Abstract
The twin-arginine translocation (Tat) pathway is responsible for the export of folded proteins across the cytoplasmic membrane of bacteria. Substrates for the Tat pathway include redox enzymes requiring cofactor insertion in the cytoplasm, multimeric proteins that have to assemble into a complex prior to export, certain membrane proteins, and proteins whose folding is incompatible with Sec export. These proteins are involved in a diverse range of cellular activities including anaerobic metabolism, cell envelope biogenesis, metal acquisition and detoxification, and virulence. The Escherichia coli translocase consists of the TatA, TatB, and TatC proteins, but little is known about the precise sequence of events that leads to protein translocation, the energetic requirements, or the mechanism that prevents the export of misfolded proteins. Owing to the unique characteristics of the pathway, it holds promise for biotechnological applications.
Keywords: Tat, translocase, protein transport, pathogenesis, biotechnology
INTRODUCTION
The twin-arginine translocation (Tat) pathway serves the unique and remarkable role of transporting folded proteins across energy-transducing membranes (133). It is widespread within the microbial world and in plants. Homologs of the genes that encode the transport apparatus occur in archaea, bacteria, chloroplasts, and plant mitochondria (135). In bacteria, the Tat pathway catalyzes the export of proteins from the cytoplasm across the inner/cytoplasmic membrane (IM). In chloroplasts, the Tat components are found in the thylakoid membrane and direct the import of proteins from the stroma. The Tat pathway acts separately from the general secretory (Sec) pathway, which transports proteins in an unfolded state (123).
Tat: twin-arginine translocation
IM: inner/cytoplasmic membrane
Sec: general secretory pathway
cpTat: chloroplast Tat/ΔpH pathway
Translocation: movement of proteins across a membrane
Signal peptide: N-terminal extension of a protein that targets it for translocation
TM: transmembrane
The discovery of the Tat pathway dates to the early 1990s, when it was noticed that a subset of polypeptides in chloroplasts could be translocated independently of ATP hydrolysis and instead relied solely on the proton gradient (26, 86). For this reason, it was initially designated the ΔpH pathway but more recently has been termed the cpTat pathway (chloroplast Tat/ΔpH pathway). In 1995 Creighton et al. (28) presented the first evidence that the cpTat pathway enables the translocation of prefolded proteins. Shortly thereafter, Berks (11) observed that a group of bacterial periplasmic proteins containing various cofactors share a unique type of signal peptide containing a consensus “double arginine” motif also found in substrates of the chloroplast pathway. The existence of a bacterial pathway analogous to the one in chloroplasts was thus established; it was initially termed mtt (membrane targeting and translocation) (132) and then later Tat (twin-arginine translocation) (114).
The minimal set of components required for Tat translocation in Escherichia coli consists of three integral membrane proteins, one from each of the TatA, TatB, and TatC families (Tha4, Hcf106, and cpTatC in chloroplasts, respectively). However, in other bacteria the composition of the Tat system is surprisingly varied. A TatB protein does not seem to be essential for export, as some genomes only encode a single TatA and TatC protein (e.g., Rickettsia prowazekii and Staphylococcus aureus). Some gram-positive bacteria and archaea contain multiple tatC genes as well as multiple tatA/B genes. For example, Bacillus subtilis has two tatC genes (and three tatA genes) (75), each of which seems to constitute separate, substrate-specific Tat systems (73, 74, 99).
TatC is the most conserved of the Tat proteins and sequence conservation is particularly strong within the transmembrane (TM) domains (17, 22). In a phylogenetic tree of TatC, the cyanobacterial TatC proteins cluster with the chloroplast TatC proteins, highlighting the likely evolutionary origin of chloroplasts. TatA and TatB are similar in terms of structure and sequence, and therefore it is difficult to delineate TatA from TatB homologs in some organisms. Phylogenetic analysis indicates that TatA homologs cluster more tightly than TatB proteins, suggesting that TatB may have diverged in sequence more rapidly than TatA (135). It appears that an early gene duplication event gave rise to tatA and tatB, but since then only tatA has been duplicated (e.g., to give tatE in E. coli). The functional significance of the duplication of tatA in E. coli is unclear because tatE is expressed at a low level (71) and its deletion does not affect the export of reporter fusions to any of the known Tat signal peptides (D. Tullman-Ercek, M. P. DeLisa, Y. Kawarasaki, P. Iranpour, B. Ribnicky, et al., unpublished data).
The degree to which organisms utilize the Tat machinery varies widely. In some organisms, the Tat pathway appears to be employed for only a handful of proteins (74). On the other hand, the soil-dwelling gram-positive bacterium Streptomyces coelicolor is predicted to have 129 Tat substrates, although only a small number of these have been confirmed experimentally (80). Halophilic archaea such as Halobacterium sp. NRC-1 and Haloferax volcanii are predicted to utilize Tat for the export of >90% of their secreted proteins (41, 107). A functional Tat pathway is not required for the growth of E. coli, other γ-proteobacteria, or B. subtilis under most conditions; however, it is essential for the viability of H. volcanii even in rich media (41). At present the Tat pathway is the subject of intense study aimed at addressing a number of intriguing mechanistic questions, understanding its role in pathogenicity, and exploiting its ability to export folded proteins for biotechnological applications. Biochemical studies suggest a highly dynamic picture with the Tat proteins participating in various multimeric membrane complexes that are thought to interact transiently during the translocation cycle. However, the precise steps in this process remain a mystery, including the mechanism by which the proton motive force (pmf) provides the energy for vectorial protein transfer across the IM.
Recent findings have shown that Tat export is essential for the establishment of bacterial infection in animals and plants. Because Tat proteins are not found in mammals, in principle the inactivation of Tat export with selective small-molecule inhibitors might lead to new antimicrobial agents with low human toxicity. Finally, the Tat pathway may provide a means for the secretion of complex recombinant proteins for biotechnological purposes. The expression of secreted proteins in E. coli has so far relied on the Sec pathway (51). However, several proteins are not compatible with Sec export and their production could benefit by directing them to the Tat apparatus. Similarly, utilization of the Tat pathway holds promise for improving the display and screening of protein libraries for the isolation of novel receptors or enzymes.
pmf: proton motive force
Translocon: machinery through which proteins are translocated
TAT SUBSTRATE PROTEINS
Signal Peptides
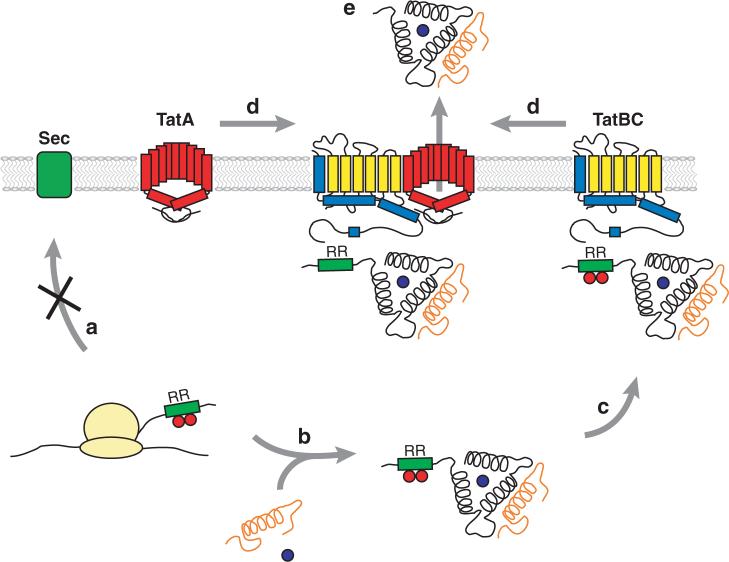
With the exception of proteins that are exported across membranes via specialized systems (e.g., type I secretion), a signal peptide is necessary in order to initiate export (133). Signal peptides consist of three domains: the positively charged N-terminal domain (or n-region), the hydrophobic domain (h-region), and the C-terminal domain (c-region) (Figure 1). Sequence analysis revealed that signal peptides capable of targeting the Tat translocon contain the consensus motif Ser/Thr-Arg-Arg-X-Phe-Leu-Lys (where X is any polar amino acid). The nearly invariant twin-arginine dipeptide gave rise to the pathway's name (11, 29). In addition, the h-region of Tat signal peptides is typically less hydrophobic than that of Sec-specific signal peptides owing to the presence of more glycine and threonine residues (29). Tat signal sequences tend to be longer than their Sec counterparts (primarily because of an extended n-region), with average lengths of 38 and 24 amino acids, respectively (29).
Figure 1.
Features of a typical Tat leader peptide, ssTorA, from E. coli. The n-region is shown in pink, the h-region in blue, and the c-region in yellow. The consensus Tat motif is boxed. The vertical dashed line indicates the site of cleavage by signal peptidase I. Charged residues in the c-region and mature proteins are marked.
TMAO: trimethylamine N-oxide
DMSO: dimethyl sulfoxide
The first arginine of the Arg-Arg dipeptide in the consensus motif can be substituted with Lys, and the second arginine can be substituted with Gln or Asn as well as Lys, with varying efficiencies (23, 37, 66, 122). The substitution of both arginines with lysines blocks the export of physiological Tat substrate proteins (122). However, the heterologous protein colicin V, when fused to a mutant Tat signal peptide containing a Lys-Lys sequence, could still be exported to the periplasm via the Tat pathway (67). To date, only two natural Tat substrates are known to lack the arginine dipeptide: the prepropenicillin amidase of E. coli (containing an Arg-Asn-Arg) (64) and tetrathionate reductase from Salmonella enterica (Lys-Arg) (60). The other residues within the consensus motif are more amenable to amino acid substitutions. For example, the Phe and Leu residues can be substituted with other highly hydrophobic amino acids with little effect on export, although efficiency does drop upon substitution with less hydrophobic amino acids, such as Tyr (122). Also, neither the Ser nor the Lys within the consensus is essential for Tat targeting (122).
In E. coli, a subset of Tat signal peptides exhibits a high degree of export pathway selectivity while others are promiscuous and can direct export through either the Sec or the Tat translocon (15, 38, 66; D. Tullman-Ercek, M. P. DeLisa, Y. Kawarasaki, P. Iranpour, B. Ribnicky, et al., unpublished data). Earlier it had been proposed that the presence of a positive charge in the c-region serves to avoid misdirecting the precursor polypep-tide to the Sec translocon (15, 29). However, many Tat signal sequences lack this charge. A recent analysis of 29 predicted E. coli Tat signal peptides revealed that Tat selectivity is imparted by the overall charge of the c-region together with the N terminus of the mature protein. A net charge of +2 or higher resulted in Tat specificity while an overall charge of −1 or lower resulted in Sec and Tat promiscuity (D. Tullman-Ercek, M. P. DeLisa, Y. Kawarasaki, P. Iranpour, B. Ribnicky, et al., unpublished data). Because export is post-translational, it should not be surprising that a property encoded by the mature polypeptide is important in routing the protein to the secretory apparatus.
The Nature of Proteins Exported via Tat
Bioinformatic analysis of signal peptides has been used extensively for the prediction of proteins that utilize the Tat apparatus (10, 42, 107). However, as with any computational method, predictions have to be interpreted with caution (73).
Initially the Tat pathway was thought to have evolved for the translocation of complex redox proteins (11). For example, in Thermoplasma acidophilum and Thermoplasma volcanicum all predicted Tat substrates are believed to be cofactor-containing proteins (63). Similarly, in E. coli about 6% of all secreted proteins are predicted to be Tat-dependent and the majority are cofactor-containing redox proteins (95, 107, 121). They include hydrogenases, formate dehydrogenases, nitrate reductases, trimethylamine N-oxide (TMAO) reductases, and dimethyl sulfoxide (DMSO) reductases, all of which function in anaerobic respiration (50). For many of these enzymes the synthesis and maturation of their cofactors occur in the cytoplasm and they must be incorporated in a manner concomitant with folding (12, 95), rendering them incompatible with the Sec pathway. Remarkably, not only can the Tat pathway accommodate folded proteins, but it can also discriminate against misfolded proteins (38, 110, 112).
Some cofactor-binding sites, such as those for flavin adenine dinucleotide or copper, are also found in proteins that are exported through the Sec pathway. Periplasmic copper proteins are intriguing because those with a single-copper binding domain are Sec dependent. Preference seems to shift toward Tat export for substrates with additional and more complex copper binding sites, such as the E. coli multi-copper oxidase CueO (12, 68, 122). Unlike the many Tat substrates involved in anaerobic growth, CueO is required for copper homeostasis during aerobic growth (68, 120).
At least eight proteins in E. coli seem to require export via the Tat pathway yet do not have a signal sequence of their own. Instead, they form a multimeric complex with another protein that contains a Tat signal sequence (“hitchhiker mechanism”) (106). Thus far, all the proteins predicted to be exported in this way are also redox proteins and include hydrogenases −1 and −2, formate dehydrogenases, and DMSO reductases. The prototypical example of this class of proteins is the hydrogenase-2 subunit HybC, which is transported with its partner HybO in a process mediated by the signal peptide of the latter polypeptide (106).
The Tat pathway is capable of translocating integral membrane proteins. In fact, the multisubunit hydrogenases −1 and −2 mentioned above are membrane bound via a TM helical domain at the C terminus (C-tail) of one of the subunits. Targeting to the IM requires both the C-tail and the N-terminal Tat signal peptide (56). It is unclear whether the TM region is inserted laterally (e.g., from the translocon) or from the periplasm after export. However, the process is independent of YidC (homologous to the mitochondrial Oxa1p), which is required for the biogenesis of most other IM proteins in bacteria, mitochondria, and chloroplasts (56, 136). Note that membrane anchoring is not limited to substrates exported as multimers (e.g., E. coli hydrogenase-2 subunit HybA) (56).
In addition to C-terminal TM α-helices, other means of membrane tethering have been found in Tat substrates of the cpTat pathway but so far have not been observed in bacteria (84, 124). For example, the Rieske protein of the cytochrome b6/f complex that is exported via the cpTat pathway uses an un-cleaved signal sequence rather than a C-tail for membrane anchoring (84). There is also some evidence to suggest that the Tat pathway is capable of translocating lipoproteins that are attached to the membrane via lipidation of the N terminus (41, 80). Signal peptides containing the Tat consensus motif as well as the lipoprotein signal peptidase cleavage site have been identified in a variety of species, including Legionella pneumophila, Streptomyces lividans, S. coelicolor, and H. volcanii (32, 41, 80, 118). Two such signal peptides from S. coeli-color directed a reporter protein to the IM in S. lividans in a Tat-dependent manner (80). Similarly, Dilks et al. (41) demonstrated that the predicted Tat lipoprotein halocyanin-1 from H. volcanii is targeted to the IM in a Tat-dependent manner. However, definitive biochemical evidence for protein lipidation was not reported in any of the studies discussed above.
Only a quarter of the putative Tat substrates in L. pneumophila or the halophilic archaeon Halobacterium sp. NRC-1 appear to be redox proteins (32, 107). In Agrobacterium tumefaciens there is a significant number of predicted Tat-dependent periplasmic ligand binding proteins as well as an assortment of other enzymes, such as guanylate kinase and epoxide hydrolase, that do not seem to require cofactors (43). Similarly, the majority of the predicted Tat substrates from S. coelicolor do not fall into one of the groups discussed above (80, 118). Sequence analysis has shown that virtually all the homologs of alkaline phosphatase D and phospholipase C, neither of which are redox proteins, have typical Tat signal peptides (42). Even in E. coli, SufI, a prototypical Tat substrate of unknown function, and the periplasmic amidases AmiA and AmiC are neither exported as multimers nor contain cofactors. The genes amiA and amiC are particularly important for cell physiology. Inactivation of either gene or mislocalization of the corresponding protein due to the impairment of Tat export results in the formation of long chains of cells that are sensitive to the anionic detergent sodium dodecyl sulfate (13, 69). Interestingly, microarray analysis revealed that the change in expression of 50% of the E. coli genes whose expression is affected by a deletion of tatC can be attributed to this cell envelope defect (68). Amidases are predicted to be Tat substrates in a variety of other bacteria, though knockouts do not always result in such severe pleiotropic membrane defects (20, 24, 118).
GFP: green fluorescent protein
Why have amidases and other proteins evolved to utilize the Tat pathway? After all, the transit times for export are considerably longer compared with export through Sec and the energetic cost may be greater (3, 4, 122). The most widely accepted hypothesis is that these proteins may fold too quickly within the cytoplasm to be compatible with the Sec pathway. However, it was recently demonstrated that the cotranslational signal recognition particle (SRP) pathway is capable of exporting soluble proteins that exhibit rapid in vitro folding kinetics, such as the disulfide-bond-forming enzyme DsbA (62, 119). Along these lines it would be interesting to determine whether proteins such as AmiA, AmiC, and SufI can be targeted to the periplasm via an SRP-dependent process.
An alternative hypothesis is that certain Tat proteins may have to fold prior to export because they cannot reach their biologically active state within the periplasmic milieu. For example, green fluorescent protein (GFP) can be translocated via both the Sec and Tat pathways but is only fluorescent in the periplasm when exported by the Tat pathway (45, 111, 125). This finding suggests that some Tat substrates may be incompatible with folding in the periplasmic space. Along these lines it is also conceivable that the Tat pathway is in fact used for the export of slowly folding proteins. The periplasm presents a relatively harsh environment, and proteins that fail to reach their native state quickly are targeted for degradation by DegP (109). By allowing folding to proceed in the cytoplasm, slowly folding proteins may attain a protease-resistant conformation prior to being exposed to the periplasmic milieu.
Protein folding in the cytoplasm also offers access to chaperones such as GroEL that require ATP hydrolysis for their action. Interactions with GroEL/ES, DnaK/J/GrpE, and other cytoplasmic chaperone networks may be required to prevent aggregation or assist in the assembly of certain Tat substrates. Rose et al. (107) postulated that for H. volcanii and other halophilic archaea the high salt conditions of the extracellular milieu could lead to extensive aggregation, and for this reason proteins of all types are first folded in the cytoplasm and then exported via the Tat pathway. However, the halophile Salinibacter ruber survives in high salt conditions, yet this bacterium does not show such extreme dependence on the Tat pathway (41). In any event, engagement of the Tat pathway by protein substrates is presumed to be dictated by folding considerations, but whether this is due to slowly or rapidly folding rates or the availability of appropriate chaperones is an open issue.
MECHANISTIC CONSIDERATIONS
The TatABC Proteins
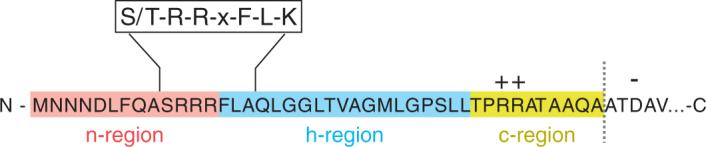
TatC is the largest and most highly conserved Tat component. The E. coli TatC is a 258-amino-acid polytopic membrane protein with six TM helices and both the N and C termini located on the cytoplasmic side of the membrane (9, 77) (Figure 2). The cytoplasmic side of TatC seems to be particularly important for function. Several residues within cytoplasmic loops are completely or functionally conserved across prokaryotes, chloroplasts, and plant mitochondria, and site-directed mutagenesis of a number of these residues has revealed that they are important or essential for TatC function (5, 8, 22). In vitro biochemical studies have revealed that TatC serves as the initial docking site for Tat signal peptides. Signal peptides from bacteria and chloroplasts interact with TatC either alone or in a complex with TatB (1, 27, 34). It is likely that more than one TatC monomer is required for each targeting event, since two mutant versions of TatC that alone blocked Tat-dependent transport support transport when expressed together (22). The interaction between TatC and the signal peptide in vitro requires the twin-arginine motif (1, 27). Recently, it was shown that export-defective signal peptides, in which the twin-arginine dipeptide had been substituted with twin lysines, could be rescued by mutations in TatC, further underscoring its role in signal peptide docking (E. M. Strauch & G. Georgiou, unpublished data).
Figure 2.
The predicted structure and topology of the E. coli Tat components. Predicted helical regions are shown as boxes.
The E. coli TatB protein consists of 171 amino acids, with an N-terminal TM helix followed by a short hinge region leading to a predicted amphipathic helix that may lie along the cytoplasmic side of the IM (35). The region of TatB following the amphipathic helix is predicted to remain helical in nature to approximately residue 75, after which TatB is considered largely unstructured (Figure 2). This latter region of TatB was shown through truncation analysis to be nonessential for Tat transport. However, once TatB is truncated to the amphipathic helix, activity is lost (79).
A number of conserved residues within TatB are important for transport activity. Site-directed mutagenesis highlighted the importance of the hinge region between the TM and cytoplasmic amphipathic helices (57). Interestingly, the importance of the hinge region may be substrate dependent (6, 8). It has been suggested that the most critical parts of TatB for transport lie within the cytoplasmic portions of the protein (79); mutations located in the TM domain have not yet been shown to have an effect on transport (6, 8, 57; P. A. Lee, N. P. Greene, G. Buchanan, P. J. Bond, G. Orriss, et al. unpublished data).
MBP: maltose binding protein
TatB was shown in vitro to contact the entire length of the signal peptide and also the mature protein more than 20 residues past the signal peptide cleavage site (1). Such interactions were seen only when TatC was present, suggesting that substrate targeting involves a series of sequential interactions, with TatC forming the primary recognition site before the substrate is transferred to TatB. In turn TatB could be considered a mediator between TatC and TatA, contacting the substrate after initial recognition by TatC and then potentially involved in transfer to a complex consisting mainly of the TatA protein prior to translocation (1, 115).
TatB is essential for the transport of endogenous substrates in E. coli (116). Nonetheless, a small but significant amount of transport of colicin V has been observed in a tatB deletion strain, suggesting that TatB may not always be essential for transport (67). Recently, a low level of Tat-dependent transport of maltose binding protein (MBP) was shown to occur when tatA and tatC were expressed from a plasmid in a strain deleted for all known tat genes (16). This construct did not support transport of the endogenous substrate TMAO reductase (TorA). However, upon mutagenesis of the tatAC genes, mutants that allowed significant transport of both MBP and TorA were isolated. Interestingly, all the mutations resulted in substitutions with methionine, serine, or aspartate and fell only within the first six residues of the TatA protein (16). The E. coli TatB contains a highly conserved aspartate in this region, and in some TatB homologs that do not contain this aspartate there is either a serine or methionine residue in the vicinity. Furthermore, the TatA protein from R. prowazekii, which has a Tat system that consists of only TatA and TatC, has both a methionine and a serine at positions three and four, respectively. These observations suggest that residues in this region are essential for at least one of the functions of the TatB protein (when it is present). These residues are thought to lie on the periplasmic side of the IM and thus are unlikely to play a role in substrate recognition by TatB.
The TatA protein is the most abundant of the Tat components and is estimated to be present at around 20 times more than TatB and TatC (71, 115). As previously mentioned, the TatA protein is predicted to have a structure similar to TatB, with an N-terminal TM domain followed by a short hinge region leading to an amphipathic helical region (extending to around residue 43) and an unstructured soluble C-terminal tail (Figure 2). TatA is shorter than TatB by 89 amino acids and contains shorter cytoplasmic helical and soluble domains. TatA is thought to function at a late stage in translocation and likely forms the major constituent of the Tat pore itself (1, 52, 85).
The TM domain of TatA is critical for export and is important for interactions with TatB (8, 30, 79). Upon removal of the TM domain TatA becomes soluble (35). There is some evidence that the cytoplasmic parts of TatA may have functional interactions with the IM because TatA assumes a helical nature that is observed only in the presence of membrane lipids (101). Both the hinge region and the cytoplasmic helical region have functional significance. The majority of the loss-of-function mutants isolated from a random library of tatA fall within these regions (58). Mutation of Phe at position 39 near the end of the cytoplasmic helical region resulted in a TatA protein that acts as a dominant-negative mutant, blocking transport even in the presence of wild-type TatA (57, 58).
A dual topology of the E. coli TatA has been proposed on the basis of topological analysis with fusions to the latter parts of TatA (53). Furthermore, TatA homologs in B. subtilis and H. volcanii are present in a soluble cytoplasmic form (41, 100). Thus, it is possible that during the transport cycle the cytoplasmic portion of TatA may insert into the IM or even cross into the periplasmic compartment.
The stoichiometry of the TatABC components is critical for export function. Overexpression of tatB results in complete loss of Tat transport (116). The overexpression of tatA has a less severe but nonetheless significant effect on translocation (116). In contrast, high expression of TatC can relieve saturation of the Tat pathway caused by the high-level expression of nonphysiological substrates such as GFP fusions (36). A general increase in the protein flux through the Tat pathway is seen only when tatABC are overexpressed in an operon together (2, 115, 134).
The Translocation Pore
Several lines of evidence suggest that TatB and TatC interact with a 1:1 stoichiometry to form a functional complex. TatB and TatC from E. coli purify predominantly as a 350- to 600-kDa membrane complex containing multiple copies of the two proteins in equimolar amounts (19, 34, 90). In addition, a fusion protein consisting of the C terminus of TatB fused to the N terminus of TatC supports Tat transport (19). Upon removal of the TM domain of TatB from this TatBC fusion construct, Tat transport activity is lost but the formation of the complex is not affected, although its interactions with TatA are weakened (82). Furthermore, TatC is unstable in the absence of TatB (116), suggesting a role for TatB in stabilizing the TatBC complex.
A second membrane complex of ∼600 kDa containing TatA and TatB has also been purified (18, 34, 115), and the two proteins can be coimmunoprecipitated (18). Chemical cross-linking has revealed that TatB forms homodimers (35, 57). Recent cysteine scanning and cross-linking experiments with TatB have shown that TatB self-interactions are mediated through both the TM domain and the early portion of the cytoplasmic amphipathic helix (P. A. Lee, N. P. Greene, G. Buchanan, P. J. Bond, G. Orriss, et al., unpublished data). Cross-linked species containing up to five TatB polypeptides were observed, suggesting that TatB may exist not only as dimers but also in higher-order oligomers. TatA also forms at least homotetramers in the E. coli IM (35, 57, 101). When expressed in the absence of TatBC, TatA forms multiple complexes ranging in size from 100 to 600 kDa (34, 35, 89, 101).
In E. coli, small amounts of a third Tat complex have been purified. This complex contained TatA, TatB, and TatC but with a large molar excess of TatA (34). The ratio of the Tat proteins in this complex more closely resembles that in the whole cell, as calculated from translational fusions or quantitative Western blotting analysis (71, 115).
The isolation of different Tat protein complexes, together with experimental evidence from both the bacterial and cpTat pathways, has led to the idea of a modular and highly dynamic system whereby the Tat proteins exist as separate complexes in the resting state and come together to form a complete translocation pore upon substrate binding (1, 85). Such a mechanism is depicted in Figure 3. Complexes of varying sizes may form pores that match the different sizes of folded Tat substrate proteins and thus minimize the space not occupied by the translocated protein, in turn restricting the formation of water-filled regions that can serve as conduits for ion leakage.
Figure 3.
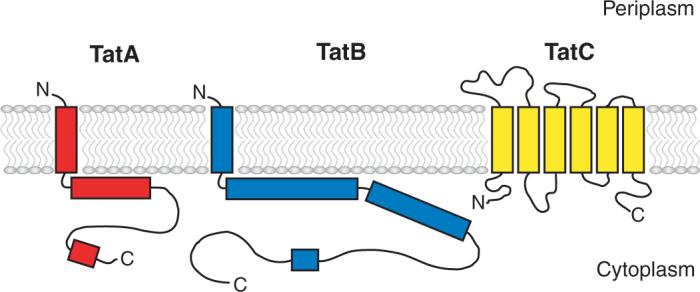
Model of Tat targeting and transport. In the membrane TatA is depicted in red, TatB in blue, and TatC in yellow. (a) Upon emerging from the ribosome the preprotein must avoid targeting to other pathways such as Sec, which is aided by the characteristics of the signal peptide and mature protein (29; D. Tullman-Ercek, M. P. DeLisa, Y. Kawarasaki, P. Iranpour, B. Ribnicky, et al., unpublished data) and/or the binding of Tat-specific chaperones (red circles) (70, 129). (b) After folding, any cofactors and/or additional subunits are added prior to targeting to the TatBC receptor complex (c) (1, 27, 34). (d) The proton motive force drives the formation of an active translocase and the substrate is transported through a pore consisting mainly of TatA (1, 52, 85). (e) Upon removal of the signal peptide the mature protein is released on the periplasmic side of the membrane.
The first structural view of the Tat translocon was obtained by Sargent et al. (115) using negative stain electron microscopy. Pore-like structures derived from a purified TatAB complex that contains an approximate 20 times molar excess of TatA to TatB and no detectible TatC were observed. The external diameter of these structures was 155 to 160 Å, with an internal diameter of around 65Å containing one to two density features, possibly forming a gate to the pore. Single-particle electron microscopy was also used to obtain low-resolution structures of TatABC complexes from three different bacteria expressed in E. coli. Similar structures were observed for the TatABC proteins from the different species, indicating structural conservation. These structures were suggested to be protein-lined pores or cavities but were not large enough to be the translocation pore itself (90).
A higher-resolution three-dimensional image of a Tat complex was recently obtained using random canonical tilt electron microscopy (Figure 4). A complex consisting almost entirely of TatA forms pore-like structures of various sizes that appear gated or blocked on the cytoplasmic side of the IM (52).
Figure 4.
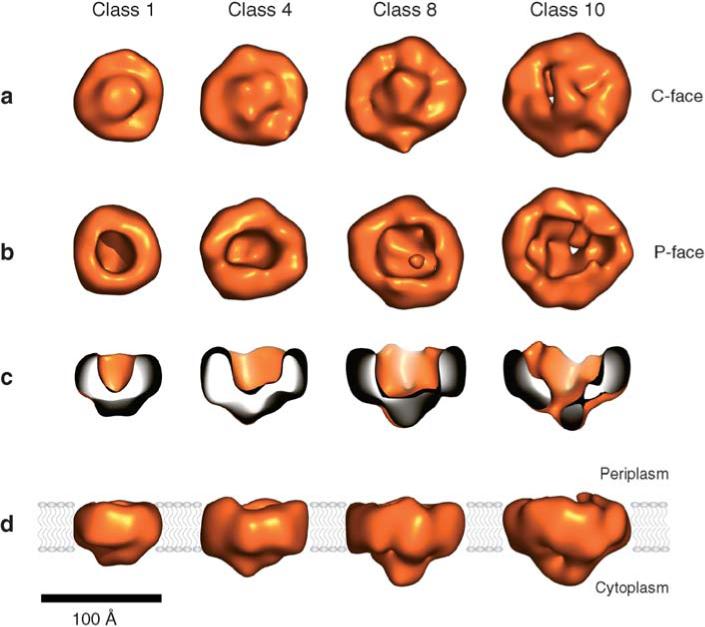
Three-dimensional density maps of TatA complexes. (a) TatA complexes viewed from the closed end of the channel proposed to be at the cytoplasmic side of the membrane (C-face). (b) TatA complexes viewed from the open end of the channel proposed to be at the periplasmic side of the membrane (P-face). (c) Side views of TatA. The front half of each molecule has been cut away to reveal internal features. (d) Views of TatA parallel to the membrane plane. The proposed position of the lipid bilayer is indicated in blue. (Scale bar, 100 Å.) Reproduced with permission from Reference 52 © 1993−2005 by The National Academy of Sciences of the United States of America, all rights reserved.
The above structural data provide support for the idea that the TatA protein forms the channel through which substrates are translocated across the IM. However, to gain a clear understanding of the structure and function of the Tat complexes that have been isolated, it is important to obtain a structural image of the translocase in action with a signal peptide or substrate in close contact with the Tat apparatus. This may be possible with a substrate that is blocked within the pore during transport or with a mutant Tat component that would effectively block transport at the substrate receptor level. A mutant recently constructed by Di Cola & Robinson (40), which was targeted to the cpTat pathway but not completely translocated, may prove useful for this purpose.
Energetics of Tat Translocation
Originally, the chloroplast cpTat pathway was termed the ΔpH pathway, since protein translocation has no requirement for ATP as an energy source but instead relied solely on the pmf (26). Similarly, transport of Tat substrates into E. coli inner membrane vesicles (IMVs) has no requirement for ATP and translocation was blocked upon dissipation of the membrane potential (2, 134). Alder & Theg (3) calculated the energy use of Tat-dependent translocation into isolated thylakoids and showed that on average 7.9 × 104 protons (equivalent to ∼10,000 ATP molecules) were released from the gradient per protein translocated. For comparison, Sec-dependent translocation is less energetically costly and has been estimated to require one ATP molecule per 20 amino acyl residues of the substrate translocated (130). A reduction in Tat-dependent transport was observed in the presence of D2O relative to H2O, suggesting that proton transfer is involved at some stage, perhaps via a H+/protein counterflux principle (87).
The requirement for the pmf under physiological conditions was challenged by recent in vivo data. Finazzi et al. (46) concluded that Tat translocation becomes pH sensitive only when probed in vitro, possibly owing to the loss of some critical intracellular factors. Di Cola et al. (39) examined cpTat translocation in vivo in tobacco protoplasts in the presence of valinomycin, which dissipates the membrane potential, and/or nigericin, which dissipates both the membrane potential and the ΔpH. The authors concluded that the initial stages of Tat transport are not dependent on either component of the pmf but also pointed out that it is not possible to conclude from these data whether or not the pmf is required at other points in the translocation process. Further exploration of the energy requirements of Tat transport is required, particularly of what factors lead to the discrepancies observed under the different experimental conditions.
IMV: inner membrane vesicle
REMPs: redox enzyme maturation proteins
Folding Quality Control
It is well established that the Tat pathway exports proteins that are folded prior to transport. It is also apparent that unfolded proteins are not compatible with transport (38, 110), implying that somewhere between protein synthesis and export there must be steps in which some sort of quality control is exerted on the folding state of the substrate. This is particularly important for a number of Tat substrates that require cofactor insertion or oligomerization with partner proteins in the cytoplasm prior to export.
Cytoplasmic chaperone-like proteins that function in the assembly of a number of cofactor and/or multimeric Tat substrates prevent targeting to the translocase until assembly is complete. The genes encoding these proteins are often found in the same operon as their corresponding Tat substrate proteins. These types of chaperones have recently been termed redox enzyme maturation proteins (REMPs) (129). It is not clear whether all Tat-dependent redox proteins have a corresponding REMP. However, homologs of the E. coli REMPs are found in many bacteria (129). Expression of the apparently REMP-less Tat substrate PhsABC from S. enterica LT2 in E. coli led to the production of active thiosulfate reductase (59). This finding suggests that either a REMP is not required for the export of PhsABC or that E. coli REMPs compensate for the lack of the native chaperone.
One well-studied group of REMPs is involved in the maturation of Tat substrates that are transported as multimeric complexes with only one of the subunits bearing a Tat signal peptide. For example, HyaE and HybE are chaperones required for the assembly of the hydrogenase components HyaA-HyaB and HybO-HybC, respectively. Only HyaA and HybO contain a Tat signal sequence, and therefore the respective dimers have to form in the cytoplasm prior to export (106). HyaE interacts with the HyaA precursor but not with the mature form and does not interact with HyaB (44). The HybE protein interacts with the precursor forms of both HybO and HybC (prior to removal of its C-terminal extension) but not with the mature form of each protein (44). HybE blocks the export of HybO without its hitchhiker partner HybC; without HybE the chaperone-mediated quality control mechanism is lost and transport of HybO is allowed prior to formation of the HybOC dimer (70).
Perhaps the most well-known E. coli REMPs are DmsD, required for the pre-export maturation of DMSO reductase (DmsA) (103), and TorD, which is required for the efficient maturation of TorA (98). DmsD and TorD belong to a family of structurally related chaperones that show high specificity for their cognate molybdoenzyme substrates (65, 70, 103). TorD has been purified in both monomeric and dimeric forms and crystallized as a dimer from Shewanella massilia (127, 128). HyaE, HybE, DmsD, and YcdY may also form similar dimeric structures (44, 113), suggesting that dimerization could be a general feature of E. coli REMPs.
DmsD was first identified owing to its ability to bind to the Tat signal peptide of DmsA (93). DmsD also binds to TatB and TatC in the IM, indicating that it may have a direct role in Tat-dependent transport (96). TorD binds the TorA signal peptide (70) and is required for the correct insertion of the molybdopterin guanine dinucleotide cofactor into TorA (98). In addition, Jack et al. (70) presented evidence that TorD can also function as a HyaE/HybE-type REMP. Replacement of the signal peptide of HybO with that of TorA results in export and correct localization of HybO without its partner protein HybC, which is found mislocalized in the cytoplasm. Upon overexpression of torD the HybOC dimer is correctly localized and hydrogenase activity is restored. In this case it is not clear whether TorD monitors the folding and assembly of the substrate proteins, or alternatively, by binding to the TorA signal peptide, whether it simply retards the export kinetics sufficiently to allow the HybO-HybC complex to form. Recently, it was also shown that TorD has an affinity for GTP, which is enhanced by signal peptide binding. This observation led Sargent and coworkers (55) to propose that nucleotide binding and release might regulate the interaction between the signal peptide and TorD.
DeLisa et al. (38) showed that a number of disulfide bond-containing proteins could be translocated via the Tat pathway only if oxidative protein folding and disulfide bond formation could take place in the cytoplasm. A trxB gor mutant strain (14), which rendered the normally reducing environment of the cytoplasm more oxidative, was employed for this study. Tat-dependent export of alkaline phosphatase (PhoA), a homodimer containing two structural disulfide bonds per polypeptide chain, was observed only in trxB gor mutants but not in the parental strain where PhoA could not fold prior to export. The requirement for oxidation in the cytoplasm for Tat export-competence was not dependent on the signal peptide fused to PhoA, arguing against the involvement of Tat-specific chaperones. On the basis of these observations, DeLisa et al. (38) proposed that a proofreading mechanism that discriminates between folded and unfolded proteins, allowing the export of only the former, must be an inherent property of the Tat translocon. Similarly, cytochrome c is also compatible for Tat transport only if the heme cofactor had been inserted, a process that occurs during folding in the cytoplasm (110). Recently, Richter & Brüser (105) reported that reduced PhoA fused to a Tat signal peptide binds to the TatBC complex resulting in a defect in the proton gradient, probably caused by the attempt to transport unfolded PhoA. They proposed that the inability of the Tat translocon to export unfolded substrates was not due to folding quality control but instead to “structural incompatibility with the translocation mechanism.” However, this seems to be a semantic distinction: The existence of a step in the translocation process that is incompatible with features found in any unfolded polypeptide de facto constitutes a folding quality control mechanism.
Recently, translocation reversal of two proteins targeted to the cpTat pathway was reported in vivo. It was concluded that the proteins traversed the membrane far enough for signal peptide cleavage to take place, but then they were somehow rejected by the translocase and returned to the stroma (40). This suggests that even at a late stage the Tat pathway can somehow sense and reject an incompatible substrate. The signal or mechanism for this rejection is not clear. No significant cpTat-dependent retrograde transport has been reported for an endogenous substrate protein in its natural setting and there is yet no evidence for a similar process in bacteria or archaea. Also, the involvement of the translocase in this phenomenon has not been demonstrated directly.
Other Proteins Involved in Tat Export
Reconstitution of the Tat pathway in vitro results in a low level of protein export. Even in IMVs obtained from cells overexpressing the tatABC genes from a T7 promoter, export is only 20% of the level seen in intact cells (2, 134), suggesting the involvement of additional factors in the Tat pathway. The data supporting the involvement of proteins other than TatABC and substrate-specific chaperones in Tat export are limited. The phage shock protein PspA was isolated from a genomic library as a protein that, when overproduced, relieves the saturation of the Tat pathway caused by high substrate levels (36). PspA is involved in the maintenance of the membrane integrity and/or pmf, suggesting an indirect role in Tat transport (31). In another study, two uncharacterized peripheral membrane proteins were reported to cross-link to the TorA signal peptide in IMVs (72).
ROLE IN PATHOGENESIS
Mutations in the tat genes have pleiotropic effects in E. coli including (a) filamentation and changes in outer membrane permeability due to the mislocalization of AmiA and AmiC, leading to the impairment of biofilm formation (68, 121); (b) inability to use various electron acceptors for growth under anaerobic conditions; and (c) defects in iron acquisition and copper homeostasis (68). Such growth defects would be expected to compromise the ability of pathogenic bacteria to colonize their host organisms. Indeed, in the food-borne pathogen E. coli 0157:H7, the Tat system is indirectly important for virulence (102). Similarly, a tat deletion strain of A. tumefaciens was unable to induce tumor formation in plant tissue (43).
The Tat pathway is specifically involved in the export of a variety of proteins that are directly involved in virulence. In the opportunistic human pathogen Pseudomonas aeruginosa two secreted virulence factors (both phospholipases) are mislocalized in tat mutants (131). Phospholipases are involved in cleavage of phospholipids found in mammalian erythrocytes (94). The two Pseudomonas enzymes were transported across the outer membrane by the type II secretion system (131), previously thought to be used only by proteins that use the Sec pathway (25). The Tat pathway of P. aeruginosa is also involved in the transport of several other virulence factors (91). Disruption of the Tat pathway abolishes virulence of pathogenic pseudomonads including P. aeruginosa and the plant pathogens Pseudomonas syringae pv. tomato DC3000 and pv. maculicola ES4326 (21, 24, 91).
Recently, attention has been focused on the Tat system of L. pneumophila, a water-borne pathogen that is the causative agent of Legionnaires’ disease. Although the Tat pathway is important for biofilm formation, replication within the host, growth under iron-limiting conditions, and secretion of phospholipase C activity, no Tat substrate directly involved in virulence of L. pneumophila has been identified so far (33, 108). The Tat pathway is also thought to be required for Helicobacter pylori and other gut bacteria to become established in their host organism (92).
The Tat pathway is directly involved in antibiotic resistance mechanisms of mycobacteria. Mycobacterium tuberculosis and Mycobacterium smegmatis tat mutants were unable to transport active β-lactamases (83). In fact, tatC may be an essential gene for optimal growth in these bacteria (83, 117). Recently, using β-lactamase as a reporter for Tat transport, McDonough et al. (83) demonstrated that the phospholipase C proteins (PlcA and PlcB), important for the virulence of M. tuberculosis (83, 104), are exported via the Tat pathway.
BIOTECHNOLOGICAL APPLICATIONS
The translocation of proteins from the bacterial cytoplasm is an important process in biotechnology. Export into the bacterial periplasm or into the growth medium is employed in preparative protein expression (51) and for protein engineering applications. Sec-dependent signal peptides, e.g., OmpA, PhoA, PelB, have been widely used to direct the export of biotechnologically relevant polypeptides. However, it has been known for 20 years that many heterologous proteins, as well as some native bacterial cytoplasmic proteins, fail to be exported when fused to Sec-dependent signal peptides partly because of folding considerations (88). Other proteins are compatible with transport but once in the periplasm are subjected to extensive degradation before they can reach their more stable, native conformation. Yet a another group of proteins, exemplified by GFP (45), are unable to fold properly once exported from the cytoplasm. Translocation of such proteins in a functional form might be achieved by routing them through the Tat pathway.
Tinker et al. (126) used the Tat pathway for the export of GFP fused to the A subunit of cholera toxin or the heat-labile toxins LT1 and LTIIb. Concomitant export of the respective B subunits via the Sec apparatus resulted in the formation of active and fluorescent holotoxin fusions in the periplasm. Other proteins of commercial interest have been translocated via the Tat pathway in E. coli and in S. lividans (49, 76).
For many exported proteins, folding is thermodynamically coupled to disulfide bond formation. The export of disulfide-containing proteins via the Tat pathway can occur only in trxB gor mutants, discussed above, or in other strains that allow the formation of disulfide bonds in the cytoplasm (38). Kim et al. (78) reported that a truncated form of tissue plasminogen activator containing nine disulfide bonds fused to a Tat leader peptide and expressed in a trxB gor E. coli strain resulted in a higher yield of active enzyme in the periplasm compared with a construct containing a Sec signal peptide. While the above results are encouraging, it is not yet clear whether Tat export can result in high yields of periplasmic protein, as desired for preparative expression purposes. In E. coli, even native Tat substrate proteins saturate the export process when overexpressed from strong promoters (36, 68, 134). Optimization of the growth conditions or coexpression of REMPs, PspA, or the TatABC proteins partially alleviates the saturation of the export apparatus (7, 36, 81). Still, protein yields for Tat-exported proteins are substantially lower than those obtained via Sec export, which often exceed 1 g liter−1. Therefore, additional fine tuning of cell physiology and fermentation conditions are required before Tat export can be utilized for protein manufacturing.
The ability of the Tat pathway to export only folded polypeptides is of considerable importance in protein engineering. Recently, a clever technique was developed for the selection of polypeptides that fold into a stable and soluble conformation (47). The efficiency with which Tat-exported β-lactamase chimeras localize in the periplasm, and thus confer ampicillin resistance, depends on the folding characteristics of the fusion partner polypeptide. Growth on increasing concentrations of ampicillin was employed to select for protein variants that exhibited higher stability and were less prone to aggregation. Screening of a library of random mutants of the Alzheimer's amyloid beta peptide (Aβ42) led to the isolation of several clones displaying greatly enhanced solubility (47). This method holds promise for the generation of well-expressed and soluble mutants of recombinant proteins for biochemical characterization and structural analysis studies.
Display on filamentous phage or on bacteria (48, 61) is widely used for the screening of combinatorial protein libraries. Both phage and bacterial display require translocation of the polypeptide from the cytoplasm, and this is typically accomplished using a Sec signal peptide. However, for the reasons discussed above, certain proteins cannot be exported via the Sec pathway and therefore cannot be displayed. Even if a protein is compatible with Sec export, mutant forms of the protein, such as those that arise during the construction of libraries by random mutagenesis, may exhibit altered folding characteristics that might prevent their display. The Tat pathway could be used to mediate the export and display of such proteins and thus expand the diversity of sequences within the library.
Recombinant polypeptides are typically displayed on filamentous phage by fusing them to the N terminus of the p3 phage protein, which is transported into the periplasmic space and resides transiently in the IM before it is incorporated into the growing virion. For unknown reasons, export of p3 with a Tat signal peptide results in low phage titers (D. Tullman-Ercek & G. Georgiou, unpublished data). However, display without significant loss of infectivity is achieved when a Tat-exported polypeptide is fused to half of a coiled-coil and coexpressed with a Sec-exported p3 fused to the second half of the coiled-coil (97). In this manner, the two polypeptides associate after they have been exported into the periplasm by different routes, thus allowing efficient display. For bacterial display purposes, proteins can be translocated via the Tat pathway and then be tethered to the IM, for example, by fusion to the C-terminal tail of HybO (56). Such periplasmically anchored polypeptides can be screened for binding to fluorescently labeled ligands using the anchored periplasmic expression (APEx) approach (54). For APEx, proteins tethered to the IM are made accessible to fluorescent ligands by making spheroplasts, which can then be analyzed by flow cytometry.
SUMMARY POINTS.
The Tat pathway transports a wide variety of prefolded proteins across bacterial and plant chloroplast membranes.
The Tat pathway is required for virulence of some bacteria.
The transport mechanism involves high-molecular-weight complexes formed by the TatABC proteins that comprise the translocon.
No detailed model of the transport process has emerged so far.
The exact energy requirements of the system are somewhat controversial.
The Tat pathway holds promise for biotechnology applications particularly in protein expression and engineering.
ACKNOWLEDGMENTS
We are grateful to Prof. Tracy Palmer and Eva Strauch for communicating results prior to publication. Support was provided by grants from the Foundation for Research and NIH 1 R01 GM069872.
Footnotes
RELATED RESOURCE
Scott JR. 2006. Synthesis and function of cell surface structures in gram-positive bacteria. Annu. Rev. Microbiol. 60:In press
LITERATURE CITED
- 1.Alami M, Luke I, Deitermann S, Eisner G, Koch HG, et al. Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol. Cell. 2003;12:937–46. doi: 10.1016/s1097-2765(03)00398-8. [DOI] [PubMed] [Google Scholar]
- 2.Alami M, Trescher D, Wu LF, Muller M. Separate analysis of twin-arginine translocation (Tat)-specific membrane binding and translocation in Escherichia coli. J. Biol. Chem. 2002;277:20499–503. doi: 10.1074/jbc.M201711200. [DOI] [PubMed] [Google Scholar]
- 3.Alder NN, Theg SM. Energetics of protein transport across biological membranes. A study of the thylakoid DeltapH-dependent/cpTat pathway. Cell. 2003;112:231–42. doi: 10.1016/s0092-8674(03)00032-1. [DOI] [PubMed] [Google Scholar]
- 4.Alder NN, Theg SM. Energy use by biological protein transport pathways. Trends Biochem. Sci. 2003;28:442–51. doi: 10.1016/S0968-0004(03)00167-1. [DOI] [PubMed] [Google Scholar]
- 5.Allen SC, Barrett CM, Ray N, Robinson C. Essential cytoplasmic domains in the Escherichia coli TatC protein. J. Biol. Chem. 2002;277:10362–66. doi: 10.1074/jbc.M109135200. [DOI] [PubMed] [Google Scholar]
- 6.Barrett CM, Mathers JE, Robinson C. Identification of key regions within the Escherichia coli TatAB subunits. FEBS Lett. 2003;537:42–46. doi: 10.1016/s0014-5793(03)00068-1. [DOI] [PubMed] [Google Scholar]
- 7.Barrett CM, Ray N, Thomas JD, Robinson C, Bolhuis A. Quantitative export of a reporter protein, GFP, by the twin-arginine translocation pathway in Escherichia coli. Biochem. Biophys. Res. Commun. 2003;304:279–84. doi: 10.1016/s0006-291x(03)00583-7. [DOI] [PubMed] [Google Scholar]
- 8.Barrett CM, Robinson C. Evidence for interactions between domains of TatA and TatB from mutagenesis of the TatABC subunits of the twin-arginine translocase. FEBS J. 2005;272:2261–75. doi: 10.1111/j.1742-4658.2005.04654.x. [DOI] [PubMed] [Google Scholar]
- 9.Behrendt J, Standar K, Lindenstrauss U, Brüser T. Topological studies on the twin-arginine translocase component TatC. FEMS Microbiol. Lett. 2004;234:303–8. doi: 10.1016/j.femsle.2004.03.048. [DOI] [PubMed] [Google Scholar]
- 10.Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. Prediction of twin-arginine signal peptides. BMC Bioinform. 2005;6:167. doi: 10.1186/1471-2105-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berks BC. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 12.Berks BC, Palmer T, Sargent F. The Tat protein translocation pathway and its role in microbial physiology. Adv. Microb. Physiol. 2003;47:187–254. doi: 10.1016/s0065-2911(03)47004-5. [DOI] [PubMed] [Google Scholar]
- 13.Bernhardt TG, de Boer PA. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 2003;48:1171–82. doi: 10.1046/j.1365-2958.2003.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bessette PH, Aslund F, Beckwith J, Georgiou G. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc. Natl. Acad. Sci. USA. 1999;96:13703–8. doi: 10.1073/pnas.96.24.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blaudeck N, Kreutzenbeck P, Freudl R, Sprenger GA. Genetic analysis of pathway specificity during posttranslational protein translocation across the Escherichia coli plasma membrane. J. Bacteriol. 2003;185:2811–19. doi: 10.1128/JB.185.9.2811-2819.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blaudeck N, Kreutzenbeck P, Muller M, Sprenger GA, Freudl R. Isolation and characterization of bifunctional Escherichia coli TatA mutant proteins that allow efficient Tat-dependent protein translocation in the absence of TatB. J. Biol. Chem. 2005;280:3426–32. doi: 10.1074/jbc.M411210200. [DOI] [PubMed] [Google Scholar]
- 17.Bogsch EG, Sargent F, Stanley NR, Berks BC, Robinson C, Palmer T. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J. Biol. Chem. 1998;273:18003–6. doi: 10.1074/jbc.273.29.18003. [DOI] [PubMed] [Google Scholar]
- 18.Bolhuis A, Bogsch EG, Robinson C. Subunit interactions in the twin-arginine translocase complex of Escherichia coli. FEBS Lett. 2000;472:88–92. doi: 10.1016/s0014-5793(00)01428-9. [DOI] [PubMed] [Google Scholar]
- 19.Bolhuis A, Mathers JE, Thomas JD, Barrett CM, Robinson C. TatB and TatC form a functional and structural unit of the twin-arginine translocase from Escherichia coli. J. Biol. Chem. 2001;276:20213–19. doi: 10.1074/jbc.M100682200. [DOI] [PubMed] [Google Scholar]
- 20.Bronstein P, Marrichi M, DeLisa MP. Dissecting the twin-arginine translocation pathway using genome-wide analysis. Res. Microbiol. 2004;155:803–10. doi: 10.1016/j.resmic.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 21.Bronstein PA, Marrichi M, Cartinhour S, Schneider DJ, Delisa MP. Identification of a twin-arginine translocation system in Pseudomonas syringae pv. tomato DC3000 and its contribution to pathogenicity and fitness. J. Bacteriol. 2005;187:8450–61. doi: 10.1128/JB.187.24.8450-8461.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buchanan G, Leeuw E, Stanley NR, Wexler M, Berks BC, et al. Functional complexity of the twin-arginine translocase TatC component revealed by site-directed mutagenesis. Mol. Microbiol. 2002;43:1457–70. doi: 10.1046/j.1365-2958.2002.02853.x. [DOI] [PubMed] [Google Scholar]
- 23.Buchanan G, Sargent F, Berks BC, Palmer T. A genetic screen for suppressors of Escherichia coli Tat signal peptide mutations establishes a critical role for the second arginine within the twin-arginine motif. Arch. Microbiol. 2001;177:107–12. doi: 10.1007/s00203-001-0366-2. [DOI] [PubMed] [Google Scholar]
- 24.Caldelari I, Mann S, Crooks C, Palmer T. The Tat pathway of the plant pathogen Pseudomonas syringae is required for optimal virulence. Mol. Plant. Microbe Interact. 2006;19:200–12. doi: 10.1094/MPMI-19-0200. [DOI] [PubMed] [Google Scholar]
- 25.Cianciotto NP. Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 2005;13:581–88. doi: 10.1016/j.tim.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 26.Cline K, Ettinger WF, Theg SM. Protein-specific energy requirements for protein transport across or into thylakoid membranes. Two lumenal proteins are transported in the absence of ATP. J. Biol. Chem. 1992;267:2688–96. [PubMed] [Google Scholar]
- 27.Cline K, Mori H. Thylakoid DeltapH-dependent precursor proteins bind to a cpTatC-Hcf106 complex before Tha4-dependent transport. J. Cell Biol. 2001;154:719–29. doi: 10.1083/jcb.200105149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Creighton AM, Hulford A, Mant A, Robinson D, Robinson C. A monomeric, tightly folded stromal intermediate on the delta pH-dependent thylakoidal protein transport pathway. J. Biol. Chem. 1995;270:1663–69. doi: 10.1074/jbc.270.4.1663. [DOI] [PubMed] [Google Scholar]
- 29.Cristobal S, de Gier JW, Nielsen H, von Heijne G. Competition between Sec- and TAT-dependent protein translocation in Escherichia coli. EMBO J. 1999;18:2982–90. doi: 10.1093/emboj/18.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dabney-Smith C, Mori H, Cline K. Requirement of a Tha4-conserved transmembrane glutamate in thylakoid Tat translocase assembly revealed by biochemical complementation. J. Biol. Chem. 2003;278:43027–33. doi: 10.1074/jbc.M307923200. [DOI] [PubMed] [Google Scholar]
- 31.Darwin AJ. The phage-shock-protein response. Mol. Microbiol. 2005;57:621–28. doi: 10.1111/j.1365-2958.2005.04694.x. [DOI] [PubMed] [Google Scholar]
- 32.De Buck E, Lebeau I, Maes L, Geukens N, Meyen E, et al. A putative twin-arginine translocation pathway in Legionella pneumophila. Biochem. Biophys. Res. Commun. 2004;317:654–61. doi: 10.1016/j.bbrc.2004.03.091. [DOI] [PubMed] [Google Scholar]
- 33.De Buck E, Maes L, Meyen E, Van Mellaert L, Geukens N, et al. Legionella pneumophila Philadelphia-1 tatB and tatC affect intracellular replication and biofilm formation. Biochem. Biophys. Res. Commun. 2005;331:1413–20. doi: 10.1016/j.bbrc.2005.04.060. [DOI] [PubMed] [Google Scholar]
- 34.De Leeuw E, Granjon T, Porcelli I, Alami M, Carr SB, et al. Oligomeric properties and signal peptide binding by Escherichia coli Tat protein transport complexes. J. Mol. Biol. 2002;322:1135–46. doi: 10.1016/s0022-2836(02)00820-3. [DOI] [PubMed] [Google Scholar]
- 35.De Leeuw E, Porcelli I, Sargent F, Palmer T, Berks BC. Membrane interactions and self-association of the TatA and TatB components of the twin-arginine translocation pathway. FEBS Lett. 2001;506:143–48. doi: 10.1016/s0014-5793(01)02904-0. [DOI] [PubMed] [Google Scholar]
- 36.DeLisa MP, Lee P, Palmer T, Georgiou G. Phage shock protein PspA of Escherichia coli relieves saturation of protein export via the Tat pathway. J. Bacteriol. 2004;186:366–73. doi: 10.1128/JB.186.2.366-373.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeLisa MP, Samuelson P, Palmer T, Georgiou G. Genetic analysis of the twin arginine translocator secretion pathway in bacteria. J. Biol. Chem. 2002;277:29825–31. doi: 10.1074/jbc.M201956200. [DOI] [PubMed] [Google Scholar]
- 38.DeLisa MP, Tullman D, Georgiou G. Folding quality control in the export of proteins by the bacterial twin-arginine translocation pathway. Proc. Natl. Acad. Sci. USA. 2003. pp. 6115–20. [DOI] [PMC free article] [PubMed]; Demonstrates the existence of a folding quality control function within the Tat export machinery
- 39.Di Cola A, Bailey S, Robinson C. The thylakoid ΔpH/Δpsi are not required for the initial stages of Tat-dependent protein transport in tobacco protoplasts. J. Biol. Chem. 2005;280:41165–70. doi: 10.1074/jbc.M509215200. [DOI] [PubMed] [Google Scholar]
- 40.Di Cola A, Robinson C. Large-scale translocation reversal within the thylakoid Tat system in vivo. J. Cell Biol. 2005;171:281–89. doi: 10.1083/jcb.200502067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dilks K, Gimenez MI, Pohlschroder M. Genetic and biochemical analysis of the twin-arginine translocation pathway in halophilic archaea. J. Bacteriol. 2005;187:8104–13. doi: 10.1128/JB.187.23.8104-8113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dilks K, Rose RW, Hartmann E, Pohlschroder M. Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J. Bacteriol. 2003;185:1478–83. doi: 10.1128/JB.185.4.1478-1483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding Z, Christie PJ. Agrobacterium tumefaciens twin-arginine-dependent translocation is important for virulence, flagellation, and chemotaxis but not type IV secretion. J. Bacteriol. 2003;185:760–71. doi: 10.1128/JB.185.3.760-771.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dubini A, Sargent F. Assembly of Tat-dependent [NiFe] hydrogenases: identification of precursor-binding accessory proteins. FEBS Lett. 2003;549:141–46. doi: 10.1016/s0014-5793(03)00802-0. [DOI] [PubMed] [Google Scholar]
- 45.Feilmeier BJ, Iseminger G, Schroeder D, Webber H, Phillips GJ. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J. Bacteriol. 2000;182:4068–76. doi: 10.1128/jb.182.14.4068-4076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Finazzi G, Chasen C, Wollman FA, de Vitry C. Thylakoid targeting of Tat passenger proteins shows no pH dependence in vivo. EMBO J. 2003;22:807–15. doi: 10.1093/emboj/cdg081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fisher AC, Kim W, DeLisa MP. Genetic selection for protein solubility enabled by the folding quality control feature of the twin-arginine translocation pathway. Protein Sci. 2006. pp. 449–58. [DOI] [PMC free article] [PubMed]; Demonstrates a biotechnological application of the Tat pathway for isolation of proteins with enhanced solubility
- 48.Francisco JA, Campbell R, Iverson BL, Georgiou G. Production and fluorescence-activated cell sorting of Escherichia coli expressing a functional antibody fragment on the external surface. Proc. Natl. Acad. Sci. USA. 1993;90:10444–48. doi: 10.1073/pnas.90.22.10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gauthier C, Li H, Morosoli R. Increase in xylanase production by Streptomyces lividans through simultaneous use of the Sec- and Tat-dependent protein export systems. Appl. Environ. Microbiol. 2005;71:3085–92. doi: 10.1128/AEM.71.6.3085-3092.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gennis RB, Stewart V. Respiration. In: Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd ed Am. Soc. Microbiol.; Washington, DC: 1996. pp. 217–61.. [Google Scholar]
- 51.Georgiou G, Segatori L. Preparative expression of secreted proteins in bacteria: status report and future prospects. Curr. Opin. Biotechnol. 2005;16:538–45. doi: 10.1016/j.copbio.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 52.Gohlke U, Pullan L, McDevitt CA, Porcelli I, de Leeuw E, et al. The TatA component of the twin-arginine protein transport system forms channel complexes of variable diameter. Proc. Natl. Acad. Sci. USA. 2005. pp. 10482–86. [DOI] [PMC free article] [PubMed]; Provides the first high-resolution images of a Tat complex, possibly the translocation channel itself
- 53.Gouffi K, Gerard F, Santini CL, Wu LF. Dual topology of the Escherichia coli TatA protein. J. Biol. Chem. 2004;279:11608–15. doi: 10.1074/jbc.M313187200. [DOI] [PubMed] [Google Scholar]
- 54.Harvey BR, Georgiou G, Hayhurst A, Jeong KJ, Iverson BL, Rogers GK. Anchored periplasmic expression, a versatile technology for the isolation of high-affinity antibodies from Escherichia coli-expressed libraries. Proc. Natl. Acad. Sci. USA. 2004;101:9193–98. doi: 10.1073/pnas.0400187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hatzixanthis K, Clarke TA, Oubrie A, Richardson DJ, Turner RJ, Sargent F. Signal peptide-chaperone interactions on the twin-arginine protein transport pathway. Proc. Natl. Acad. Sci. USA. 2005;102:8460–65. doi: 10.1073/pnas.0500737102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hatzixanthis K, Palmer T, Sargent F. A subset of bacterial inner membrane proteins integrated by the twin-arginine translocase. Mol. Microbiol. 2003. pp. 1377–90. [DOI] [PubMed]; Demonstrates the YidC-independent integration of Tat-dependent membrane proteins
- 57.Hicks MG, de Leeuw E, Porcelli I, Buchanan G, Berks BC, Palmer T. The Escherichia coli twin-arginine translocase: conserved residues of TatA and TatB family components involved in protein transport. FEBS Lett. 2003;539:61–67. doi: 10.1016/s0014-5793(03)00198-4. [DOI] [PubMed] [Google Scholar]
- 58.Hicks MG, Lee PA, Georgiou G, Berks BC, Palmer T. Positive selection for loss-of-function tat mutations identifies critical residues required for TatA activity. J. Bacteriol. 2005;187:2920–25. doi: 10.1128/JB.187.8.2920-2925.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hinsley AP, Berks BC. Specificity of respiratory pathways involved in the reduction of sulfur compounds by Salmonella enterica. Microbiology. 2002;148:3631–38. doi: 10.1099/00221287-148-11-3631. [DOI] [PubMed] [Google Scholar]
- 60.Hinsley AP, Stanley NR, Palmer T, Berks BC. A naturally occurring bacterial Tat signal peptide lacking one of the ‘invariant’ arginine residues of the consensus targeting motif. FEBS Lett. 2001;497:45–49. doi: 10.1016/s0014-5793(01)02428-0. [DOI] [PubMed] [Google Scholar]
- 61.Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nat. Biotechnol. 2005;23:1105–16. doi: 10.1038/nbt1126. [DOI] [PubMed] [Google Scholar]
- 62.Huber D, Boyd D, Xia Y, Olma MH, Gerstein M, Beckwith J. Use of thioredoxin as a reporter to identify a subset of Escherichia coli signal sequences that promote signal recognition particle-dependent translocation. J. Bacteriol. 2005;187:2983–91. doi: 10.1128/JB.187.9.2983-2991.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hutcheon GW, Bolhuis A. The archaeal twin-arginine translocation pathway. Biochem. Soc. Trans. 2003;31:686–89. doi: 10.1042/bst0310686. [DOI] [PubMed] [Google Scholar]
- 64.Ignatova Z, Hornle C, Nurk A, Kasche V. Unusual signal peptide directs penicillin amidase from Escherichia coli to the Tat translocation machinery. Biochem. Biophys. Res. Commun. 2002;291:146–49. doi: 10.1006/bbrc.2002.6420. [DOI] [PubMed] [Google Scholar]
- 65.Ilbert M, Mejean V, Iobbi-Nivol C. Functional and structural analysis of members of the TorD family, a large chaperone family dedicated to molybdoproteins. Microbiology. 2004;150:935–43. doi: 10.1099/mic.0.26909-0. [DOI] [PubMed] [Google Scholar]
- 66.Ize B, Gerard F, Wu LF. In vivo assessment of the Tat signal peptide specificity in Escherichia coli. Arch. Microbiol. 2002;178:548–53. doi: 10.1007/s00203-002-0488-1. [DOI] [PubMed] [Google Scholar]
- 67.Ize B, Gerard F, Zhang M, Chanal A, Voulhoux R, et al. In vivo dissection of the Tat translocation pathway in Escherichia coli. J. Mol. Biol. 2002;317:327–35. doi: 10.1006/jmbi.2002.5431. [DOI] [PubMed] [Google Scholar]
- 68.Ize B, Porcelli I, Lucchini S, Hinton JC, Berks BC, Palmer T. Novel phenotypes of Escherichia coli tat mutants revealed by global gene expression and phenotypic analysis. J. Biol. Chem. 2004;279:47543–54. doi: 10.1074/jbc.M406910200. [DOI] [PubMed] [Google Scholar]
- 69.Ize B, Stanley NR, Buchanan G, Palmer T. Role of the Escherichia coli Tat pathway in outer membrane integrity. Mol. Microbiol. 2003;48:1183–93. doi: 10.1046/j.1365-2958.2003.03504.x. [DOI] [PubMed] [Google Scholar]
- 70.Jack RL, Buchanan G, Dubini A, Hatzixanthis K, Palmer T, Sargent F. Coordinating assembly and export of complex bacterial proteins. EMBO J. 2004;23:3962–72. doi: 10.1038/sj.emboj.7600409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jack RL, Sargent F, Berks BC, Sawers G, Palmer T. Constitutive expression of Escherichia coli tat genes indicates an important role for the twin-arginine translocase during aerobic and anaerobic growth. J. Bacteriol. 2001;183:1801–4. doi: 10.1128/JB.183.5.1801-1804.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jong WS, ten Hagen-Jongman CM, Genevaux P, Brunner J, Oudega B, Luirink J. Trigger factor interacts with the signal peptide of nascent Tat substrates but does not play a critical role in Tat-mediated export. Eur. J. Biochem. 2004;271:4779–87. doi: 10.1111/j.1432-1033.2004.04442.x. [DOI] [PubMed] [Google Scholar]
- 73.Jongbloed JD, Antelmann H, Hecker M, Nijland R, Bron S, et al. Selective contribution of the twin-arginine translocation pathway to protein secretion in Bacillus subtilis. J. Biol. Chem. 2002;277:44068–78. doi: 10.1074/jbc.M203191200. [DOI] [PubMed] [Google Scholar]
- 74.Jongbloed JD, Grieger U, Antelmann H, Hecker M, Nijland R, et al. Two minimal Tat translocases in Bacillus. Mol. Microbiol. 2004. pp. 1319–25. [DOI] [PubMed]; Describes two distinct Tat translocases in B. subtilis, each transporting specific substrates
- 75.Jongbloed JD, Martin U, Antelmann H, Hecker M, Tjalsma H, et al. TatC is a specificity determinant for protein secretion via the twin-arginine translocation pathway. J. Biol. Chem. 2000;275:41350–57. doi: 10.1074/jbc.M004887200. [DOI] [PubMed] [Google Scholar]
- 76.Kang DG, Lim GB, Cha HJ. Functional periplasmic secretion of organophosphorous hydrolase using the twin-arginine translocation pathway in Escherichia coli. J. Biotechnol. 2005;118:379–85. doi: 10.1016/j.jbiotec.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 77.Ki JJ, Kawarasaki Y, Gam J, Harvey BR, Iverson BL, Georgiou G. A periplasmic fluorescent reporter protein and its application in high-throughput membrane protein topology analysis. J. Mol. Biol. 2004;341:901–9. doi: 10.1016/j.jmb.2004.05.078. [DOI] [PubMed] [Google Scholar]
- 78.Kim JY, Fogarty EA, Lu FJ, Zhu H, Wheelock GD, et al. Twin-arginine translocation of active human tissue plasminogen activator in Escherichia coli. Appl. Environ. Microbiol. 2005;71:8451–59. doi: 10.1128/AEM.71.12.8451-8459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee PA, Buchanan G, Stanley NR, Berks BC, Palmer T. Truncation analysis of TatA and TatB defines the minimal functional units required for protein translocation. J. Bacteriol. 2002;184:5871–79. doi: 10.1128/JB.184.21.5871-5879.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li H, Jacques PE, Ghinet MG, Brzezinski R, Morosoli R. Determining the functionality of putative Tat-dependent signal peptides in Streptomyces coelicolor A3(2) by using two different reporter proteins. Microbiology. 2005;151:2189–98. doi: 10.1099/mic.0.27893-0. [DOI] [PubMed] [Google Scholar]
- 81.Li SY, Chang BY, Lin SC. Coexpression of TorD enhances the transport of GFP via the TAT pathway. J. Biotechnol. 2005;1224:412–21. doi: 10.1016/j.jbiotec.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 82.Mangels D, Mathers J, Bolhuis A, Robinson C. The core TatABC complex of the twin-arginine translocase in Escherichia coli: TatC drives assembly whereas TatA is essential for stability. J. Mol. Biol. 2005;345:415–23. doi: 10.1016/j.jmb.2004.10.043. [DOI] [PubMed] [Google Scholar]
- 83.McDonough JA, Hacker KE, Flores AR, Pavelka MSJ, Braunstein M. The twin-arginine translocation pathway of Mycobacterium smegmatis is functional and required for the export of mycobacterial beta-lactamases. J. Bacteriol. 2005. pp. 7667–79. [DOI] [PMC free article] [PubMed]; Demonstrates that the Tat pathway exports virulence factors in pathogenic mycobacteria
- 84.Molik S, Karnauchov I, Weidlich C, Herrmann RG, Klosgen RB. The Rieske Fe/S protein of the cytochrome b6/f complex in chloroplasts: missing link in the evolution of protein transport pathways in chloroplasts? J. Biol. Chem. 2001;276:42761–66. doi: 10.1074/jbc.M106690200. [DOI] [PubMed] [Google Scholar]
- 85.Mori H, Cline K. A twin arginine signal peptide and the pH gradient trigger reversible assembly of the thylakoid ΔpH/Tat translocase. J. Cell Biol. 2002;157:205–10. doi: 10.1083/jcb.200202048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mould RM, Robinson C. A proton gradient is required for the transport of two lumenal oxygen-evolving proteins across the thylakoid membrane. J. Biol. Chem. 1991;266:12189–93. [PubMed] [Google Scholar]
- 87.Musser SM, Theg SM. Proton transfer limits protein translocation rate by the thylakoid DeltapH/Tat machinery. Biochemistry. 2000;39:8228–33. doi: 10.1021/bi000115f. [DOI] [PubMed] [Google Scholar]
- 88.Nouwen N, de Kruijff B, Tommassen J. prlA suppressors in Escherichia coli relieve the proton electrochemical gradient dependency of translocation of wild-type precursors. Proc. Natl. Acad. Sci. USA. 1996;93:5953–57. doi: 10.1073/pnas.93.12.5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Oates J, Barrett CM, Barnett JP, Byrne KG, Bolhuis A, Robinson C. The Escherichia coli twin-arginine translocation apparatus incorporates a distinct form of TatABC complex, spectrum of modular TatA complexes and minor TatAB complex. J. Mol. Biol. 2005;346:295–305. doi: 10.1016/j.jmb.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 90.Oates J, Mathers J, Mangels D, Kuhlbrandt W, Robinson C, Model K. Consensus structural features of purified bacterial TatABC complexes. J. Mol. Biol. 2003;330:277–86. doi: 10.1016/s0022-2836(03)00621-1. [DOI] [PubMed] [Google Scholar]
- 91.Ochsner UA, Snyder A, Vasil AI, Vasil ML. Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc. Natl. Acad. Sci. USA. 2002;99:8312–17. doi: 10.1073/pnas.082238299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Olson JW, Maier RJ. Molecular hydrogen as an energy source for Helicobacter pylori. Science. 2002;298:1788–90. doi: 10.1126/science.1077123. [DOI] [PubMed] [Google Scholar]
- 93.Oresnik IJ, Ladner CL, Turner RJ. Identification of a twin-arginine leader-binding protein. Mol. Microbiol. 2001;40:323–31. doi: 10.1046/j.1365-2958.2001.02391.x. [DOI] [PubMed] [Google Scholar]
- 94.Ostroff RM, Vasil AI, Vasil ML. Molecular comparison of a nonhemolytic and a hemolytic phospholipase C from Pseudomonas aeruginosa. J. Bacteriol. 1990;172:5915–23. doi: 10.1128/jb.172.10.5915-5923.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Palmer T, Sargent F, Berks BC. Export of complex cofactor-containing proteins by the bacterial Tat pathway. Trends Microbiol. 2005;13:175–80. doi: 10.1016/j.tim.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 96.Papish AL, Ladner CL, Turner RJ. The twin-arginine leader-binding protein, DmsD, interacts with the TatB and TatC subunits of the Escherichia coli twin-arginine translocase. J. Biol. Chem. 2003;278:32501–6. doi: 10.1074/jbc.M301076200. [DOI] [PubMed] [Google Scholar]
- 97.Paschke M, Hohne W. A twin-arginine translocation (Tat)-mediated phage display system. Gene. 2005;350:79–88. doi: 10.1016/j.gene.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 98.Pommier J, Mejean V, Giordano G, Iobbi-Nivol C. TorD, a cytoplasmic chaperone that interacts with the unfolded trimethylamine N-oxide reductase enzyme (TorA) in Escherichia coli. J. Biol. Chem. 1998;273:16615–20. doi: 10.1074/jbc.273.26.16615. [DOI] [PubMed] [Google Scholar]
- 99.Pop O, Martin U, Abel C, Muller JP. The twin-arginine signal peptide of PhoD and the TatAd/Cd proteins of Bacillus subtilis form an autonomous Tat translocation system. J. Biol. Chem. 2002;277:3268–73. doi: 10.1074/jbc.M110829200. [DOI] [PubMed] [Google Scholar]
- 100.Pop OI, Westermann M, Volkmer-Engert R, Schulz D, Lemke C, et al. Sequence-specific binding of prePhoD to soluble TatAd indicates protein-mediated targeting of the Tat export in Bacillus subtilis. J. Biol. Chem. 2003;278:38428–36. doi: 10.1074/jbc.M306516200. [DOI] [PubMed] [Google Scholar]
- 101.Porcelli I, de Leeuw E, Wallis R, van den Brink-van der Laan E, de Kruijff B, et al. Characterization and membrane assembly of the TatA component of the Escherichia coli twin-arginine protein transport system. Biochemistry. 2002;41:13690–97. doi: 10.1021/bi026142i. [DOI] [PubMed] [Google Scholar]
- 102.Pradel N, Ye C, Livrelli V, Xu J, Joly B, Wu LF. Contribution of the twin arginine translocation system to the virulence of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 2003;71:4908–16. doi: 10.1128/IAI.71.9.4908-4916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ray N, Oates J, Turner RJ, Robinson C. DmsD is required for the biogenesis of DMSO reductase in Escherichia coli but not for the interaction of the DmsA signal peptide with the Tat apparatus. FEBS Lett. 2003;534:156–60. doi: 10.1016/s0014-5793(02)03839-5. [DOI] [PubMed] [Google Scholar]
- 104.Raynaud C, Guilhot C, Rauzier J, Bordat Y, Pelicic V, et al. Phospholipases C are involved in the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 2002;45:203–17. doi: 10.1046/j.1365-2958.2002.03009.x. [DOI] [PubMed] [Google Scholar]
- 105.Richter S, Brüser T. Targeting of unfolded PhoA to the Tat translocon ofEscherichia coli. J. Biol. Chem. 2005;280:42723–30. doi: 10.1074/jbc.M509570200. [DOI] [PubMed] [Google Scholar]
- 106.Rodrigue A, Chanal A, Beck K, Muller M, Wu LF. Co-translocation of a periplasmic enzyme complex by a hitchhiker mechanism through the bacterial Tat pathway. J. Biol. Chem. 1999;274:13223–28. doi: 10.1074/jbc.274.19.13223. [DOI] [PubMed] [Google Scholar]
- 107.Rose RW, Brüser T, Kissinger JC, Pohlschroder M. Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol. Microbiol. 2002;45:943–50. doi: 10.1046/j.1365-2958.2002.03090.x. [DOI] [PubMed] [Google Scholar]
- 108.Rossier O, Cianciotto NP. The Legionella pneumophila tatB gene facilitates secretion of phospholipase C, growth under iron-limiting conditions, and intracellular infection. Infect. Immun. 2005;73:2020–32. doi: 10.1128/IAI.73.4.2020-2032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ruiz N, Silhavy TJ. Sensing external stress: watchdogs of the Escherichia coli cell envelope. Curr. Opin. Microbiol. 2005;8:122–26. doi: 10.1016/j.mib.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 110.Sanders C, Wethkamp N, Lill H. Transport of cytochrome c derivatives by the bacterial Tat protein translocation system. Mol. Microbiol. 2001;41:241–46. doi: 10.1046/j.1365-2958.2001.02514.x. [DOI] [PubMed] [Google Scholar]
- 111.Santini CL, Bernadac A, Zhang M, Chanal A, Ize B, et al. Translocation of jellyfish green fluorescent protein via the Tat system of Escherichia coli and change of its periplasmic localization in response to osmotic up-shock. J. Biol. Chem. 2001;276:8159–64. doi: 10.1074/jbc.C000833200. [DOI] [PubMed] [Google Scholar]
- 112.Santini CL, Ize B, Chanal A, Muller M, Giordano G, Wu LF. A novel Sec-independent periplasmic protein translocation pathway in Escherichia coli. EMBO J. 1998;17:101–12. doi: 10.1093/emboj/17.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sarfo KJ, Winstone TL, Papish AL, Howell JM, Kadir H, et al. Folding forms of Escherichia coli DmsD, a twin-arginine leader binding protein. Biochem. Biophys. Res. Commun. 2004;315:397–403. doi: 10.1016/j.bbrc.2004.01.070. [DOI] [PubMed] [Google Scholar]
- 114.Sargent F, Bogsch EG, Stanley NR, Wexler M, Robinson C, et al. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 1998;17:3640–50. doi: 10.1093/emboj/17.13.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sargent F, Gohlke U, De Leeuw E, Stanley NR, Palmer T, et al. Purified components of the Escherichia coli Tat protein transport system form a double-layered ring structure. Eur. J. Biochem. 2001;268:3361–67. doi: 10.1046/j.1432-1327.2001.02263.x. [DOI] [PubMed] [Google Scholar]
- 116.Sargent F, Stanley NR, Berks BC, Palmer T. Sec-independent protein translocation in Escherichia coli. A distinct and pivotal role for the TatB protein. J. Biol. Chem. 1999;274:36073–82. doi: 10.1074/jbc.274.51.36073. [DOI] [PubMed] [Google Scholar]
- 117.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 118.Schaerlaekens K, Van Mellaert L, Lammertyn E, Geukens N, Anne J. The importance of the Tat-dependent protein secretion pathway in Streptomyces as revealed by phenotypic changes in tat deletion mutants and genome analysis. Microbiology. 2004;150:21–31. doi: 10.1099/mic.0.26684-0. [DOI] [PubMed] [Google Scholar]
- 119.Shimohata N, Akiyama Y, Ito K. Peculiar properties of DsbA in its export across the Escherichia coli cytoplasmic membrane. J. Bacteriol. 2005;187:3997–4004. doi: 10.1128/JB.187.12.3997-4004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Singh SK, Grass G, Rensing C, Montfort WR. Cuprous oxidase activity of CueO from Escherichia coli. J. Bacteriol. 2004;186:7815–17. doi: 10.1128/JB.186.22.7815-7817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stanley NR, Findlay K, Berks BC, Palmer T. Escherichia coli strains blocked in Tat-dependent protein export exhibit pleiotropic defects in the cell envelope. J. Bacteriol. 2001;183:139–44. doi: 10.1128/JB.183.1.139-144.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Stanley NR, Palmer T, Berks BC. The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J. Biol. Chem. 2000;275:11591–96. doi: 10.1074/jbc.275.16.11591. [DOI] [PubMed] [Google Scholar]
- 123.Stephenson K. Sec-dependent protein translocation across biological membranes: evolutionary conservation of an essential protein transport pathway (review). Mol. Membr. Biol. 2005;22:17–28. doi: 10.1080/09687860500063308. [DOI] [PubMed] [Google Scholar]
- 124.Summer EJ, Mori H, Settles AM, Cline K. The thylakoid ΔpH-dependent pathway machinery facilitates RR-independent N-tail protein integration. J. Biol. Chem. 2000;275:23483–90. doi: 10.1074/jbc.M004137200. [DOI] [PubMed] [Google Scholar]
- 125.Thomas JD, Daniel RA, Errington J, Robinson C. Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli. Mol. Microbiol. 2001;39:47–53. doi: 10.1046/j.1365-2958.2001.02253.x. [DOI] [PubMed] [Google Scholar]
- 126.Tinker JK, Erbe JL, Holmes RK. Characterization of fluorescent chimeras of cholera toxin and Escherichia coli heat-labile enterotoxins produced by use of the twin arginine translocation system. Infect. Immun. 2005. pp. 3627–35. [DOI] [PMC free article] [PubMed]; Describes Tat-exported fluorescent chimeras for use in trafficking studies and the potential for Tat-exported vaccine candidates
- 127.Tranier S, Iobbi-Nivol C, Birck C, Ilbert M, Mortier-Barriere I, et al. A novel protein fold and extreme domain swapping in the dimeric TorD chaperone from Shewanella massilia. Structure. 2003;11:165–74. doi: 10.1016/s0969-2126(03)00008-x. [DOI] [PubMed] [Google Scholar]
- 128.Tranier S, Mortier-Barriere I, Ilbert M, Birck C, Iobbi-Nivol C, et al. Characterization and multiple molecular forms of TorD from Shewanella massilia, the putative chaperone of the molybdoenzyme TorA. Protein Sci. 2002;11:2148–57. doi: 10.1110/ps.0202902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Turner RJ, Papish AL, Sargent F. Sequence analysis of bacterial redox enzyme maturation proteins (REMPs). Can. J. Microbiol. 2004;50:225–38. doi: 10.1139/w03-117. [DOI] [PubMed] [Google Scholar]
- 130.Uchida K, Mori H, Mizushima S. Stepwise movement of preproteins in the process of translocation across the cytoplasmic membrane of Escherichia coli. J. Biol. Chem. 1995;270:30862–68. doi: 10.1074/jbc.270.52.30862. [DOI] [PubMed] [Google Scholar]
- 131.Voulhoux R, Ball G, Ize B, Vasil ML, Lazdunski A, et al. Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. EMBO J. 2001;20:6735–41. doi: 10.1093/emboj/20.23.6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Weiner JH, Bilous PT, Shaw GM, Lubitz SP, Frost L, et al. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell. 1998;93:93–101. doi: 10.1016/s0092-8674(00)81149-6. [DOI] [PubMed] [Google Scholar]
- 133.Wickner W, Schekman R. Protein translocation across biological membranes. Science. 2005;310:1452–56. doi: 10.1126/science.1113752. [DOI] [PubMed] [Google Scholar]
- 134.Yahr TL, Wickner WT. Functional reconstitution of bacterial Tat translocation in vitro. EMBO J. 2001;20:2472–79. doi: 10.1093/emboj/20.10.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yen MR, Tseng YH, Nguyen EH, Wu LF, Saier MHJ. Sequence and phylogenetic analyses of the twin-arginine targeting (Tat) protein export system. Arch. Microbiol. 2002;177:441–50. doi: 10.1007/s00203-002-0408-4. [DOI] [PubMed] [Google Scholar]
- 136.Yi L, Dalbey RE. Oxa1/Alb3/YidC system for insertion of membrane proteins in mitochondria, chloroplasts and bacteria (review). Mol. Membr. Biol. 2005;22:101–11. doi: 10.1080/09687860500041718. [DOI] [PubMed] [Google Scholar]