Abstract
Polyadenylation is an important factor controlling RNA degradation and RNA quality control mechanisms. In this report we demonstrate for the first time that RNase R has in vivo affinity for polyadenylated RNA and can be a key enzyme involved in poly(A) metabolism. RNase II and PNPase, two major RNA exonucleases present in Escherichia coli, could not account for all the poly(A)-dependent degradation of the rpsO mRNA. RNase II can remove the poly(A) tails but fails to degrade the mRNA as it cannot overcome the RNA termination hairpin, while PNPase plays only a modest role in this degradation. We now demonstrate that in the absence of RNase E, RNase R is the relevant factor in the poly(A)-dependent degradation of the rpsO mRNA. Moreover, we have found that the RNase R inactivation counteracts the extended degradation of this transcript observed in RNase II-deficient cells. Elongated rpsO transcripts harboring increasing poly(A) tails are specifically recognized by RNase R and strongly accumulate in the absence of this exonuclease. The 3′ oligo(A) extension may stimulate the binding of RNase R, allowing the complete degradation of the mRNA, as RNase R is not susceptible to RNA secondary structures. Moreover, this regulation is shown to occur despite the presence of PNPase. Similar results were observed with the rpsT mRNA. This report shows that polyadenylation favors in vivo the RNase R-mediated pathways of RNA degradation.
Keywords: poly(A) polymerase I, polyadenylation, PNPase, RNase II, RNase R, rpsO mRNA
INTRODUCTION
Polyadenylation is a universal post-transcriptional RNA modification that can modulate gene expression (Sarkar 1997; Dreyfus and Régnier 2002). It is implicated in RNA quality control mechanisms and modulates RNA stability in all domains of life (Slomovic et al. 2006, 2008; Doma and Parker 2007; Houseley and Tollervey 2008). In Escherichia coli, the majority of mRNAs undergo polyadenylation in exponentially growing cells (Mohanty and Kushner 2006). Rho-independent transcriptional terminators act as poly(A) signals that are recognized by poly(A) polymerase I (PAP I, encoded by the pcnB gene), the enzyme responsible for >90% of polyadenylation (O'Hara et al. 1995; Yehudai-Resheff and Schuster 2000; Mohanty and Kushner 2006). Several types of RNA molecules, including mRNA, tRNA, and rRNA are substrates for PAP I (Cao et al. 1997; Li et al. 1998b; Mohanty and Kushner 1999; Khemici and Carpousis 2004) and poly(A) tails are also important in the control of noncoding RNAs and their regulatory pathways (Xu and Cohen 1995; Viegas et al. 2007; Andrade and Arraiano 2008; Reichenbach et al. 2008). Recently, it was demonstrated that polyadenylation can be an important factor controlling protein production (Joanny et al. 2007).
In prokaryotes, poly(A) tails generally destabilize RNA by facilitating exonucleolytic attack. Addition of a poly(A) tract to the 3′ end of RNA helps the binding of 3′–5′ exonucleases and promote RNA degradation. Rounds of polyadenylation and exonucleolytic digestion can overcome RNA secondary structures and complete decay (Coburn and Mackie 1996; Haugel-Nielsen et al. 1996; Régnier and Arraiano 2000). In yeast, the addition of small poly(A) stretches by the TRAMP complex targets RNA to degradation by the exosome, in close resemblance to the prokaryotic system (LaCava et al. 2005; Vanácová et al. 2005; Wyers et al. 2005).
Poly(A)-dependent ribonucleases play important roles in the RNA surveillance and degradation pathways. E. coli RNase II and PNPase are two major degradative exonucleases involved in the control of mRNA stability (Régnier and Arraiano 2000). Polyadenylated RNA is a preferred substrate for both enzymes (Lisitsky and Schuster 1999; Mohanty and Kushner 2000; Amblar et al. 2006; Arraiano et al. 2008). Other factors such as the RNA chaperone Hfq can promote polyadenylation (Hajnsdorf and Régnier 2000; Mohanty et al. 2004). Nevertheless, the metabolism of polyadenylated RNA is still not entirely understood. The rpsO mRNA encoding for the ribosomal protein S15 has been widely used as a model of study for polyadenylation-mediated mRNA turnover (Régnier and Hajnsdorf 1991; Hajnsdorf et al. 1995; Folichon et al. 2005). Work by Marujo et al. (2000) showed that RNase II paradoxically protects this mRNA from degradation by removing the poly(A) tails that can act as a substrate to other ribonuclease(s). Notably, PNPase is not the major enzyme involved in this poly(A)-dependent degradation. Data suggested that at least one yet unknown poly(A)-dependent ribonuclease is involved in this complex regulation (Hajnsdorf et al. 1994, 1995; Marujo et al. 2000).
An excellent candidate for this is the widely distributed RNase R. RNase R-like enzymes can be part of multiprotein complexes involved in RNA degradation (Purusharth et al. 2005; Houseley et al. 2006). This 3′–5′ exonuclease is highly effective against structured RNA and has recently been described to be important in RNA quality control, processing, and turnover (Cairrão et al. 2003; Cheng and Deutscher 2005; Oussenko et al. 2005; Andrade et al. 2006; Richards et al. 2006; Purusharth et al. 2007). There are organisms such as Mycoplasma, where RNase R is the only exonuclease present (Lalonde et al. 2007). RNase R and the catalytic subunit of eukaryotic exosomes (Rrp44/Dis3) belong to the same RNase II family of enzymes (Frazão et al. 2006; Liu et al. 2006; Barbas et al. 2008; Lorentzen et al. 2008). Rrp44/Dis3 is involved in the degradation of polyadenylated RNA (Chekanova et al. 2007; Dziembowski et al. 2007; Ibrahim et al. 2008; Schneider et al. 2007). Bacterial RNase R shows in vitro affinity for polyadenylates (Cheng and Deutscher 2002; Vincent and Deutscher 2006; Amblar et al. 2007). However, the in vivo role of RNase R on poly(A)-mediated degradation of mRNA was elusive.
In this work, we demonstrate that RNase R is the major enzyme responsible for the poly(A)-dependent degradation of the rpsO mRNA. Moreover, RNase R is shown to have a more relevant effect on rpsO mRNA stability than PNPase. In the absence of RNase R, the rpsO transcript presents longer poly(A) tails and has higher stability. These new findings highlight the role of RNase R in RNA degradation. The importance of RNase R-mediated pathways of decay in polyadenylated RNA metabolism is discussed.
RESULTS
rpsO mRNA is more efficiently degraded by RNase R than by PNPase
Exonucleolytic poly(A)-dependent degradation of rpsO mRNA becomes predominant when the primary pathway of decay mediated by RNase E is inactive. Accordingly, the rpsO mRNA is dramatically stabilized upon pcnB inactivation provided RNase E is inactive. Similar results are obtained when analyzing a mutated rpsO mRNA lacking the main RNase E cleavage site (Hajnsdorf et al. 1995; Marujo et al. 2003). For this reason the experiments below were intended to investigate the role of 3′–5′ exonucleases in the poly(A)-dependent decay were performed in RNase E deficient strains. Previous work showed that RNase II inactivation results in the destabilization of the rpsO mRNA as a consequence of 3′ end elongation of this transcript due to polyadenylation (Marujo et al. 2000). PNPase deficiency, on the other hand, did not significantly affect its stability (Hajnsdorf et al. 1994, 1995; Braun et al. 1996). We, therefore, hypothesized that RNase R, the other exonuclease involved in mRNA turnover, could be responsible for this decay.
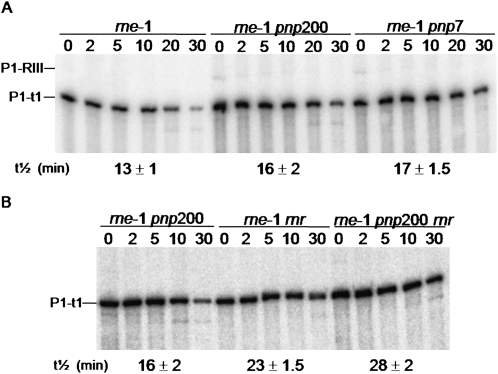
Since strains deficient for both PNPase and RNase R are not viable (Cheng and Deutscher 2003), we decided to construct a new set of multiple exonuclease mutants harboring the available pnp200 allele encoding a thermosensitive PNPasets (Yancey and Kushner 1990), to better study the roles of 3′–5′ exoribonuleases in the degradation of the rpsO mRNA (Table 1). The thermal inactivation of PNPasets is slow, but the activity of the enzyme can be significantly reduced (Yancey and Kushner 1990; Cheng and Deutscher 2003). However, this does not lead to a significant decrease in the protein levels (Supplemental Fig. S1). We found that a 30-min incubation of bacteria cultures at 44°C prior to measurement of decay rates was sufficient to observe an effect of PNPasets inactivation on rpsO mRNA stability (Fig. 1A). The rpsO mRNA was slightly more stable in the rne-1 pnp200 than in the rne-1 mutant (the half-lives were 16 min and 13 min, respectively). The pnp7 allele carries a nonsense mutation that leads to reduced levels of PNPase with a concomitant decrease in PNPase activity (Arraiano et al. 1988; Zilhão et al. 1996; Mohanty and Kushner 2003; Supplemental Fig. S1). The fact that a similar stabilization was observed under the same conditions in the rne-1 pnp7 and rne-1 pnp200 mutants suggested that the thermosensitive PNPasets encoded by pnp200 was significantly inactivated (Fig. 1). These data reinforce the earlier conclusion that PNPase has only a modest effect on the degradation of the primary rpsO mRNA (Régnier and Hajnsdorf 1991; Hajnsdorf et al. 1995; Braun et al. 1996).
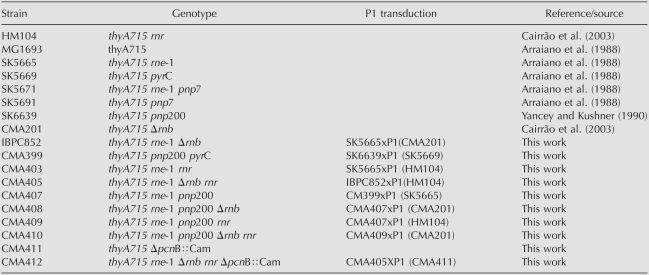
TABLE 1.
Strains used in this work
FIGURE 1.
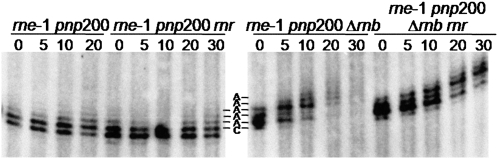
Effect of RNase R and PNPase on the degradation of the rpsO mRNA. (A) Analysis of the rpsO mRNA stability upon PNPase depletion. Total RNA from rne-1, rne-1 pnp200, and rne-1 pnp7 was extracted from exponential phase cultures grown at 30°C and incubated for 30 min at the nonpermissive temperature of 44°C, before inhibition of transcription. Two alleles coding for PNPase were tested: the pnp200 encodes the thermolabile PNPasets (Yancey and Kushner 1990) while the pnp7 is a nonsense mutant (Mohanty and Kushner 2003). The P1-t1 mRNA corresponds to the monocistronic rpsO transcript while the P1-RIII mRNA results from the processing of the rpsO-pnp bicistronic transcript (Hajnsdorf et al. 1994). An antisense riboprobe against the full-length rpsO mRNA was used. Half-lives are expressed in minutes. (B) rpsO mRNA decay in the absence of PNPase and/or RNase R. Cultures of rne-1 pnp200, rne-1 rnr, and rne-1 pnp200 rnr strains were grown at 30°C to exponential phase and shifted to 44°C; after 30 min of heat incubation, rifampicin was added and culture samples were withdrawn at times indicated. All strains were studied under the same conditions. Five micrograms of total RNA were analyzed on a 6% polyacrylamide Northern blot. Half-lives are depicted at the bottom the corresponding strain and are expressed in minutes.
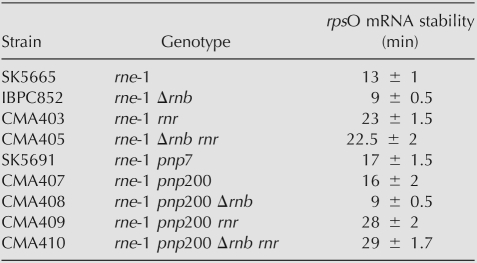
To test if the rpsO mRNA could be a substrate for RNase R, we monitored the degradation rate of this transcript in mutants for this enzyme. Notably, we found that the rpsO mRNA is significantly stabilized in the absence of RNase R. Half-life of this transcript rises from 13 min in the rne-1 strain to 23 min in the rne-1 rnr mutant (Fig. 1; Table 2). This result suggests that the rpsO mRNA is a substrate for RNase R.
TABLE 2.
rpsO mRNA stability
We then examined the effect of the combined inactivation of PNPase and RNase R. For this purpose, we extracted RNA from the rne-1 pnp200 rnr and compared its stability to that of mutants lacking only one of these exonucleases. Autoradiographs in Figure 1B show that simultaneous inactivation of the two exonucleases results in higher stabilization of the rpsO mRNA, indicating that RNase R and PNPase can both act on the turnover of this transcript. Moreover, comparison of decay rates in the rne-1, rne-1 rnr, and the rne-1 pnp200 rnr mutants (half-lives of 13, 23, and 28 min, respectively) confirms that RNase R contributes much more than PNPase to the degradation of the rpsO transcript. Accordingly, RNase R inactivation has a dramatic stabilization effect in cells deficient for RNase E and PNPase. The half-life of the rpsO mRNA rises from 16 min in the rne-1 pnp200 to 28 min in the rne-1 pnp200 rnr. A graphic representation of the rpsO mRNA decay along the time is presented in Supplemental Figure S2.
Altogether, these results indicate that RNase R and PNPase may have a cumulative effect on the control of rpsO mRNA stability, although RNase R plays a prevalent role in ruling the life span of the rpsO transcript.
RNase R is responsible for destabilization of the rpsO mRNA
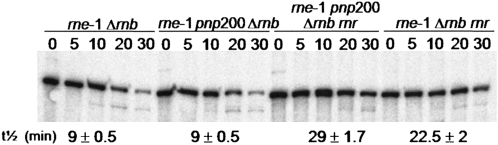
We used the multiple exonuclease mutants described above to investigate whether RNase R also accounts for the instability of the rpsO mRNA in cells lacking RNase II. As expected, this transcript is more efficiently degraded in the rne-1 Δrnb than in the rne-1 strain (Table 2). Moreover, we confirmed that additional inactivation of PNPase did not counteract this instability, as observed in the rne-1 pnp200 Δrnb mutant (Fig. 2; Hajnsdorf et al. 1994, 1995; Marujo et al. 2000). This greatly contrasts with the stabilization of the rpsO transcript observed upon RNase R depletion in the rne-1 Δrnb genetic background. Half-lives of this transcript increases from 9 min in the rne-1 Δrnb strain to 22.5 min in the rne-1 rnr Δrnb mutant. These data strongly reinforce the conclusion that RNase R mostly accounts for the exonucleolytic degradation of the rpsO mRNA. Moreover, the fact that the rpsO mRNA is further stabilized upon inactivation of PNPase in strains lacking RNase R and RNase II (half-lives are 22.5 and 29 min in the rne-1 rnr Δrnb and the rne-1 rnr Δrnb pnp200, respectively) confirms that PNPase may also contribute to the rapid degradation of this transcript in the combined absence of RNase II and RNase R.
FIGURE 2.
The RNase R counteracts the extensive degradation of the rpsO mRNA observed in an rne-1 pnp rnb mutant. Exponential phase growing cultures of the rne-1 Δrnb, rne-1 pnp200 Δrnb, rne-1 pnp200 Δrnb rnr, and rne-1 Δrnb rnr strains were shifted from 30°C to 44°C, and after 30 min of heat incubation, transcription was blocked by addition of rifampicin. The rpsO mRNA stability is indicated in minutes.
Taken together, these results indicate that RNase R is the main exonuclease responsible for the extended degradation of the rpsO mRNA in RNase II deficient cells.
RNase R degrades the oligoadenylated rpsO mRNA
Destabilization of the rpsO mRNA consecutive to RNase II inactivation was attributed to the elongation of the 3′-oligo(A) extension, which may favor the action of the exonucleases (Marujo et al. 2000). We therefore postulated that oligoadenylated molecules are RNase R substrates and checked whether the polyadenylation status affects the sensitivity of the rpsO mRNA to RNase R. In order to determine the length of these poly(A) tails, the rpsO transcripts were cleaved at a unique site by RNase H targeted through a methylated chimeric RNA–DNA oligonucleotide, and 3′ fragments were then analyzed by Northern blotting (Marujo et al. 2000). This method allowed us to detect single adenylation or nibbling events at the 3′ end of the transcript.
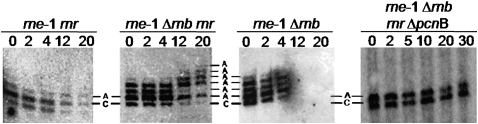
In the presence of RNase II, the rne-1 pnp200 and rne-1 pnp200 rnr strains only show addition of 2–3 adenosine (A) nucleotides (Fig. 3) downstream from the CA terminal nucleotides of rpsO primary transcripts (Folichon et al. 2005). However, inactivation of RNase II results in the increasing elongation of these molecules (Marujo et al. 2000) by at least 5 nucleotides (nt), as observed in the rne-1 pnp200 Δrnb mutant, and poly(A) tails are visible until 10 min after addition of rifampicin (Fig. 3). Nevertheless, we notice that when using the rne-1 pnp200ts Δrnb rather than the rne-1 pnp7 rnb500ts mutant (Marujo et al. 2000) we could not detect the same level of elongation, probably due to residual PNPasets activity left after heat inactivation. As expected, this progressive elongation of the rpsO transcripts (Fig. 3) is correlated with the enhanced degradation of the full-length mRNA (Fig. 2). Data from the rne-1 pnp200 Δrnb strain confirms that the oligoadenylated mRNAs arising upon RNase II inactivation are degraded by an exonuclease that is not PNPase. Strikingly, a similar experiment performed in the rne-1 pnp200 Δrnb rnr mutant shows that removal of RNase R results in poly(A) tails markedly more stable that become progressively longer as time passes, being clearly visible even 30 min after the transcriptional arrest (Fig. 3). Moreover, this 3′-end elongation is now reflected in the higher stabilization of the primary rpsO mRNA (Fig. 2).
FIGURE 3.
RNase R removes the adenosine residues added by PAP I at the 3′ end of the rpsO transcripts upon RNase II inactivation. All strains carry the pnp200 allele and were exposed to thermal inactivation for 30 min prior to addition of rifampicin. Sample time points were withdrawn at times indicated. Ten micrograms of RNA extracted from rne-1 pnp200, rne-1 pnp200 rnr, rne-1 pnp200 Δrnb, and rne-1 pnp200 Δrnb rnr strains were mixed with the chimeric DNA/RNA CrpsO oligonucleotide, which binds ∼50 nt upstream of the terminal transcription residues of the rpsO transcripts and targets them to RNase H cleavage. The mixture was then incubated with RNase H. 3′ terminal fragment of rpsO transcripts were resolved on a 10% polyacrylamide gel and identified by hybridization with a riboprobe specific to the 3′ end of the rpsO mRNA. A run-off transcript matching the nonadenylated full-length rpsO was treated in the same conditions (not shown) (Marujo et al. 2000). RNA fragments that end at the transcriptional termination C residue were identified and are indicated (Left panel/right panel: corresponds, respectively, to the second and first band observed counting from the bottom). Longer RNA fragments correspond to RNAs harboring additional adenosine residues, indicated by the letter A.
Stabilization of oligoadenylated RNA species following RNase R inactivation is consistent with the hypothesis above that RNase R is an important poly(A)-dependent exonuclease acting on the rpsO mRNA turnover in vivo. Moreover the length of RNA stabilized in the absence of RNase R suggests that this enzyme recognizes the transcript elongated of four to six adenosines downstream from the CA terminal nucleotides.
The absence of RNase II favors RNA degradation by RNase R
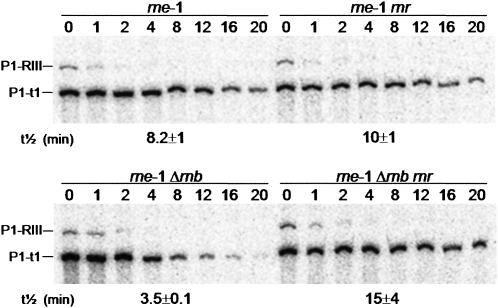
The experiments above were all performed in cells that were grown for 30 min at 44°C, in order to inactivate thermosensitive PNPasets encoded by pnp200. We decided to look at the role of RNase R under the less stringent conditions previously used to study the degradation pathways of the rpsO transcripts (Hajnsdorf et al. 1994, 1995; Marujo et al. 2000). The minor role of PNPase in exonucleolytic degradation of the rpsO mRNA prompted us to use strains wild-type for this ribonuclease. Therefore, cells were grown at 30°C and transcription was inhibited immediately after the temperature shift to 44°C, which inactivates the thermosensitive RNase E. Northern blot analysis of total RNA confirms that the RNase II deficiency destabilizes of the rpsO mRNA (half-lives are 8.2 and 3.5 min in the rne-1 and the rne-1 Δrnb strains, respectively) (Fig. 4). In the same conditions, RNase R deficiency causes only a slight increase of rpsO mRNA stability in cells containing RNase II. Half-lives are 8.2 and 10.5 min in the wild-type and the rnr strains, respectively. However, we determined that further inactivation of RNase R in cells lacking RNase II results in a more than fourfold stabilization of this mRNA (from 3.5 to 15 min), which is about twice higher than that observed after 30 min incubation at 44°C (Table 2).
FIGURE 4.
RNase II protects more efficiently the rpsO mRNA from RNase R-mediated degradation if incubation at high temperature is short. To inactivate the thermosensitive RNase Ets, cell cultures were grown at 30°C and shifted to 44°C with immediate addition of rifampicin to blockage of transcription, and samples were withdrawn at times indicated. Total RNA from cells carrying both (rne-1), only one (rne-1 rnr and rne-1 Δrnb), or any (rne-1 Δrnb rnr) of the hydrolytic exonucleases RNase II and RNase R were analyzed on 6% polyacrylamide Northern blots. rpsO mRNA was detected using an antisense riboprobe. P1-t1 corresponds to the monocistronic rpsO transcript while the P1-RIII results from the processing of the rpsO-pnp bicistronic transcript.
Experiments performed under conditions avoiding the longer incubation at 44°C confirm that the poly(A)-dependent decay of the rpsO mRNA cannot be efficiently mediated by PNPase. Furthermore, they confirm that in the absence of RNase E, RNase R is a main player in rpsO mRNA decay and that its activity is very efficiently counteracted by RNase II (Fig. 4; Supplemental Fig. S3).
RNase R level is increased at high temperature
The data above indicate that RNase R has a more pronounced effect on the rpsO transcript degradation when cells have been incubated 30 min at 44°C (an approximately twofold stabilization) than when RNA decay is measured immediately after the temperature shift (a 1.2-fold stabilization). This suggests that RNase R-mediated degradation pathways are more active after a long incubation at high temperature.
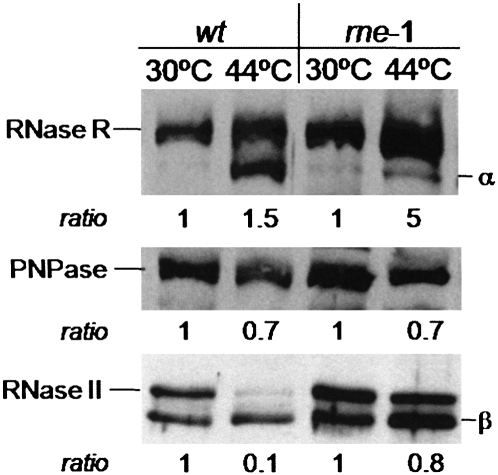
We hypothesized that this change of contribution of RNase R to the degradation of the rpsO mRNA may reflect a modification in intracellular RNase R level. Western blot assays were performed to compare RNase R present in the wild-type and rne-1 cells grown at 30°C and shifted to 44°C for 30 min. The levels of RNase II and PNPase were also analyzed in the same conditions (Fig. 5). Heat treatment was shown to affect the levels of these exonucleases, as shown in protein extract from the wild type. Unlike PNPase and particularly RNase II that show decreased levels, RNase R is slightly more abundant in wild-type cells incubated at high temperature (Fig. 5). In addition, upon heat treatment and RNase Ets inactivation (rne-1 protein extract), RNase R levels are highly increased (at least a fivefold induction) while the levels of the other RNA exonucleases show only a minor variation (Fig. 5). The findings are consistent with a previous report showing that RNase R expression is post-transcriptionally regulated by RNase E (Cairrão and Arraiano 2006). Therefore, increased intracellular levels of RNase R in cells lacking functional RNase E may help explain the more marked effect of this exonuclease in the degradation of the rpsO mRNA upon prolonged heat treatment.
FIGURE 5.
RNase R is a heat-shock protein whose levels are highly regulated by RNase E. Wild-type (wt) and RNase E-deficient (rne-1) cells were grown at 30°C and shifted to 44°C for 30 min. Samples were withdrawn immediately before and after the temperature shift. Equal amounts of protein extracts were analyzed on SDS-PAGE. Nitrocellulose membranes were incubated with both RNase II and RNase R or RNase II and PNPase antibodies. The detection of RNase II was used to validate equal protein loading in the different gels, where the same protein extracts were analyzed. Only the corresponding section of interest from different membranes is shown here. Typical results corresponding to total membranes are shown in Supplemental Figure S4. In each strain, samples collected after the incubation at 44°C are compared to the samples at 30°C. The amount of protein from cultures at 30°C detected in each strain was set at 1. Quantification of the protein bands obtained in this representative experiment were obtained by scanning densitometry using the IMAGEQUANT program, and the relative amount of each exonuclease is depicted at the bottom of the corresponding section of each membrane. (α, β) Nonspecific cross-hybridization with RNase R and RNase II antibodies, respectively.
In addition to heat shock treatment, RNase R levels are also increased in cold shock (Cairrão et al. 2003) and stationary phase (Andrade et al. 2006) and other stress conditions (Chen and Deutscher 2005). In all these conditions, increasing amounts of RNase R lead to consequences in the transcripts regulated by this enzyme. RNase R seems thus to have a primordial role in the cellular adaptation to new stressful environments, probably by degradation of unnecessary RNAs.
Poly(A) tails arising from RNase II inactivation are primarily recognized by RNase R
The data above suggest that oligo(A) tails of the rpsO transcript are primarily recognized by RNase R. A possible explanation is that RNase R can initiate decay in oligo(A) tails that are too short to be recognized by PNPase. In agreement with this hypothesis, the short extensions of 4–6 adenosines detected downstream from the CA transcription termination sites in the rne-1 pnp200 Δrnb rnr and rne-1 pnp200 Δrnb strains (Fig. 3) are also present in rne-1 Δrnb rnr, and rne-1 Δrnb cells containing PNPase (Fig. 6). Longer poly(A) tails appear upon stabilization of the full-length rpsO mRNA in the rne-1 Δrnb rnr mutant (Fig. 4). The failure to detect these oligoadenylated molecules in the rne-1 Δrnb mutant when RNase R is present suggests that they are RNase R targets. Conversely, when RNase II is present we cannot observe these elongated molecules, as seen on the rne-1 rnr mutant strain (Fig. 6). Lack of 3′ extensions in a rne-1 Δrnb rnr ΔpcnB mutant confirmed that poly(A) polymerase I accounts for the polyadenylation of the rpsO transcript.
FIGURE 6.
3′ terminal nucleotide analysis of the rpsO mRNA. All strains carry the rne-1 allele and were exposed to thermal inactivation following immediate addition of rifampicin. Sample time points were withdrawn at times indicated after the blockage of transcription. Ten micrograms of RNA extracted from rne-1 rnr; rne-1 Δrnb; rne-1 Δrnb rnr, and rne-1 Δrnb rnr ΔpcnB were treated with RNase H and the chimeric DNA/RNA oligonucleotide CrpsO as described in Material and Methods. RNAs fragments whose terminal nucleotide matches the transcriptional terminal C residue are identified. Increasing bands correspond to elongated RNAs that vary one nucleotide in size from the neighborhood bands, corresponding to a post-transcriptional added adenosine (A) residue. Cells that carry the pcnB deletion do not present this elongated rpsO mRNA species. A specific 3′ rpsO riboprobe was used to detect these transcripts.
The results above indicate that RNase II inactivation allows the synthesis of poly(A) tails that are efficiently recognized by RNase R, thus promoting the RNase R-mediated degradation of the rpsO mRNA. Moreover, our data also show that degradation of oligoadenylated rpsO transcripts by RNase R takes place in the presence of PNPase.
DISCUSSION
Exonucleolytic poly(A)-dependent degradation of rpsO mRNA becomes predominant when RNase E is inactive (Marujo et al. 2003). In this report we have shown that RNase R is the main enzyme involved in the poly(A) pathway of degradation of this transcript, in the absence of RNase E. A model for the degradation of the rpsO mRNA based on the competition for the 3′end of the RNA by PAP I and the exonucleases is shown in Figure 7. Polyadenylation (by PAP I) provides a single-stranded region downstream from termination hairpins allowing exonuclease binding and RNA degradation. RNase II removes the oligo(A) extensions very efficiently, but it fails to degrade the full-length mRNA, as it cannot overcome the RNA hairpin (Marujo et al. 2000; Folichon et al. 2005). The increasing poly(A) tails arising in the absence of RNase II favor the action of other 3′–5′ exonucleases promoting RNA decay (Marujo et al. 2000; Folichon et al. 2005).
FIGURE 7.
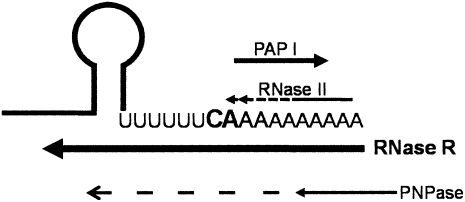
Model of the poly(A)-dependent metabolism of the rpsO mRNA. The 3′ end of the rpsO transcript has a transcriptional termination hairpin followed by a short linear sequence. The major transcriptional termination nucleotides (CA) are in bold. RNase II removes very efficiently the nucleotides added by PAP I downstream from this sequence but does not proceed through the mRNA body (Marujo et al. 2000; Folichon et al. 2005). When RNase II is inactive, longer poly(A) tails arise promoting the degradation of the rpsO mRNA. Polyadenylation extends the short 3′ linear region, providing a toehold for exonucleases. RNase R and PNPase may degrade this transcript, although with different specificities. RNase R requires a less extensive 3′ end linear region to efficiently bind to RNA than PNPase. Addition of two nucleotides after the (U)6CA fragment are enough to initiate RNase R degradation. This becomes advantageous in the competition with PNPase for the access to the 3′ end of the rpsO mRNA. The growing poly(A) tails favor RNase R degradation. In the presence of poly(A) tails RNase R can easily overcome the RNA secondary structure and completely degrade the rpsO mRNA. RNase R is therefore established as a major poly(A)-dependent exonuclease involved in the degradation of the rpsO mRNA. Polyadenylated RNAs can be attacked by RNase II and RNase R preventing the appearance of longer poly(A) tails that can act as suitable substrate to PNPase. Only if RNase II and RNase R are inactive, longer poly(A) tails might arise that can either be shortened (arrow) or possibly promote RNA degradation by PNPase (dotted arrow).
PNPase and RNase R can both degrade this transcript, although with different specificities (Fig. 7). In vitro assays suggest that PNPase may require a longer linear sequence than RNase R in order to bind and degrade RNA (McLaren et al. 1991; Vincent and Deutscher 2006; Amblar et al. 2007). Therefore RNase R seems to have an advantage in the competition with PNPase for the access to the 3′ end of the RNA. Furthermore, efficient binding of the RNA allows the complete degradation by RNase R, as this exonuclease is not susceptible to RNA secondary structures (Vincent and Deutscher 2006). The short tails of 5–6 adenosines added by PAP I (only visible after RNase R inactivation) expand the single-stranded region downstream from the RNA termination hairpin to a linear sequence of about 13 nt in length. Notably, a 10–14-nt linear sequence at the 3′ end of RNA provides an optimal toehold for RNase R (Vincent and Deutscher 2006). Hence, we provide the first in vivo evidence that the single-stranded 3′ end necessary for the binding and degradation by RNase R can be provided through polyadenylation. In addition, the degradative activities of RNase II and RNase R prevent the appearance of such longer polyadenylated species and consequently impair PNPase activity on the rpsO mRNA degradation. Only poly(A) tails escaping this degradation may become a good substrate to PNPase. This helps explain why PNPase is shown to have a more relevant role in the degradation of this transcript only in the absence of RNase R. However, we do not rule out that RNase R can also degrade the rpsO transcript in a poly(A)-independent manner, even though poly(A) tails definitely favor degradation by RNase R. It is possible that “breathing” of the hairpin may result in the relaxation of the secondary structure, providing an optimal linear region to serve as toehold for RNase R without prior requirement of polyadenylation.
PNPase was so far characterized as the main exonuclease involved in the poly(A) metabolism, either acting as a free enzyme or in the degradosome (Coburn and Mackie 1998; Blum et al. 1999; Lisitsky and Schuster 1999). However, PNPase was shown to play a modest role in the degradation of the rpsO mRNA. Here we report that in the absence of RNase E and consequent induction of RNase R levels, this transcript is much more efficiently degraded by RNase R rather than PNPase, and an rnr mutant shows RNAs harboring longer poly(A) tails. These elongated RNA molecules can only accumulate because they are not efficiently degraded in the absence of RNase R. Moreover, this is shown to occur in the presence of PNPase. The activity of other exonucleases present in the cell may be preventing the occurrence of longer poly(A) tails in rnr mutants; however, unlike RNase R, they probably cannot overcome the termination hairpin and fail to degrade the mRNA (Li et al. 1998a,b; Vincent and Deutscher 2006).
Interestingly, double mutants for PNPase and RNase II or PNPase and RNase R are not viable (Donovan and Kushner 1986; Cheng and Deutscher 2003; this work) showing that these enzymes cannot completely replace each other. RNase R levels are increased in response to changes in environmental conditions. In a double mutant rnb ts pnp mutant, mRNAs are stabilized and cells end up dying in rich media (Donovan and Kushner 1986). However, this fact is not observed in minimal media. It is tempting to suggest that up-regulation of RNase R levels can potentially alleviate the accumulation of mRNAs, overcoming the lethal phenotype of the rnb pnp mutant. In the set of experiments to inactivate PNPasets, cells were incubated for 30 min at 44°C, and this resulted in the up-regulation of RNase R. As observed, increasing levels of RNase R probably favor the degradation of this transcript; however, the up-regulation of RNase R levels is not an essential condition for the poly(A)-dependent degradation of this transcript by this enzyme (Fig. 6). This confirms that RNase R plays a major role in poly(A)-dependent RNA decay.
Poly(A)-mediated degradation of RNA molecules plays very important roles in the RNA quality control systems and modulation of RNA stability (Régnier and Arraiano 2000; Dreyfus and Régnier 2002; Li et al. 2002). RNA polyadenylation is a tightly regulated cellular process (Mohanty and Kushner 1999; Binns and Masters 2002; Jasiecki and Wegrzyn 2003). Previous work described the involvement of an additional ribonuclease in the poly(A) metabolism, but its identity remained elusive (Hajnsdorf et al. 1994, 1995; O'Hara et al. 1995; Khemici and Carpousis 2004). Our results refined the poly(A)-dependent degradation pathways of mRNA, including RNase R as an important player. In fact, the RNase R/PAP I pathway may be more generalized, as a 3′ rpsT mRNA fragment whose degradation is highly dependent on polyadenylation (Coburn and Mackie 1998) is also stabilized (a threefold) in an rnr single mutant (Fig. 8). PNPase is also shown to be important in this decay, as previously described (Coburn and Mackie 1998; Khemici and Carpousis 2004). This suggests that polyadenylated fragments can be degraded by RNase R and PNPase while oligoadenylated full-length transcripts are mainly degraded by RNase R.
FIGURE 8.
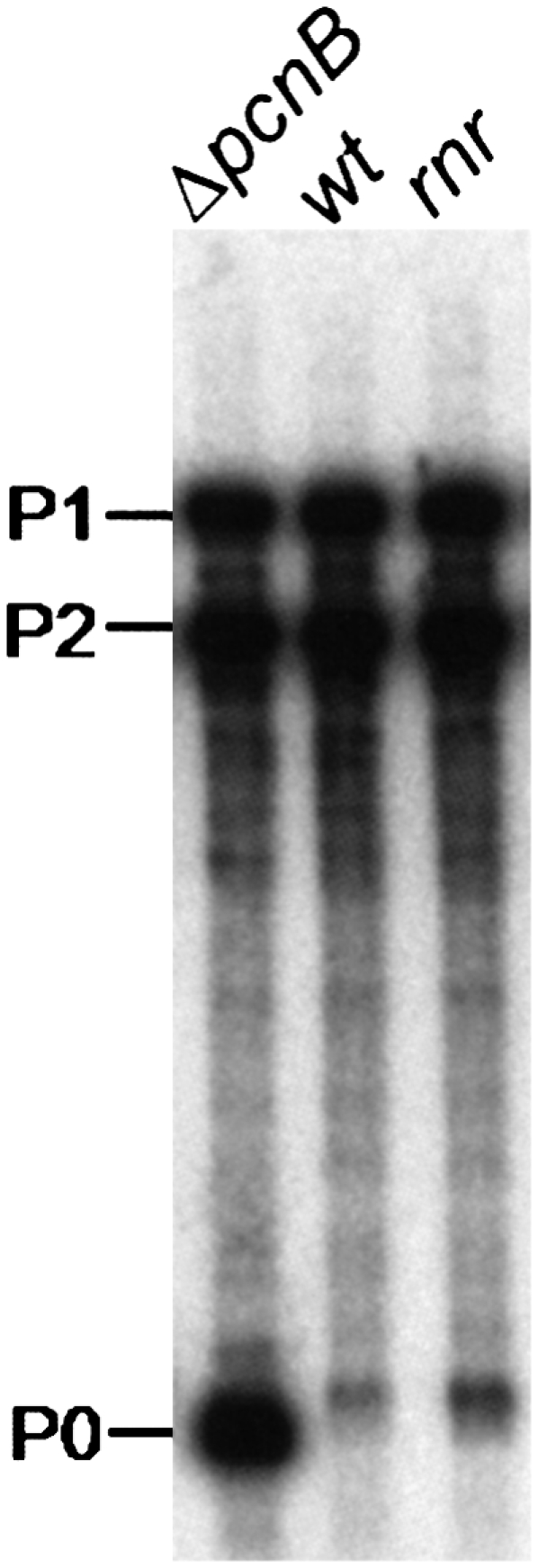
Steady-state level of an rpsT mRNA fragment is affected by RNase R. Cultures of MG1693 (wt), SK7988 (ΔpcnB), and HM104 (rnr) were grown at 30°C to exponential phase. Ten micrograms of total RNA from each strain were extracted and resolved on a 6% polyacrylamide-urea gel. A riboprobe corresponding to the full-length rpsT transcript was used to detect rpsT mRNAs. P1 (447 nt) and P2 (356 nt) mRNAs derived from tandem promoters within the rpsT gene, while P0 represents a major 147-nt degradative intermediate whose degradation is highly dependent on polyadenylation (described in Coburn and Mackie 1998). The size difference on this transcript between the different strains analyzed reflects the presence or absence of a short oligo(A) extension at the 3′ end.
All these results highlight the importance of bacterial RNase R in the post-transcriptional control of gene expression, establishing an important parallel with its eukaryotic counterparts present in exosomes, which are key players involved in the metabolism of polyadenylated RNA.
MATERIALS AND METHODS
Bacterial strains
The strains used in this work are indicated in Table 1. All experiments were performed in the E. coli MG1693 (wild type) and its derivative isogenic strains. When required, mutations were transferred from different genetic backgrounds by P1vir transduction. pnp200 cells were always used as the receptor strain, as transduction of pnp200 allele is problematic (Yancey and Kushner 1990). The rne-1 has no antibiotic resistance associated. Therefore, we used a two-step transduction protocol to pass the rne-1 mutation to the desired genetic background (Arraiano et al. 1988). The rne-1 gene is cotransducible with the adjacent pyrC gene, essential for uracil metabolism. The first step was to transduce pnp200 cells with P1(pyrC∷Tet) lysate and select for tetracycline resistant colonies on LB plates. Isolated colonies were then selected in minimal media (M9) supplemented or not with uracil. Cells deficient in uracil metabolism (cells carrying the pyrC∷TetR mutation) were then transduced with P1(rne-1) containing the wild-type pyrC gene. Rne-1 cells were tetracycline sensitive and auxotrofic for uracil metabolism (pyrC+). Additional confirmation of the rne-1 allele insertion was done by testing the thermosensitivity of these cells at 44°C.
Current available pcnB mutants were either TetR or KanR, which did not allow the selection of an rne-1 Δrnb∷Tet rnr∷Kan pcnB strain due to incompatibility in antibiotic markers. The new ΔpcnB∷Cam strain was constructed using a one-step inactivation system of chromosomal genes (Datsenko and Wanner 2000). After transformation with the PCR fragment containing the ΔpcnB deletion into MG1693 strain, chloramphenicol-resistant colonies were isolated. Transformants were reisolated on selective plates and mutation reintroduced into MG1693. The deletion was confirmed by PCR. In addition, miniprep analysis of wild-type and MG1693ΔpcnB∷Cam strains transformed with pUC19 confirmed the copy number reduction of this plasmid in the mutant strain (March et al. 1989). Primers used in this work are listed in Supplemental Table S1 and were purchased from STAB VIDA.
Growth conditions
Bacteria were grown on an orbital water shaker bath at 30°C in Luria-Bertani (LB) medium supplemented with thymine (50 μg/mL). When required, antibiotics were present at the following concentrations: chloramphenicol, 50 μg/mL, kanamycin, 50 μg/mL; tetracycline, 20 μg/mL; ampicillin, 150 μg/mL. Growth proceeded up to an OD650nm of 0.30. In steady-state experiments, culture samples were immediately collected at this stage. In decay experiments, cell cultures were then shifted to 44°C and rifampicin was added to stop transcription, either immediately or 30 min after heat shock.
RNA extraction and Northern blot analysis
Total RNA was extracted as previously described (Marujo et al. 2000). Five micrograms of total RNA from each strain were used to analyze the full-length P1-t1 rpsO mRNA on a 6% polyacrylamide-urea gel. The probe was an antisense RNA complementary to the rpsO transcript, extending from the translation initiation codon to the transcription terminator t1 of rpsO, transcribed in vitro from pEHa4 linearized at the SalI site of the polylinker (Hajnsdorf et al. 1994). Ten micrograms of total RNA were resolved on a 10% polyacrylamide-urea gel to analyze the 3′ rpsO RNA fragments generated by RNase H. Detection was performed using an antisense RNA probe complementary to the 3′ extremity of rpsO transcript between nucleotides 368 and 420 (Marujo et al. 2000). Ten micrograms of total RNA were used to analyze rpsT transcripts on a 6% polyacrylamide-urea Northern blot. An antisense RNA probe matching the full-length rpsT RNA was used to detect rpsT expression. RNA was electrotransferred onto Hybond-N+ (GE Healthcare) membrane using TAE 1× and was UV cross-linked to the membrane. Riboprobes were generated by in vitro transcription with T7 or SP6 RNA Polymerase (Promega) in the presence of [α-32P]UTP (Perkin Elmer). Riboprobes were purified on G50 Microspin columns (GE Healthcare) or fenol:chloroform extracted and ethanol precipitated overnight at −20°C.
Site-directed cleavage of rpsO mRNA by RNase H
This experiment was basically carried out as previously described (Marujo et al. 2000). Ten micrograms of total RNA and 30 ng of the CrpsO chimeric oligonucleotide (Supplemental Table S1) were mixed in 5 μL, successively incubated at 95°C for 5 min and at 50°C for 15 min, and then slowly cooled to 37°C. One unit of RNase H (USB) and 2 units of RNasin (Promega) were then added to the reaction mixture in a final volume of 10 μL and reaction was carried out for 1 h at 37°C. The reaction was stopped by addition of 15 μL of loading buffer, and RNA fragments were analyzed on 10% polyacrylamide-urea Northern blots. The 2′-O-methyl RNA–DNA chimeric oligonucleotide CrpsO is complementary to the 3′ nucleotide region of the rpsO mRNA between residues 352 and 370. A full-length rpsO mRNA was obtained by in vitro transcription with SP6 RNA Polymerase (Promega) and used as a control, allowing the identification of the fragments ended at the termination transcriptional nucleotide C.
RNA half-life determination
Quantification of full-length RNA was done after PhosphorImager scanning on STORM 860 using the IMAGEQUANT program (Molecular Dynamics). The half-lives of RNA were determined by linear regression using the logarithm of the percentage of RNA remaining versus time, considering the amount of RNA at 0 min as 100%. At least two independent RNA extractions from each strain were tested.
Western blot analysis
Wild-type and rne-1 cells were grown at 30°C in LB medium up to OD650 ∼0.3. Samples (30°C fraction) were collected at this stage, and the remaining culture was shifted to a water bath at 44°C. Thirty minutes after the heat shock, a new sample culture (44°C fraction) was collected. For Western blotting 5 or 10 μg of total protein extracts (quantified by the Bradford assay) were separated on SDS-PAGE gels and the blotting was performed as described (Andrade et al. 2006). At least three independent cultures of each strain were tested in the same conditions. Membranes were incubated with antibodies raised against RNase R, PNPase, and/or RNase II, as stated in Figure 5 and Supplemental Figures S1 and S2, and detection was made using the ECL system (GE Healthcare). After exposure of the blots to Biomax-MR film (Kodak), quantification of the intensity of bands was estimated by the scanning densitometry IMAGEQUANT program (Molecular Dynamics).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Sidney Kushner for providing the pnp200 strain and A.J. Carpousis for providing PNPase antibodies. We are grateful to Jacques Le Derout and Vânia Pobre for technical assistance and Paulo Marujo for helpful discussions. J.M.A. is the recipient of a FCT Doctoral Scholarship (Portugal). This work was supported by Fundação para a Ciência e Tecnologia (FCT), Portugal, the Centre National de la Recherche Scientifique (UPR9073), Paris 7 University (France), and a bilateral cooperation between Portugal and France.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1197309.
REFERENCES
- Amblar M., Barbas A., Fialho A.M., Arraiano C.M. Characterization of the functional domains of Escherichia coli RNase II. J. Mol. Biol. 2006;360:921–933. doi: 10.1016/j.jmb.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Amblar M., Barbas A., Gomez-Puertas P., Arraiano C.M. The role of the S1 domain in exoribonucleolytic activity: Substrate specificity and multimerization. RNA. 2007;13:317–327. doi: 10.1261/rna.220407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade J.M., Arraiano C.M. PNPase is a key player in the regulation of small RNAs that control the expression of outer membrane proteins. RNA. 2008;14:543–551. doi: 10.1261/rna.683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade J.M., Cairrão F., Arraiano C.M. RNase R affects gene expression in stationary phase: Regulation of ompA. Mol. Microbiol. 2006;60:219–228. doi: 10.1111/j.1365-2958.2006.05092.x. [DOI] [PubMed] [Google Scholar]
- Arraiano C.M., Yancey S.D., Kushner S.R. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J. Bacteriol. 1988;170:4625–4633. doi: 10.1128/jb.170.10.4625-4633.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arraiano C.M., Barbas A., Amblar M. Characterizing Ribonucleasesin vitro: Examples of synergies between biochemical and structural analysis. In: Maquat L.M., Arraiano C.M., editors. Methods in enzymology. Vol. 447. Academic Press; New York: 2008. [DOI] [PubMed] [Google Scholar]
- Barbas A., Matos R.G., Amblar M., López-Viñas E., Gomez-Puertas P., Arraiano C.M. New insights into the mechanism of RNA degradation by ribonuclease II: Identification of the residue responsible for setting the RNase II end product. J. Biol. Chem. 2008;283:13070–13076. doi: 10.1074/jbc.M709989200. [DOI] [PubMed] [Google Scholar]
- Binns N., Masters M. Expression of the Escherichia coli pcnB gene is translationally limited using an inefficient start codon: A second chromosomal example of translation initiated at AUU. Mol. Microbiol. 2002;44:1287–1298. doi: 10.1046/j.1365-2958.2002.02945.x. [DOI] [PubMed] [Google Scholar]
- Blum E., Carpousis A.J., Higgins C.F. Polyadenylation promotes degradation of 3′-structured RNA by the Escherichia coli mRNA degradosome in vitro. J. Biol. Chem. 1999;274:4009–4016. doi: 10.1074/jbc.274.7.4009. [DOI] [PubMed] [Google Scholar]
- Braun F., Hajnsdorf E., Régnier P. Polynucleotide phosphorylase is required for the rapid degradation of the RNase E-processed rpsO mRNA of Escherichia coli devoid of its 3′ hairpin. Mol. Microbiol. 1996;19:997–1005. doi: 10.1046/j.1365-2958.1996.440971.x. [DOI] [PubMed] [Google Scholar]
- Cairrão F., Arraiano C.M. The role of endoribonucleases in the regulation of RNase R. Biochem. Biophys. Res. Commun. 2006;343:731–737. doi: 10.1016/j.bbrc.2006.03.040. [DOI] [PubMed] [Google Scholar]
- Cairrão F., Cruz A., Mori H., Arraiano C.M. Cold shock induction of RNase R and its role in the maturation of the quality control mediator SsrA/tmRNA. Mol. Microbiol. 2003;50:1349–1360. doi: 10.1046/j.1365-2958.2003.03766.x. [DOI] [PubMed] [Google Scholar]
- Cao G.J., Kalapos M.P., Sarkar N. Polyadenylated mRNA in Escherichia coli: Modulation of poly(A) RNA levels by polynucleotide phosphorylase and ribonuclease II. Biochimie. 1997;79:211–220. doi: 10.1016/s0300-9084(97)83508-0. [DOI] [PubMed] [Google Scholar]
- Chekanova J.A., Gregory B.D., Reverdatto S.V., Chen H., Kumar R., Hooker T., Yazaki J., Li P., Skiba N., Peng Q., et al. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131:1340–1353. doi: 10.1016/j.cell.2007.10.056. [DOI] [PubMed] [Google Scholar]
- Chen C., Deutscher M.P. Elevation of RNase R in response to multiple stress conditions. J. Biol. Chem. 2005;280:34393–34396. doi: 10.1074/jbc.C500333200. [DOI] [PubMed] [Google Scholar]
- Cheng Z.F., Deutscher M.P. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J. Biol. Chem. 2002;277:21624–21629. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]
- Cheng Z.F., Deutscher M.P. Quality control of ribosomal RNA mediated by polynucleotide phosphorylase and RNase R. Proc. Natl. Acad. Sci. 2003;100:6388–6393. doi: 10.1073/pnas.1231041100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z.F., Deutscher M.P. An important role for RNase R in mRNA decay. Mol. Cell. 2005;17:313–318. doi: 10.1016/j.molcel.2004.11.048. [DOI] [PubMed] [Google Scholar]
- Coburn G.A., Mackie G.A. Differential sensitivities of portions of the mRNA for ribosomal protein S20 to 3′-exonucleases dependent on oligoadenylation and RNA secondary structure. J. Biol. Chem. 1996;271:15776–15781. doi: 10.1074/jbc.271.26.15776. [DOI] [PubMed] [Google Scholar]
- Coburn G.A., Mackie G.A. Reconstitution of the degradation of the mRNA for ribosomal protein S20 with purified enzymes. J. Mol. Biol. 1998;279:1061–1074. doi: 10.1006/jmbi.1998.1842. [DOI] [PubMed] [Google Scholar]
- Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma M.K., Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–680. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- Donovan W.P., Kushner S.R. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc. Natl. Acad. Sci. 1986;83:120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus M., Régnier P. The poly(A) tail of mRNAs: Bodyguard in eukaryotes, scavenger in bacteria. Cell. 2002;111:611–613. doi: 10.1016/s0092-8674(02)01137-6. [DOI] [PubMed] [Google Scholar]
- Dziembowski A., Lorentzen E., Conti E., Séraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- Folichon M., Marujo P.E., Arluison V., Le Derout J., Pelligrini O., Hajnsdorf E., Régnier P. Fate of mRNA extremities generated by intrinsic termination: Detailed analysis of reactions catalyzed by ribonuclease II and poly(A) polymerase. Biochimie. 2005;87:819–826. doi: 10.1016/j.biochi.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Frazão C., McVey C.E., Amblar M., Barbas A., Vonrhein C., Arraiano C.M., Carrondo M.A. Unraveling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature. 2006;443:110–114. doi: 10.1038/nature05080. [DOI] [PubMed] [Google Scholar]
- Hajnsdorf E., Régnier P. Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc. Natl. Acad. Sci. 2000;97:1501–1505. doi: 10.1073/pnas.040549897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnsdorf E., Steier O., Coscoy L., Teysset L., Régnier P. Roles of RNase E, RNase II and PNPase in the degradation of the rpsO transcripts of Escherichia coli: Stabilizing function of RNase II and evidence for efficient degradation in an ams pnp rnb mutant. EMBO J. 1994;13:3368–3377. doi: 10.1002/j.1460-2075.1994.tb06639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnsdorf E., Braun F., Haugel-Nielsen J., Régnier P. Polyadenylylation destabilizes the rpsO mRNA of Escherichia coli . Proc. Natl. Acad. Sci. 1995;92:3973–3977. doi: 10.1073/pnas.92.9.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugel-Nielsen J., Hajnsdorf E., Régnier P. The rpsO mRNA of Escherichia coli is polyadenylated at multiple sites resulting from endonucleolytic processing and exonucleolytic degradation. EMBO J. 1996;15:3144–3152. [PMC free article] [PubMed] [Google Scholar]
- Houseley J., Tollervey D. The nuclear RNA surveillance machinery: The link between ncRNAs and genome structure in budding yeast? Biochim. Biophys. Acta. 2008;1779:239–246. doi: 10.1016/j.bbagrm.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Houseley J., LaCava J., Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Ibrahim H., Wilusz J., Wilusz C.J. RNA recognition by 3′-to-5′ exonucleases: The substrate perspective. Biochim. Biophys. Acta. 2008;1779:256–265. doi: 10.1016/j.bbagrm.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasiecki J., Wegrzyn G. Growth-rate dependent RNA polyadenylation in Escherichia coli . EMBO Rep. 2003;4:172–177. doi: 10.1038/sj.embor.embor733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joanny G., Le Derout J., Bréchemier-Baey D., Labas V., Vinh J., Régnier P., Hajnsdorf E. Polyadenylation of a functional mRNA controls gene expression in Escherichia coli . Nucleic Acids Res. 2007;35:2494–2502. doi: 10.1093/nar/gkm120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khemici V., Carpousis A.J. The RNA degradosome and poly(A) polymerase of Escherichia coli are required in vivo for the degradation of small mRNA decay intermediates containing REP-stabilizers. Mol. Microbiol. 2004;51:777–790. doi: 10.1046/j.1365-2958.2003.03862.x. [DOI] [PubMed] [Google Scholar]
- LaCava J., Houseley J., Saveanu C., Petfalski E., Thompson E., Jacquier A., Tollevervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Lalonde M.S., Zuo Y., Zhang J., Gong X., Wu S., Malhotra A., Li Z. Exoribonuclease R in Mycoplasma genitalium can carry out both RNA processing and degradative functions and is sensitive to RNA ribose methylation. RNA. 2007;13:1957–1968. doi: 10.1261/rna.706207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Pandit S., Deutscher M.P. 3′ exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli . Proc. Natl. Acad. Sci. 1998a;95:2856–2861. doi: 10.1073/pnas.95.6.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Pandit S., Deutscher M.P. Polyadenylation of stable RNA precursors in vivo . Proc. Natl. Acad. Sci. 1998b;95:12158–12162. doi: 10.1073/pnas.95.21.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Reimers S., Pandit S., Deutscher M.P. RNA quality control: Degradation of defective transfer RNA. EMBO J. 2002;21:1132–1138. doi: 10.1093/emboj/21.5.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisitsky I., Schuster G. Preferential degradation of polyadenylated and polyuridinylated RNAs by the bacterial exoribonuclease polynucleotide phosphorylase. Eur. J. Biochem. 1999;261:468–474. doi: 10.1046/j.1432-1327.1999.00285.x. [DOI] [PubMed] [Google Scholar]
- Liu Q., Greimann J.C., Lima C.D. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Lorentzen E., Basquin J., Tomecki R., Dziembowski A., Conti E. Structure of the active subunit of the yeast exosome core, Rrp44: Diverse modes of substrate recruitment in the RNase II nuclease family. Mol. Cell. 2008;29:717–728. doi: 10.1016/j.molcel.2008.02.018. [DOI] [PubMed] [Google Scholar]
- March J.B., Colloms M.D., Hart-Davis D., Oliver I.R., Masters M. Cloning and characterization of an Escherichia coli gene, pcnB, affecting plasmid copy number. Mol. Microbiol. 1989;3:903–910. doi: 10.1111/j.1365-2958.1989.tb00239.x. [DOI] [PubMed] [Google Scholar]
- Marujo P.E., Hajnsdorf E., Le Derout J., Andrade R., Arraiano C.M., Régnier P. RNase II removes the oligo(A) tails that destabilize the rpsO mRNA of Escherichia coli . RNA. 2000;6:1185–1193. doi: 10.1017/s135583820000073x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marujo P.E., Braun F., Haugel-Nielsen J., Le Derout J., Arraiano C.M., Régnier P. Inactivation of the decay pathway initiated at an internal site by RNase E promotes poly(A)-dependent degradation of the rpsO mRNA in Escherichia coli . Mol. Microbiol. 2003;50:1283–1294. doi: 10.1046/j.1365-2958.2003.03753.x. [DOI] [PubMed] [Google Scholar]
- McLaren R.S., Newbury S.F., Dance G.S., Causton H.C., Higgins C.F. mRNA degradation by processive 3′-5′ exoribonucleases in vitro and the implications for prokaryotic mRNA decay in vivo . J. Mol. Biol. 1991;221:81–95. [PubMed] [Google Scholar]
- Mohanty B.K., Kushner S.R. Analysis of the function of Escherichia coli poly(A) polymerase I in RNA metabolism. Mol. Microbiol. 1999;34:1094–1108. doi: 10.1046/j.1365-2958.1999.01673.x. [DOI] [PubMed] [Google Scholar]
- Mohanty B.K., Kushner S.R. Polynucleotide phosphorylase, RNase II and RNase E play different roles in the in vivo modulation of polyadenylation in Escherichia coli . Mol. Microbiol. 2000;36:982–994. doi: 10.1046/j.1365-2958.2000.01921.x. [DOI] [PubMed] [Google Scholar]
- Mohanty B.K., Kushner S.R. Genomic analysis in Escherichia coli demonstrates differential roles for polynucleotide phosphorylase and RNase II in mRNA abundance and decay. Mol. Microbiol. 2003;50:645–658. doi: 10.1046/j.1365-2958.2003.03724.x. [DOI] [PubMed] [Google Scholar]
- Mohanty B.K., Kushner S.R. The majority of Escherichia coli mRNAs undergo post-transcriptional modification in exponentially growing cells. Nucleic Acids Res. 2006;34:5695–5704. doi: 10.1093/nar/gkl684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty B.K., Maples V.F., Kushner S.R. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli . Mol. Microbiol. 2004;54:905–920. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- O'Hara E.B., Chekanova J.A., Ingle C.A., Kushner Z.R., Peters E., Kushner S.R. Polyadenylylation helps regulate mRNA decay in Escherichia coli . Proc. Natl. Acad. Sci. 1995;92:1807–1811. doi: 10.1073/pnas.92.6.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oussenko I.A., Abe T., Ujiie H., Muto A., Bechhofer D.H. Participation of 3′-to-5′ exoribonucleases in the turnover of Bacillus subtilis mRNA. J. Bacteriol. 2005;187:2758–2767. doi: 10.1128/JB.187.8.2758-2767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purusharth R.I., Klein F., Sulthana S., Jäger S., Jagannadham M.V., Evguenieva-Hackenberg E., Ray M.K., Klug G. Exoribonuclease R interacts with endoribonuclease E and an RNA helicase in the psychrotrophic bacterium Pseudomonas syringae Lz4W. J. Biol. Chem. 2005;280:14572–14578. doi: 10.1074/jbc.M413507200. [DOI] [PubMed] [Google Scholar]
- Purusharth R.I., Madhuri B., Ray M.K. Exoribonuclease R in Pseudomonas syringae is essential for growth at low temperature and plays a novel role in the 3′ end processing of 16 and 5 S ribosomal RNA. J. Biol. Chem. 2007;282:16267–16277. doi: 10.1074/jbc.M605588200. [DOI] [PubMed] [Google Scholar]
- Reichenbach B., Maes A., Kalamorz F., Hajnsdorf E., Görke B. The small RNA GlmY acts upstream of the sRNA GlmZ in the activation of glmS expression and is subject to regulation by polyadenylation in Escherichia coli . Nucleic Acids Res. 2008;36:2570–2580. doi: 10.1093/nar/gkn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Régnier P., Arraiano C.M. Degradation of mRNA in bacteria: Emergence of ubiquitous features. Bioessays. 2000;22:235–244. doi: 10.1002/(SICI)1521-1878(200003)22:3<235::AID-BIES5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Régnier P., Hajnsdorf E. Decay of mRNA encoding ribosomal protein S15 of Escherichia coli is initiated by an RNase E-dependent endonucleolytic cleavage that removes the 3′ stabilizing stem and loop structure. J. Mol. Biol. 1991;217:283–292. doi: 10.1016/0022-2836(91)90542-e. [DOI] [PubMed] [Google Scholar]
- Richards J., Mehta P., Karzai A.W. RNase R degrades non-stop mRNAs selectively in an SmpB-tmRNA-dependent manner. Mol. Microbiol. 2006;62:1700–1712. doi: 10.1111/j.1365-2958.2006.05472.x. [DOI] [PubMed] [Google Scholar]
- Sarkar N. Polyadenylation of mRNA in prokaryotes. Annu. Rev. Biochem. 1997;66:173–197. doi: 10.1146/annurev.biochem.66.1.173. [DOI] [PubMed] [Google Scholar]
- Schneider C., Anderson J.T., Tollervey D. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol. Cell. 2007;27:324–331. doi: 10.1016/j.molcel.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic S., Portnoy V., Liveanu V., Schuster G. RNA polyadenylation in prokaryotes and organelles: Different tails tell different tales. Crit. Rev. Plant Sci. 2006;25:65–77. [Google Scholar]
- Slomovic S., Portnoy V., Yehudai-Resheff S., Bronshtein E., Schuster G. Polynucleotide phosphorylase and the archaeal exosome as poly(A)-polymerases. Biochim. Biophys. Acta. 2008;1779:247–255. doi: 10.1016/j.bbagrm.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Vanácová S., Wolf J., Martin G., Blank D., Dettwiler S., Friedlein A., Langen H., Keith G., Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viegas S.C., Pfeiffer V., Sttika A., Silva I.J., Vogel J., Arraiano C.M. Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 2007;35:7651–7664. doi: 10.1093/nar/gkm916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent H.A., Deutscher M.P. Substrate recognition and catalysis by the exoribonuclease RNase R. J. Biol. Chem. 2006;281:29769–29775. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- Wyers F., Rougemaille M., Badis G., Rousselle J.C., Dufour M.E., Boulay J., Régnault B., Devaux F., Namane A., Séraphin B., et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Xu F., Cohen S.N. RNA degradation in Escherichia coli regulated by 3′ adenylation and 5′ phosphorylation. Nature. 1995;374:180–183. doi: 10.1038/374180a0. [DOI] [PubMed] [Google Scholar]
- Yancey S.D., Kushner S.R. Isolation and characterization of a new temperature-sensitive polynucleotide phosphorylase mutation in Escherichia coli K-12. Biochimie. 1990;72:835–843. doi: 10.1016/0300-9084(90)90193-k. [DOI] [PubMed] [Google Scholar]
- Yehudai-Resheff S., Schuster G. Characterization of the E. coli poly(A) polymerase: Nucleotide specificity, RNA-binding affinities and RNA structure dependence. Nucleic Acids Res. 2000;28:1139–1144. doi: 10.1093/nar/28.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilhão R., Cairrão F., Régnier P., Arraiano C.M. PNPase modulates RNase II expression in Escherichia coli: Implications for mRNA decay and cell metabolism. Mol. Microbiol. 1996;20:1033–1042. doi: 10.1111/j.1365-2958.1996.tb02544.x. [DOI] [PubMed] [Google Scholar]