Abstract
Amyotrophic lateral sclerosis (ALS) is a motor neuron disease that leads to loss of motor function and early death. About 5% of cases are inherited, with the majority of identified linkages in the gene encoding copper, zinc-superoxide dismutase (SOD1). Strong evidence indicates that the SOD1 mutations confer dominant toxicity on the protein. To provide new insight into mechanisms of ALS, we have generated and characterized a model for familial ALS in Drosophila with transgenic expression of human SOD1. Expression of wild type or disease-linked (A4V, G85R) mutants of human SOD1 selectively in motor neurons induced progressive climbing deficits. These effects were accompanied by defective neural circuit electrophysiology, focal accumulation of human SOD1 protein in motor neurons, and a stress response in surrounding glia. However, toxicity was not associated with oligomerization of SOD1 and did not lead to neuronal loss. These studies uncover cell-autonomous injury by SOD1 to motor neurons in vivo, as well as non-autonomous effects on glia, and provide the foundation for new insight into injury and protection of motor neurons in ALS.
Amyotrophic lateral sclerosis (ALS)2 is a progressive neurodegenerative disease of the motor system (1-3). It is characterized by loss of muscle function secondary to dysfunction and death of both upper and lower motor neurons. Most cases occur sporadically, whereas a small percentage are inherited. About one-fifth of occurrences of familial ALS (fALS) are linked to dominantly inherited mutations in the gene encoding copper, zinc-superoxide dismutase (SOD1) (4). SOD-linked fALS resembles the sporadic disease, although some inherited cases show earlier disease onset or a particularly fast or slow rate of progression (2, 5). Uncovering how mutations in SOD1 ultimately lead to the dysfunction and death of motor neurons may shed light on how ALS develops and progresses in patients with both sporadic and familial disease.
SOD1 is an enzyme found in the cytoplasm of all cells, comprising as much as 1% of total cellular protein (6). Over 100 missense mutations affecting many different residues, an insertion, a deletion, and a truncation of SOD1 have been linked to development of motor neuron disease (1, 2). Mutations confer a dominant toxicity on the protein, rather than a loss of function. Enzymatic activity of various mutant forms of hSOD1 ranges from low to unchanged to high (7). Notably, loss of function of SOD1 in mice does not cause motor dysfunction, but expression of mutant SOD1 in mice or rats causes a late-onset progressive motor neuron disease that mimics the human disease (8-11). The precise nature of the toxic function gained by mutant SOD1 remains elusive, but studies of the pathology and modifiers of SOD-linked ALS in mice continue to yield important clues that will aid in the treatment of human fALS, and potentially, of sporadic ALS.
Although it is the motor neurons that are dying, mutant SOD1 protein is found in all cells. Early studies focused on addressing damage done to motor neurons by mutant SOD1 and how this related to cell-specific dysfunction and death. Directing mutant SOD1 expression to all (12) or most neurons (13), however, does not cause motor neuron disease in mice. Even up-regulating neuronal-specific mutant SOD1 expression in the background of a ubiquitous-expression mouse model has no effect on disease onset, severity, or progression. Conversely, directing mutant SOD1 expression to astrocytes does not induce motor neuron disease (14). Thus, attention has turned to elucidating the contribution of multiple cell types to the dysfunction and death of motor neurons in ALS. This has become particularly fundamental in light of recent work showing that decreasing the amount of mutant SOD1 in motor neurons affects disease onset (5). The importance of other cells is emphasized by studies reporting both support and damage by surrounding cells, including other neurons and microglia (5, 15).
Drosophila has proven to be instrumental in modeling various neurodegenerative diseases, including polyglutamine expansion diseases, α-synuclein-linked Parkinson disease, and tauopathies (16-18). Modeling dominant SOD-linked ALS in flies may provide a valuable tool for studying mechanisms of ALS and other motor neuron degenerative situations. Here we describe a model for SOD-linked fALS in Drosophila with expression of WT or human disease-linked mutants of hSOD1 directed to motor neurons. These studies uncover both cell-autonomous and cell-non-autonomous cellular responses and provide the foundation for new insight into mechanisms that contribute to loss of motor neuron integrity in ALS.
EXPERIMENTAL PROCEDURES
Drosophila Stocks and Transgenic Flies—The GAL4-UAS expression system was used to direct expression of transgenes to particular cell types. For motor neuron-specific expression, the D42 driver line was used (19, 20). For eye expression, the gmr-GAL4 driver line was used. The human SOD1 gene was amplified and cloned from transgenic flies bearing UAS-hSOD1 (HS1 flies, gift of Dr. Gabrielle Boulianne (21)). A missense mutation encoding the amino acid substitution K75R was corrected by site-directed mutagenesis to obtain a cDNA that matched the canonical hSOD1 open reading frame (CAG46542). Mutations corresponding to fALS A4V and G85R were introduced using site-directed mutagenesis (QuikChange II site-directed mutagenesis kit, Stratagene, La Jolla, CA). A cDNA encoding dSOD1 was obtained by amplification from larval cDNA, and the sequence was verified (Flybase ID FBgn0003462). WT and mutant SOD1 cDNAs were subcloned into the pUAST vector. Transgenic flies were made by germline transformation of w1118 embryos using standard procedures (WT, G85R, and A4V in-house, dSOD1 by Genetic Services Inc., Cambridge, MA). For G85R, four independent insertions were recombined to bring its expression level closer to that of WT and A4V. The hSOD1 and dSOD1 lines were generated in the same laboratory genetic background. Independent insertions of the hSOD1, A4V, and dSOD1 were tested with similar results. In some studies, a chromosome III insertion of UAS-eGFP (22) was used as a negative control transgene expressing an unrelated protein. Positive control flies bearing truncated spinocerebellar ataxia 3 with an expanded polyglutamine tract (UAS-SCA3-trQ78) are described (23).
Western Analysis—To determine transgene expression levels, adult progeny were homogenized in Laemmli buffer (Bio-Rad Laboratories) for protein analysis at 0-1, 28, or 49 days. Samples were separated by polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (12.5% Ready gels or Criterion gels, TransBlot membranes; Bio-Rad Laboratories) for Western analysis. Antibodies used included: NCL-SOD mouse monoclonal antibody to hSOD1 (1:500, Novocastra Laboratories Ltd., Newcastle upon Tyne, UK) for WT, G85R, and A4V; rabbit polyclonal hSOD1 antibody sc-11407 (FL154, 1:350), Santa Cruz Biotechnology, Santa Cruz, CA) for WT, A4V, and dSOD1; and mouse monoclonal antibody E7 ascites (1:1000, Developmental Studies Hybridoma Bank (DSHB), Iowa City, IA) to detect β-tubulin. Horseradish peroxidase-conjugated secondary antibodies (1:4000, Jackson ImmunoResearch Laboratories Inc., West Grove, PA) were used in combination with ECL chemiluminescence detection reagent (GE Healthcare, Buckinghamshire, UK) to visualize the immunoreactive bands. Non-saturated bands were quantified using ImageJ (National Institutes of Health) and expressed as a ratio to the internal reference β-tubulin.
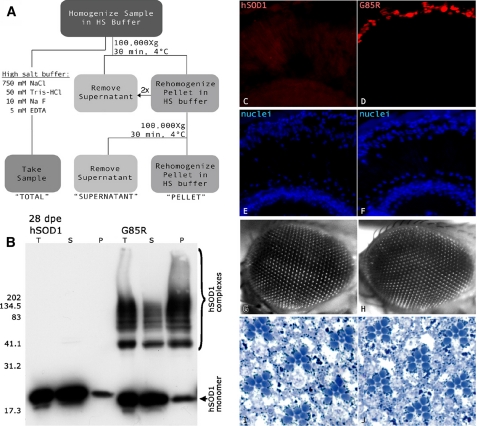
For protein fractionation studies, whole male flies at 0, 28, or 49 days were homogenized in 100 μl of cold high salt buffer (750 mm NaCl, 50 mm Tris, 10 mm NaF, 5 mm EDTA), a 25-μl sample was removed (see Fig. 4, “TOTAL”), and then the remaining sample was washed twice with an excess of high salt buffer at 100,000 × g for 30 min at 4 °C (Beckman OptimaMax, Beckman Coulter). The final pellet was homogenized in 100 μl of high salt buffer (see Fig. 4, “PELLET”). Each of the fractions was diluted 3:1 with Laemmli protein sample buffer and analyzed by Western immunoblot as above.
FIGURE 4.
G85R accumulates in high salt-resistant foci in retinal cells but does not cause degeneration. A, solubility assay for ionic fractionation of hSOD1 in homogenates of flies expressing WT or G85R under control of the retinal promoter gmr-GAL4. Proteins were separated in high salt buffer to break ionic bonds and then centrifuged at high speed to isolate soluble and insoluble species. B, total (T), supernatant (S), and pellet (P) fractions were separated by SDS-PAGE and probed for hSOD1. At 28 days, G85R but not wild type hSOD1 formed high molecular weight complexes that were found in both soluble and insoluble fractions. Both WT and G85R monomer were found in the pellet. dpe, days post eclosion of adult. C and D, cryosections of 28-day eyes from flies expressing WT or G85R expressed by the gmr-gal4 driver. Red fluorescence decorates hSOD1, and blue fluorescence labels nuclear DNA. G85R, but not WT, forms large foci recognized by hSOD1 antibody in the distal retina, just internal to the lens. E-J, nuclear arrays (E and F), external eye (G and H), and internal ommatidial structure (I and J) appear normal in eyes of 45-day flies expressing WT or G85R.
Motor Function—The ability of the flies to climb repeatedly was determined using a counter-current device placed vertically (24). Male flies (0-3 days) were separated into cohorts of 20-30 individuals that were tested together under ambient light conditions once a week over the next 28 days. During testing, flies were given 15 s to climb into a tube before being tapped down and moved to the next tube of the apparatus. After six total chances to climb upwards, the distribution of the flies in the tubes was noted, and the climbing index was determined as the proportion of times the flies climbed out of the six trials. In each of three experiments, 5-7 cohorts of each genotype (a minimum of 130 flies) were tested. Climbing indices were analyzed for differences due to transgene by age with analysis of variance using JMPin statistical software (SAS, Cary, NC) to determine that statistical significance was present (p < 0.0001). Then the Dunnett's post hoc analysis was used to compare each value with the control value at each time point; statistical significance was defined as p < 0.002.
Lifespan—A total of 200-400 male flies in cohorts of 20 individuals were observed in three independent experiments. Every 3rd day, the number of dead flies per cohort was recorded. Every 6th day, the living flies were transferred to new food vials. Lifespan data were analyzed using JMPin software (SAS). Differences were considered statistically significant if the log rank p > chi square was <0.0001.
Motor Neuron Counts—Flies expressing hSOD1 or the control protein together with nuclear GFP (stock 4775 (w1118; P{UAS-GFP.nls}14), Drosophila Stock Center, Bloomington, IN) were dissected at 0, 28, and 49 days. Thirty digital sections through paraformaldehyde-fixed thoracic ganglia were captured by confocal microscopy. GFP-positive nuclei in the region bordering T1 and T2 were counted in ImageJ (National Institutes of Health). Average cell counts, normalized to the average in control thoracic ganglia, were compared by t test, and significant difference was set at p < 0.05.
Electrophysiology—The methods of Tanouye and Wyman (25) and Martinez et al. (26) were followed to characterize the electrophysiological status of motor neurons expressing hSOD1. Briefly, flies were mounted on a glass slide with dental wax. Sharp glass microelectrodes (25 megaohms, filled with 3 m KCl) were used to record intracellularly from the respective indirect flight muscles (tergotrochanteral muscles (TTMs) and dorsal longitudinal muscles (DLMs)). The giant fiber neurons were stimulated with a sharp tungsten electrode placed inside the compound eye and in the cervical connective (1-4 V, 120-μs duration). To facilitate microelectrode access to the muscle, a small incision was made along the upper edge of the scutella. A reference electrode was inserted in the abdomen with a sharp tungsten electrode. Electrical stimulus was generated by a stimulus isolator commanded by the Master 8 stimulator (A.M.P.I., Jerusalem, Israel). The Axon Patch 2000 amplifier (Axon Instruments/Molecular Devices, Union City, CA) was set at the current clamp mode and used to detect and amplify electrical signals generated in the indirect flight muscles. The average age of the flies used for giant fiber physiology was 10 and 55 days old; for each genotype, however, 1-2 flies aged to 49 days or up to 60 days were also used. Within this age range, the following frequencies showed consistent results for each genotype.
Immunohistochemistry—At least 10 males of each genotype were dissected at 0-3, 28, or 49 days in each of at least three experiments. Flies expressing GFP were typically used as negative controls. Flies expressing SCA3-trQ78 were used in parallel to verify immunostaining of ubiquitin- and chaperone-positive inclusions in motor neurons. For analysis of hSOD1 accumulation, hSOD1 was visualized using rabbit polyclonal antibody SOD-100 (1:200, Nventa Biopharmaceuticals Corp., San Diego, CA), or the mouse monoclonal antibody NCL-SOD (1:50, Novocastra Laboratories Ltd.). Rabbit polyclonal antibody PW8765 (TBP7-27, 1:100) was used to visualize the proteasome-associated molecule Rpt3/TBP7/S6B (Biomol International, Plymouth Meeting, PA). Ubiquitin was identified using mouse monoclonal antibody NB300-130E (1:50, Novus Biologicals, Littleton, CO), and anti-Hsc/Hsp70 was labeled with SPA-822 (1:50, Nventa Biopharmaceuticals Corp.) or rat monoclonal anti-Hsp70 7F8 (1:50, gift of Dr. Susan Lindquist). Neuronal nuclei were visualized using rat monoclonal antibody 7E8A10 anti-Elav (1:75, Developmental Studies Hybridoma Bank), and glia were identified using the mouse monoclonal 8D12 anti-Repo (1:50, Developmental Studies Hybridoma Bank). Alexa Fluor-conjugated secondary antibodies were used in all immunofluorescence studies: chicken anti-mouse conjugated to Alexa Fluor 647, goat anti-mouse conjugated to Alexa Fluor 488, goat anti-rabbit conjugated to Alexa Fluor 488 or Alexa Fluor 555, and goat anti-rat conjugated to Alexa Fluor 555 (1:200, Molecular Probes, Eugene, OR). For analysis of hSOD1 accumulation, each stack of 30 confocal images representing an individual fly was categorized according to uniform standards (absent, mild, moderate, or severe). Each stack was also categorized according to intensity and breadth of the hsp70 signal by predetermined standards (absent, low, moderate, and strong). Chi square analysis within genotypes and within ages was performed using the JMPin statistical software (SAS) to identify differences due to age and UAS transgene (p < 0.001).
RESULTS
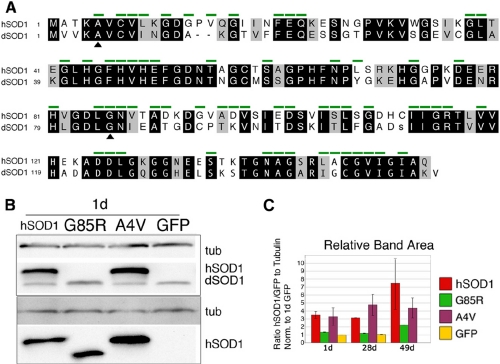
Expression of hSOD1 but Not dSOD1 in Motor Neurons Causes Progressive Motor Dysfunction—To develop a model for fALS, we generated flies bearing UAS transgenes of WT, A4V, and G85R mutant forms of hSOD1. A4V is a common mutation and is associated with fast disease progression (27). In mice, G85R causes rapid disease with low expression levels (28). We also used dSOD1 because although they are highly related, hSOD1 and dSOD1 differ at 49/153 residues (Fig. 1A). Expression was directed selectively to motor neurons using the D42 motor neuron driver, which is expressed from larval development through adult lifespan (20, 21). Western analysis confirmed expression of SOD1 in motor neurons over at least ∼50 days, allowing detailed examination of potential phenotypes over this extended time course (Fig. 1, B and C). The levels of A4V and WT hSOD1 were consistently higher than that of G85R. As observed in mouse tissue (28), G85R ran with faster mobility.
FIGURE 1.
Drosophila model of SOD-linked fALS employs motor neuron-specific expression of wild type hSOD1, A4V, and G85R mutant proteins. A, comparison of hSOD1 and dSOD1. Green bars identify residues that are mutated in SOD-linked fALS. Identities are in black, and similarities are in gray (BLOSUM 62 matrix). B, expression of transgenes in young flies with D42 motor neuron driver. Western blots were probed for an internal reference tubulin (tub, E7 ascites) and with antibodies that detect both hSOD1 and dSOD1 (top: FL154) or that detect only hSOD1 (bottom: NCL-SOD). C, relative expression of hSOD1 transgenes in young (1 day), middle-aged (28 days), and old (49 days) flies with D42 motor neuron driver, from Western blots also probed for tubulin (E7 ascites), GFP, and an hSOD1 antibody that cross-reacts to fly dSOD1 (NCL-SOD or FL154). Data were expressed as a relative ratio of immunoblot reactivity of the antibody staining of antibodies to hSOD1 (or antibodies to GFP) to antibodies to tubulin and normalized to the signal for 1-day GFP from the same Western blot.
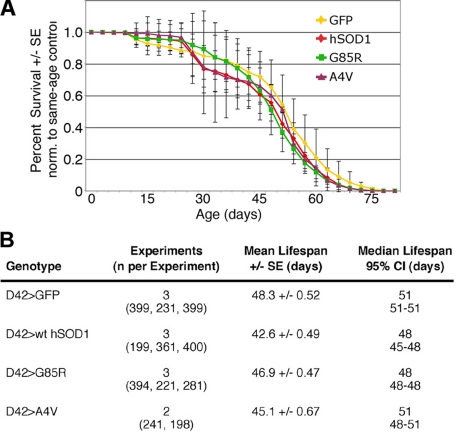
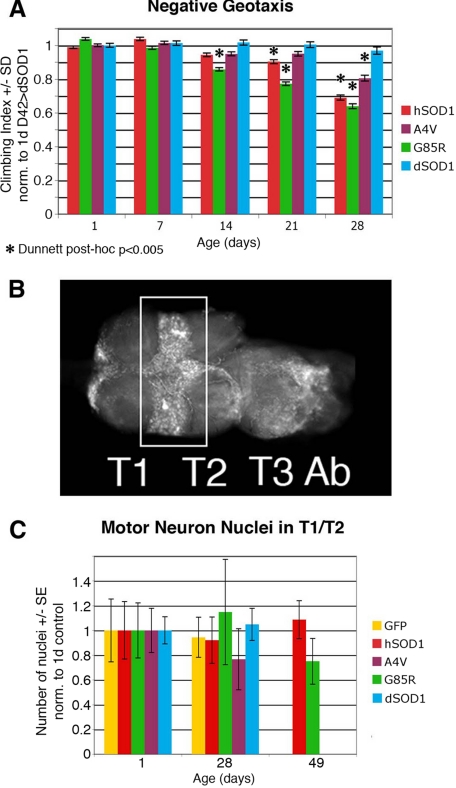
Gross observation revealed no paralysis or obvious lack of activity in flies expressing any form of hSOD1 or dSOD1 in motor neurons. Comparison of the survival of flies expressing hSOD1 to flies expressing GFP indicated that survivorship curves were similar (Fig. 2). We then examined motor function using a negative geotaxis climbing assay. When compared with flies expressing dSOD1, flies expressing hSOD1 or mutant forms in motor neurons showed typical strong climbing activity within the first week. However, flies expressing WT or mutant hSOD1 showed progressive loss of climbing when compared with dSOD1 controls, starting at 14 days (G85R) or 21 days (WT) (Fig. 3A). This finding indicates that hSOD1 may have an intrinsic toxicity to motor neurons in Drosophila. A surprising finding was that loss of climbing was not limited to mutant forms of hSOD1 but was also seen with the WT protein. Given that over 100 amino acid substitutions in hSOD1 are associated with fALS and that hSOD1 and dSOD1 differ at many residues, including some mutated in SOD-linked fALS (Fig. 1A), WT hSOD1 may be recognized as a toxic mutant form of SOD1 in Drosophila. This view was further reinforced by electrophysiological studies (see below).
FIGURE 2.
Lifespan of flies expressing WT hSOD1, A4V, or G85R hSOD1. A, survival curves, and B, details of lifespan analysis for each genotype, from multiple experimental points. The effect of hSOD1 on lifespan is known to differ between males and females and is dependent on genetic background (64). The hSOD1 transgenes used here were generated in a similar genetic background, and we do not see an extension of lifespan with hSOD1. CI, confidence interval.
FIGURE 3.
Motor neuron expression of hSOD1 induces a reduction in climbing activity without gross loss of motor neuron nuclei. A, climbing activity was compromised in flies expressing mutant or WT hSOD1 relative to flies expressing dSOD1 (blue bars). G85R showed a deficit from 14 days onwards (green bars), WT showed a deficit from 21 days (red bars), and A4V showed a deficit at 28 days (purple bars). Bars represent climbing indices for genotypes normalized to the 1-day climbing index of dSOD1 controls, ± S.E. from at least three experiments. B, the number of motor neurons was determined by counting nuclei in the T1/T2 border (rectangular selection) in confocal stacks of whole-mounted thoracic ganglia. Shown is a thoracic ganglion of genotype D42/UAS-GFP. Ab, abdominal ganglion. the number of labeled nuclei detected in the T1/T2 border was not different at any time point when compared with controls (gold bars) or between time points for flies expressing dSOD1, WT, A4V, or G85R hSOD1 (blue, red, purple, and green bars, respectively) in motor neurons (analysis of variance, p > 0.05). The cell number is normalized to 1-day controls within each experiment; average ± S.E. from at least two experiments (5-10 flies each) is presented.
No Apparent Loss of Motor Neurons—The motor dysfunction and lifespan effects on human ALS patients are due to dysfunction and loss of motor neurons. We therefore determined whether there was gross loss of motor neurons in the flies by counting nuclei in the T1/T2 region of the thoracic ganglia (Fig. 3B), an area we determined to have a large number of motor neuron somata using DiI labeling of leg muscles (data not shown). This analysis revealed no detectable loss of neuronal nuclei over time (Fig. 3C). Therefore, large scale motor neuron loss appeared not to occur; rather, climbing loss may reflect motor dysfunction.
Biochemical Oligomerization of hSOD1 Is Not Linked to Neuronal Loss or Dysfunction—Mutant hSOD1 in mice or humans often forms insoluble species or inclusions in cells (29, 30). High salt/high speed fractionation with SDS-PAGE was used to examine the solubility of hSOD1 in Drosophila motor neurons from 1-49 days. However, the solubility profile of WT and mutant hSOD1 in motor neurons did not differ from GFP or dSOD1 at any age tested (data not shown). Thus, a change in hSOD1 solubility in motor neurons did not accompany motor dysfunction. When hSOD1 was expressed in the eyes of adult flies, however, mutant G85R formed high molecular weight complexes, but WT hSOD1 did not (Fig. 4, A-D). No anatomical degeneration of the retina occurred with either form of the protein (Fig. 4, E-J). Considering their similar deleterious effects on climbing and giant fiber physiology (below), these results suggest that biochemical hSOD1 insolubility appeared dissociable from neuronal toxicity in flies.
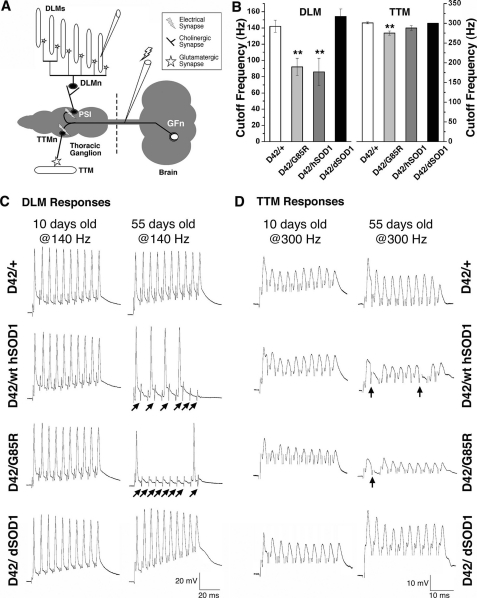
Synaptic Transmission along the Giant Fiber Motor Pathway Is Abnormal—Early signs of motor neuron disease in humans include muscle weakness and diminished motor nerve conduction. Analysis of the expression pattern of the D42 driver showed expression in motor neurons of the giant fiber system, a well defined neuronal circuit in Drosophila (25). Flies expressing hSOD1 and dSOD1 in motor neurons were therefore assessed for reduced or abnormal signaling at the neuromuscular junctions of the giant fiber system (Fig. 5A). Synaptic physiology of indirect flight muscles was compared between flies expressing WT and mutant hSOD1 to driver line controls (D42/+) and flies expressing dSOD1. The DLMs showed normal and robust responses at 10 days, with maximum following frequencies of at least 140 Hz in both experimental and control animals (Fig. 5, B and C). At 55 days, when control flies (D42/+ and D42/dSOD1) were still able to follow high frequency stimulation, flies expressing WT or G85R displayed repeated failures (Fig. 5C, arrows), with the average cut-off frequency dropping to 85-92 Hz (Fig. 5B). Recordings from the TTMs showed similar cut-off following frequencies between experimental and control flies at 10 days and a small but significant reduction in the following frequency for flies expressing G85R at 55 days (Fig. 5, B and D; p < 0.05), with a slight reduction in flies with WT hSOD1.
FIGURE 5.
hSOD1 induces age-dependent electrophysiological defects in the giant fiber neural circuit. A, schematic illustration of the giant fiber pathway responsible for jump-flight escape behavior. The giant fiber neuron (GFn) is located in the brain and descends to the thoracic ganglion, where it excites the motor neuron (TTMn) that innervates the TTM via an electrical synapse (marked with a lightning bolt). GFn also excites the peripherally synapsing interneuron (PSI) via an electrical synapse, which in turn excites five motor neurons (DLMn) innervating DLMs. Both DLM and TTM motor neurons synapse with their respective muscles via glutamatergic synapses. For illustrational purposes only, the DLMn is shown outside the thoracic ganglion. B, histograms of the average cutoff frequency in DLM (left panel) and TTM (right panel) in 55-day flies. DLM in control flies (D42/+ and dSOD1) was able to follow a train of 10 stimuli up to ∼140-Hz stimulation of the GFn, whereas DLM in flies expressing WT or G85R was only capable of following up to 80-90 Hz. Although TTM in the control flies followed up to 300 Hz, the ability of the TTM to follow high frequency stimulation was compromised in flies expressing WT. (**, p < 0.05) and slightly reduced in G85R. n ≥ 5 independent flies for each genotype. C, representative responses of DLM following 140-Hz stimulation of the GFn in control flies (top panels), flies expressing WT hSOD1 (second panels), flies expressing G85R (third panels), and those expressing dSOD1 (bottom panels). The muscle responded normally to each stimulus at 10 days but failed to follow each stimulus when aged (55 days) in the experimental flies. The arrows indicate failed responses. D, representative responses of TTM following 300-Hz stimulation of the GFn in control flies (top panels), flies expressing WT (second panels), flies expressing G85R (third panels), and those expressing dSOD1 (bottom panels). The muscle responded normally to each stimulus at 10 days, but experimental flies showed minor failures to a train of 10 stimuli at older time points (55 days). Arrows indicate failed responses. For each genotype and age group presented, n = 5.
These results showed that the giant fiber circuit is functional in older flies upon low frequency stimulation but defective with high frequency stimulation, revealing that synaptic transmission becomes progressively defective in flies expressing hSOD1. The DLM pathway was more sensitive than the TTM pathway to hSOD1 damage. DLM motor neurons mediate wing depression during the escape response, whereas the TTMs initiate leg extension of the mesothoracic leg extensor (25, 31). Although the giant fiber circuit does not directly govern climbing, these observations indicate that abnormalities in synaptic transmission or excitability may be common in motor systems of hSOD1-expressing flies.
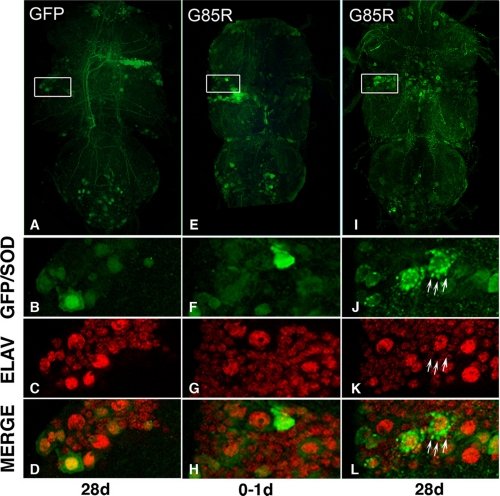
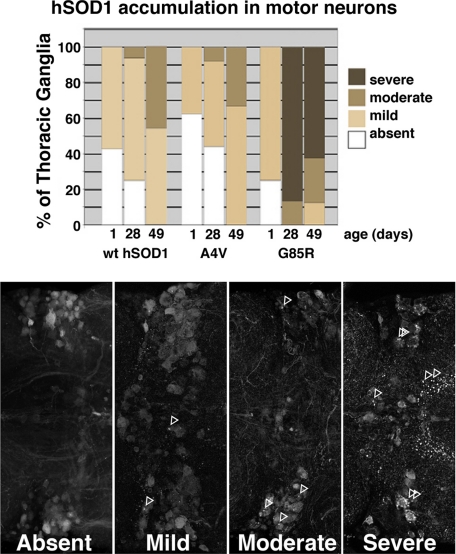
hSOD1 Progressively Accumulates in Motor Neuron Somata and Processes—Visible protein accumulations that can be ubiquitinated or associated with chaperones are hallmarks of many diseases including ALS, as well as hSOD1 inclusions in motor neurons (32-35). Given that hSOD1 conferred neuronal dysfunction, we examined motor neurons in the thoracic ganglia for evidence of pathology. Strikingly, both WT and mutant hSOD1 protein accumulated in foci within motor neurons. Foci were apparent as early as 1 day and increased with age both in number within individual cells and in total number of cells with foci (Figs. 6 and 7). At later time points, accumulations were visible not only within cell bodies but also within neuronal axons. Foci formation in young flies was similar for WT and mutant hSOD1 (1 day: absent to mild), and foci formation in old flies was similar for WT and A4V (28 and 49 days: absent to mild/moderate) and was much greater for G85R (28 and 49 days: moderate to strong) (Fig. 7). Foci were specific to hSOD1 as the control protein GFP did not form similar focal accumulations (data not shown). We also co-immunostained for ubiquitin, TBP7 (a proteasomal cofactor), and the chaperone Hsp70. None of these antibodies highlighted hSOD1-positive foci in motor neurons (data not shown). This indicates that these accumulations do not appear to represent misfolded protein recognized by ubiquitin-proteasomal or chaperone systems, although their appearance suggests a gradual overwhelming of the neuronal capacity to properly handle hSOD1 protein.
FIGURE 6.
hSOD1 accumulates in foci with age in motor neurons. The accumulation of hSOD1 into foci in the thoracic ganglion of animals expressing G85R with the D42 driver at young (0-1 days) and old (28 days) ages, when compared with animals expressing GFP only (left) is shown. Green, GFP or hSOD1 immunostaining; red, the neuronal nuclear marker Elav, in whole-mount thoracic ganglia. B-D, details of C showing homogenous GFP fluorescence in a 28-day fly. F-H, details of G showing homogenous immunoreactivity for G85R hSOD1 at 0-1 days. J-L, details of K showing striking focal accumulation of G85R at 28 days, with many foci in a single cell (arrows).
FIGURE 7.
Quantitative analysis of hSOD1 foci accumulation in motor neurons. Top, analysis of WT, A4V, and G85R hSOD1 accumulation with time. WT and mutant forms of hSOD1 accumulated into foci progressively with age (chi square p < 0.001). Bottom, whole-mount thoracic ganglia immunolabeled for hSOD1 to illustrate classification of focal protein accumulation. Arrows denote SOD-positive foci in motor neurons and neuropil. Only the T1-T2 border is shown here, but immunofluorescence in the entire thoracic ganglia was used to make the determination. Absent, SOD immunofluorescence was uniform and smooth. Mild, SOD immunofluorescence was mostly smooth and uniform, a few cells exhibited focal accumulations, and not more than one focus was observed per cell. Moderate, some smooth immunofluorescence was visible, and many cells contained at least one focal accumulation. Severe, the vast majority of visible immunofluorescence was present in foci, and most cells contained large numbers of accumulations.
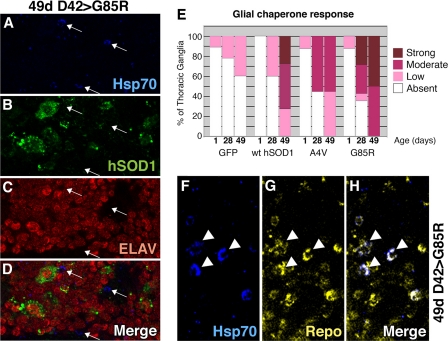
Expression of hSOD1 in Motor Neurons Produces a Stress Response in Glia—These studies, however, did uncover a chaperone response that was not present within the motor neurons themselves but rather in nearby cells. We noted that animals expressing hSOD1 showed an increase in immunostaining for Hsp70 that was not coincident with hSOD1 foci (Fig. 8, A-D). Although there was a minimal chaperone signal at 1 day, flies expressing hSOD1 showed increased immunoreactivity for Hsp70 at 28 and 49 days for mutant proteins and 49 days for the WT protein (Fig. 8E). The stress response was specific to SOD1 since expression of GFP had minimal effects, and up-regulation of Hsp70 was also seen upon expression of dSOD1 in motor neurons at 28 and 49 days (data not shown).
FIGURE 8.
Expression of SOD1 in motor neurons is associated with a stress response in glia. A-D, confocal images of a thoracic ganglion from a fly expressing G85R in motor neurons, stained for Hsc/Hsp70 (blue), hSOD1 (green), and Elav (neurons, red). Hsc/hsp70 immunoreactivity was often seen near, but not overlapping with, hSOD1 and Elav (arrows). E, WT hSOD1 induced mild to strong expression of hsc/hsp70 protein at 49 days, whereas both A4V and G85R induced immunostaining at 28 days, which was increased at 49 days. Differences when compared with control at each time point and differences due to age within genotype are statistically significant (p < 0.0001). F-H, the chaperone signal was in glia, not motor neurons. Hsp70 signal (blue) overlapped with the glial cell marker Repo (yellow). Arrowheads highlight examples of cells that immunostain strongly for both Hsc/Hsp70 and Repo.
The Hsp70 response in cells not visibly expressing the toxic protein was a striking feature of SOD1 expression in motor neurons. In contrast, neuronal expression of toxic polyglutamine protein induces dramatic up-regulation of Hsp70 in neurons (Fig. 9) (36, 37)). In the SOD situation, the Hsp70 immunostaining appeared to partially or completely fill small round cells. This suggested that it may be localized to glia, the other major cell type of the nervous system. We co-stained thoracic ganglia from flies expressing G85R with Hsp70 and the glial marker Repo (Fig. 8, F-H). These studies revealed that not all glia contained Hsp70, but all cells positive for Hsp70 were also positive for Repo, indicating that expression of SOD1 in motor neurons was inducing a chaperone response in glia.
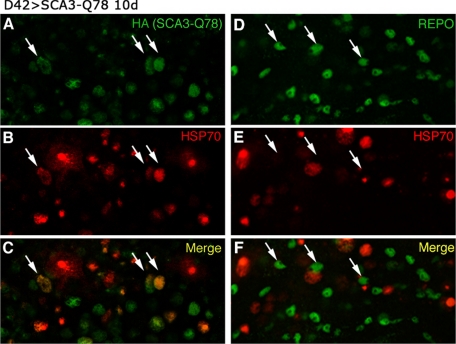
FIGURE 9.
Expression of Hsp70 in motor neurons is associated with a neuronal stress response upon polyglutamine protein expression. Shown are confocal images of thoracic ganglia from flies demonstrating expression of SCA3tr-Q78 in motor neurons, stained for Hsc/Hsp70 (red) and polyglutamine protein (A and C, green) or glia with Repo (D and F, green). Neuronal expression induces robust Hsc/Hsp70 immunoreativity in neurons (here and in Refs. 36 and 37), with a minimal response in Repo-positive cells. HA, hemagglutinin.
DISCUSSION
We present a model for SOD-linked fALS in Drosophila that displays motor dysfunction, a defining feature of the human disease. This effect in flies was accompanied by failure in high frequency synaptic transmission, focal accumulation of hSOD1 in motor neurons, and up-regulation of heat shock protein in glia. This work suggests that SOD can cause cell-autonomous damage to motor neurons, and highlight that expression of hSOD1 selectively in motor neurons induces a change in glia.
Drosophila Reveals an in Vivo Toxicity of hSOD1 to Motor Neurons—Our data indicate that a motor neuron-restricted expression pattern conferred behavioral compromise in climbing ability. This suggests that hSOD1 may have an intrinsic toxicity to motor neurons, which can be defined in the Drosophila system. Like typical mouse models of SOD1 toxicity (38), we used robust expression of hSOD1. Previous models in mice have demonstrated a dependence of toxicity on widespread tissue expression, specifically with the genes under control of the endogenous hSOD1 enhancer/promoter elements (8-10). Several studies with mice have reported no toxicity with neuron-restricted expression; Lino et al. (13) directed expression to motor neurons and sensory neurons using the Thy1 promoter, and Pramatarova et al. (12) directed expression to all neurons using the neurofilament light chain promoter. Moreover, Lino et al. (13) showed that the addition of Thy1-driven expression to hSOD1-driven expression had no enhancement effect, undermining a long held view that SOD1 toxicity is primarily cell-autonomous. This idea was expanded when Clement et al. (15) demonstrated in chimeric mice that motor neurons can display ALS-like pathology when they are not expressing the mutant protein themselves but rather are surrounded by other cell types that are expressing the mutant protein. Furthermore, they observed that motor neurons expressing mutant hSOD1 are devoid of pathology when in proximity to other cells not containing the mutant protein. Our model, on the other hand, provides an approach to define toxic properties of hSOD1 specifically in motor neurons that can lead to a motor deficit.
Although such studies in the mouse suggest that motor neurons may not be the primary site of damage in ALS models, reducing mutant hSOD1 expression in motor neurons delays disease onset (5). This finding indicates that SOD1 in motor neurons may indeed be playing a critical role. This is further substantiated in studies reporting deleterious effects in cultured motor neurons expressing mutant hSOD1 (39-42). Our findings of toxicity of hSOD1 in the fly support a role for cell-autonomous damage to motor neurons by hSOD1 as the deficits are seen with expression restricted to motor neurons. Within motor neurons, we see progressive accumulation of hSOD1, both in the somata surrounding the nucleus, as well as in neurites. These focal accumulations may both cause and result from hindrances in trafficking and axonal transport or insufficient protein degradation. It is known that disruption of anterograde and retrograde axonal movement of synaptic proteins and neurotrophic entities can negatively affect neuronal function. The p150glued mutation in dynactin-1, which severely disrupts axonal transport, causes a progressive, late onset motor phenotype in mice (43-46). Mice expressing mutant SOD1 also have compromised axonal transport (47-49). Our flies display electrophysiological defects reflective of impaired motor neuron function, indicating that the fly may provide a sensitive system for the detection of subtle motor neuron defects caused by hSOD1 and disease-linked forms. Despite a progressive motor phenotype, we did not detect a change in numbers of neuronal nuclei, excluding widespread loss of cells. The electrical features of the motor pathway also indicated that it could function fine at low activity levels, suggesting that synapses may be the primary site of dysfunction of SOD1 flies. Death of motor neurons and other aspects of the progression phase of the disease may be related to the effects of SOD1 in other cells, such as astrocytes or microglia. Alternatively, cell death may occur at a much later stage, possibly beyond the lifespan of flies, as it takes 8 months for motor neurons in the G85R transgenic mice to begin to die (28).
In our studies, WT hSOD1 imparted toxicity nearly on a par with either A4V or G85R mutant forms; WT hSOD1 even showed a tendency to accumulate in foci, a feature generally expected of a mutant but not normal hSOD1. Recent data, however, suggest that wild type as well as mutant forms of SOD1 can take on abnormal conformations that are disease-associated (50, 51). We hypothesize that WT hSOD1 may function as a conformational mutant protein in the context of Drosophila neurons for the following reasons. Toxicity can be conferred onto hSOD1 by any one of more than a hundred distinct amino acid substitutions, which implies an exquisite dependence upon conformation. This raises the possibility that any sequence other than the wild type Drosophila SOD1 conformation in the context of the SOD1 protein may appear abnormal to the fly. Although Drosophila SOD1 and hSOD1 are very similar in sequence, and hSOD1 can even functionally replace the Drosophila gene (21), the enzymes do differ in many amino acids, including locations where mutations occur that are associated with fALS. Importantly, overexpression of dSOD1 does not mimic the effects of hSOD1 expression in the fly. We note that this finding also fails to support the idea that SOD1 toxicity may be related to dismutase activity of the enzyme as both dSOD1 and hSOD1 would presumably result in the overabundance of hydrogen peroxide, yet there was selective toxicity of hSOD1.
Toxic protein-induced dysfunction in neurons is often associated with abnormal protein accumulation. This may serve as a protective measure undertaken by the cell to sequester a toxic entity, or it may signal disruption of the normal capacity of a cell for protein handling, degradation, folding, or trafficking. In ALS, inclusions consisting mainly of neurofilaments are regularly observed in affected tissue (52). SOD-linked fALS is known to be associated with accumulation of mutant SOD in motor neurons. Indeed, some mutant hSOD1 proteins form aggregated or cross-linked complexes in vitro (34, 53). In Drosophila, we observed accumulation of hSOD1 in motor neurons that increased with age. Initially, foci were rare and exclusively seen in cell bodies. At later ages, they increased in frequency and were observed both in somata and processes. This points to a gradual overwhelming of the neuronal capacity to properly process hSOD1, including flagging for degradation, trafficking to the proteasome, or degradation. We have not ruled out that the cell may be actively sequestering hSOD1, but since the foci in flies were not ubiquitinated, these foci may be early signs of a problem with handling hSOD1 protein.
A Glial Response to Motor Neurons Expressing hSOD1—Affected tissues in neurodegenerative diseases often exhibit the induction of a chaperone stress response. Heat shock protein induction has been noted in a mouse model with widespread mutant SOD1 expression (32), and chaperones have been found co-localized with SOD1 inclusions in motor neurons (32, 34, 35). The heat shock protein immunoreactivity we observed in fly thoracic ganglion did not overlap with hSOD1 staining. Rather, it was present exclusively in cells that are positive for the glial-specific marker protein Repo. Thus, in the fly model, the motor neurons contained the toxic protein, but the glia appeared to initiate a stress response.
It is unlikely that exogenous SOD1 induced a stress response due to SOD1 expression in glia themselves since the D42 motor neuron driver is specific, and we did not detect SOD1 in glia by immunofluorescence using a variety of primary antibodies, despite robust SOD1 levels. Leaky expression due to the genomic insertion sites of the transgenes could result in glial expression of the exogenous proteins, although analysis of flies lacking the GAL4 driver revealed no detectable hSOD1 protein. Furthermore, expanded polyglutamine protein in flies with the same motor neuron driver is only observed in neurons. Thus, this appears an interesting and distinct feature of SOD1 expression when compared with polyglutamine toxicity. The role of a stress response in disease is underscored by recent findings that endoplasmic reticulum stress may also contribute in mouse models (54). A current and important issue in the understanding of mechanisms of protein toxicity is the nature of the toxic species, as well as where and by what means the toxic protein acts to induce pathogenesis (55, 56). Current data favor smaller oligomers that are difficult to see or detect in vivo.
The glial chaperone up-regulation may be a reaction to the toxic protein or a signal secondary to effects of SOD1 in motor neurons. Motor neuron expression of dSOD1, but not of a pathogenic polyglutamine protein by the same driver, also resulted in a glial response, indicating that the response occurs with SOD1. In our studies, flies with greater chaperone induction showed more severe indicators of motor dysfunction. For example, G85R was associated with both the greatest chaperone up-regulation and the most severe phenotypes including climbing deficit, reduction in survival, and protein accumulation. Thus, the degree of stress response in glia may serve as a measure of neuronal dysfunction or a measure of the extent to which glia are attempting to combat problems in motor neurons. A neuron in distress may broadcast its dysfunction in various ways that are detectable to glia: for example, fibroblast growth factor release (57). Potentially, the glia are initiating a stress response to better provide support to neurons or to better cope with surroundings that have become less hospitable. Some studies have shown that Hsp70 can be released from glia and muscles (58) and affect neuronal viability (59). Thus, chaperone up-regulation in glia may be affecting motor neurons in a positive manner. It is also possible that in the absence of a glial stress response, the motor phenotypes would be even worse. Recent data suggest that up-regulation of Hsp70 is beneficial to disease progression in mouse models of ALS (60) and that astrocytes play a key role in causing toxicity to motor neurons (61).
The fly provides a rich system, complementary to mouse and others, for addressing human disease mechanisms. Molecular pathways of fundamental biology function appear sufficiently conserved to define the foundation for novel therapeutic approaches (reviewed in Refs. 16-18). Some of the most striking examples include the effects of chaperones, polymorphisms of which are risk factors for Parkinson disease (62), and the role of histone deacetylase inhibitors on polyglutamine toxicity, compounds that are now in clinical trials for Huntington disease (63). This fly model expressing human SOD1 displays some fundamental features of sporadic ALS and SOD-linked familial ALS disease. The SOD1 flies show progressive motor dysfunction, coupled with electrophysiological defects and abnormal accumulation of the protein. These effects may present early changes as no gross loss of motor neurons was detected. However, the effects on glia suggest that features of non-autonomous cellular interactions may be studied and defined in this system. These findings provide a foundation for further examination of hSOD1 damage to motor neurons and a genetic framework in which to approach neuron-glial interactions that may contribute to disease.
Supplementary Material
Acknowledgments
We thank G. Boulianne and S. Lindquist for sharing reagents.
This work was supported, in whole or in part, by National Institutes of Health Grant AG09215 from the NIA (to N. M. B.) and National Institutes of Health Grant ES014441 from the NIEHS (to B. Z.). This work was also supported by the Systems and Integrated Biology Training Grant and the Developmental Biology Training Grant (to M. R. W.) and a grant from the David and Lucile Packard Foundation (to N. M. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This article was selected as a Paper of the Week.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: ALS, amyotrophic lateral sclerosis; fALS, familial ALS; SOD1, copper, zinc-superoxide dismutase; hSOD1, human SOD1; dSOD1, Drosophila SOD1; WT, wild type; GFP, green fluorescent protein; TTM, tergotrochanteral muscle; DLM, dorsal longitudinal muscle; GFn, giant fiber neuron.
References
- 1.Boillee, S., Vande Velde, C., and Cleveland, D. W. (2006) Neuron 52 39-59 [DOI] [PubMed] [Google Scholar]
- 2.Valentine, J. S., Doucette, P. A., and Zittin Potter, S. (2005) Annu. Rev. Biochem. 74 563-593 [DOI] [PubMed] [Google Scholar]
- 3.Bradley, W. G. (1987) Muscle Nerve 10 490-502 [DOI] [PubMed] [Google Scholar]
- 4.Rosen, D. R., Siddique, T., Patterson, D., Figlewicz, D. A., Sapp, P., Hentati, A., Donaldson, D., Goto, J., O'Regan, J. P., Deng, H. X., Rahmani, S., Krizus, A., McKenna-Yasek, D., Cayabyab, A., Gaston, S. M., Berger, R., Tanzi, R. E., Halperin, J. J., Herzfeldt, B., Van Den Bergh, R., Hung, W.-Y., Deng, G., Mulder, D. W., Smyth, C., Laing, N. G., Soriano, E., Pericak-Vance, M. A., Haines, J., Rouleau, G. A., Gusella, J. S., Horvitz, H. R., and Brown, R. H., Jr. (1993) Nature 362 59-62 [DOI] [PubMed] [Google Scholar]
- 5.Boillee, S., Yamanaka, K., Lobsiger, C. S., Copeland, N. G., Jenkins, N. A., Kassiotis, G., Kollias, G., and Cleveland, D. W. (2006) Science 312 1389-1392 [DOI] [PubMed] [Google Scholar]
- 6.Pardo, C. A., Xu, Z., Borchelt, D. R., Price, D. L., Sisodia, S. S., and Cleveland, D. W. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 954-958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borchelt, D. R., Lee, M. K., Slunt, H. S., Guarnieri, M., Xu, Z. S., Wong, P. C., Brown, R. H., Jr., Price, D. L., Sisodia, S. S., and Cleveland, D. W. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 8292-8296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gurney, M. E., Pu, H., Chiu, A. Y., Dal Canto, M. C., Polchow, C. Y., Alexander, D. D., Caliendo, J., Hentati, A., Kwon, Y. W., Deng, H. X., Chen, W., Zhai, P., Sufit, R. L., and Siddique, T. (1994) Science 264 1772-1775 [DOI] [PubMed] [Google Scholar]
- 9.Ripps, M. E., Huntley, G. W., Hof, P. R., Morrison, J. H., and Gordon, J. W. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 689-693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong, P. C., Pardo, C. A., Borchelt, D. R., Lee, M. K., Copeland, N. G., Jenkins, N. A., Sisodia, S. S., Cleveland, D. W., and Price, D. L. (1995) Neuron 14 1105-1116 [DOI] [PubMed] [Google Scholar]
- 11.Nagai, M., Aoki, M., Miyoshi, I., Kato, M., Pasinelli, P., Kasai, N., Brown, R. H., Jr., and Itoyama, Y. (2001) J. Neurosci. 21 9246-9254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pramatarova, A., Laganiere, J., Roussel, J., Brisebois, K., and Rouleau, G. A. (2001) J. Neurosci. 21 3369-3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lino, M. M., Schneider, C., and Caroni, P. (2002) J. Neurosci. 22 4825-4832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong, Y. H., Parsadanian, A. S., Andreeva, A., Snider, W. D., and Elliott, J. L. (2000) J. Neurosci. 20 660-665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clement, A. M., Nguyen, M. D., Roberts, E. A., Garcia, M. L., Boillee, S., Rule, M., McMahon, A. P., Doucette, W., Siwek, D., Ferrante, R. J., Brown, R. H., Jr., Julien, J. P., Goldstein, L. S., and Cleveland, D. W. (2003) Science 302 113-117 [DOI] [PubMed] [Google Scholar]
- 16.Bilen, J., and Bonini, N. M. (2005) Annu. Rev. Genet. 39 153-171 [DOI] [PubMed] [Google Scholar]
- 17.Marsh, J. L., and Thompson, L. M. (2006) Neuron 52 169-178 [DOI] [PubMed] [Google Scholar]
- 18.Shulman, J. M., Shulman, L. M., Weiner, W. J., and Feany, M. B. (2003) Curr. Opin. Neurol. 16 443-449 [DOI] [PubMed] [Google Scholar]
- 19.Phillips, J. P., Tainer, J. A., Getzoff, E. D., Boulianne, G. L., Kirby, K., and Hilliker, A. J. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 8574-8578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeh, E., Gustafson, K., and Boulianne, G. L. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 7036-7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parkes, T. L., Elia, A. J., Dickinson, D., Hilliker, A. J., Phillips, J. P., and Boulianne, G. L. (1998) Nat. Genet. 19 171-174 [DOI] [PubMed] [Google Scholar]
- 22.Mollereau, B., Wernet, M. F., Beaufils, P., Killian, D., Pichaud, F., Kuhnlein, R., and Desplan, C. (2000) Mech. Dev. 93 151-160 [DOI] [PubMed] [Google Scholar]
- 23.Warrick, J. M., Paulson, H. L., Gray-Board, G. L., Bui, Q. T., Fischbeck, K. H., Pittman, R. N., and Bonini, N. M. (1998) Cell 93 939-949 [DOI] [PubMed] [Google Scholar]
- 24.Benzer, S. (1967) Proc. Natl. Acad. Sci. U. S. A. 58 1112-1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanouye, M. A., and Wyman, R. J. (1980) J. Neurophysiol. 44 405-421 [DOI] [PubMed] [Google Scholar]
- 26.Martinez, V. G., Javadi, C. S., Ngo, E., Ngo, L., Lagow, R. D., and Zhang, B. (2007) Dev. Neurobiol. 67 778-791 [DOI] [PubMed] [Google Scholar]
- 27.Rosen, D. R., Bowling, A. C., Patterson, D., Usdin, T. B., Sapp, P., Mezey, E., McKenna-Yasek, D., O'Regan, J., Rahmani, Z., Ferrante, R. J., Brownstein, M. J., Kowall, N. W., Beal, M. F., Horvitz, H. R., and Brown, R. H., Jr. (1994) Hum. Mol. Genet. 3 981-987 [DOI] [PubMed] [Google Scholar]
- 28.Bruijn, L. I., Becher, M. W., Lee, M. K., Anderson, K. L., Jenkins, N. A., Copeland, N. G., Sisodia, S. S., Rothstein, J. D., Borchelt, D. R., Price, D. L., and Cleveland, D. W. (1997) Neuron 18 327-338 [DOI] [PubMed] [Google Scholar]
- 29.Johnston, J. A., Dalton, M. J., Gurney, M. E., and Kopito, R. R. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 12571-12576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsumoto, G., Stojanovic, A., Holmberg, C. I., Kim, S., and Morimoto, R. I. (2005) J. Cell Biol. 171 75-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trimarchi, J. R., and Schneiderman, A. M. (1995) J. Comp. Physiol. 176 355-364 [DOI] [PubMed] [Google Scholar]
- 32.Liu, J., Shinobu, L. A., Ward, C. M., Young, D., and Cleveland, D. W. (2005) J. Neurochem. 93 875-882 [DOI] [PubMed] [Google Scholar]
- 33.Ross, C. A., and Poirier, M. A. (2004) Nat. Med. 10 (suppl.) S10-S17 [DOI] [PubMed] [Google Scholar]
- 34.Shinder, G. A., Lacourse, M. C., Minotti, S., and Durham, H. D. (2001) J. Biol. Chem. 276 12791-12796 [DOI] [PubMed] [Google Scholar]
- 35.Watanabe, M., Dykes-Hoberg, M., Culotta, V. C., Price, D. L., Wong, P. C., and Rothstein, J. D. (2001) Neurobiol. Dis. 8 933-941 [DOI] [PubMed] [Google Scholar]
- 36.Chan, H. Y., Warrick, J. M., Gray-Board, G. L., Paulson, H. L., and Bonini, N. M. (2000) Hum. Mol. Genet. 9 2811-2820 [DOI] [PubMed] [Google Scholar]
- 37.Warrick, J. M., Chan, H. Y., Gray-Board, G. L., Chai, Y., Paulson, H. L., and Bonini, N. M. (1999) Nat. Genet. 23 425-428 [DOI] [PubMed] [Google Scholar]
- 38.Turner, B. J., and Talbot, K. (2008) Prog. Neurobiol. 85 94-134 [DOI] [PubMed] [Google Scholar]
- 39.Bruening, W., Roy, J., Giasson, B., Figlewicz, D. A., Mushynski, W. E., and Durham, H. D. (1999) J. Neurochem. 72 693-699 [DOI] [PubMed] [Google Scholar]
- 40.Durham, H. D., Roy, J., Dong, L., and Figlewicz, D. A. (1997) J. Neuropathol. Exp. Neurol. 56 523-530 [DOI] [PubMed] [Google Scholar]
- 41.Pasinelli, P., Borchelt, D. R., Houseweart, M. K., Cleveland, D. W., and Brown, R. H., Jr. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 15763-15768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rabizadeh, S., Gralla, E. B., Borchelt, D. R., Gwinn, R., Valentine, J. S., Sisodia, S., Wong, P., Lee, M., Hahn, H., and Bredesen, D. E. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 3024-3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eaton, B. A., Fetter, R. D., and Davis, G. W. (2002) Neuron 34 729-741 [DOI] [PubMed] [Google Scholar]
- 44.LaMonte, B. H., Wallace, K. E., Holloway, B. A., Shelly, S. S., Ascano, J., Tokito, M., Van Winkle, T., Howland, D. S., and Holzbaur, E. L. (2002) Neuron 34 715-727 [DOI] [PubMed] [Google Scholar]
- 45.Levy, J. R., and Holzbaur, E. L. (2006) Int. J. Dev. Neurosci. 24 103-111 [DOI] [PubMed] [Google Scholar]
- 46.Levy, J. R., Sumner, C. J., Caviston, J. P., Tokito, M. K., Ranganathan, S., Ligon, L. A., Wallace, K. E., LaMonte, B. H., Harmison, G. G., Puls, I., Fischbeck, K. H., and Holzbaur, E. L. (2006) J. Cell Biol. 172 733-745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borchelt, D. R., Wong, P. C., Becher, M. W., Pardo, C. A., Lee, M. K., Xu, Z. S., Thinakaran, G., Jenkins, N. A., Copeland, N. G., Sisodia, S. S., Cleveland, D. W., Price, D. L., and Hoffman, P. N. (1998) Neurobiol. Dis. 5 27-35 [DOI] [PubMed] [Google Scholar]
- 48.Gonzalez de Aguilar, J. L., Gordon, J. W., Rene, F., Lutz-Bucher, B., Kienlen-Campard, P., and Loeffler, J. P. (1999) Eur. J. Neurosci. 11 4179-4187 [DOI] [PubMed] [Google Scholar]
- 49.Zhang, B., Tu, P., Abtahian, F., Trojanowski, J. Q., and Lee, V. M. (1997) J. Cell Biol. 139 1307-1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gruzman, A., Wood, W. L., Alpert, E., Prasad, M. D., Miller, R. G., Rothstein, J. D., Bowser, R., Hamilton, R., Wood, T. D., Cleveland, D. W., Lingappa, V. R., and Liu, J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 12524-12529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rakhit, R., Robertson, J., Vande Velde, C., Horne, P., Ruth, D. M., Griffin, J., Cleveland, D. W., Cashman, N. R., and Chakrabartty, A. (2007) Nat. Med. 13 754-759 [DOI] [PubMed] [Google Scholar]
- 52.Strong, M. J., Kesavapany, S., and Pant, H. C. (2005) J. Neuropathol. Exp. Neurol. 64 649-664 [DOI] [PubMed] [Google Scholar]
- 53.Stathopulos, P. B., Rumfeldt, J. A., Scholz, G. A., Irani, R. A., Frey, H. E., Hallewell, R. A., Lepock, J. R., and Meiering, E. M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 7021-7026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nishitoh, H., Kadowaki, H., Nagai, A., Maruyama, T., Yokota, T., Fukutomi, H., Noguchi, T., Matsuzawa, A., Takeda, K., and Ichijo, H. (2008) Genes Dev. 22 1451-1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haass, C., and Selkoe, D. J. (2007) Nat. Rev. Mol. Cell. Biol. 8 101-112 [DOI] [PubMed] [Google Scholar]
- 56.Muchowski, P. J., and Wacker, J. L. (2005) Nat. Rev. Neurosci. 6 11-22 [DOI] [PubMed] [Google Scholar]
- 57.Cassina, P., Pehar, M., Vargas, M. R., Castellanos, R., Barbeito, A. G., Estevez, A. G., Thompson, J. A., Beckman, J. S., and Barbeito, L. (2005) J. Neurochem. 93 38-46 [DOI] [PubMed] [Google Scholar]
- 58.Guzhova, I., Kislyakova, K., Moskaliova, O., Fridlanskaya, I., Tytell, M., Cheetham, M., and Margulis, B. (2001) Brain Res. 914 66-73 [DOI] [PubMed] [Google Scholar]
- 59.Robinson, M. B., Tidwell, J. L., Gould, T., Taylor, A. R., Newbern, J. M., Graves, J., Tytell, M., and Milligan, C. E. (2005) J. Neurosci. 25 9735-9745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gifondorwa, D. J., Robinson, M. B., Hayes, C. D., Taylor, A. R., Prevette, D. M., Oppenheim, R. W., Caress, J., and Milligan, C. E. (2007) J. Neurosci. 27 13173-13180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagai, M., Re, D. B., Nagata, T., Chalazonitis, A., Jessell, T. M., Wichterle, H., and Przedborski, S. (2007) Nat. Neurosci. 10 615-622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu, Y. R., Wang, C. K., Chen, C. M., Hsu, Y., Lin, S. J., Lin, Y. Y., Fung, H. C., Chang, K. H., and Lee-Chen, G. J. (2004) Hum. Genet. 114 236-241 [DOI] [PubMed] [Google Scholar]
- 63.Marx, J. (2005) Science 310 43-45 [DOI] [PubMed] [Google Scholar]
- 64.Spencer, C. C., Howell, C. E., Wright, A. R., and Promislow, D. E. (2003) Aging Cell 2 123-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.