Summary
The homeodomain transcription factor Nkx2-1 plays key roles in the developing telencephalon, where it regulates the identity of progenitor cells in the medial ganglionic eminence (MGE) and mediates the specification of several classes of GABAergic and cholinergic neurons. Here we have investigated the postmitotic function of Nkx2-1 in the migration of interneurons originating in the MGE. Experimental manipulations and mouse genetics show that downregulation of Nkx2-1 expression in postmitotic cells is necessary for the migration of interneurons to the cortex, whereas maintenance of Nkx2-1 expression is required for interneuron migration to the striatum. Nkx2-1 exerts this role in the migration of MGE-derived interneurons by directly regulating the expression of a guidance receptor, Neuropilin-2, which enables interneurons to invade the developing striatum. Our results demonstrate a novel role for the cell-fate determinant Nkx2-1 in regulating neuronal migration by direct transcriptional regulation of guidance receptors in postmitotic cells.
Introduction
During development of the nervous system, migrating neurons and axons are guided to their final destination by the coordinated activity of multiple extracellular cues that act on specific membrane receptors to steer their movement in the right direction (Dickson, 2002; Tessier-Lavigne and Goodman, 1996). Because neuronal guidance typically involves a complex set of instructions even in the simplest organisms, adoption of a specific program of migration requires that neurons respond to guidance cues in a highly regulated pattern, both in time and space. Transcriptional regulation is a key determinant in this process, as it ultimately represents one of the primary mechanisms controlling the repertoire of receptors expressed by neurons (Butler and Tear, 2007; Polleux et al., 2007).
Transcription factors regulating neuronal fate specification typically function before the last division of progenitor cells, while guidance decisions are made at later stages of development (Jessell, 2000). Thus, it is likely that transcription factors expressed in postmitotic neurons are responsible for activating specific migration and axon guidance programs. In the vertebrate spinal cord, for example, the expression of a specific combination of transcription factors in postmitotic motoneurons appears to encode their axon trajectory and final targeting (Kania et al., 2000; Sharma et al., 2000; Sharma et al., 1998; Thaler et al., 2004). Interestingly, some of the same transcription factors that regulate axon guidance also play a major role in the early specification of different neuronal pools, suggesting that the same transcription factors may carry on different functions depending on the cellular context (Garcia and Jessell, 2008; Müller et al., 2003; Shirasaki and Pfaff, 2002). In addition to demonstrating the function of transcription factors in the regulation of axon guidance, studies over the past few years have begun to identify possible candidate genes that would function as downstream effectors of these factors. For example, it has been suggested that Lim1 expression in LMC neurons may regulate the expression of the receptor tyrosine kinase EphA4, which is essential for the final targeting of their axonal projections to the limb mesenchyma (Kania and Jessell, 2003). Similarly, the role of the transcription factor Zic2 in regulating midline crossing by retinal axons appears to involve the regulation of another member of the Eph family, EphB1 (García-Frigola et al., 2008; Lee et al., 2008; Williams et al., 2004).
Since migrating neurons and growing axons are instructed towards their final destination by similar guidance molecules (Bagri and Tessier-Lavigne, 2002; Brose and Tessier-Lavigne, 2000), it seems conceivable that equivalent mechanisms regulate the expression of guidance receptors in both migrating neurons and axons. However, the function of specific transcription factors in the migration and positioning of neurons is still very limited (McEvilly et al., 2002; Sugitani et al., 2002; Ge et al., 2006; Hand et al., 2005; Le et al., 2007). In the developing telencephalon, the medial ganglionic eminence (MGE) is the origin of interneurons that migrate tangentially to the striatum and cerebral cortex (Lavdas et al., 1999; Marín et al., 2000; Sussel et al., 1999; Wichterle et al., 1999; Wichterle et al., 2001). We have previously shown that sorting of striatal and cortical interneurons to their respective target territories depends on neuropilin/semaphorin interactions (Marín et al., 2001). Here we have investigated the transcriptional mechanisms regulating this process. We found that downregulation of postmitotic Nkx2-1 expression is a necessary event for the migration of interneurons to the cortex, whereas Nkx2-1 expression is required for interneuron migration to the striatum. Forced Nkx2-1 expression in MGE-derived cells prevents interneuron migration to the cortex, whereas loss of Nkx2-1 function reduces the number of interneurons that accumulate in the striatum. Nkx2-1 exerts this role in the migration of MGE-derived interneurons by directly regulating the expression of Neuropilin2 (Nrp2), the receptor of Semaphorin-3F (Sema3F). Our results demonstrate that direct transcriptional regulation of guidance receptors in postmitotic neurons is an essential mechanism in neuronal migration.
Results
Interneurons migrating to the cortex rapidly down-regulate Nkx2-1 expression
Nkx2-1 is one of the earliest genes expressed in the mouse forebrain (Sussel et al., 1999). At embryonic day (E) 13.5, the peak of interneuron generation in the mouse, Nkx2-1 is strongly expressed by cells in several progenitor domains of the subpallium, including the MGE, the preoptic area and part of the septum, whereas it is absent from the lateral ganglionic eminence (Figure 1A). At this stage, Nkx2-1 expression was also observed in many postmitotic neurons derived from the MGE, such as striatal interneurons (Figures 1A and 1C) (Marín et al., 2000). In contrast, Nkx2-1 expression was not detected in cortical interneurons at this or any other embryonic stages (Figure 1A and 1B, and data not shown), even though many of them originate in the MGE (Lavdas et al., 1999; Sussel et al., 1999; Wichterle et al., 1999; Wichterle et al., 2001). There are two possible hypotheses that would explain the differential expression of Nkx2-1 in postmitotic striatal and cortical interneurons. One possibility is that MGE progenitors producing cortical interneurons never express Nkx2-1. Alternatively, Nkx2-1 might be expressed by MGE progenitors giving rise to both striatal and cortical interneurons, but this latter population would rapidly down-regulate Nkx2-1 expression soon after leaving the MGE. To test these alternative hypotheses, we first quantified the percentage of cells expressing Nkx2-1 located within 100 µm of the ventricle in the MGE of E13.5 embryos. Virtually all cells present in the ventricular zone (VZ) of the MGE expressed Nkx2-1 at this stage (Figures 1D and 1D’; 99.2 ± 0.4% of Propidium Iodine cells, average ± s.e.m.). We next quantified the number of progenitor cells expressing Nkx2-1 in the VZ of the MGE in E13.5 embryos. In agreement with previous reports (Xu et al., 2005), we found that almost every cell in the MGE that incorporated the S-phase marker BrdU at this stage also expressed Nkx2-1 (Figures 1E and 1E’, 96.9 ± 1.1% of BrdU cells, average ± s.e.m.), reinforcing the view that all MGE progenitors express this transcription factor.
Figure 1. Cortical interneurons downregulate Nkx2-1 protein expression after leaving the MGE progenitor zone.

(A) Coronal section through the brain of an E13.5 mouse embryo showing Nkx2-1 protein expression in the medial ganglionic eminence (MGE), striatum (Str), globus pallidus (GP), preoptic area (POA) and septum (Se).
(B and C) Higher magnification images of the areas boxed in (A).
(D–E’) MGE ventricular zone (VZ) from an E13.5 embryo depicting MGE cells stained with Nkx2-1 and propidium iodine (PI) (D–D’), and MGE progenitors stained S-phase marker BrdU and Nkx2-1 (E–E’).
(F–H) Expression of Nkx2-1 protein in GFP-expressing cells after transplantation of GFP-expressing progenitors in the MGE. As illustrated in the schematic diagram (F’) and the high magnification images (G and H), the majority of migrating cells had undetectable levels of Nkx2-1 (green dots), many cells expressed low levels of Nkx2-1 (yellow dots with green circle), and a small number of cells expressed high levels of Nkx2-1 protein (yellow dots). The numbers depicted in the schematic (F’) describe the location of cells shown in (G) and (H).
H, hippocampus; LGE, lateral ganglionic eminence; LV, lateral ventricle; NCx, neocortex. Asterisk, transplant.
Scale bars equal 200 µm (A and F), 50 µm (B, C, G and H), and 10 µm (D–E’).
To specifically test whether MGE-derived interneurons down-regulate Nkx2-1 expression while migrating towards the cortex, we performed experiments in which a piece of E13.5 MGE VZ from a green fluorescence protein (GFP)-expressing transgenic mouse brain was transplanted into the same region of wild-type host slices. After 18 h in culture, we analyzed the expression of Nkx2-1 in GFP migrating cells (Figures 1F and 1F’; n = 3 experiments, 132 GFP cells analyzed). The majority of GFP-expressing cells derived from the transplant were found in a corridor deep to the striatal mantle through which most interneurons reach the cortex (Flames et al., 2004), and they did not contain detectable levels of Nkx2-1 protein (Figures 1F’ and 1G; 67.0 ± 14.3% of GFP cells, average ± s.e.m.). About 22% of the GFP-expressing cells were also located in the corridor and contained traces of Nkx2-1 (Figures 1F’ and 1G; 22.2 ± 14.8% of GFP cells, average ± s.e.m.), suggesting that these cells might be undergoing down regulation of this protein. Finally, a small proportion of GFP-expressing cells was located in the ventrolateral aspect of the prospective striatum and expressed high levels of Nkx2-1 protein (Figures 1F’ and 1H; 10.9 ± 4.3% of GFP cells, average ± s.e.m.). These experiments suggest that MGE-derived interneurons migrating towards the cortex rapidly down regulate the expression of Nkx2-1 after leaving the MGE, whereas those interneurons migrating to the striatum maintain the expression of this transcription factor.
Nkx2-1 expression prevents the migration of MGE-derived cells towards the cortex
Our previous observations raised the intriguing possibility that down regulation of Nkx2-1 in MGE-derived interneurons might be necessary to acquire a cortical migratory fate. To test this hypothesis, we forced the expression of Nkx2-1 in MGE progenitor cells through focal electroporation in E13.5 slices (Figure 2A). In control experiments, Gfp-expressing cells migrated to the cortex following their normal route (> 90 cells per cortex in 22/22 slices; Figures 2C and 2H), and only a minority of cells was found in the striatum. In contrast, in slices co-electroporated with Gfp and Nkx2-1, the majority of MGE-derived cells accumulated in the basal ganglia, and only occasional cells were found in the cortex (< 20 cells per cortex in 22/22 slices; Figures 2D and 2H). To exclude the possibility that overexpression of Nkx2-1 prevents interneuron migration rather than changing their direction, we electroporated small MGE explants with either Gfp alone or Gfp and Nkx2-1 and cultured them in three-dimensional matrigel matrices. After 36 h in culture, cells were found to migrate a similar distance in the two experimental conditions (data not shown; see also Figures 6B and 6D), suggesting that expression of Nkx2-1 in postmitotic interneurons does not impair cell migration but appears to specifically disrupt their target selection.
Figure 2. Nkx2-1 overexpression in MGE-derived interneurons prevents their migration to the cortex.

(A and B) Schematic diagrams of the focal electroporation experiment and the Nkx2-1 (372 amino acids) constructs used in these experiments.
(C–G) Migration of MGE-derived cells electroporated with Gfp (C) or with Gfp and Nkx2-1 (D), Nkx2-1ΔTN (E), Nkx2-1ΔCt (F) or Nkx2-1A35T (G). Arrowheads point to cells that have reached the cortex. Dotted lines indicate the limits of the organotypic slices.
(H) Schematic representation of the migratory routes adopted by MGE-derived cells electroporated with Gfp and Gfp + Nkx2-1A35T (green arrow) or with Gfp Nkx2-1, Nkx2-1ΔTN or Nkx2-1ΔCt (black dotted arrow).
GP, globus pallidus; H, hippocampus; HD, homeodomain; LGE, lateral ganglionic eminence; MGE, medial ganglionic eminence; NCx, neocortex; NK2-SD, NK2 specific domain; PCx, piriform cortex; POA, preoptic area; Str, striatum; TN, Tinman motif.
Scale bar equals 200 µm.
Nkx2-1 belongs to the NK-2 class of homeodomain (HD) transcription factors (Harvey, 1996). In addition to the HD, two other peptide domains are conserved within the NK-2 class of transcription factors. The short TN domain (Tinmam motif), located at the N-terminal region, has been suggested to underlie the function of the NK-2 proteins as repressors during neural patterning events in the ventral neural tube (Muhr et al., 2001). The function of the NK2 specific domain (NK2-SD) is not fully understood. In Nkx2-2, NK2-SD has been shown to act as an intra-molecular inhibitor of a transcriptional activator domain located at the C-terminus of the protein (Watada et al., 2000). To investigate the mechanism through which Nkx2-1 may regulate the migration of MGE-derived cells, we performed a structure-function analysis in organotypic slices. We first evaluated the role of the TN motif by electroporating a truncated form of Nkx2-1 missing the first 20 amino acids (Nkx2-1ΔTN; Figure 2B). Analysis of slices electroporated with Gfp and Nkx2-1ΔTN revealed that MGE-derived cells also failed to reach the cortex in the absence of the TN motif (< 20 cells per cortex in 17/17 slices; Figures 2E and 2H). To investigate if a putative C-terminus activator domain plays a role in the migration of MGE-derived cells, we used a truncated form of Nkx2-1 missing 4 amino acids of the NK2-SD domain and the remaining C-terminal of the protein (Nkx2-1ΔCt; Figure 2B). Analysis of slices electroporated with Gfp and Nkx2-1ΔCt showed that MGE-derived cells also failed to reach the cortex in the absence of the C-terminus activator domain and an intact NK-SD motif (< 20 cells per cortex in 11/11 slices; Figures 2F and 2H). Finally, we assessed the role of the HD in this process by performing a single amino acid substitution in the position 35 of the Nkx2-1 HD (Nkx2-1A35T; Figure 2B). This mutated form of Nkx2-1 binds to DNA target sequences with 50-fold less affinity than wild type Nkx2-1 (Xiang et al., 1998). Analysis of slices electroporated with Gfp and Nkx2-1A35T revealed that MGE-derived cells expressing this construct were able to migrate towards the cortex like in control slices (> 90 cells per cortex in 14/15 slices; Figures 2G and 2H). Similar results were obtained when a different point mutation in the HD (Nkx2-1Y54M) was used (> 90 cells per cortex in 18/19 slices; data not shown). Of note, the A35T replacement does not appear to decrease the stability of the protein (which would have explained the absence of a phenotype), since Gfp/Nkx2-1A35T-expressing cells migrating towards the cortex have detectable levels of Nkx2-1 (Figure S1). Altogether, these experiments revealed that the mechanism through which Nkx2-1 prevents MGE-derived cells from migrating towards the cortex requires an intact HD and does not rely on interactions involving the TN and the C-terminus activator/SD domains.
Loss of postmitotic Nkx2-1 function decreases the number of striatal interneurons
If Nkx2-1 regulates the sorting of cortical and striatal interneurons, loss of Nkx2-1 function in postmitotic cells should lead to a reduction in the number of MGE-derived cells that accumulate in the striatum. To test this hypothesis, we bred mice carrying floxed alleles of the Nkx2-1 locus (Kusakabe et al., 2006) with transgenic mice in which Cre recombinase is expressed under the control of Lhx6 (Fogarty et al., 2007), a LIM-HD transcription factor expressed by MGE-derived neurons that drives recombination almost exclusively in postmitotic cells (Figure S2). To verify the efficiency and specificity of recombination, we analyzed the distribution of cells expressing Nkx2-1 protein in the subpallium of E15.5 control and Lhx6-Cre;Nkx2-1Fl/Fl mutant embryos. As expected, we found that expression of Nkx2-1 in the MGE VZ was not affected whereas the number of Nkx2-1 expressing cells throughout the subpallial mantle, including the developing striatum, was dramatically reduced in Lhx6-Cre;Nkx2-1Fl/Fl mutant embryos (Figure S3). The presence of Nkx2-1 mRNA in some striatal cells suggests that either they have yet to recombine the two Nkx2-1 alleles, or that a fraction of striatal interneurons may not express Lhx6 or, at least, the Lhx6-Cre transgene (Figure S3).
To assess the impact of postmitotic loss of Nkx2-1 function in the development of striatal interneurons, we analyzed the expression of several markers for striatal interneurons, such as Lhx6, Lhx7 (also known as Lhx8), Er81 and Somatostatin (Sst) (Kawaguchi et al., 1995; Marín et al., 2000; Stenman et al., 2003). We found that the striatum of E15.5 Lhx6-Cre;Nkx2-1Fl/Fl mutant embryos contained significantly fewer Lhx6-, Lhx7-, Er81- and Sst-expressing neurons than controls (n = 3; Figure 3). We next analyzed the distribution of interneurons in the striatum of postnatal day 25 control and Lhx6-Cre;Nkx2-1Fl/Fl mutant mice. Quantification of the number of neurons expressing Choline acetyltranferase (ChAT), Parvalbumin (PV) and SST, markers of the three main classes of mature striatal interneurons (Kawaguchi et al., 1995), revealed that the striatum of P25 Lhx6-Cre;Nkx2-1Fl/Fl mutant mice contained significantly fewer ChAT- and PV-expressing interneurons than controls (n = 3; Figures 4G–I), while the number of SST-expressing cells did not differ (n = 3; Figure 4).
Figure 3. Reduced numbers of striatal interneurons after postmitotic loss of Nkx2-1function.
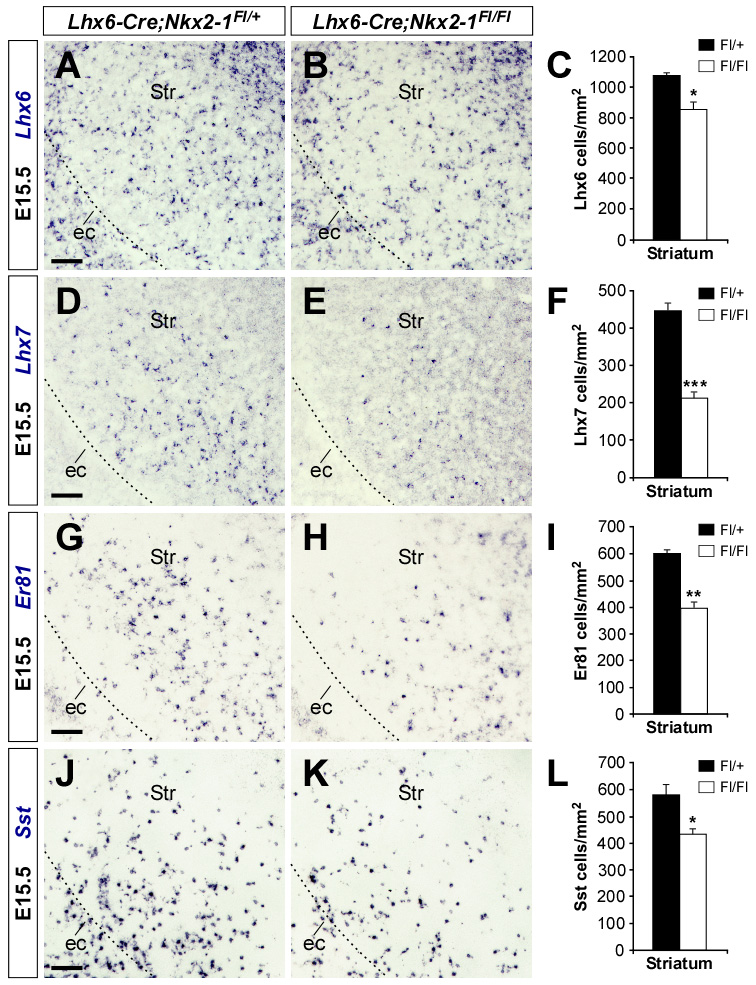
(A, B, D, E, G, H, J and K) Coronal sections through the telencephalon of E15.5 control (A, D, G and J) and Lhx6-Cre;Nkx2-1Fl/Fl mutant (B , E, H and K) embryos showing Lhx6 (A and B), Lhx7 (D and E), Er81 (G and H) and Sst (J and K) mRNA expression. (C, F, I and L) Quantification of the number of Lhx6, Lhx7, Er81 and Sst-expressing cells in the striatum of E15.5 control and Lhx6-Cre;Nkx2-1Fl/Fl mutant embryos. Histograms show average ± s.e.m. 1083.96 ± 23.47 (Lhx6 control); 862.07 ± 49.01 (Lhx6 mutant); 452.32 ± 21.78 (Lhx7 control); 212.02 ± 18.68 (Lhx7 mutant); 601.60 ± 12.74 (Er81 control); 397.05 ± 23.84 (Er81 mutant); 579.03 ± 39.17 (Sst control); 432.18 ± 20.03 (Sst mutant). *** p < 0.001, ** p < 0.01 and * p < 0.05, t-test.
ec, external capsule; Str, striatum.
Scale bar equals 100 µm.
Figure 4. Loss of Nkx2-1 function decreases the number of interneurons in the postnatal striatum.
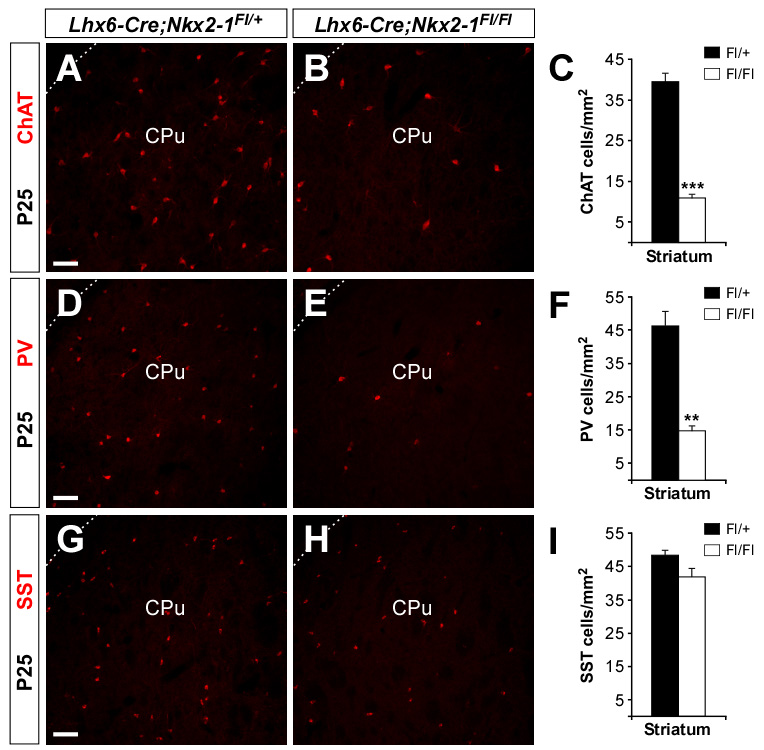
(A, B, D, E, G and H) Coronal sections through the striatum of P25 control (A, D and G) and Lhx6-Cre;Nkx2-1Fl/Fl mutant (B , E and H) mice showing ChAT (A and B), PV (D and E) and SST (G and H) expression.
(C, F and I) Quantification of the number of ChAT, PV and SST-expressing cells in the striatum of P25 control and Lhx6-Cre;Nkx2-1Fl/Fl mutant mice. Histograms show average ± s.e.m. 39.42 ± 2.26 (ChAT control); 10.94 ± 1.04 (ChAT mutant); 46.32 ± 4.52 (PV control); 14.71 ± 1.77 (PV mutant); 48.23 ± 1.65 (SST control); 41.73 ± 2.81 (SST mutant). *** p < 0.001 and ** p < 0.01, t-test.
CPu, caudate putamen.
Scale bar equals 100 µm.
The previous results were consistent with the hypothesis that loss of Nkx2-1 function prevents the migration of MGE-derived interneurons to the striatum. There were, however, alternative possibilities to explain these results. For example, loss of Nkx2-1 could lead to increased cell death in striatal interneurons. Quantification of the density of apoptotic cells (marked by cleaved Caspase3 expression) revealed no differences in the MGE and striatum of E13.5 control and Lhx6-Cre;Nkx2-1Fl/Fl mutant embryos (n = 3; MGE: 77.2 ± 3.3% [control] and 70.2 ± 4.4% [mutant]; striatum: 49.0 ± 0.7% [control] and 45.5 ± 5.7% [mutant]; average ± s.e.m). Similar results were observed at E12.5 and E14.5, data not shown), suggesting that this was not the case.
Nkx2-1 could also be required to induce or maintain the expression of striatal interneuron markers, and so the observed decrease in the number of striatal interneurons could merely reflect a failure in the expression of those genes. This seems unlikely, because similar numbers of Lhx6 and Lhx7-expressing cells were found throughout the telencephalon outside the striatum in control and mutant mice (Figure S4). However, to completely rule out this possibility, we performed two additional series of experiments. First, we bred a Cre-reporter line (Rosa-LoxP-STOP-LoxP-YFP, also known as Rosa-EYFP mice; Srinivas et al., 2001) into the mutant background to obtain Lhx6-Cre;Nkx2-1Fl/+;Rosa-YFP (control) and Lhx6-Cre;Nkx2-1Fl/Fl;Rosa-YFP (mutant) embryos. In these mice, Cre-mediated recombination in Lhx6-expressing cells leads to their permanent labeling with EYFP through the ubiquitous Rosa promoter. Consistent with the hypothesis that MGE-derived interneurons fail to migrate to the striatum in the absence of Nkx2-1, the striatum of Lhx6-Cre;Nkx2-1Fl/Fl;Rosa-YFP mutants at P0 contained fewer YFP cells than controls (n = 3; Figure 5A–C). This deficit was readily detectable as early as E13.5 (n = 3; Figures S5).
Figure 5. Tracing experiments reveal less interneurons invading the striatum after postmitotic loss of Nkx2-1 function.
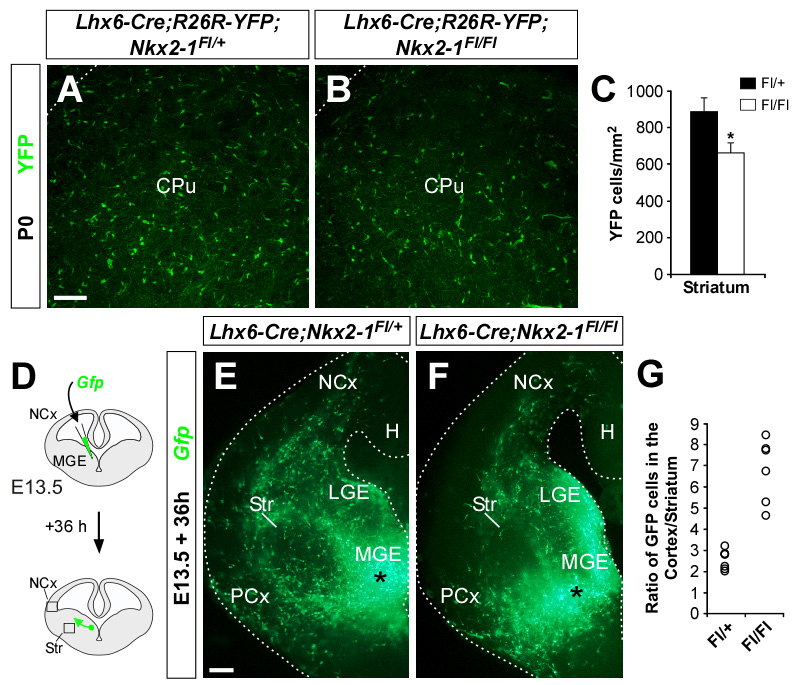
(A and B) Coronal sections through the striatum of P0 Lhx6-Cre;Nkx2-1Fl/+;Rosa-YFP control (A) and Lhx6-Cre;Nkx2-1Fl/Fl;Rosa-YFP mutant (B) mice showing YFP expression. YFP is also detected in scattered blood vessels, as previously reported (Fogarty et al., 2007).
(C) Quantification of the number of YFP-expressing cells in the striatum of P0 Lhx6-Cre;Nkx2-1Fl/+;Rosa-YFP control and Lhx6-Cre;Nkx2-1Fl/Fl;Rosa-YFP mutant mice. Histograms show average ± s.e.m. 889.31 ± 79.03 (YFP control); 664.12 ± 58.40 (YFP mutant). * p < 0.05, t-test.
(D) Schematic diagram of the focal electroporation experiment. The number of Gfp-expressing cells was counted in a fixed volume of the cortex and striatum (black boxes) and the ratio between these populations was determined for each slice
(E–F) Migration of MGE-derived cells in E13.5 Lhx6-Cre;Nkx2-1Fl/+;Rosa-YFP control (E) and Lhx6-Cre;Nkx2-1Fl/Fl;Rosa-YFP mutant (F) slices. Occasionally, Gfp-expressing cells accumulated in the piriform cortex of mutant slices. Dotted lines indicate the limits of the organotypic slices.
(G) Ratio of Gfp-expressing cells in the quantified region of the cortex and striatum for each individual E13.5 Lhx6-Cre;Nkx2-1Fl/+;Rosa-YFP control and Lhx6-Cre;Nkx2-1Fl/Fl;Rosa-YFP mutant slices. Average ± s.e.m. 2.55 ± 0.41 (control); 6.80 ± 1.22 (mutant). *** p < 0.001, t-test. Number of Gfp-expressing cells in the striatum (30.38 ± 4.03, control; 12.20 ± 3.80, mutant. *** p < 0.001, t-test) and cortex (76.80 ± 12.76, control; 84.20 ± 21.20, mutant) of electroporated slices.
CPu, caudate putamen; H, hippocampus; LGE, lateral ganglionic eminence; MGE, medial ganglionic eminence; NCx, neocortex; PCx, piriform cortex; Str, striatum.
Scale bar equals 100 µm (A and B) and00 µm (E and F).
In a second series of experiments, we directly assessed the migration of MGE-derived interneurons by focally electroporating a plasmid encoding Gfp (Figure 5D). In control experiments, the ratio between the number of Gfp-expressing cells in an equal volume of cortex and striatum was approximately 3:1 (n = 6 slices; Figure 5E and 5G). In contrast, in slices obtained from Lhx6-Cre;Nkx2-1Fl/Fl mutant embryos, this proportion increased to approximately 7:1 due to a reduction in the number of Gfp-cells that migrated to the striatum, while the number of cortical Gfp-cells remained similar to controls (n = 6 slices; Figure 5F and 5G). In conclusion, our experiments demonstrate that postmitotic Nkx2-1 function is required for the migration of MGE-derived interneurons to the developing striatum.
Nkx2-1 expression in postmitotic cells suppresses Sema3A/3F-mediated repulsion of MGE-derived interneurons
Nkx2-1 might control the sorting of MGE-derived cortical and striatal interneurons through regulating the expression of specific receptors for guidance factors in those cells. Our previous work has shown that Neuropilin-1 (Nrp1) and Neuropilin-2 (Nrp2), the binding receptors for the repulsive molecules Semaphorin 3A (Sema3A) and Semaphorin 3F (Sema3F), respectively, are expressed by MGE-derived cortical interneurons, but not by striatal interneurons (Marín et al., 2001). Because the developing striatum expresses both Sema3A and Sema3F, expression of Nrp1 and Nrp2 in cortical interneurons prevents their entry into the striatum, channeling them towards the cortex (Marín et al., 2001). We thus hypothesized that postmitotic Nkx2-1 may participate in the sorting of cortical and striatal interneurons by regulating the expression of the receptors for Sema3A and/or Sema3F. To test this idea, we electroporated E13.5 MGE explants with plasmids encoding Gfp or Nkx2-1-IRES-Gfp and cultured them along with aggregates of COS cells expressing either DsRed or DsRed and Sema3A/3F in matrigel matrices (Figure 6A). As expected from our previous work (Marín et al., 2001), we found that Sema3A/3F exerted a potent chemorepulsive effect over Gfp-expressing MGE-derived cells (n = 12; Figures 6B, 6C and 6H). In contrast, Sema3A/3F-expressing COS cells did not repel MGE-derived cells expressing Nkx2-1 (n = 12; Figures 6D, 6E and 6H). To determine if the Nkx2-1 HD mediates the suppression of Sema3A/3F-mediated chemorepulsion, we confronted Nkx2-1A35T-electroporated MGE explants with COS cells aggregates. As in controls, MGE-derived cells expressing the HD mutation Nkx2-1A35T were repelled by COS cells expressing Sema3A/3F (n = 11; Figures 6F, 6G and 6H). In sum, these results demonstrate that expression of Nkx2-1 in migrating MGE-derived cells renders them insensitive to Sema3A/3F chemorepulsion through an Nkx2-1 HD-dependent mechanism.
Figure 6. Postmitotic Nkx2-1 expression suppresses Sema3A/3F-mediated repulsion in MGE-derived interneurons.
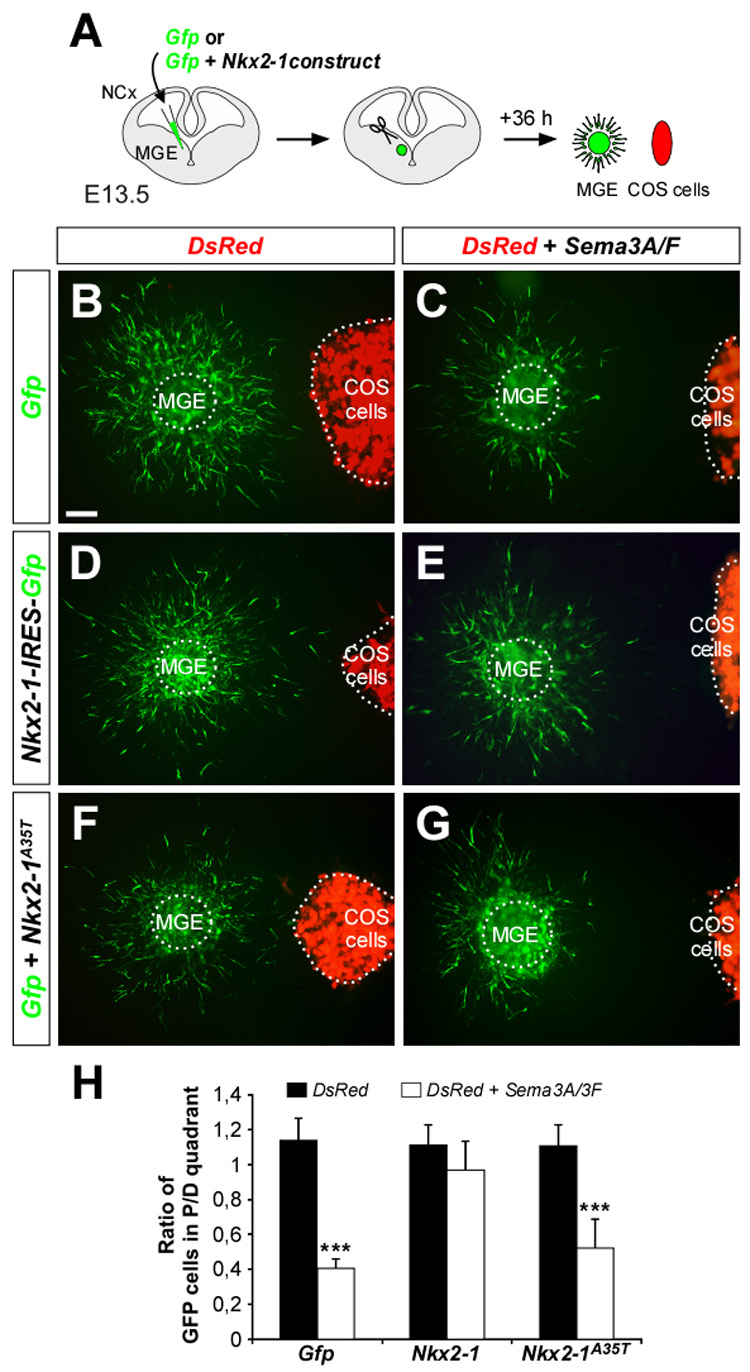
(A) Schematic diagram of the experimental paradigm used in MGE/COS co-culture confrontation assays. E13.5 slices were focally electroporated with Gfp, Nkx2-1-IRES-Gfp or Gfp + Nkx2-1A35T. MGE explants were dissected from the electroporated region, confronted to DsRed or DsRed + Sema3A/3F-transfected COS cells aggregates and cultured in Matrigel® matrices.
(B–G) Migration of Gfp (B and C) Nkx2-1-IRES-Gfp (D and E), and Gfp + Nkx2-1A35T (F and G) electroporated MGE-derived cells in response to mock-transfected (B, D and F) or Sema3A/3F-transfected (C, E and G) COS cells aggregates.
(H) Quantification of co-culture confrontation assays. P and D, proximal and distal quadrants, respectively. Histograms show average ± s.e.m. 1.13 ± 0.12 (Gfp MGE cells, mock-COS cells); 0.40 ± 0.05 (Gfp MGE cells, Sema3A/3F- COS cells); 1.11 ± 0.12 (Nkx2-1IRES-Gfp MGE cells, mock-COS cells); 0.97 ± 0.16 (Nkx2-1IRES-Gfp MGE cells, Sema3A/3F -COS cells); 1.10 ± 0.12 (Gfp + Nkx2-1A35T MGE cells, mock-COS cells); 0.52 ± 0.17 (Gfp + Nkx2-1A35T MGE cells, Sema3A/3F -COS cells). *** p < 0.001, t-test.
MGE, medial ganglionic eminence; NCx, neocortex.
Scale bar equals 50 µm.
Nkx2-1 represses Nrp2 expression in migrating MGE-derived interneurons
The previous results are consistent with the hypothesis that Nkx2-1 represses the expression of receptors for Sema3A and/or Sema3F in MGE-derived migrating neurons. To directly test this, we developed an in vitro assay in which we could specifically isolate RNA from migrating MGE-derived neurons (Figure 7A). MGE explants were electroporated with Gfp or Nkx2-1-IRES-Gfp plasmids and migrating cells were collected after 48 h in culture. A limitation of this experimental approach is that, although the population of cells is highly enriched in migratory neurons, the proportion of electroporated cells was relatively low (~30%; Figures 7B–C’). Despite this caveat, gene expression analysis using semi-quantitative RT-PCR revealed a dramatic increase in the expression of Nkx2-1 in migrating MGE-derived cells expressing Nkx2-1-IRES-Gfp compared to those expressing the control plasmid (n = 3, Figure 7D). Compared to controls, we also detected a mild reduction in the expression of Nrp1 and a prominent decrease in the expression of Nrp2 transcripts in migrating neurons expressing Nkx2-1-IRES-Gfp (Figure 7D). In contrast, we did not detect significant differences in the expression of PlexinA3 and PlexinA4 (Figure 7D), signaling components of the receptor complexes for Sema3A and Sema3F (Yaron et al., 2005). Similarly, the expression of the GABA synthesizing enzyme Gad67 or the transcription factor Lhx6, both present in cortical and striatal interneurons, did not differ between Gfp and Nkx2-1-IRES-Gfp expressing cells. We next used quantitative RT-PCR to precisely determine the influence of Nkx2-1 in the transcription of Nrp1 and Nrp2 in migrating MGE-derived cells. In these experiments, we could confirm that Nrp2 expression was reduced in Nkx2-1-IRES-Gfp-expressing MGE-derived cells (n = 6; Figure 7E). In contrast, although we consistently found reduced levels of Nrp1 expression in Nkx2-1-IRES-Gfp-expressing MGE-derived cells compared to controls, these differences were not statistically significant (n = 6; Figure 7E). Together with our previous findings of reduced sensitivity to semaphorin signaling, these results strongly suggest a role for Nkx2-1 in the transcriptional repression of, at least, the Nrp2 receptor.
Figure 7. Nkx2-1 represses Neuropilin-2 expression in MGE-derived cells.
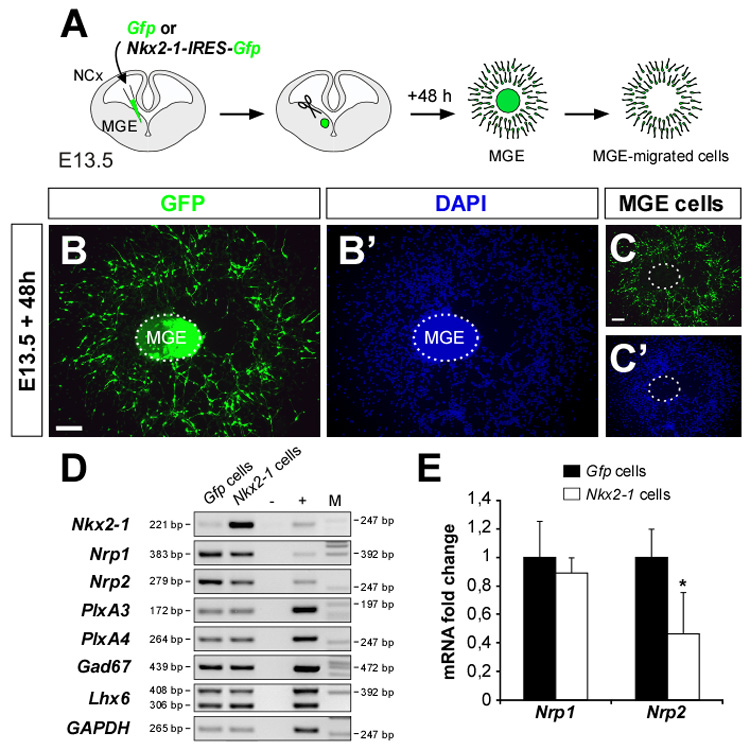
(A) Schematic diagram of the experimental paradigm used to isolate RNA from migrating MGE-derived cells.
(B–C’) A Gfp-electroporated MGE explant stained with DAPI after 48 h in culture, before (B and B’) and after (C and C’) removing the explant core, which contains progenitor cells.
(D) Semi-quantitative RT-PCR analysis comparing gene expression in Gfp- and Nkx2-1-electroporated MGE-derived cells. Negative (-, all reagents except cDNA) and positive (+, E14.5 MGE cDNA) controls were included in each run. Amplicon and molecular marker (M) base pairs (bp) are shown at the left and right sides of the panels, respectively. The Lhx6 gene has two transcripts: Lhx6 or Lhx6.1a (408 bp) and Lhx6.1b (306 bp). GAPDH was used as loading control.
(E) Quantitative RT-PCR analysis for Neuropilin-1 and Neuropilin-2 expression in Gfp-and Nkx2-1-electroporated MGE-derived cells. Histograms show average ± s.e.m. 1.00 ± 0.25 (Gfp cells, Nrp1); 0.89 ± 0.11 (Nkx2-1 cells, Nrp1); 1.00 ± 0.19 (Gfp cells, Nrp2); 0.46 ± 0.29 (Nkx2-1 cells, Nrp2). * p < 0.05, t-test.
MGE, medial ganglionic eminence; NCx, neocortex.
Scale bar equals 50 µm.
Nkx2-1 directly represses the transcription of Nrp2
Nkx2-1 could inhibit the expression of Nrp2 in MGE-derived migrating interneurons by directly interacting with the Nrp2 promoter. To test this, we first examined whether Nkx2-1 protein directly binds to Nrp2 regulatory sequences in vivo. A phylogenetic footprinting analysis of the 20 kb sequence upstream from the Nrp2 transcription initiation site reveal two putative Nrp2 regulatory regions containing two adjacent Nkx2-1 binding sequences (Francis-Lang et al., 1992), which we designated as Nrp2-region2 (from -21375 bp 5’-CTTGC-3’ to -21086 bp 5’-GTGCT-3’) and Nrp2-region1 (from -327 bp 5’-CCGGA-3’ to -68 bp 5’-GGGGA-3’) (Figure 8A and Figure S6). Chromatin immunoprecipitation (ChIP) analyses demonstrated that Nkx2-1 binds to the Nrp2-region1 in E13.5 MGE-derived cells, while it does not seem to complex with the Nrp2-region2 (n = 3; Figures 8B and 8C). To demonstrate that Nkx2-1 represses the expression of Nrp2 through regulatory sequences located in the Nrp2-region1, we cloned this 260 bp DNA fragment upstream of a luciferase reporter plasmid containing a c-fos minimal promoter (Figure 8D). Co-transfection of HEK293 cells with the reporter plasmid and full-length Nkx2-1 produced a significant transcriptional repression of luciferase activity (n = 5; Figure 8D). In contrast, the mutated Nkx2-1A35T did not repress luciferase gene expression (Figure 8D). These results indicate that Nkx2-1 transcriptional repression of Nrp2 requires the direct binding of Nkx2-1 to, at least, the Nrp2-region1 regulatory sequence, and that the integrity of the Nkx2-1 HD motif is essential for this activity.
Figure 8. Nkx2-1 binds the Neuropilin-2 promotor in vivo and regulates its expression.
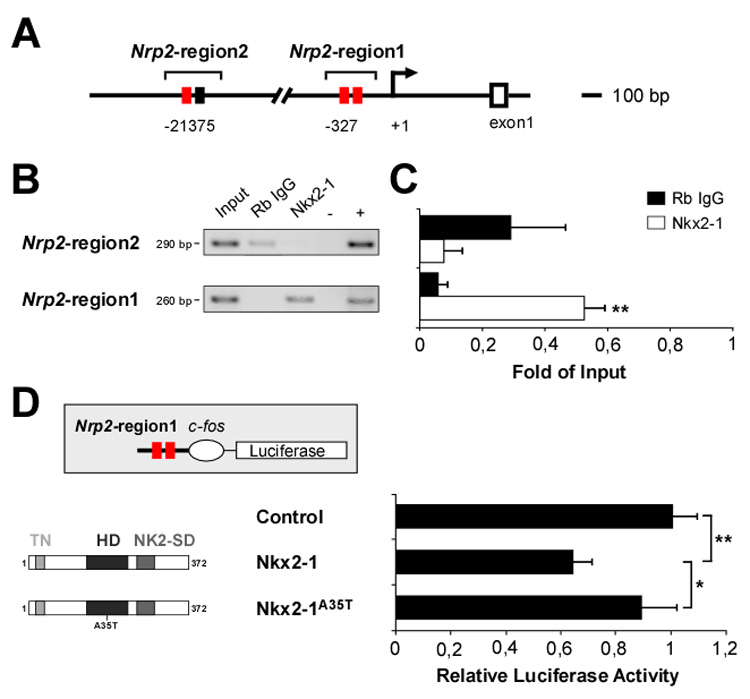
(A) Putative Nkx2-1 DNA binding sites [red and black boxes indicate 8/9 and 6-base pairs (bp) consensus sequences, respectively] in Nrp2-region2 (from -21375 bp 5’-CTTGC-3’ to -21086 bp 5’-GTGCT-3’) and Nrp2-region1 (from -327 bp 5’-CCGGA-3’ to -68 bp 5’-GGGGA-3’) of the Neuropilin-2 locus.
(B) ChIP assays were performed using E13.5 MGE cells and a non-specific rabbit anti-IgG (Rb IgG) or a polyclonal antibody against Nkx2-1. Input chromatin represents 1% of the total chromatin. Negative (-, all reagents except DNA) and positive (+, E13.5 mouse genomic DNA) controls were included in each run. Nrp2-region 2 and Nrp2-region 1 amplicon size (bp, base pairs) are indicated.
(C) The intensity of each PCR band was quantified and normalized against the input band. Histograms show average ± s.e.m. For Nrp2-region2: 0.29 ± 0.17 (Rb IgG) and 0.07 ± 0.06 (Nkx2-1). For Nrp2-region1: 0.06 ± 0.02 (Rb IgG) and 0.52 ± 0.06 (Nkx2-1). ** p < 0.01, t-test.
(D) A luciferase reporter plasmid containing the Nrp2-region1 sequence upstream of the c-fos minimal promoter driving luciferase (pGL3-Nrp2-cfos-Luc) was co-transfected with either mock, Nkx2-1 or Nkx2-1A35T expression vectors. For each condition, the relative luciferase activity corresponds to the ratio of normalized activities from the promoter-luciferase (pGL3-Nrp2-cfos-Luc) and empty-luciferase (pGL3-cfos-Luc) reporter vectors. Histograms show average ± s.e.m. 1.00 ± 0.09 (control), 0.64 ± 0.07 (Nkx2-1), and 0.89 ± 0.13 (Nkx2-1A35T). ** p < 0.01, * p < 0.05, one-way ANOVA followed by Tukey’s post-test.
Discussion
During development, neurons are instructed to migrate and project to specific regions of the brain in a process that is tightly regulated by a variety of guidance cues. A fundamental question that remains to be clarified is how distinct neuronal populations respond selectively to guidance. Transcription factors are thought to play a major role in regulating the differential expression of guidance receptors during cell migration and axon guidance (Butler and Tear, 2007; Guthrie, 2007; Polleux et al., 2007), but it is presently unclear how they exert their influence. Using gain and loss of function approaches, our results demonstrate that the Nkx2-1 transcription factor is uniquely required for the differential migration of cortical and striatal GABAergic interneurons. Postmitotic interneurons expressing Nkx2-1 become insensitive to semaphorin signaling and migrate towards the striatum, whereas those that lack Nkx2-1 expression migrate to the cortex. ChIP and luciferase assays revealed that Nkx2-1 fulfills this function, at least in part, by directly repressing the expression of Nrp2, a receptor for class III semaphorins. Our results therefore demonstrate that postmitotic transcriptional mechanisms play an important role in neuronal migration by directly regulating the repertoire of guidance receptors expressed by migrating neurons.
Transcriptional control of telencephalic interneuron migration
The medial ganglionic eminence (MGE) gives rise to many cortical GABAergic interneurons while concurrently generating most striatal interneurons (Lavdas et al., 1999; Marín et al., 2000; Sussel et al., 1999; Wichterle et al., 1999), but the mechanisms controlling the segregation of these two neuronal populations remain poorly understood. We have previously shown that Nrp1 and Nrp2, the binding receptors for the striatal repulsive molecules Sema3A and Sema3F (Bagri and Tessier-Lavigne, 2002; Kruger et al., 2005), respectively, are expressed by MGE-derived cortical interneurons and absent from MGE-derived striatal interneurons (Marín et al., 2001). Loss of neuropilin function increases the number of interneurons migrating to the striatum and simultaneously decreases the number of cells reaching the cortex. Since the final destination of tangentially migrating interneurons (striatum or cortex) is determined by the expression of semaphorin receptors, we have investigated the nature of the factors controlling this process.
Our experiments demonstrate that postmitotic Nkx2-1 controls the segregation of MGE-derived cells by regulating neuropilin/semaphorin interactions. The maintenance or down-regulation of Nkx2-1 expression in migrating cells is linked to their final destination: striatal interneurons maintain Nkx2-1 expression, whereas cortical cells rapidly downregulate Nkx2-1 mRNA and protein. Moreover, when MGE-derived cells were forced to express Nkx2-1, they failed to reach the cortex and accumulated in the basal telencephalon. In contrast, loss of postmitotic Nkx2-1 function resulted in a reduction in the number of MGE-derived interneurons that populate the striatum. There are two possible caveats in the latter experiment. First, the timing of Cre recombination leaves a very short period between the loss of Nkx2-1 expression (which happens almost exclusively in cells negative for Ki67, Figure S2) and the selection of a target territory. Thus, by the time interneurons lose Nkx2-1 in Lhx6-Cre;Nkx2-1Fl/Fl mutant embryos, many may have already arrived to the striatum. Second, Lhx6 or the Lhx6-Cre transgene are not expressed by all striatal interneurons, and therefore some of them maintain Nkx2-1 expression (Figure S3). Despite these limitations, many MGE-derived interneurons failed to reach the striatum in the absence of Nkx2-1 postmitotic function, suggesting that Nkx2-1 is an important regulator of this process. One question that remains unresolved is the destiny of Nkx2-1-deficient striatal interneurons. Quantification of the number of Lhx6-expressing interneurons in the cortex of E15.5 control and Nkx2-1 conditional mutant embryos revealed no differences (n = 3; Figure S7). However, since cortical interneurons greatly outnumber striatal interneurons, it is very unlikely that rerouting of some striatal cells to the cortex could be easily perceived. Alternatively, loss of Nkx2-1 function may not be enough to redirect all interneurons to the cortex and many cells could have disperse through other subpallial regions, avoiding the striatum. Consistent with view, Lhx7-expressing cells were never observed in the cortex of Lhx6;Nkx2.1Fl/Fl mutant embryos (data not shown).
Expression of Nkx2-1 rendered MGE-migrating cells insensitive to the Sema3A/3F chemorepulsion. This result directly implicates Nkx2-1 function in suppressing the responsiveness of MGE-derived cells to semaphorin signaling. Nkx2-1 suppresses the response of MGE-derived cells to class III semaphorins by directly controlling the expression of, at least, Nrp2. Nkx2-1 directly binds in vivo to a region of the Nrp2 promoter containing two Nkx2-1 specific binding sites located in close proximity (~151 base pairs). The interaction of Nkx2-1 with this short sequence was sufficient to repress transcription in vitro, reinforcing the notion that Nkx2-1 directly suppresses the expression of Nrp2 in migrating MGE-derived interneurons. Nkx2-1 regulates the transcription of other genes by interacting with clustered binding sites (Bohinski et al., 1994) and since this transcription factor binds DNA as a monomer, these repeated consensus sequences might represent a unique arrangement for Nkx2-1 binding site recognition. Our experiments failed to show a conclusive relationship between Nkx2-1 and Nrp1. It should be noted, however, that the relatively low efficiency of Nkx2-1 over-expression in postmitotic migrating interneurons (~ 30%) might have precluded the identification of additional targets genes, such as Nrp1.
Nkx2-1 transcriptional activity depends on the interactions of its HD motif with specific DNA target sequences (Harvey, 1996; Damante et al., 1994). Other highly conserved domains of the protein, such as the TN motif and the NK2-specific domain (SD), could further modulate Nkx2-1 transcriptional activity (Muhr et al., 2001; Watada et al., 2000). Our experiments demonstrate that Nkx2-1 regulates the segregation of MGE-derived cells through transcriptional interactions involving specific residues of the HD (Ala35 and Tyr45). These amino acid residues are needed to suppress the responsiveness to semaphorins and are involved in the interactions between Nkx2-1 and the Nrp2 regulatory sequence. In contrast, neither the TN nor the C-terminus/SD domains appear to regulate the migration of MGE-derived cells to the cortex. Apart from binding to specific DNA sequences, the Nkx2-1 HD has been shown to regulate transcriptional activity by interacting with other transcription factors or through post-translational modifications (Minoo et al., 2007; Yang et al., 2004). Thus, in addition to conferring binding specificity to the Nrp2-region1 promoter, it is possible that specific amino acid residues of the Nkx2-1 HD establish additional interactions that mediate the repression of Nrp2 expression in MGE-derived migrating cells.
The transcriptional regulation of telencephalic interneuron migration is likely to involve additional factors. For example, the Dlx1/2 transcription factors appear also to repress Nrp2 in the developing forebrain (Le et al., 2007), although the functional consequences of this regulation for the migration of MGE-derived cells have not been examined. In addition to neuropilin/semaphorin interactions, it is likely that Nkx2-1 may control other guidance systems in MGE-derived neurons. Of note, migrating neurons expressing Nkx2-1 do not simply migrate through the striatum as they fail to sense semaphorins, but indeed they seem to be attracted to that location. This result suggests that the striatum may also contain an attractive factor for MGE-derived interneurons, although the molecular nature of this activity remains unknown. Alternatively, Nkx2-1 may confer striatal interneurons with sensitivity for a cortical chemorepulsive cue, which will prevent migrating MGE-derived interneurons expressing Nkx2-1 from entering the cortex.
A question that remains to be elucidated is the mechanism regulating the postmitotic expression of Nkx2-1 in migrating MGE-derived neurons. Sonic hedgehog (Shh) signaling induces and maintains Nkx2-1 expression in MGE progenitors during development (Ericson et al., 1995; Shimamura et al., 1995; Xu et al., 2005), and supports the expansion of progenitors through neurogenesis (Machold et al., 2003). However, the analysis of Dlx5/6Cre;SmoFl/Fl conditional mutant mice suggests that Shh does not control the expression of Nkx2-1 in striatal postmitotic interneurons (Xu et al., 2005). Future work should address this issue, as it seems critical for understanding how appropriate numbers of different inhibitory populations are generated during development.
Multiple roles for Nkx2-1 in the development of telencephalic interneurons
Previous studies have revealed that Nkx2-1 is critical for the development of the mammalian subpallium. The emerging idea, however, is that Nkx2-1 plays diverse roles in closely related cells depending on their relative stage of differentiation. During early patterning of the telencephalon, Nkx2-1 regulates the specification of the MGE and preoptic area (POA) progenitor cells (Corbin et al., 2003; Sussel et al., 1999). In the absence of Nkx2-1 function, the MGE and POA progenitors are re-specified to a more dorsal fate, similar to that of LGE progenitors. This transformation leads to a dramatic reduction in the number of cells derived from those structures, such as GABAergic cortical and striatal interneurons, as well as projection neurons of the globus pallidus and other basal forebrain structures (Sussel et al., 1999).
In addition to its master role in ventral subpallial identity, Nkx2-1 also instructs the selection of specific fates by controlling the expression of differentiation genes in subpallial-derived neurons, such as the LIM-HD transcription factors Lhx6 and Lhx7 (Sussel et al., 1999). As shown for Lhx6, Nkx2-1 appears to control the expression of these genes through direct transcriptional regulation (Du et al., 2008). Lhx6 and Lhx7 are essential regulators of the fate of several types of GABAergic and cholinergic neurons derived from Nkx2-1-expressing progenitors (Fragkouli et al., 2005; Mori et al., 2004; Wonders and Anderson, 2006; Zhao et al., 1999), reinforcing the idea that Nkx2-1 expression prior to cell cycle exit influences the fate of MGE-derived cells.
Our results demonstrate that Nkx2-1 plays an additional role in postmitotic MGE-derived cells by controlling the repertoire of guidance receptors expressed by migrating interneurons. As for the induction of the cell fate determinant Lhx6 (Du et al., 2008), the postmitotic function of Nkx2-1 in neuronal migration is achieved through the direct transcriptional repression of a guidance receptor, Nrp2. Thus, the cellular context in which Nkx2-1 operates at different stages of differentiation greatly influences the functional outcome of its transcriptional activity. The mechanisms conferring Nkx2-1 with time and context-dependent transcriptional specificity remain to be elucidated.
Experimental Procedures
Mouse lines
Wild-type mice and GFP-expressing transgenic mice (Hadjantonakis et al., 2002) maintained in a CD1 background were used for expression analysis and tissue culture experiments. Lhx6-Cre (Fogarty et al., 2007), Rosa-EYFP (Srinivas et al., 2001) and Nkx2-1Fl/Fl (Kusakabe et al., 2006) mice were maintained in a mixed C57Bl/6 × 129/SvJ × CBA background. Animals were kept under Spanish, UK and EU regulation.
DNA constructs
A cDNA encoding Nkx2-1 (accession number NM_009385) was used. The truncated constructs Nkx2-1ΔTN-IRES-Gfp (deletion [Δ] of amino acids 1–20) and Nkx2-1ΔCt (Δ amino acids 271–372) were generated by PCR. The single-amino acid substitutions Nkx2-1A35T and Nkx2-1Y54M were prepared using the QuickChange II XL Kit (Stratagene). All constructs (Gfp, Nkx2-1-IRES-Gfp, Nkx2-1ΔTN-IRES-Gfp, Nkx2-1ΔCt, Nkx2-1A35T and Nkx2-1Y54M) were inserted into the pCAGGS chicken β-actin promoter expression vector.
In vitro focal electroporation
E13.5 organotypic coronal slice cultures from wild type or Gfp transgenic embryos were obtained as described previously (Anderson et al., 1997). Expression vectors were electroporated at a concentration of 1 µg/µl and mixed in a 0.9/1.5 ratio when co-electroporated. Expression vectors were focally injected into the MGE and embryonic slice cultures were electroporated as previously described (Flames et al., 2004).
In situ hybridization and immunohistochemistry
For in situ hybridization, brains were fixed overnight in 4% paraformaldehyde in PBS (PFA). Twenty µm frozen sections were hybridized with digoxigenin-labeled probes as described before (Flames et al., 2007). Immunohistochemistry was performed on: culture slices, MGE explants in matrigel® pads or 20 µm cryostat sections. Slices, explants and embryos were fixed in 4% PFA at 4°C from 2–6 h. The following primary antibodies were used: rat anti-BrdU (1/100, Accurate),chicken anti-GFP (1/1000, Aves Labs), rabbit anti-Nkx2-1 (1/2000, Biopat), rabbit anti-PV (1/3000, Swant), goat anti-ChAT(1/100, Chemicon), rat anti-SST (1/200, Chemicon), rabbit anti-cleaved Caspase 3 (1/150, Cell Signaling) and rabbit anti- Ki67 (1/500, Novocastra). The following secondary antibodies were used: donkey anti-rat 488, goat anti-chicken 488, donkey anti-rabbit 555, donkey anti-goat 555 (Molecular Probes) and donkey anti-rat Cy3 (Jackson Laboratories). The immunofluorescence detection of EYFP was performed using an anti-GFP antibody. DAPI (Sigma) and Propidium Iodine (Molecular Probes) were used for fluorescent nuclear counterstaining.
Quantification
For the quantification of interneurons in E13.5, E15.5, P0 and P25 control and Lhx6-Cre;Nkx2-1Fl/Fl mutant brains, the outline of the striatum or cortex at rostral, intermediate and caudal levels was delineated in 20 or 60 µm sections, different interneuron markers (Lhx6, Lhx7, Er81, Sst, ChAT, PV and SST) or YFP-expressing cells were counted for three different brains from each genotype and the cell density (number of cells per mm2) was calculated. In the transplantation experiments, the intensity of Nkx2-1 fluorescence in Gfp-expressing cells was quantified and classified into high or low levels when presenting >90% or <30%, respectively, of the Nkx2-1 fluorescence intensity found in MGE progenitors (considered as 100%).
MGE explants cultures
MGE explants were dissected out from organotypic slices after electroporation. For gene-expression analysis, MGE explants were cultured on glass coverslips coated with poly-L-Lysine and laminin in Neurobasal medium containing 0.3% methylcellulose (Sigma). In coculture experiments, MGE explants were confronted with COS7 cells aggregates expressing DsRed alone or DsRed and Sema3A/3F in Matrigel matrix (Beckton-Dickinson) as described previously (Flames et al., 2004). Sema3A and Sema3F cDNA sequences used have been described elsewhere (Marín et al., 2001). For quantification, each explant was divided into 4 quadrants, the number of Gfp-expressing cells was quantified in the proximal and distal quadrant (in relation to the COS7 cells) and the proximal/distal ratio was calculated.
Semi-quantitative and quantitative RT-PCR
Total RNA from MGE-derived cells was extracted with TRIzol according to the manufacturer’s instructions (Invitrogen). RNA (150 ng) was treated with DNaseI RNase-free (Fermentas) for 30 min at 37°C prior to reverse transcription into single-stranded cDNA using SuperScriptII Reverse Transcriptase and Oligo(dT)12–18 primers (Invitrogen) for 1 hour at 42°C. For semi-quantitative PCR, 2 µl of cDNA, the appropriate primers (Figure S8) and recombinant Taq DNA polymerase (Invitrogen) were used. For Nrp1 and Nrp2, a pre-amplification PCR step (multiplex) was performed using 10 µl of cDNA and a primer mix containing GAPDH, Nrp1 and Nrp2 multiplex primers. PCR products were analyzed by electrophoresis on a 2% agarose gel. Quantitative (q) PCR was carried out in an Applied Biosystems 7300 real-time PCR unit using the Platinum SYBR Green qPCR Supermix UDG with ROX (Invitrogen), 5 µl of cDNA and the appropriate primers (Figure S8). Each independent sample was assayed in duplicate. Gene expression levels were normalized using GAPDH.
Phylogenetic footprinting analysis
In silico analysis of the predicted 5’ flanking region of the Mus musculus Nrp2 locus (accession number NT_039170) was performed using Vista and University California Santa Cruz (UCSC) Genome Browsers. Putative Nkx2-1 binding sites match between 6 to 9 nucleotides the described Nkx2-1 consensus sequence 5’-CCACTC/GAAGTG-3’.
Chromatin Immunoprecipitation assay
Chromatin Immunoprecipitation (ChIP) assay was performed using the EZ ChIP Kit (Upstate) according to the manufacturer’s instructions. Briefly, mouse E13.5 MGE cells (1–2×107) were cross-linked with 1% PFA for 25 minutes at room temperature. Sonication of cells in SDS lysis buffer on ice (Bioruptor, Diagenode; 200W potency; 40 seconds on, 20 seconds off; 5×5 minutes) generated soluble chromatin fragments between 150–400 bp. Chromatin was immunoprecipitated with 5–6 µg of rabbit anti-Nkx2-1 (Biopat) and rabbit anti-immunoglobulin G (IgG) antibodies. Immunoprecipitated DNA sequences were analyzed by PCR using primer pairs spanning the Nrp2-region2 and Nrp2-region1 (Figure S6). PCR products were analyzed by electrophoresis on a 2% agarose gel.
Promoter luciferase assay
HEK293 cells were co-transfected with 200 ng of a luciferase reporter plasmid (pGL3-cfos-Luc or pGL3-Nrp2-cfos-Luc), 8 ng of CMVβgal and 200 ng of an empty (mock), full-length Nkx2-1 or Nkx2-1A35T expression vector using Fugene reagent (Roche). After one day in culture, cells were collected and assayed for luciferase and β-galactosidase activity using the Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Each independent sample was assayed in duplicate and luciferase activities were normalized using β-galactosidase activity.
Supplementary Material
Acknowledgements
We thank M. Bonete, T. Gil, M. Pérez, M. Grist and A. Rubin for excellent technical assistance, V. Pachnis, M. Tessier-Lavigne and J.M. Ortiz for plasmids, A. Barco for the q-PCR equipment, C. García-Frigola and J. Galcerán for advise on ChIP and luminescence experiments, respectively, and A. Nagy and S. Srinivas for Gfp and Rosa-EYFP mice, respectively. We are grateful to J. Galcerán and members of the Marín and Rico labs for stimulating discussions and critically reading this manuscript. This work was supported by grants from Spanish Ministry of Education and Science BFU2005-04773/BMC and CONSOLIDER CSD2007-00023, Fundació “la Caixa”, the European Commission through STREP contract number 005139 (INTERDEVO) and the EURYI program (to OM), NINDS and NIMH (to SAA), and UK Medical Research Council and European Research Council (to NK). SNP was supported by a predoctoral fellowship from the Foundation for Science and Technology (POCI 2010/FSE), Portugal.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- Bagri A, Tessier-Lavigne M. Neuropilins as Semaphorin receptors: in vivo functions in neuronal cell migration and axon guidance. Adv. Exp. Med. Biol. 2002;515:13–31. [PubMed] [Google Scholar]
- Bohinski RJ, Di Lauro R, Whitsett JA. The lung-specific surfactant protein B gene promoter is a target for thyroid transcription factor 1 and hepatocyte nuclear factor 3, indicating common factors for organ-specific gene expression along the foregut axis. Mol. Biol. Cell. 1994;14:5671–5681. doi: 10.1128/mcb.14.9.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brose K, Tessier-Lavigne M. Slit proteins: key regulators of axon guidance, axonal branching, and cell migration. Curr. Opin. Neurobiol. 2000;10:95–102. doi: 10.1016/s0959-4388(99)00066-5. [DOI] [PubMed] [Google Scholar]
- Butler SJ, Tear G. Getting axons onto the right path: the role of transcription factors in axon guidance. Development. 2007;134:439–448. doi: 10.1242/dev.02762. [DOI] [PubMed] [Google Scholar]
- Corbin JG, Rutlin M, Gaiano N, Fishell G. Combinatorial function of the homeodomain proteins Nkx2.1 and Gsh2 in ventral telencephalic patterning. Development. 2003;130:4895–4906. doi: 10.1242/dev.00717. [DOI] [PubMed] [Google Scholar]
- Damante G, Fabbro D, Pellizzari L, Civitareale D, Guazzi S, Polycarpou-Schwartz M, Cauci S, Quadrifoglio F, Formisano S, Di Lauro R. Sequence-specific DNA recognition by the thyroid transcription factor-1 homeodomain. Nucleic Acids Res. 1994;22:3075–3083. doi: 10.1093/nar/22.15.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson BJ. Molecular mechanisms of axon guidance. Science. 2002;298:1959–1964. doi: 10.1126/science.1072165. [DOI] [PubMed] [Google Scholar]
- Du T, Xu Q, Ocbina PJ, Anderson SA. NKX2.1 specifies cortical interneuron fate by activating Lhx6. Development. 2008;135:1559–1567. doi: 10.1242/dev.015123. [DOI] [PubMed] [Google Scholar]
- Ericson J, Muhr J, Placzek M, Lints T, Jessell TM, Edlund T. Sonic hedgehog induces the differentiation of ventral forebrain neurons: a common signal for ventral patterning within the neural tube. Cell. 1995;81:747–756. doi: 10.1016/0092-8674(95)90536-7. [DOI] [PubMed] [Google Scholar]
- Flames N, Long JE, Garratt AN, Fischer TM, Gassmann M, Birchmeier C, Lai C, Rubenstein JL, Marín O. Short- and long-range attraction of cortical GABAergic interneurons by neuregulin-1. Neuron. 2004;44:251–261. doi: 10.1016/j.neuron.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Flames N, Pla R, Gelman DM, Rubenstein JL, Puelles L, Marín O. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J. Neurosci. 2007;27:9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty M, Grist M, Gelman D, Marín O, Pachnis V, Kessaris N. Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J. Neurosci. 2007;27:10935–10946. doi: 10.1523/JNEUROSCI.1629-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkouli A, Hearn C, Errington M, Cooke S, Grigoriou M, Bliss T, Stylianopoulou F, Pachnis V. Loss of forebrain cholinergic neurons and impairment in spatial learning and memory in LHX7-deficient mice. Eur. J. Neurosci. 2005;21:2923–2938. doi: 10.1111/j.1460-9568.2005.04141.x. [DOI] [PubMed] [Google Scholar]
- Francis-Lang H, Price M, Polycarpou-Schwarz M, Di Lauro R. Cell-type-specific expression of the rat thyroperoxidase promoter indicates common mechanisms for thyroid-specific gene expression. Mol. Biol. Cell. 1992;12:576–588. doi: 10.1128/mcb.12.2.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia NV, Jessell TM. Early Motor Neuron Pool Identity and Muscle Nerve Trajectory Defined by Postmitotic Restrictions in Nkx6.1 Activity. Neuron. 2008;57:217–231. doi: 10.1016/j.neuron.2007.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Frigola C, Carreres MI, Vegar C, Mason C, Herrera E. Zic2 promotes axonal divergence at the optic chiasm midline by EphB1-dependent and -independent mechanisms. Development. 2008;135:1833–1841. doi: 10.1242/dev.020693. [DOI] [PubMed] [Google Scholar]
- Ge W, He F, Kim KJ, Blanchi B, Coskun V, Nguyen L, Wu X, Zhao J, Heng JI, Martinowich K, et al. Coupling of cell migration with neurogenesis by proneural bHLH factors. Proc. Natl. Acad. Sci. U. S. A. 2006;103:1319–1324. doi: 10.1073/pnas.0510419103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie S. Patterning and axon guidance of cranial motor neurons. Nat. Rev. Neurosci. 2007;8:859–871. doi: 10.1038/nrn2254. [DOI] [PubMed] [Google Scholar]
- Hadjantonakis AK, Macmaster S, Nagy A. Embryonic stem cells and mice expressing different GFP variants for multiple non-invasive reporter usage within a single animal. BMC Biotechnol. 2002;2:11. doi: 10.1186/1472-6750-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand R, Bortone D, Mattar P, Nguyen L, Heng JI, Guerrier S, Boutt E, Peters E, Barnes AP, Parras C, et al. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron. 2005;48:45–62. doi: 10.1016/j.neuron.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Harvey RP. NK-2 homeobox genes and heart development. Dev. Biol. 1996;178:203–216. doi: 10.1006/dbio.1996.0212. [DOI] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Kania A, Jessell TM. Topographic motor projections in the limb imposed by LIM homeodomain protein regulation of ephrin-A:EphA interactions. Neuron. 2003;38:581–596. doi: 10.1016/s0896-6273(03)00292-7. [DOI] [PubMed] [Google Scholar]
- Kania A, Johnson RL, Jessell TM. Coordinate roles for LIM homeobox genes in directing the dorsoventral trajectory of motor axons in the vertebrate limb. Cell. 2000;102:161–173. doi: 10.1016/s0092-8674(00)00022-2. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y, Wilson CJ, Augood SJ, Emson PC. Striatal interneurones: chemical, physiological and morphological characterization. Trends Neurosci. 1995;18:527–535. doi: 10.1016/0166-2236(95)98374-8. [DOI] [PubMed] [Google Scholar]
- Kruger RP, Aurandt J, Guan KL. Semaphorins command cells to move. Nat. Rev. Mol. Cell Biol. 2005;6:789–800. doi: 10.1038/nrm1740. [DOI] [PubMed] [Google Scholar]
- Kusakabe T, Kawaguchi A, Hoshi N, Kawaguchi R, Hoshi S, Kimura S. Thyroid-specific enhancer-binding protein/NKX2.1 is required for the maintenance of ordered architecture and function of the differentiated thyroid. Mol. Endocrinol. 2006;20:1796–1809. doi: 10.1210/me.2005-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavdas AA, Grigoriou M, Pachnis V, Parnavelas JG. The medial ganglionic eminence gives rise to a population of early neurons in the developing cerebral cortex. J. Neurosci. 1999;19:7881–7888. doi: 10.1523/JNEUROSCI.19-18-07881.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TN, Du G, Fonseca M, Zhou QP, Wigle JT, Eisenstat DD. Dlx homeobox genes promote cortical interneuron migration from the basal forebrain by direct repression of the semaphorin receptor neuropilin-2. J. Biol. Chem. 2007;282:19071–19081. doi: 10.1074/jbc.M607486200. [DOI] [PubMed] [Google Scholar]
- Lee R, Petros TJ, Mason CA. Zic2 regulates retinal ganglion cell axon avoidance of ephrinB2 through inducing expression of the guidance receptor EphB1. J Neurosci. 2008;28:5910–5919. doi: 10.1523/JNEUROSCI.0632-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machold R, Hayashi S, Rutlin M, Muzumdar MD, Nery S, Corbin JG, Gritli-Linde A, Dellovade T, Porter JA, Rubin LL, et al. Sonic hedgehog is required for progenitor cell maintenance in telencephalic stem cell niches. Neuron. 2003;39:937–950. doi: 10.1016/s0896-6273(03)00561-0. [DOI] [PubMed] [Google Scholar]
- Marín O, Anderson SA, Rubenstein JL. Origin and molecular specification of striatal interneurons. J. Neurosci. 2000;20:6063–6076. doi: 10.1523/JNEUROSCI.20-16-06063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín O, Yaron A, Bagri A, Tessier-Lavigne M, Rubenstein JL. Sorting of striatal and cortical interneurons regulated by semaphorin-neuropilin interactions. Science. 2001;293:872–875. doi: 10.1126/science.1061891. [DOI] [PubMed] [Google Scholar]
- McEvilly RJ, de Diaz MO, Schonemann MD, Hooshmand F, Rosenfeld MG. Transcriptional regulation of cortical neuron migration by POU domain factors. Science. 2002;295:1528–1532. doi: 10.1126/science.1067132. [DOI] [PubMed] [Google Scholar]
- Minoo P, Hu L, Xing Y, Zhu NL, Chen H, Li M, Borok Z, Li C. Physical and functional interactions between homeodomain NKX2.1 and winged helix/forkhead FOXA1 in lung epithelial cells. Mol. Biol. Cell. 2007;27:2155–2165. doi: 10.1128/MCB.01133-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Yuxing Z, Takaki H, Takeuchi M, Iseki K, Hagino S, Kitanaka J, Takemura M, Misawa H, Ikawa M, et al. The LIM homeobox gene, L3/Lhx8, is necessary for proper development of basal forebrain cholinergic neurons. Eur. J. Neurosci. 19:3129–3141. doi: 10.1111/j.0953-816X.2004.03415.x. [DOI] [PubMed] [Google Scholar]
- Muhr J, Andersso E, Persson M, Jessell TM, Ericson J. Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell. 2001;104:861–873. doi: 10.1016/s0092-8674(01)00283-5. [DOI] [PubMed] [Google Scholar]
- Müller M, Jabs N, Lorke DE, Fritzsch B, Sander M. Nkx6.1 controls migration and axon pathfinding of cranial branchio-motoneurons. Development. 2003;130:5815–5826. doi: 10.1242/dev.00815. [DOI] [PubMed] [Google Scholar]
- Polleux F, Ince-Dunn G, Ghosh A. Transcriptional regulation of vertebrate axon guidance and synapse formation. Nat. Rev. Neurosci. 2007;8:331–340. doi: 10.1038/nrn2118. [DOI] [PubMed] [Google Scholar]
- Sharma K, Leonard AE, Lettieri K, Pfaff SL. Genetic and epigenetic mechanisms contribute to motor neuron pathfinding. Nature. 2000;406:515–519. doi: 10.1038/35020078. [DOI] [PubMed] [Google Scholar]
- Sharma K, Sheng HZ, Lettieri K, Li H, Karavanov A, Potter S, Westphal H, Pfaff SL. LIM homeodomain factors Lhx3 and Lhx4 assign subtype identities for motor neurons. Cell. 1998;95:817–828. doi: 10.1016/s0092-8674(00)81704-3. [DOI] [PubMed] [Google Scholar]
- Shimamura K, Hartigan DJ, Martínez S, Puelles L, Rubenstein JL. Longitudinal organization of the anterior neural plate and neural tube. Development. 1995;121:3923–3933. doi: 10.1242/dev.121.12.3923. [DOI] [PubMed] [Google Scholar]
- Shirasaki R, Pfaff SL. Transcriptional codes and the control of neuronal identity. Annu. Rev. Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenman J, Toresson H, Campbell K. Identification of two distinct progenitor populations in the lateral ganglionic eminence: implications for striatal and olfactory bulb neurogenesis. J. Neurosci. 2003;23:167–174. doi: 10.1523/JNEUROSCI.23-01-00167.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugitani Y, Nakai S, Minowa O, Nishi M, Jishage K, Kawano H, Mori K, Ogawa M, Noda T. Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev. 2002;16:1760–1765. doi: 10.1101/gad.978002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L, Marín O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126:3359–3370. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- Tessier-Lavigne M, Goodman CS. The molecular biology of axon guidance. Science. 1996;274:1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- Thaler JP, Koo SJ, Kania A, Lettieri K, Andrews S, Cox C, Jessell TM, Pfaff SL. A postmitotic role for Isl-class LIM homeodomain proteins in the assignment of visceral spinal motor neuron identity. Neuron. 2004;41:337–350. doi: 10.1016/s0896-6273(04)00011-x. [DOI] [PubMed] [Google Scholar]
- Watada H, Mirmira RG, Kalamaras J, German MS. Intramolecular control of transcriptional activity by the NK2-specific domain in NK-2 homeodomain proteins. Proc. Natl. Acad. Sci. U. S. A. 2000;97:9443–9448. doi: 10.1073/pnas.97.17.9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H, Garcia-Verdugo JM, Herrera DG, Alvarez-Buylla A. Young neurons from medial ganglionic eminence disperse in adult and embryonic brain. Nat. Neurosci. 1999;2:461–466. doi: 10.1038/8131. [DOI] [PubMed] [Google Scholar]
- Wichterle H, Turnbull DH, Nery S, Fishell G, Alvarez-Buylla A. In utero fate mapping reveals distinct migratory pathways and fates of neurons born in the mammalian basal forebrain. Development. 2001;128:3759–3771. doi: 10.1242/dev.128.19.3759. [DOI] [PubMed] [Google Scholar]
- Williams SE, Mason CA, Herrera E. The optic chiasm as a midline choice point. Curr. Opin. Neurobiol. 2004;14:51–60. doi: 10.1016/j.conb.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat. Rev. Neurosci. 2006;7:687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- Xiang B, Weiler S, Nirenberg M, Ferretti JA. Structural basis of an embryonically lethal single Ala -->Thr mutation in the vnd/NK-2 homeodomain. Proc. Natl. Acad. Sci. U. S. A. 1998;95:7412–7416. doi: 10.1073/pnas.95.13.7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Wonders CP, Anderson SA. Sonic hedgehog maintains the identity of cortical interneuron progenitors in the ventral telencephalon. Development. 2005;132:4987–4998. doi: 10.1242/dev.02090. [DOI] [PubMed] [Google Scholar]
- Yang L, Yan D, Bruggeman M, Du H, Yan C. Mutation of a lysine residue in a homeodomain generates dominant negative thyroid transcription factor 1. Biochemistry. 2004;43:12489–12497. doi: 10.1021/bi049283o. [DOI] [PubMed] [Google Scholar]
- Yaron A, Huang PH, Cheng HJ, Tessier-Lavigne M. Differential requirement for Plexin-A3 and -A4 in mediating responses of sensory and sympathetic neurons to distinct class 3 Semaphorins. Neuron. 2005;45:513–523. doi: 10.1016/j.neuron.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Guo YJ, Tomac AC, Taylor NR, Grinberg A, Lee EJ, Huang S, Westphal H. Isolated cleft palate in mice with a targeted mutation of the LIM homeobox gene lhx8. Proc Natl Acad Sci USA. 1999;96:15002–15006. doi: 10.1073/pnas.96.26.15002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


