Abstract
Cyclic nucleotide phosphodiesterase-8 (PDE8) is a family of cAMP-specific enzymes and plays important roles in many biological processes, including T-cell activation, testosterone production, adrenocortical hyperplasia, and thyroid function. However, no PDE8 selective inhibitors are available for trial treatment of human diseases. Here we report kinetic properties of the highly active PDE8A1 catalytic domain prepared from refolding and its crystal structures in the unliganded and 3-isobutyl-1-methylxanthine (IBMX) bound forms at 1.9 and 2.1 Å resolutions, respectively. The PDE8A1 catalytic domain has KM of 1.8 μM, Vmax of 6.1 μmol/min/mg, kcat of 4.0 s−1 for cAMP, and KM of 1.6 mM, Vmax of 2.5 μmol/min/mg, kcat of 1.6 s−1 for cGMP, thus indicating that the substrate specificity of PDE8 is dominated by KM. The structure of the PDE8A1 catalytic domain has similar topology as those of other PDE families, but contains two extra helices around Asn685-Thr710. Since this fragment is distant from the active site of the enzyme, its impact on the catalysis is unclear. The PDE8A1 catalytic domain is insensitive to the IBMX inhibition (IC50 = 700 μM). The unfavorable interaction of IBMX in the PDE8A1-IBMX structure suggests an important role of Tyr748 in the inhibitor binding. Indeed, the mutation of Tyr748 to phenylalanine increases the PDE8A1 sensitivity to several non-selective or family-selective PDE inhibitors. Thus, the structural and mutagenesis studies provide not only insight into the enzymatic properties, but also guidelines for design of PDE8 selective inhibitors.
Adenosine and guanosine 3′,5′-cyclic monophosphates (cAMP and cGMP) are the second messengers that mediate the response of cells to a wide variety of hormones and neurotransmitters and modulate many metabolic processes (1–5). Phosphodiesterases (PDEs) are the sole enzymes hydrolyzing these cyclic nucleotides and thus play pivotal roles in the physiological processes involving the nucleotide signaling pathway. Human genome contains 21 PDE genes that are categorized into 11 families (6–9). Alternative mRNA splicing of these genes produces over 100 isoforms of PDE proteins. Molecules of PDEs can be divided into a variable regulatory domain at the N-terminus and a conserved catalytic domain at the C-terminus.
Family selective inhibitors of PDEs have been widely studied as therapeutics for treatment of various human diseases, including cardiotonics, vasodilators, smooth muscle relaxants, antidepressants, antiasthmatics, and agents for improvement of learning and memory (10–17). A well known example is the PDE5 inhibitor sildenafil (Viagra) that has been approved for treatment of both male erectile dysfunction and pulmonary hypertension (10,18). Among PDE inhibitors, 3-isobutyl-1-methylxanthine (IBMX) is commonly used for characterization of enzymatic properties. IBMX is a non-selective inhibitor for most PDE families. However, an uncategorized PDE enzyme that was purified from the rat liver homogenate is insensitive to the IBMX inhibition (19). For its preference to cAMP over cGMP, this rat protein is probably the first report on a fragment of PDE8.
Human genome expresses two PDE8 subfamilies (PDE8A and PDE8B), both of which are cAMP-specific and have KM of 40–150 nM for cAMP and >100 μM for cGMP (20–23). Isoforms of PDE8 distribute in various human tissues and are abundant in testis (24–27). PDE8 has been shown to be involved in regulation of T-cell activation (28), chemotaxis of activated lymphocytes (29), modulation of testosterone production in Leydig cell (30), and potentiation of biphasic insulin response to glucose (31). Recently, the H305P mutation of PDE8B1 is reported to associate with micronodular adrenocortical hyperplasia (32) and PDE8B gene variants are associated with thyroid-stimulating hormone levels and thyroid function (33). Molecules of PDE8 contain a Per-ARNT-Sim (PAS) domain that is a structural motif and an environmental protein sensor involved in many biological processes such as response to oxygen partial pressure and redox signaling (34, 35). PDE8 was reported to bind IκBβ, a regulatory protein of transcription factor NF-κB (36), presumably in a mode that the PAS domain of PDE8 competes with NF-κB for IκBβ binding.
Although PDE8 plays important roles in the physiological processes, the molecular basis has not been fully understood. Neither structures of any PDE8 fragments nor PDE8 selective inhibitors have been reported. The lack of structural information on PDE8 is apparently due to the difficulty of protein purification. While the catalytic domains of eight PDE families have been expressed and their crystal structures have been determined (37), preparation of large quantity of PDE8 has not been easy and the purified proteins in literature typically have low catalytic activity (20–23). For example, the C-terminal 545 amino acid fragment of PDE8A that was expressed in the baculovirus system had Vmax of 0.15 μmol/min/mg (20), which is at least 10 times worse than those of other PDE families. Thus, finding an alternative and effective way to produce large quantity of active PDE8 is essential for structural study. Reported here are the refolding of the PDE8A1 catalytic domain, the kinetic characterization of the refolded PDE8A1, and the crystal structures of PDE8A1 in the unliganded and IBMX-bound forms. The structures suggest a critical role of Tyr748 in the inhibitor selectivity of PDE8. The Y748F mutation showed increased sensitivity of the PDE8A catalytic domain to many of non-selective and family-selective PDE inhibitors.
Experimental Procedures
Subcloning of the PDE8A catalytic domain
The Expressed Sequence Tag cDNA clone of PDE8A1 (GenBank #AF332653) was purchased from American Type Culture Collection (category number 10325182). The cDNA fragment for expression of the catalytic domain of PDE8A1 (residues 480–820) was amplified by PCR and subcloned into vector pET15b. The following oligonucleotide primers that contain the restriction sites of NdeI and XhoI were used for expression of desired genes: 5′-GTGCCGCGCGGCAGCCATATGGCTAGCTCCCTTGATGATGTCCCAC and 3′-GGATCCTCGAGTTACTTCATTTCGTCCAGTCCTTTC. The amplified cDNAs of PDE8A1 and the expression vector pET15b were digested by the restriction enzymes, purified with agarose gel, and ligated by T4 DNA ligase.
The plasmid pET15b-PDE8A1 was transferred into E. coli strain BL21 (CodonPlus) for overexpression. The cell containing the vector pET15b-PDE8A1 was grown in a modified 2xYT culture medium (16g trypton, 10g yeast extracts, and 5g NaCl per liter) that was autoclaved before addition of 0.4% glucose, 100mg ampicillin, and 20mg chloramphenicol. After the cell was grown at 37°C to A600 = 0.7, 0.1 mM isopropyl β-D-thiogalactopyranoside (IPTG) was added to induce the overexpression at room temperature for overnight. The Y748F mutant of PDE8A1 was constructed by the standard protocol of site-directed mutagenesis and overexpressed in the similar conditions as the wild type PDE8A1.
Protein refolding and purification
Since the wild type PDE8A1 (480–820) and its mutant were expressed as inclusion body, refolding was performed to obtain the soluble and active PDE8A1 proteins. About 10 grams of frozen cell from 2 liter culture were suspended in 40 ml of the extraction buffer and past through French Press three times at 1200 psi to homogenize it. The extraction buffer was 20 mM Tris.base, pH 7.5, 50 mM NaCl, 1 mM β-mercaptoethanol (β-ME), and 1 mM EDTA. The homogenate was centrifuged at 15,000 rpm in a JA20 rotor for 30 min. The pellet of PDE8A was dissolved in 10 ml buffer of 6 M guanidine, 0.1 M Tris·HCl pH 8.0 under shaking at room temperature for 3 hours. The dissolved mixture was centrifuged at 15,000 rpm for 20 min.
The supernatant was loaded into a Ni-NTA column (φ=2.5 cm, 25 ml of QIAGEN agarose beads). The column was washed with 100 ml buffer of 8 M urea, 0.1 M Tris·HCl pH 8.0, and then eluted with the same buffer plus 0.5 M arginine. The eluted fractions of the PDE8A1 catalytic domain were combined and added with 10 mM β-ME and 10 mM DTT. The refolding was initiated by addition of 30 μg/ml protein to a buffer of 0.5 M Tris·HCl pH 7.0, 20 mM MgCl2, 20 mM MnCl2, 20 μM ZnSO4, 0.7 M arginine, 30% glycerol, 10 mM NaCl, 1 mM KCl, and 10 mM DTT at 4°C for three days. The protein concentration was estimated by the absorption of A280 (1.097 unit of A280 = 1 mg/ml), as calculated by program ProtParam (38).
The refolded PDE8A1 was mixed with 15 grams of hydroxyapatite HTP GEL bead (Bio-Rad) that was presoaked in water. After stirred at room temperature for 15 min, the suspension was filtered with a filter paper. The bead was re-suspended in 100 ml of 20 mM Tris·HCl pH 8.0, 50 mM NaCl, 1 mM β-ME and packed into a column. The column was washed with 50 ml of the same buffer and eluted with 100ml buffer of 20mM Tris·HCl pH 8.0, 100 mM KH2PO4, and 1 mM β-ME. The combined fractions were dialyzed against 0.5 liter of 20 mM Tris pH 7.5, 50 mM NaCl, and 1 mM β-ME, twice, 1 hour and overnight.
To remove the His-tag, 2.5 mM CaCl2 and 1 μg/ml thrombin (Haematologic Tech. Inc.) was added for digestion at 25°C for 1 hour. The digested PDE8A1 was loaded into a Q-sepharose column (GE Healthcare) that was pre-equilibrated with a buffer of 50 mM NaCl, 20 mM Tris·HCl pH 7.5, 1 mM β-ME, and 1 mM MgCl2. The column was washed with 200 ml of 20 mM Tris·HCl pH 7.5, 100 mM NaCl, 1 mM β-ME, 1 mM EDTA and then PDE8A1 was eluted out with the same buffer but 300 mM NaCl. After concentrated to about 10 ml, the protein was loaded into Sepharyl S-300 column (GE Healthcare) and eluted with a buffer of 20 mM Tris·HCl pH 7.5, 50 mM NaCl, 1 mM β-ME, 1 mM MgCl2. The protein was finally concentrated to 10 mg/ml. A typical batch of the refolding and purification yielded about 10 mg PDE8A1 from 2 liters of the refolding buffer. The protein was estimated to have purity greater than 95%, as shown in the SDS gel.
The subcloning, expression, refolding, and purification of the PDE8A fragment (residue 205–820) were carried out in the similar condition as those for the PDE8A catalytic domain (480–820). The refolding buffer for the PDE8 fragment was 50 mM Tris.HCl, pH 7.0, 1.25 M urea, 1 M Arg, 40 mM MnCl2, 10 mM NaCl, 1 mM KCl, and 10 mM DTT.
Enzymatic assay
The enzymatic activities were assayed by using 3H-cAMP or 3H-cGMP as substrates, as previously reported (39). The catalytic domain of PDE8A1 was incubated with a reaction mixture of 20 mM Tris·HCl, pH 7.5, 4 mM MnCl2, 1 mM DTT, 3H-cAMP or 3H-cGMP (20000–40000 cpm/assay) at 24°C for 15 min. The reaction was terminated by addition of 0.2 M ZnSO4 and 0.12 M Ba(OH)2. The reaction product 3H-AMP or 3H-GMP was precipitated out by BaSO4 while unreacted 3H-cAMP or 3H-cGMP remained in supernatant. After centrifugation, the radioactivity in the supernatant was measured in a liquid scintillation counter. The turn over was controlled at hydrolysis of <30% substrate under a suitable enzyme concentration (0.004 to 0.085 μg/ml PDE8A1). Twelve concentrations of cAMP or cGMP in a range of 0.04 to 50 μM were used to obtain the kinetic parameters.
The enzymatic properties were analyzed by the steady state kinetics (40). The non-linear regression of the Michealis-Menten equation, as well as Eadie-Hofstee plots were analyzed to obtain the values of KM, Vmax, and kcat. For measurement of IC50, ten concentrations of inhibitors were used at the substrate concentration of <1/10 KM and the suitable enzyme concentration. All measurements were repeated three times.
Crystallization and structure determination of PDE8A
The crystal of the unliganded catalytic domain of PDE8A1 (480–820) was grown by vapor diffusion of 10 mg/ml PDE8A1 against a well buffer of 100 mM Na cacodylate (pH 6.5), 15% isopropanol, 30% ethylene glycol, 10% PEG3350 at 4°C. The unliganded crystal has the space group C2221 with a = 76.3, b = 132.7, and c = 101.5 Å. The PDE8A1-IBMX crystal was grown by the hanging drop against the same buffer for the unliganded PDE8A1, except for 8% PEG3350. The PDE8A1-IBMX crystal has the space group P21 with cell dimensions of a = 76.3, b = 101.6, c = 76.6 Å and β = 119.9°. The diffraction data were collected on beamline X29 in Brookhaven National Laboratory and processed by program HKL (41). The unliganded PDE8A1 structure was solved by the molecular replacement program AMoRe (42), with PDE4D2 as the initial model. The structure of PDE8A1-IBMX was solved by using the unliganded PDE8A1 as the model. The atomic models were rebuilt by program O (43) against the electron density maps that were improved by the density modification package of CCP4. The structures were refined by CNS (Table 1, 44).
Table 1.
Statistics on diffraction data and structure refinement
| Data collection | Unliganded PDE8A | PDE8A-IBMX |
|---|---|---|
| Space group | C2221 | P21 |
| Unit cell (a, b, c, Å, β) | 76.3, 132.7, 101.5 | 76.3, 101.6, 76.6, 119.9° |
| Resolution (Å) | 1.9 | 2.1 |
| Unique reflections | 36,345 | 52,808 |
| Fold of redundancy | 7.4 | 4.0 |
| Completeness (%) | 89.0 (45.8)* | 90.7 (59.6) |
| Average I/σ | 15.2 (2.8)* | 13.0 (2.9) |
| Rmerge | 0.054 (0.26)* | 0.043 (0.23) |
| Structure Refinement | ||
| R-factor | 0.236 | 0.221 |
| R-free | 0.265 (10%)‡ | 0.256 (10%) |
| Resolution (Å) | 30–1.9 | 30–2.1 |
| Reflections | 35,445 | 52,075 |
| RMS deviation for | ||
| Bond (Å) | 0.005 | 0.006 |
| Angle | 1.1° | 1.1° |
| Average B-factor (Å2) | ||
| Protein | 47.9 (2744)§ | 51.0 (5488) |
| IBMX | 75.5(32)§ | |
| Waters | 40.2 (64) § | 41.5 (134) |
| Zn | 34.9 (1) | 40.2 |
| Mg | 36.2 (1) | 38.4 |
The numbers in parentheses are for the highest resolution shell.
The percentage of reflections omitted for calculation of R-free.]
The number of atoms in the crystallographic asymmetric unit.
RESULTS
Kinetic properties of the refolded PDE8A1
When 4 mM MnCl2 is used as the catalytic ion, the refolded PDE8A1 catalytic domain has KM of 1.8 ± 0.1 μM, Vmax of 6.1 ± 0.1 μmol/min/mg, and kcat of 4.0 ± 0.1 s−1 for cAMP, and KM of 1.6 ± 0.1 mM, Vmax of 2.5 ± 0.3 μmol/min/mg, and kcat of 1.6 ± 0.2 s−1 for cGMP. When 10 mM Mg2+ was used as the catalytic ion, the refolded PDE8A1 catalytic domain has KM of 7.0 ± 0.1 μM, Vmax of 4.5 ± 0.1 μmol/min/mg, and kcat of 2.9 ± 0.1 s−1 for cAMP, and KM of 1.5 ± 0.1 mM, Vmax of 0.6 ± 0.1 μmol/min/mg, and kcat of 0.4 s−1 for cGMP. The specificity constant of (kcat/KM)cAMP/(kcat/KM)cGMP was 1680 and 2200 respectively for the Mg2+ and Mn2+catalyses, thus confirming the early conclusion that PDE8 is cAMP-specific (20–23). These kinetic parameters of the refolded PDE8A catalytic domain are comparable with those of other cAMP-specific PDE families. For example, the full length PDE4D2 and the catalytic domain of PDE7A1 have the KM values of 1.5 and 0.2 μM, kcat of 3.9 and 1.6 s−1 for cAMP, and the specificity constants (kcat/KM)cAMP/(kcat/KM)cGMP of 500 and 4000, respectively (39). Thus, the substrate specificity of PDE4, PDE7 and PDE8A is determined by the apparent affinity constant KM. A notable difference among these three cAMP-specific PDEs is that the kcat of PDE8A for cGMP is about 2.5-fold lower than that for substrate cAMP, in contrast to the comparable or even higher kcat for cGMP than those for cAMP in the cases of full length PDE4D2 and the PDE7A1 catalytic domain (39). Although the 2.5-fold difference is marginal, kcat might have somewhat contribution to the substrate specificity of PDE8A. The above kinetic parameters of the PDE8A catalytic domain show that the manganese is slightly more efficient than magnesium and might thus imply that manganese is the catalytic metal ion in cells, yet to be further studied.
An unusual observation is that KM of 1.8 μM for the catalytic domain of PDE8A1 is 12–45 fold larger than the KM values of 40–150 nM for the full length PDE8A and PDE8B, as reported by four research groups (20–23). The KM disagreement is in contrast to our early observations that the full length and catalytic domains of other PDE families have comparable KM values, as shown in the cases of PDE4 (39, 45), PDE7 (39, 46), PDE9 (47), and PDE10 (48, 49). To study if the refolding process generates non-native conformation and thus false kinetic behaviors, the soluble fraction of the PDE8A1 catalytic domain from the E. coli expression was purified and assayed for its enzymatic properties. The PDE8A1 that was naturally folded in the E. coli cell has KM of 1.5 ± 0.2 μM that is well compared with KM of 1.8 μM for the refolded protein. In addition, the crystal structure of the refolded PDE8A1 has the similar topology as those of other PDE families, suggesting the biologically relevant conformation. Therefore, the different KM values between the isolated catalytic domain and the full-length PDE8A1 likely reflect their intrinsic kinetic behaviors and the regulation of the catalytic activity of PDE8 by the PAS domain. This argument is supported by our progress on the preparation of the PDE8A1 fragment (residue 205–820) that contains both PAS and catalytic domain. This fragment has KM of 0.28 ± 0.01 μM and kcat of 1.1 ± 0.01 s−1 when cAMP is used as the substrate and 4 mM MnCl2 is the catalytic ion. The KM of 280 nM for the PDE8A (205–820) fragment is 2–7 fold different from the values of 40–150 nM for the full length PDE8s in literature (20–23), in comparison to 10–45 fold difference between the catalytic domain and full-length PDE8. Therefore, the comparison of the KM values of the catalytic domain, the fragment (205–820), and the full-length PDE8 suggests that the PAS domain impacts the substrate binding and the catalysis, possibly via an allosteric mode. This hypothesis is consistent with the early observation that the full-length PDE8A had an activity about 5-fold higher than its catalytic domain and binding of the partner protein IκBβ further stimulated about 2-fold catalytic activity (36).
Structure of the PDE8A1 catalytic domain
Almost all the residues in the PDE8A1 catalytic domain (residues 480–820) were traced without ambiguity in both crystal structures, except for a few of the N- and C-terminal residues. The crystallographic asymmetric units contain one molecule in the unliganded PDE8A1 crystals, but two in the PDE8A1-IBMX complex. The two molecules in PDE8A1-IBMX apparently assemble into a dimer that is superimposable with the dimeric catalytic domain of PDE4D2. However, it is not clear if the PDE8A1 dimer in the crystal is biologically relevant. The catalytic domain of PDE8A1 contains 18 α-helices (Fig. 1), in comparison with 16 α-helices in the most other PDE families (37). Structural superposition of PDE8A-IBMX over other PDEs yielded RMS deviations of 0.94 Å for 291 comparable residues of PDE4D2-IBMX, 1.27 Å for 294 residues of PDE7A1-IBMX, and 1.37 Å for 254 residues of PDE5A1-sildenafil. This comparison indicates the similar topology of the PDE families. The H-loop or The603-Asn620 of PDE8A1, which is an integrated portion of the active site, is well superimposed over the corresponding Ser208-Asn224 of PDE4D2-IBMX (Fig. 1), as well as most other PDE families (37). However, the H-loop of PDE5A1 has been shown to exist in five different conformations upon inhibitor binding (50, 51). While the conformational changes of the H-loop in PDE5A1 clearly suggest its role in discrimination of inhibitors, it remains puzzle how the H-loop impacts the catalysis in the other PDE families.
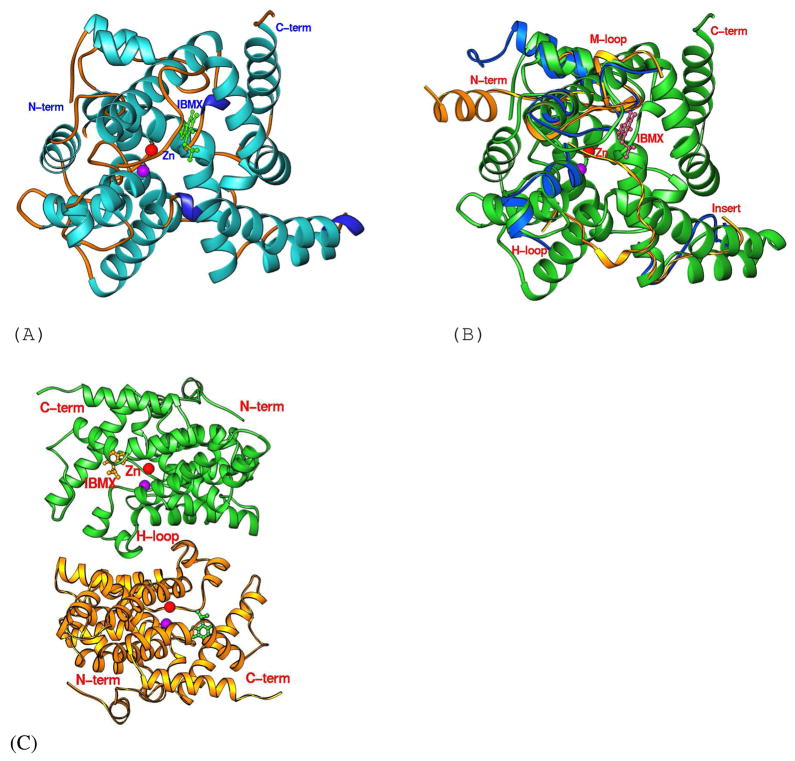
Fig. 1.
The PDE8A1 structure. (A) Ribbon diagram of the PDE8A1 catalytic domain. The purple ball represents the second divalent metal that needs to be identified. (B) Superposition of PDE8A1 (green ribbons) over PDE4D2 (blue ribbons) and PDE5A1-sildenafil (golden ribbons). The comparable portions of the PDE4D2 and PDE5A1 structures were omitted. The H-loop of PDE8A1 is similar to that of PDE4D2, but different from that of PDE5A1. (C) Dimer of PDE8A1 catalytic domain. The H-loops (residues 604-620) form the dimer interface. (D) Sequence alignment between PDE4D2 and PDE8A1.
In spite of the overall structure similarity, three regions of PDE8A1 show significant differences in the position and conformation from other PDE families. First, the N-terminal helix H1 of PDE8A1 is not comparable with other PDE families and helix H2 is superimposable with those of PDE4 and PDE7, but not PDE5 (Fig. 1). Thus, the PDE8 structure supported our early proposal (37) that the core catalytic domain starts from helix H3 or the residue around Pro512 of PDE8A1 (Pro117 of PDE4D2). Second, the PDE8A1 loop of Asn685 to Thr710 contains two α-helices and a 310-helix, and has an insert of more than 10 residues in comparison with other PDE families (Fig. 1). This loop is distant from the active site and thus unlikely plays an important role in the recognition of substrates and inhibitors. Third, the M-loop, which is defined as residues of Phe749-Ser773 of PDE8A1 or Phe341-Ser364 of PDE4D2 (50, 52), shows significant conformational variation and positional difference (Fig. 1). For example, the Cα atom of Pro764 of PDE8A1 is 3.8 Å away from the corresponding Met357 of PDE4D2 and the Cα atom of Val763 of PDE8A1 has a distance of 7.5 Å from Pro801 of PDE5A1. Since the M-loop is a component of the active site and is involved in interaction with inhibitors and substrate (52), the conformational difference of the M-loop implies its role in selective recognition of substrates and inhibitors.
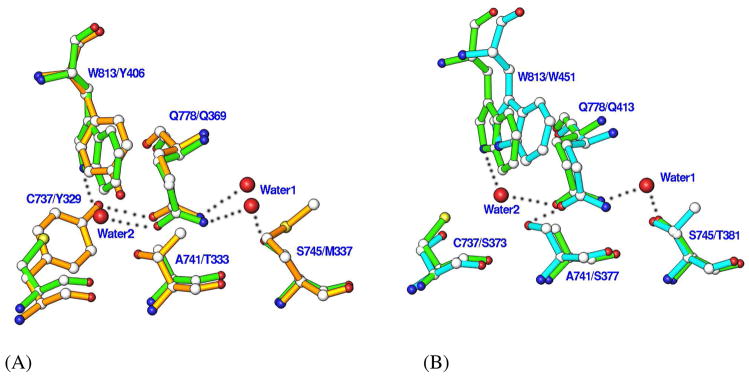
While the PDE8A active site closely resembles those of cAMP-specific PDE4 and PDE7, there are some subtle but significant differences among these structures. One notable difference is associated with the invariant glutamine (Gln778 in PDE8A1) that has been thought to play an important role in the inhibitor binding and the substrate recognition (37, 53). Gln778 of PDE8A1 occupies similar position and has the same conformation as the glutamines of PDE4 and PDE7. However, its side chain forms no hydrogen bonds with PDE8A residues, but two water molecules that respectively bind to Ser745 and Trp813 (Fig. 2). In comparison, Gln369 of PDE4D2 interacts with Tyr329 and a water molecule while Gln413 in PDE7A1 forms a hydrogen bond with Ser377 (Fig. 2). Thus, the side chain of Gln778 is more flexible than Gln369 of PDE4D2 and Gln413 of PDE7A1, and may be capable of switching its conformation. This observation is negative evidence for the hypothesis of “glutamine switch”, in which the invariant glutamine was assumed to have a fixed orientation in the cAMP- or cGMP-specific PDE families (53).
Fig. 2.
Superposition of the active site residues of the cAMP-specific PDE families. (A) Comparison between PDE8A1 (green) and PDE4D2 (golden). Dotted lines represent the hydrogen bond. The residues are labeled in a format of PDE8A1/PDE4D2. (B) Comparison between PDE8A1 (green) and PDE7A1 (cyan). The residues are labeled in a format of PDE8A1/PDE7A1.
IBMX binding at the active site of PDE8A1
Since no PDE8 selective inhibitors are available at the present, dipyridamole and IBMX were used for the structural studies to reveal inhibitor binding. Dipyridamole is a non-selective inhibitor for most PDE families and has IC50 of 0.5–1.5 μM for PDE5 (54) and 4–9 μM for PDE8 (20, 21). The co-crystal of the PDE8A1 catalytic domain with dipyridamole showed that dipyridamole occupies the active site of PDE8 in a similar pattern to the inhibitor binging in other PDE families. However, the conformation of dipyridamole could not be unambiguously determined in the crystal for unclear reason (unpublished data).
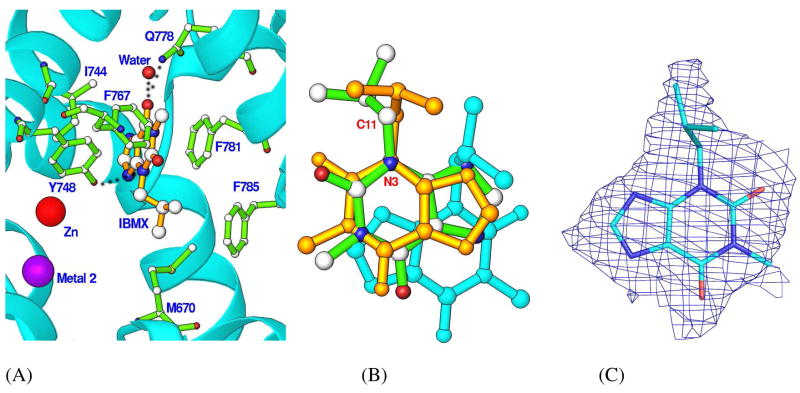
IBMX is a non-selective inhibitor for most PDE families with IC50 of 2–50 μM (55), but does not effectively inhibit PDE8 activity at concentration up to 200 μM (20–23). This is confirmed by the IC50 of 698 ± 29 μM for the refolded catalytic domain of PDE8A1. Nevertheless, we hope, based on our experience in the co-crystallization of IBMX with PDE9, another IBMX-insensitive PDE family (47), whose PDE8A-IBMX crystal revealed meaningful information on the inhibitor binding. Indeed, the co-crystal of PDE8A1 with 2 mM IBMX showed IBMX binding to the active site of PDE8A1 (Fig. 3). The electron density in the (2Fo-Fc) and (Fo-Fc) maps clearly revealed the stack of the xanthine ring of IBMX against Phe781 of PDE8A1, while the isobutyl group of IBMX has weaker electron density and is flexible (Fig. 3). In addition, IBMX forms a hydrogen bond with each of the side chains of Tyr748 and Gln778, and makes van der Waals’ interactions with residues Met670, Ile744, Tyr748, Phe767, Phe781, and Phe785.
Fig. 3.
IBMX binding to PDE8A1. (A) The interaction of IBMX with the active site residues of PDE8A1. The oxygen of the side chain of Tyr748 forms a hydrogen bond with N3 of IBMX as marked with the dotted line, but has an energetically unfavorable distance of 2.9 Å to C11 of IBMX (the atoms are marked in Fig. 3B). (B) The superposition of IBMXs of PDE8A1 (green sticks), PDE4D2 (golden sticks), and PDE5A1 (cyan sticks). (C) The (Fo-Fc) map for IBMX, which was contoured at 2.5 sigmas.
The binding of IBMX does not significantly change the conformation of PDE8A1, as shown by the very small RMS deviation (0.24 Å) for the structural superposition between the unliganded PDE8A1 and PDE8A1-IBMX. This is consistent with the observations of no significant conformational changes induced by IBMX binding in the most other PDE families, except for PDE5 (39, 47, 56). In addition, two characteristic features of the IBMX binding are conserved in the reported PDE structures: the stack against a phenylalanine and the hydrogen bond with the invariant glutamine (Gln778 in PDE8A1). However, the orientation and position of IBMX show some changes in the structures (Fig. 3). These differences might not be significant, but reflect the fact that the binding pockets of PDEs are much larger than the size of IBMX so as to allow various orientations.
Tyr748 is an important residue for selectivity of PDE8 inhibitors
The structure of the PDE8A-IBMX complex reveals that the side chain oxygen of Tyr748 forms a hydrogen bond with N3 of IBMX, but also has a distance of 2.9 Å from the neighboring carbon C11 (Fig. 3). Since the van der Waal force has typically 3.5 Å for an O-C interaction, we believed that this unfavorable interaction between the tyrosine and the carbon is a factor making IBMX bound weakly. In fact, Tyr748 is typically a phenylalanine in most PDE families, which forms a hydrophobic environment for the inhibitor stack against Phe781. Thus, a polar oxygen atom in this environment will weaken the hydrophobic interaction. This might be the reason why so many PDE inhibitors bind PDE8 poorly. On the other hand, this Tyr748 could serve as one of discriminating residues to enhance PDE8 selectivity.
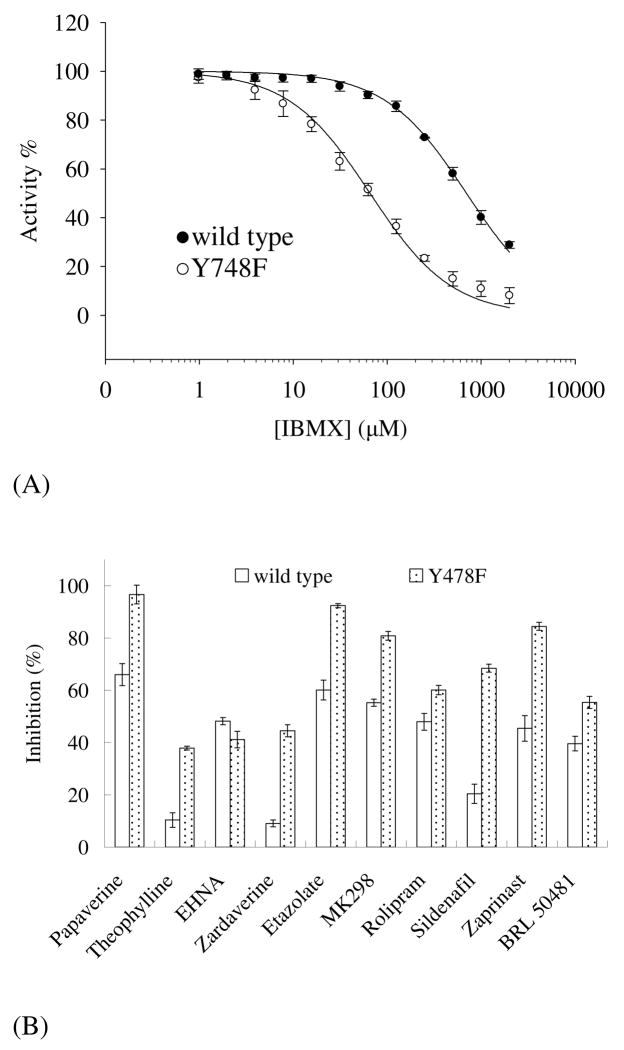
To test this hypothesis, Tyr748 was mutated to phenylalanine that exists in all other PDE families except for PDE8 and PDE9. The Y748F mutant has KM of 0.7 ± 0.1 μM, kcat of 4.3 ± 0.1 s−1, and Vmax of 6.6 ± 0.2 μmol/min/mg. Thus, the catalytic efficiency of the mutant, kcat/KM is about 2-fold better than that of the wild type PDE8A1. In addition, the Y748F mutant of PDE8A1 has IC50 of 65.8 ± 5.0 μM for IBMX (Fig. 4), which is about 10-fold better than the IC50 of 698 μM for the wild type PDE8A1. However, the Y748F mutant has IC50 of 4.1 ± 0.4 μM for dipyridamole, which is comparable with 6.0 ± 0.4 μM for the wild type PDE8A1. We also measured the inhibitory potency of the following PDE inhibitors to elucidate the impact of the mutation: non-selective inhibitors papaverine and theophylline, PDE2 inhibitor EHNA (57), PDE3/4 dual-selective zardaverine (58), PDE4 inhibitors etazolate, rolipram, and MK298 (59, 60), PDE5 inhibitors sildenafil and zaprinast (10), and PDE7 inhibitor BRL50481 (61). As shown in Fig. 4, the most inhibitors except for EHNA demonstrated some degree of improved inhibitory effects on the Y748F mutant. Taking these data together, we believe that Tyr748 is likely a residue useful for development of PDE8 selective inhibitors.
Fig. 4.
Inhibition of PDE8A1. (A) Inhibition by IBMX. (B) Inhibition of the wild type PDE8A1 and its Y748F mutant by the inhibitors of papaverine and theophylline (non-selective), EHNA (PDE2), zardaverine (PDE3/4), etazolate, rolipram, and MK298 (PDE4), sildenafil and zaprinast (PDE5), and BRL50481 (PDE7). The concentrations used were 100 μM for all inhibitors and 10–20 ng/ml for PDEs. The activities of the enzymes in the absence of the inhibitors are taken as 100%.
DISCUSSION
Structure information has played a critical role in the discovery of drugs for treatment of human diseases (62). PDE8 is one out of three PDE families whose crystal structures remain to be solved (37). The unavailability of PDE8 structures is apparently due to the difficulty on preparation of large quantity of the active enzyme. Although several research groups have reported the expression of PDE8A and 8B in the baculovirus systems, the purified proteins often have low catalytic activities (20–23). For example, the Vmax of 0.15 μmol/min/mg for a 545-residue fragment of PDE8A (20) is about 40-fold less active than our PDE8A catalytic domain (Vmax of 6.1 μmol/min/mg). A fragment of C-terminal 584 amino acids of PDE8B that was expressed in E. coli had Vmax of 0.14 nmol/min/mg (22), which is over four magnitudes slower than that of our refolded PDE8A catalytic domain. Therefore, the procedure for the expression and refolding of the PDE8A catalytic domain in this paper is valuable for not only the basic studies, but also design of PDE8 inhibitors.
PDE8 plays important roles in many physiological processes such as modulation of testosterone production (28–33). However, no PDE8 selective inhibitors are available for trial treatment of any diseases. PDE8 is insensitive to many PDE inhibitors, including IBMX, rolipram, sildenafil, and zaprinast (20–23). The crystal structure of PDE8A-IBMX reveals unique characteristic conformation of the PDE8 active site and identified Tyr748 as a key factor responsible for the insensitivity of PDE8 to most PDE inhibitors. The Y748F mutation makes PDE8A more sensitive to many of PDE inhibitors (Fig. 4), thus supporting the speculation that Tyr748 is an important residue for design of PDE8 inhibitors.
Acknowledgments
We thank beamline X29 at NSLS for collection of the diffraction data.
This work was supported in part by NIH GM59791 to HK, the 985 project of Science Foundation of Sun Yat-sen University (JC), and the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health (HR).
Abbreviation
- PDE
hosphodiesterase
- cAMP
adenosine 3′,5′-cyclic monophosphate
- cGMP
guanosine 3′,5′-cyclic monophosphate
- IBMX
3-isobutyl-1-methylxanthine
- Per
Period clock protein
- ARNT
Aryl hydrocarbon receptor nuclear translocator
- Sim
Single-minded protein
- PAS
Per-ARNT-Sim
Footnotes
The atomic coordinates and structure factors have been deposited into the Protein Data Bank with entry codes of 3ECM and 3ECN
References
- 1.Antoni F. Molecular diversity of cyclic AMP signaling. Front Neuroendocrin. 2000;21:103–132. doi: 10.1006/frne.1999.0193. [DOI] [PubMed] [Google Scholar]
- 2.Zaccolo M, Movsesian MA. Molecular diversity of cyclic AMP signaling. Circ Res. 2007;100:1569–1578. doi: 10.1161/CIRCRESAHA.106.144501. [DOI] [PubMed] [Google Scholar]
- 3.De Felice FG, Wasilewska-Sampaio AP, Barbosa AC, Gomes FC, Klein WL, Ferreira ST. Cyclic AMP enhancers and Abeta oligomerization blockers as potential therapeutic agents in Alzheimer’s disease. Curr Alzheimer Res. 2007;4:263–271. doi: 10.2174/156720507781077287. [DOI] [PubMed] [Google Scholar]
- 4.Piper M, van Horck F, Holt C. The role of cyclic nucleotides in axon guidance. Adv Exp Med Biol. 2007;621:134–143. doi: 10.1007/978-0-387-76715-4_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320:949–953. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender AT, Beavo JA. Cyclic nucleotide phosphodiesterases: molecular regulation to clinical use. Pharmacol Rev. 2006;58:488–520. doi: 10.1124/pr.58.3.5. [DOI] [PubMed] [Google Scholar]
- 7.Lugnier C. Cyclic nucleotide phosphodiesterase (PDE) superfamily: a new target for the development of specific therapeutic agents. Pharmacol Ther. 2006;109:366–398. doi: 10.1016/j.pharmthera.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Omori K, Kotera J. Overview of PDEs and their regulation. Circ Res. 2007;100:309–327. doi: 10.1161/01.RES.0000256354.95791.f1. [DOI] [PubMed] [Google Scholar]
- 9.Conti M, Beavo J. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: Essential components in cyclic nucleotide signaling. Ann Rev Biochem. 2007;76:481–511. doi: 10.1146/annurev.biochem.76.060305.150444. [DOI] [PubMed] [Google Scholar]
- 10.Rotella DP. Phosphodiesterase 5 inhibitors: current status and potential applications. Nature Rev Drug Discovery. 2002;1:674–682. doi: 10.1038/nrd893. [DOI] [PubMed] [Google Scholar]
- 11.Schrör K. The pharmacology of cilostazol. Diabetes Obes Metab. 2002;4:S14–S19. doi: 10.1046/j.1463-1326.2002.0040s2s14.x. [DOI] [PubMed] [Google Scholar]
- 12.Lipworth BJ. Phosphodiesterase-4 inhibitors for asthma and chronic obstructive pulmonary disease. Lancet. 2005;365:167–175. doi: 10.1016/S0140-6736(05)17708-3. [DOI] [PubMed] [Google Scholar]
- 13.Castro A, Jerez MJ, Gil C, Martinez A. Cyclic nucleotide phosphodiesterases and their role in immunomodulatory responses: advances in the development of specific phosphodiesterase inhibitors. Med Res Rev. 2005;25:229–244. doi: 10.1002/med.20020. [DOI] [PubMed] [Google Scholar]
- 14.Houslay MD, Schafer P, Zhang KY. Keynote review: phosphodiesterase-4 as a therapeutic target. Drug Discov Today. 2005;10:1503–1519. doi: 10.1016/S1359-6446(05)03622-6. [DOI] [PubMed] [Google Scholar]
- 15.Blokland A, Schreiber R, Prickaerts J. Improving memory: a role for phosphodiesterases. Curr Pharm Des. 2006;12:2511–2523. doi: 10.2174/138161206777698855. [DOI] [PubMed] [Google Scholar]
- 16.Menniti FS, Faraci WS, Schmidt CJ. Phosphodiesterases in the CNS: targets for drug development. Nat Rev Drug Discov. 2006;5:660–670. doi: 10.1038/nrd2058. [DOI] [PubMed] [Google Scholar]
- 17.Gales BJ, Gales MA. Phosphodiesterase-5 inhibitors for lower urinary tract symptoms in men. Ann Pharmacother. 2008;42:111–115. doi: 10.1345/aph.1K422. [DOI] [PubMed] [Google Scholar]
- 18.Galie N, Ghofrani HA, Torbicki A, Barst RJ, Rubin LJ, Badesch D, Fleming T, Parpia T, Burgess G, Branzi A, Grimminger F, Kurzyna M, Simonneau G. Sildenafil citrate therapy for pulmonary arterial hypertension. N Engl J Med. 2005;353:2148–2157. doi: 10.1056/NEJMoa050010. [DOI] [PubMed] [Google Scholar]
- 19.Lavan BE, Lakey T, Houslay MD. Resolution of soluble cyclic nucleotide phosphodiesterase isoenzymes, from liver and hepatocytes, identifies a novel IBMX-insensitive form. Biochem Pharmacol. 1989;38:4123–4136. doi: 10.1016/0006-2952(89)90694-1. [DOI] [PubMed] [Google Scholar]
- 20.Fisher DA, Smith JF, Pillar JS, St Denis SH, Cheng JB. Isolation and characterization of PDE8A, a novel human cAMP-specific phosphodiesterase. Biochem Biophys Res Commun. 1998;246:570–577. doi: 10.1006/bbrc.1998.8684. [DOI] [PubMed] [Google Scholar]
- 21.Soderling SH, Bayuga SJ, Beavo JA. Cloning and characterization of a cAMP-specific cyclic nucleotide phosphodiesterase. Proc Natl Acad Sci USA. 1998;95:8991–8996. doi: 10.1073/pnas.95.15.8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayashi M, Matsushima K, Ohashi H, Tsunoda H, Murase S, Kawarada Y, Tanaka T. Molecular cloning and characterization of human PDE8B, a novel thyroid-specific isozyme of 3′,5′-cyclic nucleotide phosphodiesterase. Biochem Biophys Res Commun. 1998;250:751–756. doi: 10.1006/bbrc.1998.9379. [DOI] [PubMed] [Google Scholar]
- 23.Gamanuma M, Yuasa K, Sasaki T, Sakurai N, Kotera J, Omori K. Comparison of enzymatic characterization and gene organization of cyclic nucleotide phosphodiesterase 8 family in humans. Cell Signal. 2003;15:565–574. doi: 10.1016/s0898-6568(02)00146-8. [DOI] [PubMed] [Google Scholar]
- 24.Wang P, Wu P, Egan RW, Billah MM. Human phosphodiesterase 8A splice variants: cloning, gene organization, and tissue distribution. Gene. 2001;280:183–194. doi: 10.1016/s0378-1119(01)00783-1. [DOI] [PubMed] [Google Scholar]
- 25.Hayashi M, Shimada Y, Nishimura Y, Hama T, Tanaka T. Genomic organization, chromosomal localization, and alternative splicing of the human phosphodiesterase 8B gene. Biochem Biophys Res Commun. 2002;297:1253–1258. doi: 10.1016/s0006-291x(02)02371-9. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi T, Gamanuma M, Sasaki T, Yamashita Y, Yuasa K, Kotera J, Omori K. Molecular comparison of rat cyclic nucleotide phosphodiesterase 8 family: unique expression of PDE8B in rat brain. Gene. 2003;319:21–31. doi: 10.1016/s0378-1119(03)00809-6. [DOI] [PubMed] [Google Scholar]
- 27.Pérez-Torres S, Cortés R, Tolnay M, Probst A, Palacios JM, Mengod G. Alterations on phosphodiesterase type 7 and 8 isozyme mRNA expression in Alzheimer’s disease brains examined by in situ hybridization. Exp Neurol. 2003;182:322–334. doi: 10.1016/s0014-4886(03)00042-6. [DOI] [PubMed] [Google Scholar]
- 28.Glavas NA, Ostenson C, Schaefer JB, Vasta V, Beavo JA. T cell activation up-regulates cyclic nucleotide phosphodiesterases 8A1 and 7A3. Proc Natl Acad Sci USA. 2001;98:6319–6324. doi: 10.1073/pnas.101131098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong H, Osmanova V, Epstein PM, Brocke S. Phosphodiesterase 8 (PDE8) regulates chemotaxis of activated lymphocytes. Biochem Biophys Res Commun. 2006;345:713–719. doi: 10.1016/j.bbrc.2006.04.143. [DOI] [PubMed] [Google Scholar]
- 30.Vasta V, Shimizu-Albergine M, Beavo JA. Modulation of Leydig cell function by cyclic nucleotide phosphodiesterase 8A. Proc Natl Acad Sci USA. 2006;103:19925–19930. doi: 10.1073/pnas.0609483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dov A, Abramovitch E, Warwar N, Nesher R. Diminished phosphodiesterase-8B potentiates biphasic insulin response to glucose. Endocrinology. 2008;149:741–748. doi: 10.1210/en.2007-0968. [DOI] [PubMed] [Google Scholar]
- 32.Horvath A, Mericq V, Stratakis CA. Mutation in PDE8B, a cyclic AMP-specific phosphodiesterase in adrenal hyperplasia. N Engl J Med. 2008;358:750–752. doi: 10.1056/NEJMc0706182. [DOI] [PubMed] [Google Scholar]
- 33.Arnaud-Lopez L, Usala G, Ceresini G, Mitchell BD, Pilia MG, Piras MG, Sestu N, Maschio A, Busonero F, Albai G, Dei M, Lai S, Mulas A, Crisponi L, Tanaka T, Bandinelli S, Guralnik JM, Loi A, Balaci L, Sole G, Prinzis A, Mariotti S, Shuldiner AR, Cao A, Schlessinger D, Uda M, Abecasis GR, Nagaraja R, Sanna S, Naitza S. Phosphodiesterase 8B gene variants are associated with serum TSH levels and thyroid function. Am J Hum Genet. 2008;82:1270–1280. doi: 10.1016/j.ajhg.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crews ST, Fan CM. Remembrance of things PAS: regulation of development by bHLH-PAS proteins. Curr Opin Genet Dev. 1999;9:580–587. doi: 10.1016/s0959-437x(99)00003-9. [DOI] [PubMed] [Google Scholar]
- 35.Wenger RH, Katschinski DM. The hypoxic testis and post-meiotic expression of PAS domain proteins. Semin Cell Dev Biol. 2005;16:547–553. doi: 10.1016/j.semcdb.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 36.Wu P, Wang P. Per-Arnt-Sim domain-dependent association of cAMP-phosphodiesterase 8A1 with IkappaB proteins. Proc Natl Acad Sci USA. 2004;101:17634–17639. doi: 10.1073/pnas.0407649101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ke H, Wang H. Crystal structures of phosphodiesterases and implications on substrate specificity and inhibitor selectivity. Curr Top Med Chem. 2007;7:391–403. doi: 10.2174/156802607779941242. [DOI] [PubMed] [Google Scholar]
- 38.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. In: The Proteomics Protocols Handbook. Walker John M., editor. Humana Press; 2005. pp. 571–607. [Google Scholar]
- 39.Wang H, Liu Y, Chen Y, Robinson H, Ke H. Multiple elements jointly determine inhibitor selectivity of cyclic nucleotide phosphodiesterases 4 and 7. J Biol Chem. 2005;280:30949–30955. doi: 10.1074/jbc.M504398200. [DOI] [PubMed] [Google Scholar]
- 40.Fersht A. Structure and mechanism in protein science. Freeman and Company; New York: 1999. pp. 103–132. [Google Scholar]
- 41.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 42.Navaza J, Saludjian P. AMoRe: an automated molecular replacement program package. Methods Enzymol. 1997;276:581–594. doi: 10.1016/S0076-6879(97)76079-8. [DOI] [PubMed] [Google Scholar]
- 43.Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 44.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 45.Bolger GB, Erdogan S, Jones RE, Loughney K, Scotland G, Hoffmann R, Wilkinson I, Farrell C, Houslay MD. Characterization of five different proteins produced by alternatively spliced mRNAs from the human cAMP-specific phosphodiesterase PDE4D gene. Biochem J. 1997;328:539–548. doi: 10.1042/bj3280539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Michaeli T, Bloom TJ, Martins T, Loughney K, Ferguson K, Riggs M, Rodgers L, Beavo JA, Wigler M. Isolation and characterization of a previously undetected human cAMP phosphodiesterase by complementation of cAMP phosphodiesterase-deficient Saccharomyces cerevisiae. J Biol Chem. 1993;268:12925–12932. [PubMed] [Google Scholar]
- 47.Huai Q, Wang H, Zhang W, Colman R, Robinson H, Ke H. Crystal structure of phosphodiesterase 9 in complex with inhibitor IBMX. Proc Natl Acad Sci USA. 2004;101:9624–9629. doi: 10.1073/pnas.0401120101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soderling SH, Bayuga SJ, Beavo JA. Isolation and characterization of a dual-substrate phosphodiesterase gene family: PDE10A. Proc Natl Acad Sci USA. 1999;96:7071–7076. doi: 10.1073/pnas.96.12.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang H, Liu Y, Hou J, Zheng M, Robinson H, Ke H. Structural insight into substrate specificity of phospodiesterase 10. Proc Natl Acad Sci, USA. 2007;104:5782–5787. doi: 10.1073/pnas.0700279104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H, Liu Y, Huai Q, Cai J, Zoraghi R, Francis SH, Corbin JD, Robinson H, Xin Z, Lin G, Ke H. Multiple conformations of phosphodiesterase-5: implications for enzyme function and drug development. J Biol Chem. 2006;281:21469–21479. doi: 10.1074/jbc.M512527200. [DOI] [PubMed] [Google Scholar]
- 51.Wang H, Ye M, Robinson H, Francis SH, Ke H. Conformational variations of both PDE5 and inhibitors provide the structural basis for the physiological effects of vardenafil and sildenafil. Mol Pharmacol. 2008;73:104–110. doi: 10.1124/mol.107.040212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huai Q, Wang H, Sun Y, Kim HY, Liu Y, Ke H. Three dimensional structures of PDE4D in complex with roliprams and implication on inhibitor selectivity. Structure. 2003;11:865–873. doi: 10.1016/s0969-2126(03)00123-0. [DOI] [PubMed] [Google Scholar]
- 53.Zhang KY, Card GL, Suzuki Y, Artis DR, Fong D, Gillette S, Hsieh D, Neiman J, West BL, Zhang C, Milburn MV, Kim SH, Schlessinger J, Bollag G. A glutamine switch mechanism for nucleotide selectivity by phosphodiesterases. Mol Cell. 2004;15:279–286. doi: 10.1016/j.molcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 54.Wang P, Wu P, Myers JG, Stamford A, Egan RW, Billah MM. Characterization of human, dog and rabbit corpus cavernosum type 5 phosphodiesterases. Life Sci. 2001;68:1977–1987. doi: 10.1016/s0024-3205(01)00989-4. [DOI] [PubMed] [Google Scholar]
- 55.Beavo JA, Rogers NL, Crofford OB, Hardman JG, Sutherland EW, Newman EV. Effects of xanthine derivatives on lipolysis and on adenosine 3′,5′-monophosphate phosphodiesterase activity. Mol Pharmacol. 1970;6:597–603. [PubMed] [Google Scholar]
- 56.Huai Q, Liu Y, Francis SH, Corbin JD, Ke H. Crystal structures of phosphodiesterases 4 and 5 in complex with inhibitor IBMX suggest a conformation determinant of inhibitor selectivity. J Biol Chem. 2004;279:13095–13101. doi: 10.1074/jbc.M311556200. [DOI] [PubMed] [Google Scholar]
- 57.Podzuweit T, Nennstiel P, Muller A. Isozyme selective inhibition of cGMP-stimulated cyclic nucleotide phosphodiesterases by erythro-9-(2-hydroxy-3-nonyl) adenine. Cell Signal. 1995;7:733–778. doi: 10.1016/0898-6568(95)00042-n. [DOI] [PubMed] [Google Scholar]
- 58.Schudt C, Winder S, Müller B, Ukena D. Zardaverine as a selective inhibitor of phosphodiesterase isozymes. Biochem Pharmacol. 1991;42:153–162. doi: 10.1016/0006-2952(91)90694-z. [DOI] [PubMed] [Google Scholar]
- 59.Wang P, Myers JG, Wu P, Cheewatrakoolpong B, Egan RW, Billah MM. Expression, purification, and characterization of human cAMP-specific phosphodiesterase (PDE4) subtypes A, B, C, and D. Biochem Biophys Res Communi. 1997;234:320–324. doi: 10.1006/bbrc.1997.6636. [DOI] [PubMed] [Google Scholar]
- 60.Huai Q, Sun Y, Wang H, Macdonald D, Aspiotis R, Robinson H, Huang Z, Ke H. Enantiomer discrimination illustrated by high resolution crystal structures of type 4 phosphodiesterase. J Med Chem. 2006;49:1867–1873. doi: 10.1021/jm051273d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith SJ, Cieslinski LB, Newton R, Donnelly LE, Fenwick PS, Nicholson AG, Barnes PJ, Barnette MS, Giembycz MA. Discovery of BRL 50481 [3-(N, N-dimethylsulfonamido)-4-methyl-nitrobenzene], a selective inhibitor of phosphodiesterase 7: in vitro studies in human monocytes, lung macrophages, and CD8+ T-lymphocytes. Mol Pharmacol. 2004;66:1679–1689. doi: 10.1124/mol.104.002246. [DOI] [PubMed] [Google Scholar]
- 62.Zhou JZ. Structure-directed combinatorial library design. Curr Opin Chem Biol. 2008;12:379–385. doi: 10.1016/j.cbpa.2008.02.007. [DOI] [PubMed] [Google Scholar]