Abstract
Predicting how species distributions might shift as global climate changes is fundamental to the successful adaptation of conservation policy. An increasing number of studies have responded to this challenge by using climate envelopes, modeling the association between climate variables and species distributions. However, it is difficult to quantify how well species actually match climate. Here, we use null models to show that species–climate associations found by climate envelope methods are no better than chance for 68 of 100 European bird species. In line with predictions, we demonstrate that the species with distribution limits determined by climate have more northerly ranges. We conclude that scientific studies and climate change adaptation policies based on the indiscriminate use of climate envelope methods irrespective of species sensitivity to climate may be misleading and in need of revision.
Keywords: bioclimatic niche, global change, null models, ornithology, species distribution
As global climates warm, some species distributions are moving upward and poleward (1, 2). Predicting how individual species respond to climate change allows assessment of extinction risk and spatial planning of conservation activity (3, 4). Climate envelopes (or the climatic niche concept) are the current methods of choice for prediction of species distributions under climate change and their use is growing rapidly in many areas of ecology (5–7). However, although climate envelope methods and assumptions have been criticized as ecologically and statistically naïve (8, 9), there exists no quantitative evaluation of the importance of these criticisms.
Because it is axiomatic that climate influences species distributions (8), the climate envelope approach of matching distributions to climate is intrinsically appealing. However, the use of such simplistic models is risky on both biological and statistical grounds: there are many reasons why species distributions may not match climate, including biotic interactions (10), adaptive evolution (11), dispersal limitation (12), and historical chance (13). Although debate began before the current explosion in climate envelope studies (8, 9), there remains no quantitative information that would allow assessment of how well, or even if, species distributions match climate. Here, we quantify the match of species distributions to environment by generating synthetic species distributions that retain the spatial structure in the observed distributions but are randomly placed with respect to climate.
Ideally, the predictions of climate envelope models would be verified on an entirely independent dataset (14, 15) and some attempts have been made at this, both by prediction of the potential distribution of introduced species in new continents (16) or through backward prediction (hindcasting) of prehistoric distributions reconstructed from the fossil record (17). Unfortunately, truly independent data are generally unavailable, so the usefulness of a climate envelope model is typically measured by how well it fits a subset of the current species distribution reserved for evaluation (8). This match is usually assessed by using the area under the receiver operating curve (AUC) (18) or other goodness-of-fit measures such as Cohen's Kappa (19). Species with current distributions that are not well modeled by climate envelopes will have low goodness-of-fit scores and are considered less likely to be limited by climate. Because reported goodness-of-fit scores are often high, it is widely accepted that climate does determine many species distributions, suggesting climate envelope-based predictions of future distribution should be robust (4, 8, 19). Unfortunately, the most popular goodness-of-fit statistic (AUC) can be misleadingly high (18), but there has been no attempt to quantify how often high goodness-of-fit scores, and hence ostensibly good matches between distribution and climate, can occur by chance alone. Consequently, the degree to which species really are constrained by climate remains unresolved. Probably the main reasons why this has not been investigated to date are the conceptual and technical challenges presented by formulating null models for spatially autocorrelated patterns (20).
The development of appropriately constrained null models offers an intuitive method for assessing the scale of this problem: if climate envelopes fit real species distributions no better than they do null model distributions, we should conclude that climate envelopes are misleading (21). Alternatively, if real climate envelopes are significant improvements on the null models, we should retain confidence in their predictions. What, then, is an appropriate null model? An appropriate null model is a pattern that retains everything in the real pattern, but excludes only the factor of interest (21): in this case, climate. The null distribution implicit in the nonspatial statistical methods used in current climate envelope methodologies is that of complete spatial randomness: the match between climate and a random scatter of presence/absence across the survey area. Such a null model is clearly inappropriate beccause all species distributions show autocorrelation, potentially attributable to both intrinsic factors such as dispersal and extrinsic factors such as climate, land use, and other anthropogenic activities (20). Indeed, even neutral models predict that species distributions will be autocorrelated (22).
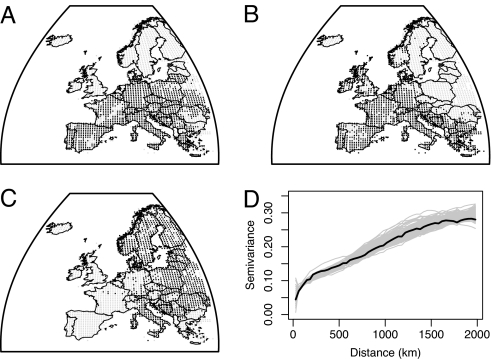
A better null distribution would be generated by simply cutting and pasting the true distribution onto a different section of the map with the new location and orientation selected at random. This would clearly be identical in autocorrelation structure to the true distribution, but any causal relationships that limited the original distribution would be broken. In practice, actual geography of the shape of continents makes this cutting and pasting impossible without imposing unreasonable restrictions on the new location: parts of the new distribution may otherwise fall into the sea. Therefore, instead of either a random scatter of presences or cutting and pasting a real distribution, the simplest appropriate null model involves the simulation of distribution patterns that have the same prevalence and the same spatial structure as actual distributions, but that have no deterministic relationship with the covariates of interest (23), in this case, climatic variables (Fig. 1).
Fig. 1.
Output from the null distribution algorithm. (A) Real distribution (Serinus serinus) with presence indicated in black, absence in gray. (B and C) Two realizations of the null distribution. (D) Semivariograms of the real distribution (black) and 99 simulations (thin gray): note that the real distribution falls entirely within the null distributions.
Results
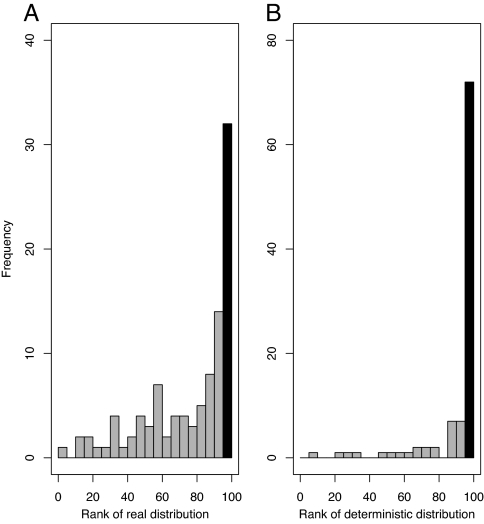
Using data on the European distribution of 100 bird species (24), we generated 99 synthetic distribution patterns for each species. For each of the 100 species, we fitted climate envelope models to both the true distribution and the 99 simulated distributions by using standard climate variables (1, 14, 19). AUC scores for the observed species distributions were similar to those in other published studies (range, 0.71–0.994; median, 0.869; supporting information (SI) Table S1) (19, 25–27), but these were ranked in the top five of the 99 simulations for only 32 species (Fig. 2A, species with significant patterns are identified in Table S1). These findings are not substantially affected by choice of significance level, preferred goodness-of-fit statistic, or choice of climate variables (number of significant patterns: range, 18–52; median, 33; Table S2).
Fig. 2.
Histograms of ranked climate envelope AUC scores of 100 distribution patterns among 99 null models. (A) 100 real species distributions. (B) 100 semideterministic patterns used in the power analysis. Black bar indicates the number of species for which the AUC score for the distribution of interest was in the top 5% of randomizations: 32 species for real species, 72 for the power analysis.
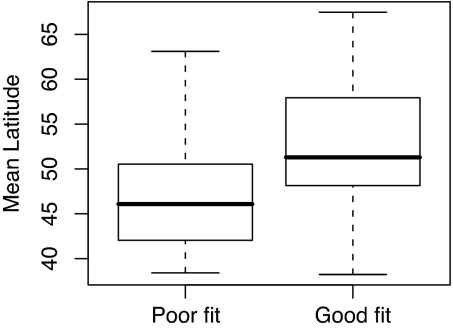
Additionally, because climate is widely believed to have stronger influence in more extreme environments (28, 29), we predicted that these 32 species would have a more northerly distribution than those that were not fitted better than null models: an effect that is clear in our data even after controlling for potentially confounding effects of the proportion of the global distribution contained within the study area (Fig. 3; F1,96 = 10.8, P = 0.001).
Fig. 3.
Boxplot of the mean latitudes of the ranges of species that were poorly or well fit by climate envelopes. The median is indicated by the black line and first and interquartile range by the box. Whiskers cover the full range of the data.
These results are potentially of great concern, but before concluding that >60% of our climate envelopes are misleading, it is important to assess the power of our null model methods to identify strongly deterministic patterns. It could be that failure to detect matches between species distributions and climate is due to insufficient information and poor statistical modeling rather than to a true absence of association. Consequently, we generated 100 simulated distributions that had the same prevalence as the 100 real species but a perfect climatic match. Using these patterns, we repeated our null model procedure, generating 99 simulations with similar spatial structure and prevalence for each pattern and fitting climate envelopes to all deterministic and simulated distributions. We found that 99 of these 100 simulated species perfectly matching climate had AUC scores ranked within the top five of the null models, demonstrating that our method correctly identifies the pattern showing a deterministic climatic signal in a sample of null European distributions. Next we generated 100 simulated species distributions with both a deterministic component and a component of added noise (the noise component comprised between 80% and 20% of the distribution) and repeated the process. Here, AUC scores for the deterministic patterns were ranked in the top five of the 99 distributions with added noise for 72 species (Fig. 2B). Full details of the power analysis are available as SI Text, but our method correctly identified patterns that were at least 50% deterministic signal in 96% of cases (Fig. S1).
Discussion
Our results are a quantitative assessment of the degree to which widely used climate envelope approaches are fit for purpose. That we use the same climate and species data that other high-profile studies have used (3, 4, 26) and find that most climate models are no better than chance associations is of considerable concern. Because our power analysis suggests that our method is highly likely to identify distribution patterns that are strongly determined by climate, we are confident that the distributions of most birds in our study are not strongly associated with the climate variables currently available.
It may be argued that assessing the statistical significance of climate envelope models is inappropriate: in their simplest form climate envelopes are widely used in ecology simply as a descriptive characterization of niche space. However, once used to make predictions, it is important that the models are falsifiable, because predictions from climate envelope models can influence decisions (7). For example, published predictions for the Scottish endemic Loxia scotica suggest that the species may loose all suitable climate space and would need to move to Iceland to escape global extinction (19). In the face of such predictions, and given scarce conservation resources, it might be rational to accept the inevitable extinction of this species. However, our results suggest that although the goodness-of-fit for our model of this species' distribution is extremely high (e.g., AUC = 0.988), this model is no better than a chance association: it is certainly not a model that should inform policy.
Does this mean that the orthodox view that macroscale bird distributions are driven by climate is wrong? There is growing evidence that a more cautious approach to predicting climate impacts is required, inspired by observations that biotic interactions can overwhelm the direct impacts of climate (30) and that climate change occurs against a complex background of land-use change and habitat fragmentation that must also be considered (31). Our observation that northern species are more likely to have distributions that are significantly determined by climate could therefore be due to the expected greater influence of abiotic factors in northern climes, but might also reflect the greater human footprint on habitats and distributions in more southern regions (32). Direct persecution of raptor species such as the red kite Milvus milvus have wiped it out across much of its native range in western Europe, whereas the distribution of agricultural specialists such as cirl bunting Emberiza cirlus must similarly have been quite different before the advent of modern agriculture (24). Other historical factors may similarly affect current distribution of European birds. Despite this inherent uncertainty, it seems likely that there are genuine biological causes of the differences between species in the degree to which their distribution is associated with climate: it has been suggested that wetland species are less well predicted by climate envelopes than terrestrial species (19), and migrant species are less likely to be affected by winter conditions than residents (33). Our data, however, do not show patterns of predictability across either wetland or terrestrial habitat preferences, or migrant status (Table S1).
By applying our methods to other taxa in different regions where larger latitudinal ranges are available it may be possible to further assess whether climatic limits are genuinely weaker than has been thought, relative to biotic interactions. Alternatively, climate effects may be hidden behind significant anthropogenic impact. Perhaps also the degree to which species distributions are subject to climatic limitation is not a global property of the distribution but varies spatially; poleward distribution limits have been suggested to be more sensitive to climate than more equatorial limits (34). It would be relatively straightforward to adapt our methods to test this hypothesis.
There are a number of possible artefactual reasons that may also explain why bioclimate envelopes are unable to detect significant climate associations. It is possible, for example, that our results are simply a consequence of the data quality currently available: climate data are based on interpolations of weather stations potentially far from the location of interest, whereas organism data may be subject to observer bias. For example, do the large holes in the distribution of Serinus serinus in eastern France and southeastern Spain (Fig. 1) represent genuine gaps that are determined by unsuitable local environmental conditions not shown by current climate surfaces, or are they simply areas with poor observer coverage? It is difficult to answer these questions at present. Furthermore, use of a 50 × 50 km grid replicates most equivalent analyses but is a blunt tool in regions such as the Alps and the Pyrenees where average climate on this scale is a poor representation of the conditions experienced within most of the square (nearly 10% of cells average over significant altitude variability). It will be interesting to see whether finer-scale analyses can improve on our current results, but, whether or not the distributions of birds are truly climate-driven, we found that by using the best available datasets and one of the best known taxonomic groups we are currently unable to build useful distribution models for many species.
Although we fitted climate envelopes that are similar in goodness-of-fit to those published in the literature, this degree of model accuracy was also possible for many null distributions that had no relationship with climate but maintained the prevalence and spatial autocorrelation of real species distributions. Because birds are perceived to be equally strongly associated with climate as other species groups and trophic levels (26), our results cast doubt on the predictions of climate envelope models for all taxa, although it seems likely that further research may reveal differences between ectotherms and endoterms (35). We therefore conclude that many, if not most, published climate envelopes may be no better than expected from chance associations alone, questioning the implications of many published studies. We recommend that future work using bioclimate envelopes take into account the likelihood that many species distributions may match climate by chance alone.
Materials and Methods
Bird Data.
Species data consisted of presence (“probable” and “confirmed” breeding records)/absence within 50 × 50 km squares for 100 native bird species taken from the European Breeding Bird Atlas west of 30°E and excluding Svalbard and the Azores (24). We chose the 100 species used in this analysis based on three criteria: first, all European endemics were included. Second, we included all species with >60% of their world distribution falling in the study area. Third, remaining species were selected at random (a list of species is provided in Table S1, together with summaries of their distribution and analysis results). Reason for inclusion was recorded as a measure of endemism to be used in later analyses.
Climate Data.
We used three climate parameters popular in avian climate envelope studies (1, 14, 19): annual growing degree days >5°C, mean temperature of the coldest month, and soil water availability. Additionally, because growing degree days and mean temperature of the coldest month are highly correlated, we used the coefficient of variation in mean monthly temperature (K), a measure of continentality. It is unlikely that all of these variables have direct effects on all bird species, but they are perceived to have strong indirect effects on birds and other taxa through effects on food availability or habitat type (1, 19). Alternative climate variables are usually strongly correlated with one or more of these variables and we found no substantial differences in our results when using two alternative combinations of climate variables also sometimes used in avian studies: (i) mean temperature of the hottest month, mean temperature of the coldest month, mean number of frosty days, and the ratio of actual to potential annual evapotranspiration, and (ii) growing degree days, mean temperature of the coldest month, seasonal variation in rainfall (coefficient of variation in monthly mean rainfall), and mean monthly rain (see section on congruence in SI Text where these datasets are referred to as climate datasets 2 and 3, respectively). Mean monthly climate variables from 1961 to 1990 were available at 0.5° resolution for the whole of the study area [dataset CRU CL 1.0 (36)] and were projected onto the 50-km bird dataset by using ordinary Kriging assuming an exponential spatial structure. Soil parameters were available globally at 1° resolution [dataset WISE.AWC (37)], were interpolated onto the 50-km squares and combined with climate variables (38).
Null Model Algorithm.
Null models of species distributions were built from real species distributions to preserve both prevalence and autocorrelation structure building on a published method (23). This method uses “clumping statistics” that measure of the conditional probability of presence in one square, given presence/absence in neighboring squares. Starting with a random scatter of the required prevalence, the algorithm repeatedly swaps pairs of squares (one with presence, the other with absence) and compares the clumping statistics of the simulation with that of the real pattern, gradually clumping squares until there is sufficient match. Sufficiency is assessed by using a semivariogram. In documented code (SI Appendix) we extend this method for larger areas and irregular data. In brief, we also use conditional probability to assess whether changes improve the match between the simulation and real datasets, but extend it from squares that are simply first neighbors to squares in 10 separate distance classes. Our algorithm continues until either convergence is reached (23), or 10,000 iterations have completed. Completing 10,000 iterations requires >1 h computer time (Dell Precision PWS690, 2.66GHz, 3GB RAM) limiting the number of null models to 99 for each species. Most null model exercises use 999 or even 9,999 simulations to generate more accurate P values but using fewer does not generate bias.
This method will result in a middomain effect whereby squares in the center of Europe will more frequently form part of the null distribution than squares around the edge of the study area (39). Theoretically, this could lead to the null distributions of species with prevalence close to 0.5 more regularly overlapping with the true distribution, perhaps making differences between null and real distributions harder to detect. This, however, is not the case in our data (detailed in the power analysis below). Although our method preserves prevalence and spatial autocorrelation, it does not explicitly preserve the extent of occurrence for null distributions. In practice we found that the area of the smallest convex polygon enclosing the entire real distribution fell within the 95% range of the null distributions for all but 14 real distributions. Because these 14 species showed no difference from the remaining species in the frequency with which the models of their distribution were better than the null distributions (χ2 = 0.24, df = 1, P = 0.63), any impact of not explicitly preserving the extent of occurrence is insignificant.
Climate Envelope Methods.
All analyses were undertaken by using R v 2.6.0 (40) (see SI Appendix for code). Climate envelope methods follow BIOMOD (41) fitting generalized additive models (GAMs), neural networks (ANNs) and generalized linear models (GLMs) to each dataset. For the real and 99 simulated distributions of each species, we fitted climate envelopes to a random selection of 70% of data and assessed the match between predicted and actual distribution in the remaining 30% of data by using AUC and Kappa (κ) scores (41). For each species, we used the rank AUC and κ values for the real distribution among the 99 simulations to assess whether the real distribution was better fitted than the simulated distributions. Because all three modeling methods and all four goodness-of-fit measures were congruent (SI Text), we focus here on ANN models and AUC scores, methods that are regularly preferred in comparative studies (26, 42).
We used logistic regression to determine the effect of the mean latitude of a species' distribution on whether or not the real species distribution was better fitted than the simulated distributions. To control for possibly confounding effects, we included a factor identifying the reason a species was included in the sample (a measure of endemism) as a nuisance variable, although this term had no effect on the results.
Power Analysis.
For each real species distribution we generated two patterns with the same prevalence as the real species, one completely determined by the climate variables, another containing an element of noise. (Full details of these methods and code for generating these patterns is provided as SI Text.) To generate deterministic patterns we randomly selected distribution limits along all climate axes and widened or narrowed the climatic limits until the required number of presences was achieved. We generated noisy patterns starting with 100 patterns wholly determined by climate as before. Next, we allowed the “species” to disperse into all squares neighboring a square with presence (generating presence in squares that are climatically unfavorable). We then eroded these larger patterns, sequentially removing presences until returning to initial prevalence. We selected presences for removal by comparing the spatial structure of the pattern with that of the real species by using conditional probabilities, generating gaps in the distributions that approximated patterns shown by real species. This is equivalent to a species with habitat requirements beyond that of climate alone and ensured that these distributions had gaps in climatically favorable areas. For each of these patterns we recorded the proportion of the original deterministic pattern that remained in the final pattern as an index of the signal-to-noise ratio. In real species distributions we have little understanding of the true signal-to-noise ratio, although for reliable prediction of future distribution from climate envelopes to be possible the signal must be strong relative to the noise (19). We used each pattern as if they were real species distributions, building 99 simulations with similar spatial structure and fitting climate envelopes to all as above.
Supplementary Material
Acknowledgments.
We thank the European Bird Census Council for access to European breeding bird distributions and R. Pakeman, J. Perez-Barberia, P. Goddard, M. Brewer, and the anonymous reviewers for their comments on the manuscript. This project was funded by the Rural and Environment Research and Analysis Directorate of the Scottish Government.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803506105/DCSupplemental.
References
- 1.Araujo MB, Pearson RG, Thuiller W, Erhard M. Validation of species-climate impact models under climate change. Glob Chang Biol. 2005;11:1504–1513. [Google Scholar]
- 2.Parmesan C, Yohe G. A globally coherent fingerprint of climate change impacts across natural systems. Nature. 2003;421:37–42. doi: 10.1038/nature01286. [DOI] [PubMed] [Google Scholar]
- 3.Huntley B, Green RE, Collingham YC, Willis SG. A Climatic Atlas of European Breeding Birds. Barcelona: Lynx Editions; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas CD, et al. Extinction risk from climate change. Nature. 2004;427:145–148. doi: 10.1038/nature02121. [DOI] [PubMed] [Google Scholar]
- 5.Nogues-Bravo D, Rodriguez J, Hortal J, Batra P, Araujo MB. Climate change, humans, and the extinction of the Woolly Mammoth. PLoS Biol. 2008;6:e79. doi: 10.1371/journal.pbio.0060079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heikkinen RK, et al. Methods and uncertainties in bioclimatic envelope modelling under climate change. Prog Phys Geogr. 2006;30:751–777. [Google Scholar]
- 7.Thuiller W, Lavorel S, Araujo MB, Sykes MT, Prentice IC. Climate change threats to plant diversity in Europe. Proc Natl Acad Sci USA. 2005;102:8245–8250. doi: 10.1073/pnas.0409902102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearson RG, Dawson TP. Predicting the impacts of climate change on the distribution of species: Are bioclimate envelope models useful? Glob Ecol Biogeogr. 2003;12:361–371. [Google Scholar]
- 9.Hampe A. Bioclimate envelope models: what they detect and what they hide. Glob Ecol Biogeogr. 2004;13:469–471. [Google Scholar]
- 10.Davis AJ, Jenkinson LS, Lawton JH, Shorrocks B, Wood S. Making mistakes when predicting shifts in species range in response to global warming. Nature. 1998;391:783–786. doi: 10.1038/35842. [DOI] [PubMed] [Google Scholar]
- 11.Thomas CD, et al. Ecological and evolutionary processes at expanding range margins. Nature. 2001;411:577–581. doi: 10.1038/35079066. [DOI] [PubMed] [Google Scholar]
- 12.Svenning JC, Skov F. Could the tree diversity pattern in Europe be generated by postglacial dispersal limitation? Ecol Lett. 2007;10:453–460. doi: 10.1111/j.1461-0248.2007.01038.x. [DOI] [PubMed] [Google Scholar]
- 13.Cotgreave P, Harvey PH. Associations among biogeography, phylogeny and bird species diversity. Biodivers Lett. 1994;2:46–55. [Google Scholar]
- 14.Elith J, et al. Novel methods improve prediction of species' distributions from occurrence data. Ecography. 2006;29:129–151. [Google Scholar]
- 15.Araujo MB, Rahbek C. How does climate change affect biodiversity? Science. 2006;313:1396–1397. doi: 10.1126/science.1131758. [DOI] [PubMed] [Google Scholar]
- 16.Roura-Pascual R, et al. Geographic potential of Argentine ants (Linepithema humile Mayr) in the face of global climate change. Proc R Soc Lond B. 2004;271:2527–2534. doi: 10.1098/rspb.2004.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Meyer E, Peterson AT, Hargrove WW. Ecological niches as stable distributional constraints on mammal species, with implications for Pleistocene extinctions and climate change projections for biodiversity. Glob Ecol Biogeogr. 2004;13:305–314. [Google Scholar]
- 18.Lobo JM, Jimenez-Valverde A, Real R. AUC: A misleading measure of the performance of predictive distribution models. Glob Ecol Biogeogr. 2008;17:145–151. [Google Scholar]
- 19.Huntley B, Collingham YC, Willis SG, Green RE. Potential impacts of climatic change on European breeding birds. PLoS ONE. 2008;3:e1439. doi: 10.1371/journal.pone.0001439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Legendre P. Spatial autocorrelation—Trouble or new paradigm. Ecology. 1993;74:1659–1673. [Google Scholar]
- 21.Gotelli NJ, Graves GR. Null Models in Ecology. Washington, DC: Smithsonian Institution Press; 1996. [Google Scholar]
- 22.Hubbell S. P. The Unified Neutral Theory of Biodiversity and Biogeography. Princeton, NJ: Princeton Univ Press; 2001. [DOI] [PubMed] [Google Scholar]
- 23.Roxburgh SH, Chesson P. A new method for detecting species associations with spatially autocorrelated data. Ecology. 1998;79:2180–2192. [Google Scholar]
- 24.Hagemeijer W. J. M., Blair M. The EBCC Atlas of European Breeding Birds: Their Distribution and Abundance. London: T. & A.D. Poyster; 1997. [Google Scholar]
- 25.Berry PM, Dawson TP, Harrison PA, Pearson RG. Modelling potential impacts of climate change on the bioclimatic envelope of species in Britain and Ireland. Glob Ecol Biogeogr. 2002;11:453–462. [Google Scholar]
- 26.Huntley B, et al. The performance of models relating species geographical distributions to climate is independent of trophic level. Ecol Lett. 2004;7:417–426. [Google Scholar]
- 27.Lawler JJ, White D, Neilson RP, Blaustein AR. Predicting climate-induced range shifts: Model differences and model reliability. Glob Chang Biol. 2006;12:1568–1584. [Google Scholar]
- 28.Brown JH, Stevens GC, Kaufmann DM. The geographic range: Size, shape, boundaries, and internal structure. Annu Rev Ecol Syst. 1996;27:597–623. [Google Scholar]
- 29.Brooker RW. Plant-plant interactions and environmental change. New Phytol. 2006;171:271–284. doi: 10.1111/j.1469-8137.2006.01752.x. [DOI] [PubMed] [Google Scholar]
- 30.Suttle KB, Thomsen MA, Power ME. Species interactions reverse grassland responses to changing climate. Science. 2007;315:640–642. doi: 10.1126/science.1136401. [DOI] [PubMed] [Google Scholar]
- 31.Sala OE, et al. Global biodiversity scenarios for the year 2100. Science. 2000;287:1770–1774. doi: 10.1126/science.287.5459.1770. [DOI] [PubMed] [Google Scholar]
- 32.Sanderson EW, et al. The human footprint and the last of the wild. Bioscience. 2002;52:891–904. [Google Scholar]
- 33.Sanderson FJ, Donald PF, Pain DJ, Burfield IJ, van Bommel FPJ. Long-term population declines in Afro-Palearctic migrant birds. Biol Conserv. 2006;131:93–105. [Google Scholar]
- 34.Thomas CD, Lennon JJ. Birds extend their ranges northwards. Nature. 1999;416:389–395. [Google Scholar]
- 35.Deutsch CA, et al. Impacts of climate warming on terrestrial ectotherms across latitude. Proc Natl Acad Sci USA. 2008;105:6668–6672. doi: 10.1073/pnas.0709472105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.New M, Hulme M, Jones P. Representing twentieth-century space-time climate variability. Part I: Development of a 1961–90 mean monthly terrestrial climatology. J Climate. 1999;12:829–856. [Google Scholar]
- 37.Batjes NH. Development of a world data set of soil water retention properties using pedotransfer rules. Geoderma. 1996;71:31–52. [Google Scholar]
- 38.Prentice IC, et al. A global biome model based on plant physiol and dominance, soil properties and climate. J Biogeogr. 1992;19:117–134. [Google Scholar]
- 39.Colwell RK, Lees DC. The mid-domain effect: Geometric constraints on the geography of species richness. Trends Ecol Evol. 2000;15:70–76. doi: 10.1016/s0169-5347(99)01767-x. [DOI] [PubMed] [Google Scholar]
- 40.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2006. [Accessed January 27, 2007]. Available at http://www.R-project.org. [Google Scholar]
- 41.Thuiller W. BIOMOD—Optimizing predictions of species distributions and projecting potential future shifts under global change. Glob Chang Biol. 2003;9:1353–1362. doi: 10.1111/gcb.12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McPherson JM, Jetz W, Rogers DJ. The effects of species' range sizes on the accuracy of distribution models: Ecological phenomenon or statistical artefact? J Appl Ecol. 2004;41:811–823. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.