Abstract
Mindin (spondin-2) is an extracellular matrix protein of unknown structure that is required for efficient T-cell priming by dendritic cells. Additionally, mindin functions as a pattern recognition molecule for initiating innate immune responses. These dual functions are mediated by interactions with integrins and microbial pathogens, respectively. Mindin comprises an N-terminal F-spondin (FS) domain and C-terminal thrombospondin type 1 repeat (TSR). We determined the structure of the FS domain at 1.8-Å resolution. The structure revealed an eight-stranded antiparallel β-sandwich motif resembling that of membrane-targeting C2 domains, including a bound calcium ion. We demonstrated that the FS domain mediates integrin binding and identified the binding site by mutagenesis. The mindin FS domain therefore represents a new integrin ligand. We further showed that mindin recognizes lipopolysaccharide (LPS) through its TSR domain, and obtained evidence that C-mannosylation of the TSR influences LPS binding. Through these dual interactions, the FS and TSR domains of mindin promote activation of both adaptive and innate immune responses.
Keywords: extracellular matrix, innate immunity, integrin, mindin (spondin-2), structure
Introduction
The extracellular matrix (ECM) consists of a complex assortment of glycoproteins and proteoglycans that are produced locally by cells in the matrix and are assembled into an organized meshwork (Kreis and Vale, 1999). The ECM serves not only as a scaffold to provide structural support for tissues but also regulates the behaviour of cells that contact it. Interactions between cells and ECM components influence cell migration, adhesion, differentiation and proliferation (Kreis and Vale, 1999). Additionally, the ECM may provide docking sites for invading microorganisms. For example, microbial pathogens use adhesins to attach to ECM proteins, such as fibronectin, collagen and thrombospondin (Patti et al, 1994). Conversely, the ECM may participate in immune defence against infections by recruiting leukocytes to sites of inflammation. Inflammatory cell recruitment is mediated by interactions between integrins and several ECM proteins, including fibronectin, vitronectin and laminin, which function as integrin ligands (Hynes, 2002; Shimaoka and Springer, 2003; Arnaout et al, 2007). ECM components may also contribute to host defence by interacting with pattern recognition molecules of the innate immune system, such as Toll-like receptor 4 (TLR4) (Okamura et al, 2001; Lasarte et al, 2007).
Mindin (also called spondin-2) is a highly conserved ECM protein that is abundantly expressed in the spleen and lymph nodes (He et al, 2004). It is a member of the mindin–F-spondin (FS) family of secreted ECM proteins that also includes mammalian FS, zebrafish mindin 1 and mindin 2, and Drosophila M-spondin (Klar et al, 1992; Higashijima et al, 1997; Umemiya et al, 1997; Feinstein et al, 1999; Manda et al, 1999; Zisman et al, 2007). Mindin and FS promote the outgrowth and adhesion of embryonic hippocampal neurons in rodents (Feinstein et al, 1999). Remarkably, mindin also functions as a pattern recognition molecule for microbial pathogens (He et al, 2004), and as an integrin ligand for inflammatory cell recruitment and T-cell priming (Jia et al, 2005; Li et al, 2006).
Mice lacking mindin are resistant to lipopolysaccharide (LPS)-induced septic shock and exhibit an impaired ability to clear bacterial infections (He et al, 2004). Macrophages and mast cells from mindin-deficient mice show defective responses to a broad spectrum of microbial stimuli. Moreover, mindin agglutinates bacteria and binds directly to LPS and lipoteichoic acid through their carbohydrate moieties. Mindin also functions as an opsonin for macrophage phagocytosis of bacteria (He et al, 2004). Mice lacking mindin exhibit defective clearance of influenza virus, whereas mindin-deficient macrophages show impaired activation following influenza infection (Jia et al, 2008). Thus, mindin is a pattern recognition molecule that is critical for initiating innate immune responses to both bacterial and viral pathogens (McDonald and Núñez, 2004; Jia et al, 2008).
Mindin-deficient mice display severely impaired recruitment of neutrophils and macrophages to inflammation sites (Jia et al, 2005). Furthermore, neutrophils directly adhere to immobilized mindin, and mindin promotes neutrophil migration in vitro. These effects are mediated through the interaction of mindin with αMβ2 and α4β1 integrins on neutrophils. Mindin–integrin interactions also have a key function in T-cell priming by dendritic cells (DCs). Mice lacking mindin have defective humoral immune responses to T-dependent, but not T-independent, antigens (Li et al, 2006). DCs from mindin-deficient mice show an impaired capacity to prime CD4+ T cells due to inefficient engagement of T lymphocytes. Additionally, these DCs have reduced levels of the Rho guanosine triphosphatases (GTPases) Rac1 and Rac2, which control DC priming capability (Benvenuti et al, 2004). Mindin regulates Rac1 and Rac2 expression by signalling through α4β1 and α5β1 integrins on DCs (Li et al, 2006).
Despite the importance of mindin in both innate and adaptive immunity, no information is available on its three-dimensional structure or ligand-binding sites. Indeed, such information is lacking for any member of the mindin–FS family of ECM proteins, each of which comprises a unique N-terminal region (termed the FS domain) with no detectable sequence homology to any known protein, and one (in the case of mindin) or more (six in the case of FS) C-terminal thrombospondin type 1 repeats (TSRs) (Higashijima et al, 1997). We determined the crystal structure of the FS domain of human mindin and demonstrated that this domain mediates integrin binding. We further localized the binding site by mutagenesis and characterized the integrin specificity of the FS domain. In addition, we showed that mindin recognizes LPS through its TSR domain.
Results
Structure determination
Initially, we attempted crystallizing full-length mindin (mindin-FL; residues 1–309), which we expressed as bacterial inclusion bodies and folded in vitro. However, the protein underwent slow degradation, and no crystals were obtained. SDS–PAGE revealed a stable product of ∼25 kDa, which amino-acid sequencing identified as an N-terminal fragment of mindin. Further characterization by mass spectrometry showed that the fragment comprised residues 1–223, which contained the FS region. Notably, mindin-FL includes a 26-residue segment between residues 223 and 250, the predicted start of the TSR domain, which contains several potential protease sites. A new expression construct was designed accordingly, and the corresponding protein (mindin-FS) crystallized readily. The structure was determined to 1.8 Å resolution by multiple wavelength anomalous dispersion (MAD) using a selenomethionine-substituted crystal (Table I). The final model contains two monomers in the asymmetric unit. However, the interface between the molecules is relatively small, in agreement with the behaviour of both mindin-FS and mindin-FL as monomers in gel filtration (not shown). The root mean squared (r.m.s.) difference in α-carbon positions for the two mindin-FS molecules in the asymmetric unit is 0.43 Å, indicating close similarity.
Table 1.
Data collection, structure determination and refinement
| Data set | ||||
|---|---|---|---|---|
| Se peak | Se inflection | Se remote | Native | |
| Data collection | ||||
| Space group | P212121 | P212121 | ||
| Unit cell (Å) | a=50.7, b=68.1, c=110.1 | a=50.6, b=68.0, c=110.1 | ||
| X-ray source | Synchrotron | CuKα | ||
| Wavelength (Å) | 0.9791 | 0.9795 | 0.9611 | 1.5418 |
| Resolution (Å) | 30.0–2.04 | 30.0–2.04 | 30.0–2.04 | 30.0–1.80 |
| Observations | 307 332 | 285 232 | 244 891 | 133 390 |
| Unique reflections | 24 768 | 24 501 | 23 837 | 39 410 |
| I/σIa | 23.5 (2.8) | 20.0 (2.1) | 15.3 (1.7) | 11.4 (3.4) |
| Completeness (%)a | 99.2 (93.3) | 98.2 (88.2) | 95.3 (74.1) | 98.5 (96.2) |
| Rmerge (%)a,b | 9.4 (39.6) | 9.4 (41.2) | 10.0 (45.8) | 5.1 (20.4) |
| Structure determination | ||||
| Resolution (Å) | 20.0–2.60 | |||
| Figure of merit | 0.51 | |||
| Structure refinement | ||||
| Resolution (Å) | 30.0–1.80 | |||
| Protein atoms | 3289 | |||
| Ca2+ (per monomer) | 1 | |||
| Ni2+ | 6 | |||
| Water molecules | 319 | |||
| Rwork (%)c | 18.9 | |||
| Rfree (%)c | 21.5 | |||
| r.m.s. deviations from ideality | ||||
| Bond lengths (Å) | 0.005 | |||
| Bond angles (deg) | 1.34 | |||
| Ramachandran plot statistics | ||||
| Most favoured (%) | 88.0 | |||
| Additionally allowed (%) | 11.5 | |||
| Generously allowed (%) | 0.5 | |||
| Disallowed (%) | 0 | |||
| Average B values (Å2) | ||||
| Protein main chain | 17.4 | |||
| Protein side chain | 21.0 | |||
| Ca2+ | 12.9 | |||
| Ni2+ | 35.1 | |||
| Water molecules | 27.5 | |||
| aValues in parentheses are statistics for the highest resolution shells. | ||||
| bRmerge=∑∣Ij–<I>∣/∑Ij, where Ij is the intensity of an individual reflection and <I> is the average intensity of that reflection. | ||||
| cRwork=∑∣∣Fo∣–∣Fc∣∣/∑∣Fo∣, where Fc is the calculated structure factor. Rfree is as for Rwork but calculated for a randomly selected 5.0% of reflections not included in the refinement. | ||||
Overall structure
On the basis of sequence alignments, it was predicted that all members of the mindin–FS family share three structural features: two N-terminal FS domains (FS1 and FS2) and one (in the case of mindin) or more TSRs (Higashijima et al, 1997). However, the structure of mindin-FS revealed that FS1 and FS2 actually form a single domain (Figure 1A and B), and that full-length mindin therefore consists of one N-terminal FS domain linked to one C-terminal TSR domain.
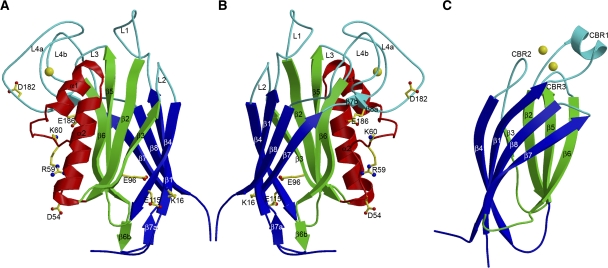
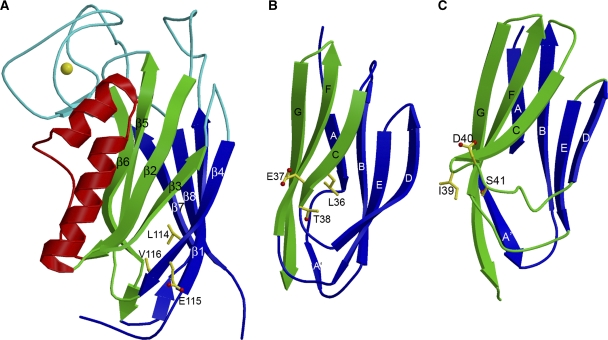
Figure 1.
Structure of the F-spondin domain of mindin and comparison with a Ca2+-dependent C2 domain. (A) Side view of mindin-FS (ribbon diagram). The two four-stranded β-sheets, β6β5β2β3 (front) and β4β1β8β7 (back), are green and blue, respectively. Helices α1 and α2 are red. The bound Ca2+ ion is yellow. Loops L1–L4 at the top of the molecule are cyan. Residues mutated to alanine to localize the integrin-binding site are drawn in ball-and-stick representation, with carbon atoms in yellow, oxygen atoms in red and nitrogen atoms in blue. (B) Mindin-FS is rotated ∼180° about the vertical axis with respect to the view in (A). (C) Side view of the C2 domain of cytosolic phospholipase A2 (PDB accession code 1RLW). The orientation is similar to that of mindin-FS in (B). The two four-stranded β-sheets, β4β1β8β7 (front) and β6β5β2β3 (back), are blue and green, respectively. The bound Ca2+ ion is yellow. Calcium-binding loops CBR1–CBR3 are cyan.
The overall fold of mindin-FS is that of an eight-stranded antiparallel β-sandwich composed of two four-stranded β-sheets, with two contiguous α-helices packed on one side of the β-sandwich sheet. Unexpectedly, a Dali structure homologue search (http://ekhidna.biocenter.helsinki.fi/dali/) identified significant similarities between the eight-stranded antiparallel β-sandwich motif of mindin and C2 domains (Figure 1C), despite the absence of sequence homology. The top hit (Z-score=5.9) was the C2 domain of phosphoinositide 3-kinase (PI3K), a lipid kinase (Walker et al, 1999). Superposition of the mindin-FS and PI3K C2 structures gave an r.m.s. deviation of 3.3 Å for 111 α-carbon atoms composing the β-sandwich motif. C2 domains (∼130 residues) are found in diverse cytosolic proteins involved in membrane trafficking and signal transduction, where they function as membrane-targeting modules through Ca2+-dependent or -independent binding to phospholipids (Rizo and Südhof, 1998; Cho and Stahelin, 2005). On the basis of differences in the connectivity of their β-strands, C2 domains have been subdivided into two types of topology variants, I and II (Nalefski and Falke, 1996). Mindin, similar to PI3K, possesses a type II topology.
The two β-sheets of mindin are composed of β-strands β1, β4, β7 and β8, and strands β2, β3, β5 and β6, respectively, using the labeling convention of type II C2 domains (Nalefski and Falke, 1996) (Figure 1). In most C2 domains, the three loops connecting β-strands β1 and β2, β3 and β4, and β5 and β6 (designated calcium-binding region 1 (CBR1), CBR2 and CBR3) form a site for binding one or more Ca2+ ions (Figure 1C), although some C2 domains, for example PI3K C2, do not bind Ca2+. Mindin contains one bound Ca2+ ion (Figure 1A and B). However, it is not located within the loops corresponding to the CBRs of C2 domains (L1, L2 and L3), but instead at a site formed mainly by the loop connecting strands β7 and β8 (L4). This loop, which contains two short β-strands (β7b and β8a), is exceptionally long (52 residues) compared with the corresponding loop of C2 domains (∼8 residues). Additionally, mindin includes two α-helices (α1 and α2), inserted between strands β2 and β3, that are not found in C2 domains (Figure 1B and C). The mindin-FS structure contains a single disulphide bond, linking Cys9 and Cys145. Because both residues are strictly conserved in FS domains (Figure 3A), a corresponding disulphide is likely present in all members of the mindin–FS family.
Calcium-binding site
The final refined structure contains one Ca2+ in each of the two mindin molecules in the asymmetric unit. Both Ca2+ ions were refined at full occupancy, with an average B value (13 Å2) less than the average main chain B value for the protein (17 Å2) (Table I), indicating high-affinity binding to mindin. In each case, the bound Ca2+ sits on a small β-sheet platform formed by strands β6, β7b and β8a (Figure 2A). It is enveloped by residues 162–166 from loop L4, resulting in nearly complete burial. Hence, the most likely role of Ca2+ in mindin is to stabilize the protein structure, particularly the conformation of the long L4 loop, rather than to mediate ligand binding, at least directly. In Ca2+-dependent C2 domains, by contrast, bound Ca2+ is available to directly coordinate the phosphoryl group of phospholipids (Cho and Stahelin, 2005).
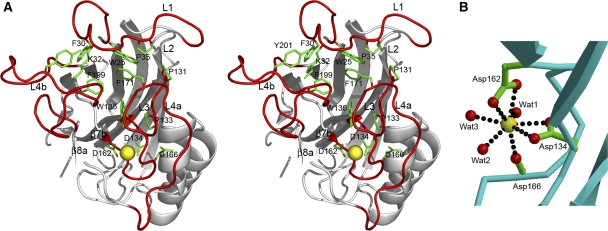
Figure 2.
Calcium-binding site of mindin-FS. (A) Stereo view of the top of mindin-FS. Loops L1, L3 and L4 are red. Conserved residues within these loops that participate in key interloop interactions, or in coordinating the Ca2+ ion (yellow), are green. (B) Coordination geometry of the Ca2+ ion. The calcium is ligated by Asp134, Asp162, Asp166, and three water molecules (Wat1–Wat3). Oxygen and carbon atoms are red and green, respectively.
The Ca2+ ion is ligated by three aspartic acid residues (Asp134, Asp162 and Asp166) and three water molecules (Figure 2B). Asp166 binds Ca2+ through its main chain carbonyl oxygen atom, whereas Asp134 and Asp162 function as bidentate ligands, each binding through its two carboxyl group oxygen atoms. Hence, the Ca2+ ion in mindin coordinates eight oxygen atoms, instead of the seven that typically define the pentagonal bipyramidal coordination geometry of Ca2+ in most Ca2+-binding proteins (Strynadka and James, 1989). Nevertheless, the coordination geometry in mindin may still be regarded as pentagonal bipyramidal, with one bidentate aspartate ligand at one of the vertices. All metal–ligand distances (2.3–2.6 Å) are within the range reported for Ca2+ ions in proteins (Strynadka and James, 1989). Moreover, sequence alignment of mindin, M-spondin and FS showed absolute conservation of all three Ca2+-coordinating aspartate residues in mindin, as well as residues 162–166 of L4 (Figure 3A), strongly suggesting that all members of the mindin–FS family bind Ca2+.
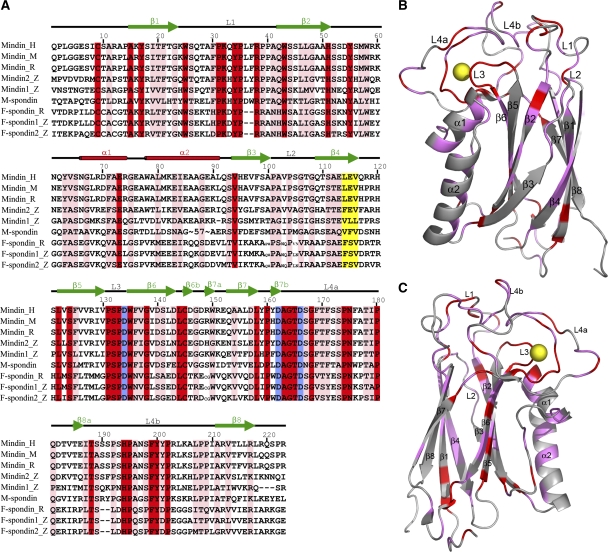
Figure 3.
Mindin–F-spondin family. (A) Structure-based sequence alignment of the F-spondin domains of mindin, M-spondin and F-spondin. Secondary structure elements are denoted by red cylinders (α-helices) and green arrow (β-strands); these are numbered sequentially. Loops located at the top of the mindin domain are labelled L1–L4. Residues that are strictly conserved are shaded red; residues that are well conserved are shaded pink. The integrin-binding triplet of mindin (LEV) is shaded yellow. Residues that coordinate the calcium ion are shaded blue. H, humans; M, mouse; R, rat; Z, zebrafish; D, Drosophila. Sequences were retrieved from SwissProt using the following accession numbers: human mindin (Q9BUD6); mouse mindin (Q8BMS2); rat mindin (Q9WV75); zebrafish mindin 1 (O42111); zebrafish mindin 2 (O42112); Drosophila M-spondin (O02092); rat F-spondin (P35446); zebrafish F-spondin 1 (O42113); zebrafish F-spondin-2 (O42114). Sequence alignments were performed with the program ClustalW (http://www.expasy.ch). (B) Distribution of conserved residues in the mindin-FS structure, based on the sequence alignment in (A). Regions containing strictly conserved residues are highlighted in red; regions with well conserved residues are highlighted in magenta. All other regions are grey. The calcium ion is yellow. Secondary structure elements are labelled according to (A). (C) Mindin-FS is rotated ∼180° about the vertical axis with respect to the view in (B).
Identification of the integrin-binding site
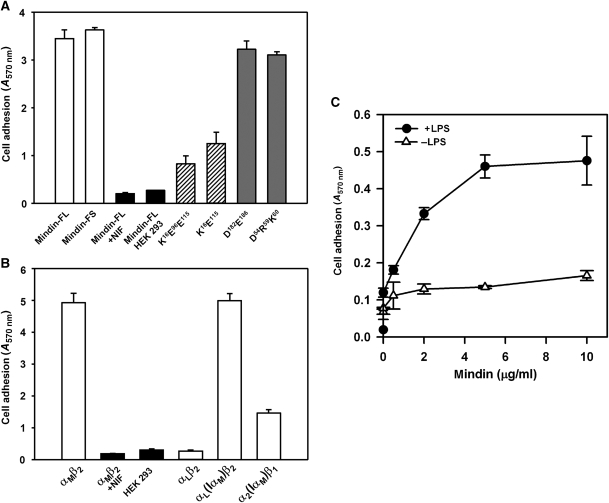
We used mutational analysis and cell adhesion assays to identify the integrin-binding site of mindin. To permit directional coupling of the protein to streptavidin-coated plastic surfaces, as well as to minimize denaturation due to immobilization, peptide tags containing biotinylation sequences were added to the C termini of mindin-FL and mindin-FS. Both immobilized proteins mediated equally strong adhesion of HEK 293 cells expressing αMβ2 integrin (αMβ2/293) (Figure 4A), which demonstrates that the integrin-binding site of mindin resides in its FS, rather than TSR, domain. Mock-transfected HEK 293 cells did not adhere to mindin (Figure 4A). Furthermore, adhesion was abolished by neutrophil inhibitory factor (NIF), an αMβ2 integrin antagonist (Muchowski et al, 1994) (Figure 4A), by the anti-αMβ2 antibody M1/70, and by the addition of EDTA (not shown), thereby confirming the specificity and cation dependency of mindin binding to αMβ2 integrin. Moreover, primary murine peritoneal neutrophils adhered to human mindin in a dose-dependent manner, which was further enhanced by cellular activation with LPS (Figure 4C).
Figure 4.
Cell adhesion to mindin. (A) αMβ2-mediated cell adhesion to mindin. A total of 2 × 106 αMβ2-expressing or mock-transfected HEK 293 cells were allowed to adhere to 24-well non-tissue culture polystyrene plates precoated with recombinant mindin-FL, mindin-FS or its different mutants. After incubation at 37°C for 30 min, unbound cells were removed by three washes with DPBS and adherent cells were quantified by staining with crystal violet, measuring absorption at 570 nm. Specificity of cell adhesion was verified using the αMβ2-specific antagonist NIF, mock-transfected cells and biotinylated IgG or ovalbumin as the substrate (not shown). Data shown are the means±s.d. of triplicate wells and are representative of three independent experiments. (B) The αMβ2-mediated cell adhesion to mindin is mediated by its αM I domain. Cell adhesion to mindin-FL by different integrin receptors, including αMβ2, αLβ2, αL(IαM)β2 and α2(IαM)β1, where the αL or α2 I domain was replaced with that of αM, were determined as above. Specificity was verified by the addition of the αMβ2-specific antagonist NIF and by using mock-transfected HEK 293 cells. Data shown are the means ±s.d. of triplicate wells and are representative of two independent experiments. (C) Neutrophil adhesion to mindin is enhanced by LPS. Murine neutrophils were added to mindin-coated 24-well plates (0.5 × 106 cells per well) in the presence or absence of 1 μg/ml LPS. After 10 min at 37°C, unbound cells were removed by washing with PBS and bound cells were quantified by crystal violet staining, measuring absorption at 570 nm.
The integrin-binding site of mindin was localized further by mutagenesis. Ligand binding to integrins is universally dependent on bivalent cations, such that Mg2+ or Mn2+ at the metal ion-dependent adhesion site (MIDAS) of the integrin directly coordinates to the carboxyl group of a glutamate or aspartate residue of the ligand (Shimaoka and Springer, 2003; Arnaout et al, 2007). However, the FS domain of mindin contains 22 glutamate or aspartate residues on its surface that could potentially ligate metals at the MIDAS. In the crystal structure, we observed that Lys16, Glu96 and Glu115 coordinated a cluster of five Ni2+ ions, which presumably originated from the crystallization buffer. As these Ni2+-binding residues should also have affinities for other bivalent cations, we constructed the triple mutant K16A/E96A/E115A. These mutations reduced adhesion of cells expressing αMβ2 integrin by ∼75% compared with wild-type mindin (Figure 4A), implicating Glu96 or Glu115 as the key metal-coordinating residue.
Glu96 is unlikely to be responsible for MIDAS binding, based on the following observations: first, whereas human mindin (the protein studied here) contains glutamate at position 96, the corresponding residue in mouse (and rat) mindin is alanine (Figure 3A). Second, mouse mindin interacts with αMβ2 integrin (Jia et al, 2005; Li et al, 2006). To directly test whether Glu115 is involved in αMβ2 binding, we constructed the double mutant K16A/E115A. As these mutations reduced adhesion of αMβ2-expressing cells to immobilized mindin to nearly the same extent as the K16A/E96A/E115 mutations (Figure 4A), we concluded that Glu115 most likely coordinates to the MIDAS of αMβ2 integrin. As controls, we introduced double (D182A/E186A) or triple (D54A/R59A/K60A) mutations on other faces of the mindin molecule (Figure 1A and B). However, these mutations did not appreciably affect adhesion (Figure 4A), confirming the specificity of the binding results.
Further examination of the human mindin sequence revealed that Glu115 is embedded within the tripeptide LEV (Figure 3A), which conforms to the consensus sequence for LDV-binding integrins (L/I-D/E-V/S/T) (Hynes, 2002; Humphries et al, 2006). Moreover, none of the other 21 solvent-accessible glutamate or aspartate residues of mindin forms part of a known integrin recognition motif, in agreement with localization of the integrin-binding site at Glu115. The LEV motif is conserved in human, mouse and rat mindin, but not in zebrafish mindin 1 (VLL), Drosophila M-spondin (VFV) or in rat or zebrafish F-spondin (FSV) (Figure 3A). Thus, within the mindin–FS family of ECM proteins, only mammalian mindins appear to be integrin ligands, at least with respect to the Glu115 site. However, zebrafish mindin 2 contains the sequence FEV at this position, which could conceivably represent an integrin-binding site in other vertebrates.
The LEV tripeptide of mindin is located at the end of strand β4 near the base of the FS domain (Figure 5A). Interestingly, this location is somewhat analogous to that of the integrin binding LET triplet of ICAM-3, which is situated at the end of β-strand C on the lower side of this immunoglobulin superfamily domain (Song et al, 2005) (Figure 5B). By contrast, the metal-coordinating IDV triplet of VCAM-1 is found in a loop connecting β-strands C and D (Shimaoka and Springer, 2003) (Figure 5C).
Figure 5.
Comparison of integrin recognition sites. (A) Mindin. The LEV tripeptide in strand β4 is drawn in ball-and-stick representation with carbon atoms in yellow and oxygen atoms in red. (B) ICAM-3 (PDB accession code 1T0P) showing the LET tripeptide in β-strand C. (C) VCAM-1 (1VCA) showing the IDS tripeptide in the loop connecting β-strands C and D.
Integrin specificity
Integrins are heterodimers composed of non-covalently associated α- and β-subunits (Hynes, 2002; Shimaoka and Springer, 2003; Arnaout et al, 2007). In mammals, 18 α-subunits and 8 β-subunits form 24 heterodimers that recognize distinct but overlapping ligands. Half of integrin α-subunits, including αM, contain inserted (I) domains (also known as A domains), which are the principal ligand-binding domains when present (Shimaoka and Springer, 2003). Although HEK 293 cells expressing αMβ2 bound strongly to mindin, cells expressing αLβ2 integrin showed no adhesion (Figure 4B). To identify the source of this differential recognition, we constructed a chimaeric integrin by replacing the I domain of αL with that of αM. As cells expressing the αL(IαM)β2 chimaera adhered to mindin as well as cells expressing αMβ2 (Figure 4B), specificity is clearly mediated through the I domains. However, other parts of the integrin αβ heterodimer also appear to contribute to mindin binding, as suggested by a chimaera in which the α2 I domain of integrin α2β1, which did not bind mindin (not shown), was replaced by that of αMβ2. In this context, the αM I domain only partially restored adhesion to mindin (Figure 4B).
Comparison with other mindin–FS family members
Structure-based sequence alignment of the FS domain of human mindin with that of other members of mindin–FS family (zebrafish mindin 1 and 2, Drosophila M-spondin, rat and zebrafish FS) showed that the N-terminal region of these proteins, corresponding to strands β1 and β2 and loop L1 in mindin, is highly conserved, as is the region from strand β5 to the middle of loop L4 (Figure 3A). By contrast, the segment that includes helices α1 and α2 and strands β3 and β4, which together form one face of the mindin molecule (Figure 1A), is considerably more variable. Thus, M-spondin contains a 57-residue insertion at a site corresponding to helix α2 of mindin, indicating significant structural differences in this region that could translate into functional differences among mindin–FS family members. Indeed, the integrin binding LEV triplet of mindin, which is absent in M- or FS, is located in strand β4, on the variable face of the FS domain (Figure 5A).
Surprisingly, the most conserved regions of sequence within the mindin–FS family correspond to loops, rather than strands or helices (Figure 3B and C): all residues in L3, and many in L1 and L4, are absolutely conserved, with most remaining residues conservatively substituted (Figure 3A). The L1, L3 and L4 loops form a cage-like structure at the top of the mindin molecule (Figure 2A). At the centre of the cage, Phe171 interacts with Trp25, Trp135, Phe199 and Pro35 to form a tight hydrophobic cluster. At one corner of the cage, Tyr201 makes hydrophobic contacts to Phe30 and Lys32, which in turn forms hydrogen bonds with Pro195, Asn197 and Tyr200. At the opposite corner, Asp134, Asp162 and Asp164 coordinate the nearly buried Ca2+ ion, as described above. This configuration of L1, L3 and L4, which is most likely to be maintained in the mindin–FS family, could imply related functions for the top of FS domains (Figure 2A), as suggested by the finding that the isolated FS domains of both mindin and FS promote outgrowth of sensory neurons in rats (Feinstein et al, 1999).
Identification of the LPS-binding region
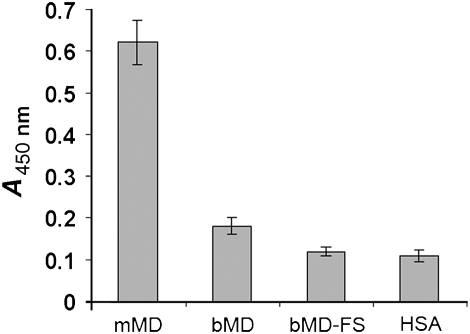
Previously, recombinant mindin produced in mammalian cells was shown to bind directly to LPS through interactions with its carbohydrate, rather than lipid A, moiety (He et al, 2004). Our finding that the FS domain of mindin most closely resembles C2 domains in its overall fold initially led us to expect that the FS domain might mediate LPS recognition, as C2 domains are membrane-targeting modules (Cho and Stahelin, 2005). However, immobilized mindin-FS did not bind LPS (Figure 6), thereby implicating the TSR domain. Accordingly, bacterially expressed mindin-FL, which contains both FS and TSR domains, was tested for LPS binding. In contrast to full-length mindin produced in HEK 293 cells, the bacterial protein showed no significant binding to LPS (Figure 6).
Figure 6.
LPS-binding capacity of different forms of mindin. Full-length mindin from HEK 293 cells (mMD), full-length mindin from E. coli (bMD), mindin FS domain alone from E. coli (bMD-FS) and human serum albumin (HSA) were coated on ELISA plate, which was washed and incubated with LPS from Salmonella typhosa (10 μg/ml) at room temperature for 1 h as described (He et al, 2004). Bound LPS was detected using mouse anti-LPS antibody. The error bars represent standard deviations of triplicate wells.
To explain this functional difference between bacterial and mammalian mindin, we analysed mindin from HEK 293 cells for possible post-translational modifications. Previous studies of the TSRs of FS, thrombospondin-1 and properdin identified two types of protein glycosylation: O-fucosylation and C-mannosylation (Hofsteenge et al, 2001; Gonzalez de Peredo et al, 2002). Although the O-fucosylation site is absent in the TSR of mindin, which has proline in place of serine/threonine at position 266, this TSR does contain a potential site for C-mannosylation at Trp257. This unusual modification involves attachment of a single α-mannopyranosyl residue to the C-2 atom of the first tryptophan in the recognition motif W-X-X-W (W257SSW in mindin) (Doucey et al, 1998).
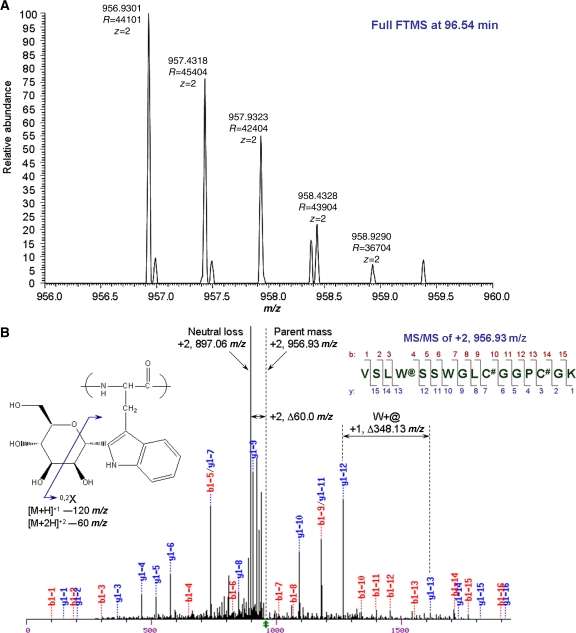
To determine whether mindin is C-mannosylated, the protein was digested with trypsin and endoproteinase Glu-C, and the resulting peptides were analysed by reversed phase liquid chromatography coupled with tandem mass spectrometry (RP-LC-MS/MS). The parent masses calculated from the peptide sequence containing the C-mannosylation recognition motif, VSLW@SSWGLC#GGPC#GK, allowed for alkylated cysteine (#=57.0215 Da) with and without mannosylated tryptophan (@=162.0528 Da). A C-mannosylated glycopeptide (VSLW@SSWGLC#GGPC#GK, +2, at m/z 956.9301 as mono isotopic mass) was observed at 96.54 min (Figure 7A). The mass of the peptide indicated two alkylated cysteines and a mannosylated tryptophan.
Figure 7.
Identification of a C-mannosylation site in mindin. (A) LC-MS/MS result of C-mannosylated glycopeptides (VSLW@SSWGLC#GGPC#GK, +2, m/z 956.9320, @=162.0528 Da and #=57.0215 Da) produced by digestion of mindin with endoproteinase Glu-C and trypsin. A full FTMS spectrum at 96.54 min of liquid chromatogram has the doubly charged C-mannosylated glycopeptides with 2.1 p.p.m. mass accuracy. (B) An MS/MS spectrum shows the fragmentation of the glycopeptides for peptide identification and a cross-ring cleavage on zero and two carbons of mannose by neutral loss.
The MS/MS spectrum of +2, at m/z 956.93, confirmed the peptide sequence (Figure 7B). In the y-ion series from the fragmentation, C-mannosylated tryptophan was assigned with 348.13 Da differences (186.08 Da (W)+162.05 Da (@)) between the y-12 ion and y-13 ion from the fragmentations. The 120 Da loss, which is the typical ring cleavage of aromatic C-glycosides (Figure 7B, inset), has been observed with loss of m/z 120 and m/z 60 for singly and doubly charged ions, respectively. The cross-ring cleavage of mannose occurred on zero and two carbon bonds (0.2A/0.2X) by neutral loss, which gave the most intense ion at m/z 897.06 and resulted in a 60 Da loss from the parent ion. Therefore, mindin produced in HEK 293 cells is C-mannosylated at Trp257.
Analysis of the entire mindin sequence using the program ExPASy (http://ca.expasy.org/tools/) identified Trp257 as the only potential glycosylation site of any type. Consistent with this result, LC-MS/MS analysis of peptides generated by digesting mindin with trypsin and chymotrypsin revealed no post-translational modifications elsewhere in the protein (74% sequence coverage; not shown). As discussed below, the C-mannosylated Trp257 residue of mindin TSR is located on the proposed ligand-binding face of TSR domains (Tan et al, 2002).
Discussion
The ECM protein mindin has multiple functions in nervous system development and innate and adaptive immunity. These include patterning axonal trajectory in the embryonic spinal cord (Feinstein et al, 1999), acting as a pattern recognition molecule for microbial pathogens (He et al, 2004; Jia et al, 2008), recruiting leukocytes to inflammation sites (Jia et al, 2005), and priming CD4+ T cells (Li et al, 2006). Of these diverse functions, inflammatory cell recruitment and T-cell priming are mediated by interactions of mindin with integrins on neutrophils and DCs, respectively (Jia et al, 2005; Li et al, 2006). We have demonstrated that mindin binds integrins through its FS domain, which therefore constitutes a new integrin ligand. Interestingly, the isolated FS domain of mindin has also been shown to facilitate the outgrowth of dorsal root ganglia neurons, although the relevant receptors have not been identified in that case (Feinstein et al, 1999). In this respect, mindin resembles FS, the founding member of the mindin–FS family, which too is involved in axon guidance (Burstyn-Cohen et al, 1999; Tzarfaty-Majar et al, 2001; Zisman et al, 2007).
The structure of the FS domain of human mindin revealed a broad surface which, based on sequence comparisons, exhibits substantial topological variability within the mindin–FS family. Importantly, this surface contains the integrin recognition motif that is unique to mammalian mindin. Mindin-FS, ICAM-1 and ICAM-3 all utilize the L/I-D/E-V/S/T triplet to bind integrins (LEV, IET and LET, respectively). In the crystal structures of ICAM-1 and ICAM-3 bound to the I domain of αL, the side chain of the central glutamate residue projects from the ligand to coordinate to the MIDAS of the I domain, whereas the side chains of the two flanking residues point towards the ligand and do not contact the integrin (Shimaoka et al, 2003; Song et al, 2005). The LEV triplet of mindin-FS is similarly disposed, with the side chain of Glu115 fully accessible to solvent, but those of Leu114 and Val116 nearly completely buried in the FS domain. Apart from the L/I-D/E-V/S/T motif, however, no residues of ICAM-1 or ICAM-3 that contact the αL I domain in the complex structures are conserved in mindin-FS, implying a different set of interactions outside the MIDAS.
Our data support a mosaic model of the mindin-binding site within αMβ2, where the αM I domain has a major function and other domains, such as the αM β-propeller or the β2 I domain, have supporting functions. Thus, mindin binding to αMβ2 is completely blocked by NIF, an αMβ2-specific antagonist, the binding site of which is located exclusively on the αM I domain (Zhang and Plow, 1997). Furthermore, substitution of the αL I domain with that of αM conferred αLβ2, an αMβ2 homologue that does not bind mindin, with full mindin-binding activity, suggesting that the I domain forms a major binding interface with mindin. However, when the αM I domain was inserted into α2, which complexes with a different β-subunit (β1) and is less homologous to αM, only partial binding was recovered. This suggests that other domains within αMβ2 may also contribute to binding, either by contacting mindin directly or by altering the conformation of the αM I domain. Our previous study showed that mindin also interacts with α4β1 integrin on neutrophils (Jia et al, 2005). As the α4 subunit lacks an I domain (Hynes, 2002), we anticipate that mindin binding to α4β1 may occur through a different mechanism than the one we describe here for αMβ2.
TSR domains (∼60 residues) have been identified in over 40 human proteins belonging to multiple families (Venter et al, 2001). Structural studies of isolated TSRs have shown that they are elongated molecules characterized by an unusual antiparallel, three-stranded fold consisting of alternating stacked layers of tryptophan and arginine residues (Tan et al, 2002; Pääkkönen et al, 2006). TSRs are found in secreted proteins or in the extracellular portion of transmembrane proteins that are involved in cell adhesion, neuronal development and immunity (Tucker, 2004). Besides LPS, molecules that bind to particular TSR modules include glycosaminoglycans, fibronectin and laminin. Mindin, which does not bind fibronectin or laminin, recognizes LPS through its carbohydrate, rather than lipid A, moiety (He et al, 2004).
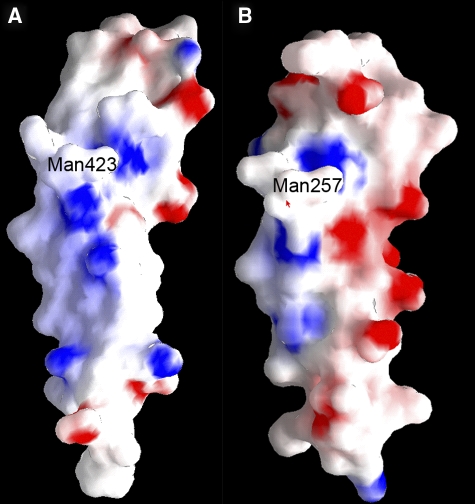
Although no structure is available for any TSR bound to a specific ligand, the second TSR module of thrombospondin-1 contains a long, positively-charged groove that is the likely recognition surface for glycosaminoglycans (Tan et al, 2002) (Figure 8A). A homology model of the mindin TSR domain displayed a similar, though less electropositive, groove (Figure 8B). Notably, the C-mannosylation site of thrombospondin-1 TSR is located along one edge of its putative glycosaminoglycan-binding groove, such that the mannose attached to Trp423, which corresponds to Trp257 of mindin TSR, could form part of the binding site (Tan et al, 2002) (Figure 8A and B). Our finding that C-mannosylated mindin from HEK 293 cells binds LPS, but that bacterially expressed mindin does not, is consistent with a role for glycosylation in ligand recognition. A role for glycosylation in TSR function is also suggested by the finding that chemically synthesized C-mannosylated peptides derived from TSRs enhanced LPS-induced signalling in macrophages (Muroi et al, 2007).
Figure 8.
Electrostatic potential surface of TSRs. (A) The proposed ligand recognition face of the second TSR module of thrombospondin-1 with a mannose (Man) attached to Trp423 (Tan et al, 2002). Solvent-accessible surfaces are coloured according to electrostatic potential, with positively charged regions in blue and negatively charged regions in red. Electrostatic surface potentials were calculated with GRASP (Nicholls et al, 1991). (B) The corresponding face of a homology model of mindin TSR. A mannose (Man) is attached to Trp257, which corresponds to Trp423 of the thrombospondin-1 TSR. Homology modelling of the mindin TSR domain was performed with the program SWISS-MODEL (http://swissmodel.expasy.org) using the NMR solution structure of the first TSR domain of F-spondin as a template (PDB accession code 1VEX) (Pääkkönen et al, 2006).
Although most of the sensors that detect and report invading microbes are associated with immune cells (e.g. TLRs, Nod-like receptors and RIG-I-like receptors), the ECM is also essential for host defence against pathogens (McDonald and Núñez, 2004). We have shown that the ECM protein mindin interacts with integrins and LPS through its FS and TSR domains, respectively. In this way, mindin promotes activation of both adaptive and innate immune responses to microbial invaders. The structural and binding studies described here provide a molecular framework for further investigating the mechanism of mindin's multiple actions in the immune system.
Materials and methods
Protein expression and purification
Gene fragments encoding full-length human mindin (mindin-FL; residues Gln1-Val309), or the FS domain alone (mindin-FS; residues Gln1-Arg223) were inserted into pET-26b (Novagen). Both proteins were expressed as inclusion bodies in Escherichia coli BL21(DE3) cells (Novagen). Inclusion bodies were dissolved in 8 M urea, 50 mM Tris–HCl (pH 8.0) and 10 mM DTT. For in vitro folding, mindin-FL or mindin-FS inclusion bodies were diluted into folding buffer containing 0.8 M L-arginine-HCl, 50 mM Tris–HCl (pH 8.0), 1 mM EDTA, 3 mM reduced glutathione and 0.9 mM oxidized glutathione to a final protein concentration of 70 mg/l. After 72 h at 4°C, the mixture was dialysed against 50 mM MES (pH 6.0). Mindin-FL and mindin-FS were purified using Superdex S-75 and Mono Q columns (GE Healthcare). Mutants were produced and purified similarly to the wild-type protein. For the production of biotinylated mindin, a tag (GSLNDIFEAQKIEWHE) was added to the C termini of both mindin-FL and mindin-FS.
The FS domain of mindin contains only two methionine residues, at positions 57 and 81. To assure sufficient anomalous signal for MAD phasing, Leu215 was mutated to methionine. For selenomethionine (SeMet) labeling, mindin-FS was expressed in methionine-auxotroph E. coli B834(DE3) cells (Novagen) grown in SelenoMet medium (Athena Enzyme Systems).
Full-length mouse mindin containing a C-terminal Myc tag was produced in transfected HEK 293 cells as described (He et al, 2004). The protein was purified from culture supernatants using a monoclonal antibody to Myc (9E10) conjugated to Sepharose.
Crystallization and data collection
Crystals of mindin-FS (native or SeMet derivative) grew in hanging drops at room temperature in 0–6% (w/v) polyethylene glycol monomethyl ether 2000, 100 mM Tris–HCl (8.8–9.3) and 10 mM NiCl2. For data collection, crystals were transferred to a cryoprotectant solution (mother liquor containing 20% (v/v) glycerol), prior to flash cooling. A native X-ray diffraction data set was collected at 100 K with a Siemens HI-STAR area detector. MAD data from a SeMet crystal were collected to 2.04-Å resolution at beamline X29 of the Brookhaven National Synchrotron Light Source. All data were indexed, integrated and scaled with the HKL2000 program (Otwinowski and Minor, 1997). Data collection statistics are presented in Table I.
Structure determination and refinement
The mindin-FS structure was determined by MAD phasing with data collected from the SeMet derivative at three different wavelengths (0.9791, 0.9795 and 0.9611 Å). Six Se positions were determined using HKL2MAP (Pape and Schneider, 2004) or SOLVE (Terwilliger, 2003), corresponding to three Se atoms per molecule in the asymmetric unit, and giving initial phases to 2.5-Å resolution with an overall figure of merit of 0.51. Density modification and automatic model building were performed with RESOLVE (Terwilliger, 2003), resulting in models for the two molecules in the asymmetric unit that were approximately 60 and 40% complete. By combining the partial structures, 80% of the model could be built, with initial Rwork of 40.4% and Rfree of 42.6% at 2.1-Å resolution.
Model building and refinement were completed using the native mindin-FS data set. Refinement was carried out using CNS1.1 (Brünger et al, 1998), including iterative cycles of simulated annealing, positional refinement and B-factor refinement, interspersed with model rebuilding into σA-weighted Fo–Fc and 2Fo–Fc electron density maps using XtalView (McRee, 1999). The final model of mindin-FS contains residues 8–218 of one monomer and 8–222 of the other, 2 calcium ions, 6 nickel ions and 319 water molecules. Refinement statistics are summarized in Table I. Stereochemical parameters were evaluated with PROCHECK (Laskowski et al, 1993). Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession code 3D34.
Cell adhesion assay
Cell adhesion to mindin was carried out as described (Li and Zhang, 2003) with minor modifications. Twenty four-well non-tissue culture polystyrene plates were coated with 100 μl of 0.2 μM streptavidin at room temperature overnight. The coated plates were blocked sequentially with 200 μl of 1% bovine serum albumin (BSA) and 200 μl of 0.5% polyvinylpyrrolidone in Dulbecco's phosphate-buffered saline (DPBS) at room temperature for 1h each. Biotinylated mindin and its different mutants (0.8 μM) or control IgG were added and the plates were incubated at room temperature for 30 min. After washing the plates with DPBS, 2 × 106 HEK 293 cells expressing different integrin receptors or their mock-transfected controls in DPBS plus 1 mM Ca2+ and 1 mM Mg2+ were incubated with 100 nM NIF, 50 μg/ml M1/70 or IgG for 15 min at 4°C and then added to each well. After 30 min at 37°C, unbound cells were removed by three washes with DPBS and adherent cells were fixed with 200 μl of 4% PFA in DPBS. The number of adherent cells was quantified by staining the plates with 0.5% crystal violet and measuring absorption at 570 nm.
LPS-binding assay
LPS binding was measured as described (He et al, 2004) with minor modifications. Purified full-length mindin from HEK 293 cells, full-length mindin from E. coli, mindin FS domain alone from E. coli or control human serum albumin were coated on ELISA plates at 2 μg/ml in 100 μl in sodium bicarbonate buffer (pH 9.6) at 4°C overnight. After washing with 0.2% Tween in PBS, the wells were blocked with 3% BSA in PBS for 1 h at room temperature. Salmonella typhosa LPS (Sigma) at 10 μg/ml was added and incubated at room temperature for 1 h. Bound LPS was detected with mouse anti-LPS polyclonal antibody followed by horseradish peroxidase-conjugated goat anti-mouse IgG antibody (Jackson Laboratory). After washing, 3,3,5,5′-tetramethyl-benzidine was added; absorbance was measured at 450 nm.
Mass spectrometry
Affinity-purified mouse mindin from HEK 293 cells (20 μg) was reduced with 25 mM DTT for 1 h at 55°C and carboxyamidomethylated with 90 mM iodoacetamide in the dark for 45 min. The modified protein was dialysed with a 7.5-kDa cutoff membrane (Millipore) against nanopure water at 4°C overnight and dried in a SpeedVac concentrator. The dried mindin was resuspended in 50 mM ammonium bicarbonate and digested with endoproteinase Glu-C (Roche) at 25°C for 20 h. The protein was further digested with trypsin (Promega) at 37°C for 20 h. The Glu-C/tryptic peptides were resuspended with 19.5 μl of mobile phase A (0.1% formic acid in water) and 0.5 μl of mobile phase B (80% acetonitrile and 0.1% formic acid in water). The sample was loaded off-line onto a nanospray tapered capillary column/emitter (360 μm × 75 μm × 15 μm) and separated through a 160 min linear gradient of increasing mobile phase B at a flow rate of ∼200 nl/min directly into the mass spectrometer.
LC-MS/MS analysis was performed on a LTQ Orbitrap XL mass spectrometer (ThermoFisher) equipped with a nanospray ion source. A full FTMS spectrum at 60 000 resolution was collected at 400–2000 m/z, followed by three data-dependent MS/MS spectra of ITMS in the most intense ion peaks from parent mass list following CID. The parent masses calculated from the peptide sequence VSLW@SSWGLC#GGPC#GK allowed for alkylated cysteine (#=57.0215 Da) with and without mannosylated tryptophan (@=162.0528 Da).
The resulting data were searched against the mouse mindin sequence (SwissProt accession number Q8BMS2), including contaminant database using the TurboSequest algorithm (BioWorks 3.3.1 SP1; ThermoFisher). DTA files were generated for spectra with a threshold of 15 ions, a TIC of 1e3 and a range of MH+ 400–6000 m/z. The SEQUEST parameters were set to allow 30.0 p.p.m. of precursor ion mass tolerance and 0.5 Da of fragment ion tolerance with monoisotopic mass. Tryptic and Glu-C peptides were allowed with up to three missed internal cleavage sites and differential modifications were allowed for alkylated cysteine and mannosylated tryptophan.
Additional sequence mapping of mindin from HEK 293 cells was carried by subjecting an SDS–PAGE gel slice containing mindin to in-gel digestion with trypsin, chymotrypsin or both enzymes combined. The resulting peptides were analysed by LC-MS/MS (Midwest Bio Services).
Acknowledgments
This study was supported by National Institutes of Health Grants AI065612 (RAM), P01 HL54710 (LZ), and AI054658 and AI061364 (Y-WH). Support for the data collected at beamline X29 of the National Synchrotron Light Source comes from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health.
References
- Arnaout MA, Goodman SL, Xiong JP (2007) Structure and mechanics of integrin-based cell adhesion. Curr Opin Cell Biol 19: 495–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuti F, Hughes S, Walmsley M, Ruf S, Fetler L, Popoff M, Tybulewicz VL, Amigorena S (2004) Requirement of Rac1 and Rac2 expression by mature dendritic cells for T cell priming. Science 305: 1150–1153 [DOI] [PubMed] [Google Scholar]
- Burstyn-Cohen T, Tzarfaty V, Frumkin A, Feinstein Y, Stoeckli E, Klar A (1999) F-spondin is required for accurate pathfinding of commissural axons at the floor plate. Neuron 23: 233–246 [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography and NMR systems: a new software suite for macromolecular structure determination. Acta Crystallogr Sec D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Cho W, Stahelin RV (2005) Membrane–protein interactions in cell signaling and membrane trafficking. Annu Rev Biophys Biomol Struct 34: 119–151 [DOI] [PubMed] [Google Scholar]
- Doucey M-A, Hess D, Cacan R, Hofsteenge J (1998) Protein C-mannosylation is enzyme-catalyzed and uses dolichyl-phosphate-mannose as a precursor. Mol Biol Cell 9: 291–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein Y, Borrell V, Garcia C, Burstyn-Cohen T, Tzarfaty V, Frumkin A, Nose A, Okamoto H, Higashijima S, Soriano E, Klar A (1999) F-spondin and mindin: two structurally and functionally related genes expressed in the hippocampus that promote outgrowth of embryonic hippocampal neurons. Development 126: 3637–3648 [DOI] [PubMed] [Google Scholar]
- Gonzalez de Peredo A, Klein D, Macek B, Hess D, Peter-Katalinic J, Hofsteenge J (2002) C-mannosylation and O-fucosylation of thrombospondin type 1 repeats. Mol Cell Proteomics 1: 11–18 [DOI] [PubMed] [Google Scholar]
- He Y-W, Li H, Zhang J, Hsu C-L, Lin E, Zhang N, Guo J, Forbush KA, Bevan MJ (2004) The extracellular matrix protein mindin is a pattern-recognition molecule for microbial pathogens. Nat Immunol 5: 88–97 [DOI] [PubMed] [Google Scholar]
- Higashijima S, Nose A, Eguchi G, Hotta Y, Okamoto H (1997) Mindin/F-spondin family: novel ECM proteins expressed in the zebrafish embryonic axis. Dev Biol 192: 211–227 [DOI] [PubMed] [Google Scholar]
- Hofsteenge J, Huwiler KG, Macek B, Hess D, Lawler J, Mosher DF, Peter-Katalinic J (2001) C-mannosylation and O-fucosylation of the thrombospondin type 1 module. J Biol Chem 276: 6485–6498 [DOI] [PubMed] [Google Scholar]
- Humphries JD, Byron A, Humphries MJ (2006) Integrin ligands at a glance. J Cell Sci 119: 3901–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO (2002) Integrins: bidirectional, allosteric signaling machines. Cell 110: 673–687 [DOI] [PubMed] [Google Scholar]
- Jia W, Li H, He Y-W (2005) The extracellular matrix protein mindin serves as an integrin ligand and is critical for inflammatory cell recruitment. Blood 106: 3854–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W, Li H, He Y-W (2008) Pattern recognition molecule mindin promotes intranasal clearance of influenza viruses. J Immunol 180: 6255–6261 [DOI] [PubMed] [Google Scholar]
- Klar A, Baldassare M, Jessell TM (1992) F-spondin: a gene expressed at high levels in the floor plate encodes a secreted protein that promotes neural cell adhesion and neurite extension. Cell 69: 95–110 [DOI] [PubMed] [Google Scholar]
- Kreis T, Vale R (eds) (1999) Guidebook to the Extracellular Matrix, Anchor, and Adhesion Proteins, 2nd edn Oxford University Press: Oxford [Google Scholar]
- Lasarte JJ, Casares N, Gorraiz M, Hervás-Stubbs S, Arribillaga L, Mansilla C, Durantez M, Llopiz D, Sarobe P, Borrás-Cuesta F, Prieto J, Leclerc C (2007) The extra domain A from fibronectin targets antigens to TLR4-expressing cells and induces cytotoxic T cell responses in vivo. J Immunol 178: 748–756 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereo chemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Li H, Oliver T, Jia W, He Y-W (2006) Efficient dendritic cell priming of T lymphocytes depends on the extracellular protein mindin. EMBO J 25: 4097–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang L (2003) The fourth blade within the β-propeller is involved specifically in C3bi recognition by integrin αMβ2. J Biol Chem 278: 34395–34402 [DOI] [PubMed] [Google Scholar]
- Manda R, Kohno T, Matsuno Y, Takenoshita S, Kuwano H, Yokota J (1999) Identification of genes (SPON2 and C20orf2) differentially expressed between cancerous and noncancerous lung cells by mRNA differential display. Genomics 61: 5–14 [DOI] [PubMed] [Google Scholar]
- McDonald C, Núñez G (2004) Mindin the fort. Nat Immunol 5: 16–18 [DOI] [PubMed] [Google Scholar]
- McRee DE (1999) XtalView/Xfit–a versatile program for manipulating atomic coordinates and electron density. J Struct Biol 125: 156–165 [DOI] [PubMed] [Google Scholar]
- Muchowski PJ, Zhang L, Chang ER, Soule HR, Plow EF, Moyle M (1994) Functional interaction between the integrin antagonist neutrophil inhibitory factor and the I domain of CD11b/CD18. J Biol Chem 269: 26419–26423 [PubMed] [Google Scholar]
- Muroi E, Manabe S, Ikezaki M, Urata Y, Sato S, Kondo T, Ito Y, Ihara Y (2007) C-mannosylated peptides derived from the thrombospondin type 1 repeat enhance lipopolysaccharide-induced signaling in macrophage-like RAW264.7 cells. Glycobiology 17: 1015–1028 [DOI] [PubMed] [Google Scholar]
- Nalefski EA, Falke JJ (1996) The C2 domain calcium-binding motif: structural and functional diversity. Protein Sci 5: 2375–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11: 281–296 [DOI] [PubMed] [Google Scholar]
- Okamura Y, Watari M, Jerud ES, Young DW, Ishizaka ST, Rose J, Chow JC, Strauss JF III (2001) The extra domain A of fibronectin activates Toll-like receptor 4. J Biol Chem 276: 10229–10233 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Pääkkönen K, Tossavainen H, Permi P, Rakkolainen H, Rauvala H, Raulo E, Kilpeläinen I., Güntert P (2006) Solution structures of the first and fourth TSR domains of F-spondin. Proteins 64: 665–672 [DOI] [PubMed] [Google Scholar]
- Pape T, Schneider TR (2004) HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J Appl Cryst 37: 843–844 [Google Scholar]
- Patti JM, Allen B, McGavin MJ, Hook M (1994) MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol 48: 585–617 [DOI] [PubMed] [Google Scholar]
- Rizo J, Südhof TC (1998) C2-domains, structure and function of a universal Ca2+-binding domain. J Biol Chem 273: 15879–15882 [DOI] [PubMed] [Google Scholar]
- Shimaoka M, Springer TA (2003) Therapeutic antagonists and conformational regulation of integrin function. Nat Rev Drug Disc 2: 703–715 [DOI] [PubMed] [Google Scholar]
- Shimaoka M, Xiao T, Liu JH, Yang Y, Dong Y, Jun CD, McCormack A, Zhang R, Joachimiak A, Takagi J, Wang JH, Springer TA (2003) Structures of the αL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation. Cell 112: 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G, Yang Y, Liu J, Casasnovas JM, Shimaoka M, Springer TA, Wang J-H (2005) An atomic resolution view of ICAM recognition in a complex between the binding domains of ICAM-3 and integrin αLβ2. Proc Natl Acad Sci USA 102: 3366–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strynadka NCJ, James MNG (1989) Crystal structures of the helix-loop-helix calcium-binding proteins. Annu Rev Biochem 58: 951–998 [DOI] [PubMed] [Google Scholar]
- Tan K, Duquette M, Liu J, Dong Y, Zhang R, Joachimiak A, Lawler J, Wang J-H (2002) Crystal structure of the TSP-1 type 1 repeats: a novel layered fold and its biological implication. J Cell Biol 159: 373–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC (2003) SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol 374: 22–37 [DOI] [PubMed] [Google Scholar]
- Tucker RP (2004) The thrombospondin type 1 repeat family. Int J Biochem Cell Biol 36: 969–974 [DOI] [PubMed] [Google Scholar]
- Tzarfaty-Majar V, Burstyn-Cohen T, Klar A (2001) F-spondin is a contact-repellent molecule for embryonic motor neurons. Proc Natl Acad Sci USA 98: 4722–4727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemiya T, Takeichi M, Nose A (1997) M-spondin, a novel ECM protein highly homologous to vertebrate F-spondin, is located at the muscle attachment sites in the Drosophila embryo. Dev Biol 186: 165–176 [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M et al. (2001) The sequence of the human genome. Science 291: 1304–1351 [DOI] [PubMed] [Google Scholar]
- Walker EH, Perisic O, Ried C, Stephens L, Williams RL (1999) Structural insights into phosphoinositide 3-kinase catalysis and signaling. Nature 402: 313–320 [DOI] [PubMed] [Google Scholar]
- Zhang L, Plow EF (1997) Identification and reconstruction of the binding site within αMβ2 for a specific and high affinity ligand, NIF. J Biol Chem 272: 17558–17564 [DOI] [PubMed] [Google Scholar]
- Zisman S, Marom K, Avraham O, Rinsky-Halivni L, Gai U, Kligun G, Tzarfaty-Majar V, Suzuki T, Klar A (2007) Proteolysis and membrane capture of F-spondin generates combinatorial guidance clues from a single molecule. J Cell Biol 178: 1237–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]