suzuki. 2005. Toward understanding the dynamics of membrane-raft-based molecular interactions. Biochim. Biophys. Acta. 1746:234–251. [DOI] [PubMed] [Google Scholar]
- Kusumi, A., Y. Sako, and M. Yamamoto. 1993. Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys. J. 65:2021–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwik, J., S. Boyle, D. Fooksman, L. Margolis, M.P. Sheetz, and M. Edidin. 2003. Membrane cholesterol, lateral mobility, and the phosphatidylinositol 4,5-bisphosphate-dependent organization of cell actin. Proc. Natl. Acad. Sci. USA. 100:13964–13969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenne, P.F., L. Wawrezinieck, F. Conchonaud, O. Wurtz, A. Boned, X.J. Guo, H. Rigneault, H.T. He, and D. Marguet. 2006. Dynamic molecular confinement in the plasma membrane by microdomains and the cytoskeleton meshwork. EMBO J. 25:3245–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, S., and T. Shoham. 2005. Protein-protein interactions in the tetraspanin web. Physiology (Bethesda). 20:218–224. [DOI] [PubMed] [Google Scholar]
- Lommerse, P.H., H.P. Spaink, and T. Schmidt. 2004. In vivo plasma membrane organization: results of biophysical approaches. Biochim. Biophys. Acta. 1664:119–131. [DOI] [PubMed] [Google Scholar]
- London, E. 2005. How principles of domain formation in model membranes may explain ambiguities concerning lipid raft formation in cells. Biochim. Biophys. Acta. 1746:203–220. [DOI] [PubMed] [Google Scholar]
- Lozahic, S., D. Christiansen, S. Manie, D. Gerlier, M. Billard, C. Boucheix, and E. Rubinstein. 2000. CD46 (membrane cofactor protein) associates with multiple beta1 integrins and tetraspans. Eur. J. Immunol. 30:900–907. [DOI] [PubMed] [Google Scholar]
- Manie, S.N., S. Debreyne, S. Vincent, and D. Gerlier. 2000. Measles virus structural components are enriched into lipid raft microdomains: a potential cellular location for virus assembly. J. Virol. 74:305–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margeat, E., A.N. Kapanidis, P. Tinnefeld, Y. Wang, J. Mukhopadhyay, R.H. Ebright, and S. Weiss. 2006. Direct observation of abortive initiation and promoter escape within single immobilized transcription complexes. Biophys. J. 90:1419–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor, S., and M. Rao. 2004. Rafts: scale-dependent, active lipid organization at the cell surface. Traffic. 5:231–240. [DOI] [PubMed] [Google Scholar]
- Melkonian, K.A., A.G. Ostermeyer, J.Z. Chen, M.G. Roth, and D.A. Brown. 1999. Role of lipid modifications in targeting proteins to detergent-resistant membrane rafts - many raft proteins are acylated, while few are prenylated. J. Biol. Chem. 274:3910–3917. [DOI] [PubMed] [Google Scholar]
- Nishimura, S.Y., S.J. Lord, L.O. Klein, K.A. Willets, M. He, Z. Lu, R.J. Twieg, and W.E. Moerner. 2006. Diffusion of lipid-like single-molecule fluorophores in the cell membrane. J. Phys. Chem. B. 110:8151–8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nydegger, S., S. Khurana, D.N. Krementsov, M. Foti, and M. Thali. 2006. Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J. Cell Biol. 173:795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odintsova, E., T.D. Butters, E. Monti, H. Sprong, G. van Meer, and F. Berditchevski. 2006. Gangliosides play an important role in the organization of CD82-enriched microdomains. Biochem. J. 400:315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, G., D. Hu, S. Ozcelik, L.K. Opresko, H.S. Wiley, and S.D. Colson. 2005. Cholesterol dictates the freedom of EGF receptors and HER2 in the plane of the membrane. Biophys. J. 89:1362–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein, E., F. Le Naour, C. Lagaudriere-Gesbert, M. Billard, H. Conjeaud, and C. Boucheix. 1996. CD9, CD63, CD81, and CD82 are components of a surface tetraspan network connected to HLA-DR and VLA integrins. Eur. J. Immunol. 26:2657–2665. [DOI] [PubMed] [Google Scholar]
- Rubinstein, E., A. Ziyyat, M. Prenant, E. Wrobel, J.P. Wolf, S. Levy, F. Le Naour, and C. Boucheix. 2006. Reduced fertility of female mice lacking CD81. Dev. Biol. 290:351–358. [DOI] [PubMed] [Google Scholar]
- Sako, Y. 2006. Imaging single molecules in living cells for systems biology. Mol. Syst. Biol. 2:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako, Y., S. Minoghchi, and T. Yanagida. 2000. Single-molecule imaging of EGFR signalling on the surface of living cells. Nat. Cell Biol. 2:168–172. [DOI] [PubMed] [Google Scholar]
- Schutz, G.J., G. Kada, V.P. Pastushenko, and H. Schindler. 2000. Properties of lipid microdomains in a muscle cell membrane visualized by single molecule microscopy. EMBO J. 19:892–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serru, V., F. Le Naour, M. Billard, D.O. Azorsa, F. Lanza, C. Boucheix, and E. Rubinstein. 1999. Selective tetraspan-integrin complexes (CD81/alpha4beta1, CD151/alpha3beta1, CD151/alpha6beta1) under conditions disrupting tetraspan interactions. Biochem. J. 340:103–111. [PMC free article] [PubMed] [Google Scholar]
- Sieber, J.J., K.I. Willig, C. Kutzner, C. Gerding-Reimers, B. Harke, G. Donnert, B. Rammner, C. Eggeling, S.W. Hell, H. Grubmuller, and T. Lang. 2007. Anatomy and dynamics of a supramolecular membrane protein cluster. Science. 317:1072–1076. [DOI] [PubMed] [Google Scholar]
- Silvie, O., E. Rubinstein, J.F. Franetich, M. Prenant, E. Belnoue, L. Renia, L. Hannoun, W. Eling, S. Levy, C. Boucheix, and D. Mazier. 2003. Hepatocyte CD81 is required for Plasmodium falciparum and Plasmodium yoelii sporozoite infectivity. Nat. Med. 9:93–96. [DOI] [PubMed] [Google Scholar]
- Silvie, O., S. Charrin, M. Billard, J.F. Franetich, K.L. Clark, G.J. van Gemert, R.W. Sauerwein, F. Dautry, C. Boucheix, D. Mazier, and E. Rubinstein. 2006. Cholesterol contributes to the organization of tetraspanin-enriched microdomains and to CD81-dependent infection by malaria sporozoites. J. Cell Sci. 119:1992–2002. [DOI] [PubMed] [Google Scholar]
- Simons, K., and G. van Meer. 1988. Lipid sorting in epithelial cells. Biochemistry. 27:6197–6202. [DOI] [PubMed] [Google Scholar]
- Stipp, C.S., T.V. Kolesnikova, and M.E. Hemler. 2001. EWI-2 is a major CD9 and CD81 partner and member of a novel Ig protein subfamily. J. Biol. Chem. 276:40545–40554. [DOI] [PubMed] [Google Scholar]
- Suzuki, K., K. Ritchie, E. Kajikawa, T. Fujiwara, and A. Kusumi. 2005. Rapid hop diffusion of a G-protein-coupled receptor in the plasma membrane as revealed by single-molecule techniques. Biophys. J. 88:3659–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K.G., T.K. Fujiwara, F. Sanematsu, R. Iino, M. Edidin, and A. Kusumi. 2007. GPI-anchored receptor clusters transiently recruit Lyn and Gα for temporary cluster immobilization and Lyn activation: single-molecule tracking study 1. J. Cell Biol. 177:717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.C., L.R. Begin, N.G. Berube, S. Chevalier, A.G. Aprikian, H. Gourdeau, and M. Chevrette. 2007. Down-regulation of CD9 expression during prostate carcinoma progression is associated with CD9 mRNA modifications. Clin. Cancer Res. 13:2354–2361. [DOI] [PubMed] [Google Scholar]
- Wieser, S., M. Moertelmaier, E. Fuertbauer, H. Stockinger, and G.J. Schutz. 2007. (Un)confined diffusion of CD59 in the plasma membrane determined by high-resolution single molecule microscopy. Biophys. J. 92:3719–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancey, P.G., W.V. Rodrigueza, E.P. Kilsdonk, G.W. Stoudt, W.J. Johnson, M.C. Phillips, and G.H. Rothblat. 1996. Cellular cholesterol efflux mediated by cyclodextrins. Demonstration of kinetic pools and mechanism of efflux. J. Biol. Chem. 271:16026–16034. [DOI] [PubMed] [Google Scholar]
- Yang, X., C. Claas, S.K. Kraeft, L.B. Chen, Z. Wang, J.A. Kreidberg, and M.E. Hemler. 2002. Palmitoylation of tetraspanin proteins: modulation of CD151 lateral interactions, subcellular distribution, and integrin-dependent cell morphology. Mol. Biol. Cell. 13:767–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.