Abstract
The endoplasmic reticulum chaperone GRP78/BIP plays a central role in the prosurvival machinery, and its enhanced expression has been implicated in drug resistance, carcinogenesis, and metastasis. E2F1, as part of an antitumor safeguard mechanism, promotes apoptosis regardless of functional p53. Using cells that are defective in p53, we show that E2F1 represses GRP78/BIP at the transcriptional level, and this requires its DNA binding domain. Analysis of human GRP78/BIP promoter reporter constructs revealed that the region between -371 and -109 of the proximal promoter contains major E2F1-responsive elements. Toward understanding the underlying mechanism of this regulation, we performed chromatin immunoprecipitation and gel shift assays, demonstrating that E2F1 directly binds to GC-rich regions in the distal GC-box and endoplasmic reticulum stress response element -126 by interfering with the binding of positive regulatory proteins Sp1 and TFII-I of the ER stress response element-binding factor complex. We further show that TFII-I, which is required for optimal stress induction of GRP78/BIP, is suppressed by E2F1 on the protein level. Finally, our studies suggest a molecular link between the inhibition of GRP78/BIP and E2F1-mediated chemosensitization of tumor cells, underscoring its relevance for cancer treatment. Together, the data provide a new mechanism for the incompletely understood tumor suppressor function of E2F1.
Resistance to chemotherapy remains a major obstacle for the treatment of malignant tumors. The complexity of drug resistance in human cancer strongly suggests the involvement of multiple pathways. One mechanism, both intrinsic and acquired, is the result of genetic alterations within cancer cells. Another mechanism may result from environmental conditions that occur naturally in solid tumors (1). Hypoxia and glucose starvation caused by poor vascularization of tumors represent physiological endoplasmic reticulum (ER)4 stress activating the unfolded protein response (2, 3). A major unfolded protein response target is GRP78 (glucose-regulated protein 78), also known as BIP, whose induction is critical for control of protein folding and assembly, targeting of misfolded proteins for proteasome degradation, ER Ca2+ binding, and regulation of the activity of ER stress transducers, such as IRE1, PERK, and ATF6, through a binding-release mechanism (4-6). GRP78/BIP also acts as an apoptotic regulator by protecting cells against ER stress-induced cell death. Overexpression of GRP78/BIP blocks cleavage of procaspase-7 and -12 in its active form, inhibits stimulation of proapoptotic proteins of the Bcl-2 family, such as BIK and BAX, and prevents cytochrome c release from the mitochondria (7). GRP78/BIP is highly up-regulated in various cancer cells and human tumors, including breast, lung, liver, prostate, colon, and gastric cancers, correlating with malignancy, metastasis, and drug resistance (8, 9). Suppression of GRP78/BIP through small interfering RNA sensitizes human cancer cells to chemotherapeutic drug-mediated cell death and inhibits tumor progression in vivo (10, 11). The intensity of GRP78/BIP expression is generally associated with survival and clinical recurrence in prostate cancer patients (8). Thus, inhibition of GRP78/BIP expression represents a novel goal for successful cancer treatment.
The ER stress-induced activation of GRP78/BIP is primarily mediated by multiple copies of the ER stress response element (ERSE) with a consensus sequence of CCAAT(N9)CCACG located upstream of the TATA element, although part of the response may also be attributed to ERSE-independent pathways (12). Interaction of NF-Y/CBF and YY1 with the two end-flanking motifs of the ERSE has been well characterized (13, 14). The inner nine-nucleotide sequence in most ERSEs, which is required for maximal stress-dependent transactivation, is GC-rich (12, 15). Sp family proteins bind the N9 region and interact with GC motifs in untreated and stress-induced cells (16). Induction of ER stress is accompanied by cleavage of p90 ATF6 to p60 ATF6, a nuclear transcription factor that interacts with NF-Y proteins (4, 17, 18). TFII-I is also induced by ER stress and interacts with ATF6 to form a part of the ERSE-protein complex (19). Previous studies showed that maximal stimulation of ERSE by ATF6 requires its interaction with TFII-I and binding to the conserved GGC sequence motif within the 9-bp region (15). Based on the data by Abdelrahim et al. (16), it is possible that Sp proteins may function to enhance the interaction of the ATF6 and TFII-I complex with basal transcription factors or other nuclear factors under ER stress conditions.
The cellular transcription factor E2F1 plays a critical role in the control of cell cycle progression by regulating the timely expression of many genes required for the transition from G1 to S phase. A key component that controls cell cycle transition through the restriction point is the retinoblastoma protein (Rb), which, when hypophosphorylated, binds to E2F1 and thereby prevents its normal function. Deregulated E2F1 activity leads to uncontrolled cell proliferation, which is one of the hallmarks of cancer. Interestingly, E2F1 can also efficiently induce apoptosis via p53-dependent and p53-independent pathways, and this is an effective mechanism for suppressing tumorigenesis. Moreover, DNA damage leads to up-regulation of E2F1 and its stabilization via direct phosphorylation by ATM, ATR, and CHK2 (checkpoint 2) kinases and by p300-mediated acetylation (20, 21). Besides direct activation of various apoptosis related genes, E2F1 sensitizes cells to apoptosis via inhibition of antiapoptotic survival signals mediated by NF-κB or Bcl-2 and its family member Mcl-1 (22-24). The E2F1-induced decrease in Bcl-2 and Mcl-1 levels occurs independently of the ARF/p53 pathway. Taken together, the frequent deregulation of E2F1 in human tumors, its apoptotic potential, and stabilization after damage suggest that E2F1 may play an important role in enhanced sensitivity of malignant cells to chemotherapy-induced cell death.
Previously, we performed a differential proteomic analysis to identify proteins associated with E2F1 activity in inducible p53-deficient cells (25). This approach revealed that E2F1 decreases the protein level of GRP78/BIP. To understand the molecular mechanism underlying the selective suppression of GRP78/BIP by E2F1 in malignant cells lacking functional p53, we investigated the core promoter region for putative transcription factor binding sites and found that repression of GRP78/BIP by E2F1 occurs at the transcriptional level via direct binding to noncanonical E2F1-like (GC-rich) sites between -324 and -311 and between -126 and -108 of the distal ERSE element, thereby interfering with DNA binding of Sp1 and TFII-I. We also demonstrate that protein expression of the nuclear transcription factor TFII-I, which is required for stress-induced transactivation of the GRP78/BIP promoter, is significantly reduced in response to E2F1 activation. Last, our data provide evidence that repression of GRP78/BIP by E2F1 is one of the underlying mechanisms of E2F1-mediated sensitization of cancer cells to chemotherapy.
EXPERIMENTAL PROCEDURES
Cell Lines, Genotoxic Treatment, and Transfections—H1299, Hep3B, and Saos-2ERE2F1 cell lines were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum and 1% penicillin G/streptomycin sulfate (Invitrogen). 4-Hydroxytamoxifen (4-OHT; Sigma) was used at a final concentration of 1 μm. For genotoxic treatment, cells were exposed to 30 μm cisplatin (Sigma). Transfections were performed with Effectene transfection reagent (Qiagen, Hilden, Germany).
Adenoviral Infection—Replication-deficient recombinant adenoviral vectors AdGFP and AdERE2F have been described previously (26). Ad-vectors were propagated in 293 cells, purified by CsCl buoyant density centrifugation, and titrated by standard plaque assay. Adenoviral infections were carried out at multiplicities of infection that allow 100% transduction of each cell line (multiplicity of infection = 10 for H1299, 20 for Hep3B).
Plasmid Construction—Expression plasmids for wild-type E2F1, the DNA binding-defective E2F1 mutant E132, and the E(-TA) mutant lacking the transactivation domain have been described previously (26). GRP78/BIP promoter fragments were amplified from chromosomal DNA isolated from HeLa cells and cloned into the TA-cloning vector (Invitrogen). Promoter fragments were subcloned into pGL3-Basic (Promega) to allow transcription of the firefly luciferase gene under their control. Primers used for amplification of the promoter fragments are shown in supplemental Table 1.
Luciferase Assay—Luciferase activity was measured 24 or 36 h post-transfection using a commercially available luciferase reporter assay system (Promega) and normalized to total protein concentration in cell extracts.
Reverse Transcription-PCR—Semiquantitative reverse transcription-PCR was performed on total RNA prepared by an RNeasy minikit (Qiagen). Following DNase I treatment, 1 μg of RNA was reverse transcribed using Omniscript RT (Qiagen) and oligo(dT). PCR amplification was performed as described (11). A minimum number of cycles was performed to obtain a clear signal within the linear amplification phase. Primer sequences are shown in supplemental Table 1.
Western Blotting—Cells were lysed in radioimmune precipitation buffer (50 mm Tris-HCl, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS), and whole cell extracts were separated on 8-12.5% SDS-polyacrylamide gels and transferred to nitrocellulose membranes (Amersham Biosciences). Membranes were incubated with the following primary antibodies: anti-GRP78 (sc-13968; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), anti-E2F1 (catalog number 554213; BD Pharmingen), anti-Sp1 (Upstate Biotechnology), anti-TFII-I (sc-9943; Santa Cruz Biotechnology), anti-ATF6 (sc-14253; Santa Cruz Biotechnology), anti-YY1 (sc-7341; Santa Cruz Biotechnology), anti-NF-YA (Upstate Biotechnology), and anti-actin (sc-1615; Santa Cruz Biotechnology). The corresponding peroxidase-labeled secondary antibody was detected using ECL Western blotting reagents (Amersham Biosciences).
Chromatin Immunoprecipitation—The chromatin immunoprecipitation assay was performed on Saos-2ERE2F1 cells grown in the presence or absence of 4-OHT. Proteins bound to DNA were cross-linked using formaldehyde at a final concentration of 1.42% for 15 min at room temperature. The reaction was stopped by adding glycine to a final concentration of 125 μm. Following sonication protein-DNA complexes were immunoprecipitated using primary antibody for E2F1 (sc-193; Santa Cruz Biotechnology), Sp1 (Upstate Biotechnology), TFII-I (sc-9943; Santa Cruz Biotechnology), or appropriate control IgG antibodies overnight at 4 °C with rotation. Immunoprecipitated chromatin was eluted from the beads in 10% Chelex100 and boiled for 10 min. The precipitated DNA was analyzed by PCR. Primers utilized are shown in supplemental Table 1. The PCR products were run on 2% agarose gels and visualized by ethidium bromide staining. Expression levels were quantitated in relative software units using a Bio-Imaging Analyzer (Fuji, Düsseldorf, Germany) with the TINA program. Data were normalized to the input after subtraction of background signals.
Electrophoretic Mobility Shift Assay—Electrophoretic mobility shift assays were carried out with nuclear extracts from Saos-2ERE2F cells using biotin end-labeled (BrightStar BioDetect kit; Ambion) double-stranded oligonucleotides corresponding to the GC-box containing element (-327 to -304) and ERSE 1 element (-126 to -108) in the GRP78/BIP promoter. Binding reactions were performed with 2 μg of nuclear extracts and labeled probe incubated in appropriate buffer for 30 min at room temperature. For competition assays, the competitor DNA was added 10 min prior to the addition of the labeled probe. The specific E2F1 binding site of the Apaf-1 promoter (-532 to -526) (27) was used as a positive control. Samples were evaluated by electrophoresis on an 8% nondenaturing acrylamide gel. The gels were blotted on nylon transfer membrane and exposed to x-ray film. Oligonucleotide sequences are shown in supplemental Table 1.
Determination of Cell Death—Treated cells were harvested with trypsin-EDTA, washed once with ice-cold phosphate-buffered saline, and incubated overnight at -20 °C with 1 ml of 70% ice-cold ethanol. Cells were washed with phosphate-buffered saline, resuspended in 400 ml of propidium iodide/RNase A solution (phosphate-buffered saline, 100 mg/ml RNase A, and 100 mg/ml propidium iodide) for 30 min, and subjected to flow cytometry using a FACSCalibur (BD Biosciences). Data were analyzed by CellQuest software. The subdiploid population was calculated as an estimate of the apoptotic cell population.
Statistical Analysis—Statistical significance was calculated by Student's t test.
RESULTS
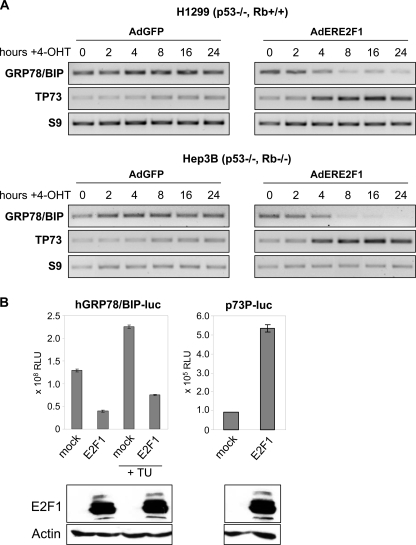
E2F1 Expression Leads to Repression of GRP78/BIP Promoter Activity—Human cancer cells often acquire mutations of the p53 tumor suppressor gene or defects in p53 apoptosis pathways. The effect of E2F1 on tumor growth, which compensates or bypasses cell death defects regardless of the p53 status, has been evaluated in several types of human cancer. To investigate the impact of E2F1 on GRP78/BIP expression in p53-defective cancer cells, H1299 and Hep3B cells were infected with AdERE2F1 at a multiplicity of infection, which allows 100% transduction. The AdERE2F1 vector, in which transgene-mediated toxicity is regulated by fusion to the tamoxifen-inducible estrogen receptor ligand-binding domain, has been shown to efficiently induce cell death of human tumor cells with defects in apoptotic pathways. Ad-vector expressing GFP was used as negative control. In both cell lines, decreased transcript levels of GRP78/BIP were observed at 4 h after E2F1 induction (Fig. 1A), compared with unchanged mRNA levels in cells lacking activated E2F1, suggesting that E2F1-mediated reduction of GRP78/BIP occurs independently of the p53 and Rb status. Repression of GRP78/BIP expression correlated with the ability of E2F1 to transactivate proapoptotic genes, such as TP73. During this early time period, we did not observe E2F1-mediated growth arrest or apoptosis (data not shown). The direct effect of E2F1 on GRP78/BIP was further investigated in Hep3B cells transfected with the plasmid hBIP-luc, which contains the proximal fragment (-371 to +2) of the human GRP78/BIP promoter. Overexpression of E2F1 resulted in a substantial inhibition of the basal promoter activity, indicating that regulation of GRP78/BIP expression by E2F1 occurs at the transcriptional level. A strong E2F1-mediated promoter repression was also detected under ER stress conditions following tunicamycin treatment. In contrast, the p73 promoter (p73P-luc (26)) was significantly activated in response to enhanced E2F1 expression (Fig. 1B).
FIGURE 1.
Repression of GRP78/BIP promoter activity by E2F1 is independent from p53 and Rb status. A, semiquantitative reverse transcription-PCR analysis of GRP78/BIP expression in p53-negative H1299 (Rb +/+) and Hep3B (Rb -/-) cells infected with Ad-vector expressing ER-E2F1. Infection by AdGFP was carried out as negative control. 16 h after infection, cells were grown in the presence of 4-OHT at the indicated times, followed by RNA isolation. p73 was used as positive control. Expression levels of RNA from the ribosomal S9 gene served as loading control. B, luciferase assay of p53-negative Hep3B cells co-transfected with 0.5 μg of GRP78/BIP promoter-luciferase reporter construct using the proximal core promoter (-371 to +2) and 0.5 μg of expression plasmid encoding E2F1 in the absence or presence of tunicamycin (TU; 0.5 μg/ml). 0.5 μg of the p73P-luc reporter construct was used as positive control. Luciferase activity (relative luciferase units; RLU) was measured 24 h after transfection. Error bars, S.D. of three independent measurements. E2F1 expression after transfection is shown in the bottom panel using actin as loading control.
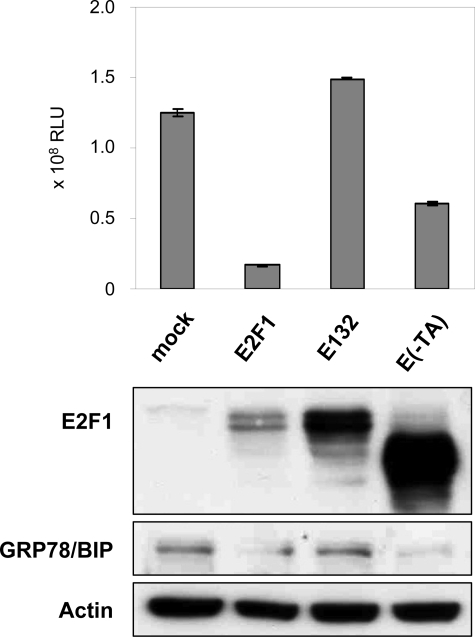
The DNA Binding Domain of E2F1 Is Involved in Full Repression of the GRP78/BIP Promoter—Previous studies using various E2F1 mutants have demonstrated that suppression of antiapoptotic mediators, such as Bcl-2 and its family member Mcl-1, by E2F1 is dependent on its DNA binding activity (22, 28, 29). To determine whether the previously mapped transactivation (TA)/Rb binding domain or the DNA binding domain of E2F1 is required for the suppressive activity on GRP78/BIP expression, the E2F1 transactivation defective mutant E(-TA) and E132 with a mutation in the DNA binding domain were tested for their ability to mediate GRP78/BIP repression (Fig. 2). Compared with the wild-type E2F1 expression plasmid, E2F1-induced repression of GRP78/BIP-luciferase activity was completely abolished by cotransfection of the plasmid encoding E2F1 without the DNA binding domain. Instead, deletion of the TA domain (residues 1-374) maintained at least 50% repression of the promoter. Western blot analysis of cell extracts confirmed that Hep3B cells overexpressing the E132 mutant showed no reduction of the GRP78/BIP protein level compared with mock-transfected cells, whereas ectopic expression of E2F1 and E(-TA) was associated with decreased expression of GRP78/BIP. These data indicate that the DNA binding domain of E2F1 is required for the full repressive effect on the GRP78/BIP promoter.
FIGURE 2.
E2F1-mediated GRP78/BIP inhibition requires the DNA binding and transactivation domain. Hep3B cells were co-transfected with 0.5 μg of GRP78/BIP reporter plasmid using the proximal core promoter region between -371 and +2 and 1 μg of expression plasmid encoding E2F1, DNA binding-defective mutant E132, E(-TA), lacking the transactivation domain, or pcDNA3.1 as mock control. Luciferase activity (RLU) was measured 36 h after transfection. Error bars, S.D. E2F1 and GRP78/BIP protein expression was verified by Western blotting. The blots were reprobed for α-actin as a loading control.
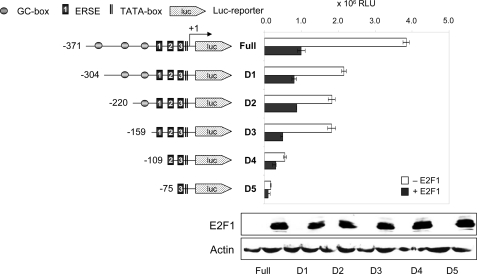
Identification of GRP78/BIP Promoter Elements Responsible for E2F1-mediated Transcriptional Repression—A unique feature shared among GRP promoters is the presence of a large number of CCAAT elements flanked by GC-rich motifs (12). The human GRP78/BIP promoter contains three ER stress response elements. However, previous studies analyzing the promoter for its response to ER stress inducers revealed that part of its activity is induced by ERSE-independent pathways. To identify the promoter region mediating responsiveness to E2F1, a panel of luciferase reporter plasmids containing the proximal -371 to +2 promoter fragment with increasing 5′-deletions was constructed by removing GC-rich and ERSE elements and tested after transient cotransfection of E2F1 in H1299 cells. As shown in Fig. 3, the main (∼2-fold) loss of basal promoter activity was observed with the constructs D1 (-371 to -304) and D4 (-159 to -109). Further deletions of the 5′-end (D1-D3) gradually affected the responsiveness to E2F1, which was nearly abrogated in the D4/D5 constructs. These findings suggest that the 296-bp region between -371 and -109 of the proximal GRP78/BIP promoter contains the major E2F1-responsive elements. Similar protein expression levels of E2F1 were confirmed by Western blotting.
FIGURE 3.
Identification of GRP78/BIP promoter elements responsible for transcriptional down-regulation by E2F1. The 5′-deletion mutants of the human GRP78/BIP promoter cloned upstream of the firefly luciferase gene are shown by nucleotide positions. Locations of the GC-boxes, ER stress elements (1-3), and TATA-box are indicated. The transcription start site of the GRP78/BIP gene is indicated by an arrow. 0.5 μg of the indicated reporter constructs either alone or together with 0.5 μg of E2F1 expression plasmid were transfected into H1299 cells. Luciferase activity (RLU) was measured 36 h after transfection. Data were obtained from three replicates. Error bars, one S.D. Protein expression levels in E2F1-transfected cells are indicated (bottom panel). Actin was used for equal loading.
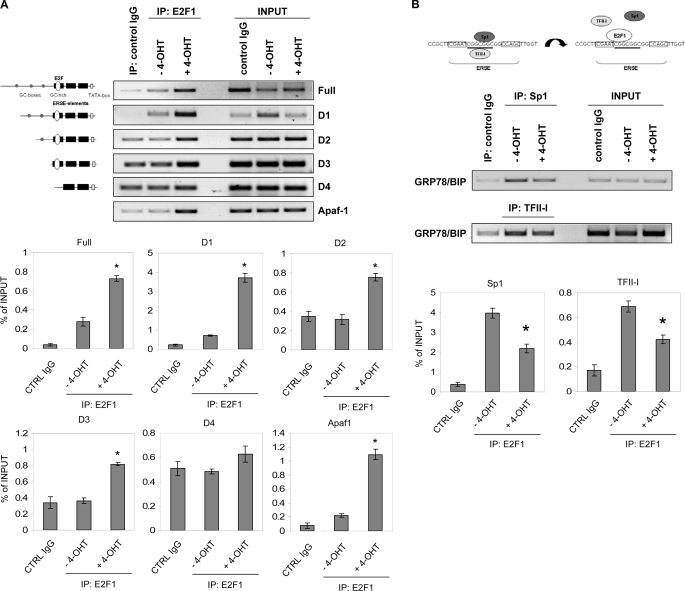
E2F1 Binds the GRP78/BIP Promoter in Vivo and Abolishes Interaction with Sp1 and TFII-I Proteins—Chromatin immunoprecipitation assays were performed to determine the in vivo binding of E2F1 to the GRP78/BIP promoter. Analysis of the entire 371-bp promoter region responsive to E2F1 revealed three GC-boxes in the -371 to -159 bp region and a GC-rich element within the three ERSE elements, which represent putative Sp1 binding sites. In addition, we have identified two DNA sequences resembling E2F binding sites, one located in close proximity (partially overlapping) to the distal GC-box (-324 to -311) and a second in the ERSE 1 element (-126 to -108) as part of the GC-rich motif. Saos-2 cells that stably express the ER-E2F1 fusion protein were used to conditionally regulate E2F1 activation through 4-OHT administration (25, 30). After synchronization of cells by serum starvation, nuclear translocation of E2F1 was induced by the addition of 1 μm 4-OHT for 24 h. DNA-protein complexes were immunoprecipitated with antibodies against E2F1 and IgG. Purified DNA was analyzed by PCR using primer pairs that amplified a 371-bp region of the GRP78/BIP promoter (full) containing the three GC-boxes and ERSEs, regions D1-D3, which revealed constitutive Luc repression in response to E2F1, D4 (-109 to +2) lacking the distal consensus ERSE sequence, or, as a positive control, the E2F1 binding region of the Apaf-1 promoter (27) (Fig. 4A). The chromatin immunoprecipitation assay showed a strong increase in E2F1 binding to full GRP78/BIP comparable with its binding to the Apaf-1 consensus sequence. Consistent with the luciferase reporter data, the presence of E2F1 on the promoter persisted through regions spanning D1-D3. Although the strongest binding of E2F1 was detectable to the 159-bp region with the ERSE elements, it was barely detectable on the D4 promoter. In contrast, there was no binding of E2F1 in untreated cells.
FIGURE 4.
E2F1 binds to the GRP78/BIP promoter in vivo by interfering with ER stress element-binding factors. A, serum-starved Saos-2 cells stably transfected with ER-E2F1 were grown in the presence or absence of 4-OHT for 24 h. Chromatin immunoprecipitation was performed using either a control IgG antibody or antibody against E2F1. PCR primers were designed to amplify the different GRP78/BIP promoter fragments spanning from -371 to +2 (full), -304 to +2 (D1), -220 to +2 (D2), -159 to +2 (D3), and -109 to +2 (D4). PCR primers for the Apaf-1 promoter were used as positive control. Input lane, 10% of total chromatin used in chromatin immunoprecipitation assay. B, chromatin immunoprecipitation was performed under the same conditions as in A using specific antibodies against Sp1 or TFII-I. PCR primers were used to amplify the GRP78/BIP promoter fragment between -220 and +2. Representative bands are indicated. Bar graphs show results from two independent experiments as relative software units cleared for background signals and normalized to input bands. Data represent the mean ± S.D. Significant differences in E2F1, Sp1, or TFII-I enrichment of chromatin in 4-OHT-treated versus untreated cells (A, p ≤ 0.005; B, p ≤ 0.01) are labeled with an asterisk (t test).
ERSEs from various ER stress-responsive genes exhibit a tripartite structure, which includes two motifs that bind NF-Y/CBF and YY1 and flank a central nonanucleotide (N9) sequence that shows some variability in different ERSEs. Previous studies have demonstrated that the GC-rich N9 motif, which binds TFII-I (15) also interacts with Sp proteins both in stressed and unstressed cells, resulting in GRP78/BIP activation (16). E2F1 is capable of regulating promoter activity through Sp1 binding sites (31, 32). In order to examine the possible role of the ERSE(N9) sequence in E2F1-mediated repression of the GRP78/BIP promoter, we analyzed whether binding of E2F1 interferes with DNA-protein interaction of the transcriptional activator Sp1 (Fig. 4B). Immunoprecipitation of chromatin-protein complexes with antibody against Sp1 and subsequent PCR amplification of the 159-bp region revealed a pronounced decrease in the intensity of the DNA-Sp1 complex upon activation of endogenous E2F1. We also observed a clearly reduced binding of TFII-I in Saos-2ERE2F1 cells treated with 4-OHT, which binds directly to the conserved GGC motif within the 9-bp region of the ERSE and is required for maximal stimulation of ERSE by ATF6 (15). Collectively, these results suggest that E2F1 exhibits its repressive effect on GRP78/BIP expression by direct interaction with the GC-rich region of the distal ERSE through interfering with the binding of positive regulatory proteins, such as Sp1 and TFII-I.
Because the distal GC-box in addition to the Sp1 binding region of the first ERSE element governed a component of E2F1-regulated GRP78/BIP repression, binding of the E2F1 protein to both regions was investigated in gel mobility shift assays using labeled oligonucleotides corresponding to the putative E2F1 sites between -327 and -304 and between -126 and -108. A comparison was made between the E2F1-like sequences from the GRP78/BIP promoter and a wild-type E2F site from the Apaf-1 promoter shown to specifically bind E2F1 (27). Consistent with the Apaf-1 E2F1 probe (Fig. 5, lane 2), both GRP78/BIP oligonucleotides formed one predominant nuclear complex, A′ (lanes 5 and 8). Band A′ of the GRP78/BIP GC-box and ERSE E2F site binding complexes was competed most efficiently by an excess (50-fold) of its cognate E2F1 site from the Apaf-1 oligonucleotide (Fig. 5, lanes 6 and 9) but not by the probe in which the E2F1 site was mutated (lanes 7 and 10), showing that both Sp1 binding sequences of the GRP78/BIP promoter directly interact with E2F1 protein. In contrast, formation of two additional faster migrating complexes, B′ and C′, with the GC-box E2F site (lane 8) was reduced neither by the specific competitor for the Apaf-1 E2F1 site (lanes 9) nor by the oligonucleotide carrying the mutated E2F motif (lane 10), assuming that both complexes do not contain E2F1. However, the fact that these bands are even enhanced in the cold competition lane 9 may argue for the binding of additional, as yet unidentified, proteins when E2F1 is absent. No binding was observed in the absence of nuclear extract (lane 1).
FIGURE 5.
The GRP78/BIP promoter E2F1-like sequences in the distal GC-box and ERSE 1 element compete for nuclear binding with the specific Apaf-1 promoter E2F1 site. Biotin-labeled Apaf-1 E2F1 sequence (lanes 2-4) and GRP78/BIP Sp1 binding sequences -324 to -311 (distal GC-box; lanes 5-7) and -126 to -108 (GC-rich element of ERSE 1; lanes 8-10) were incubated with Saos-2ERE2F1 cell nuclear extracts and a 50-fold molar excess of cold double-stranded specific competitor for the Apaf-1 E2F1 site (lanes 3, 6, and 9) or an equimolar amount of mutant Apaf-1 E2F1 site competitor (lanes 4, 7, and 10). The arrow indicates a specific band competed by double-stranded cognate competitor. The faster migrating complexes B′ and C′ with the GC-box E2F site are not E2F1-specific. Lane 1, binding without nuclear extract.
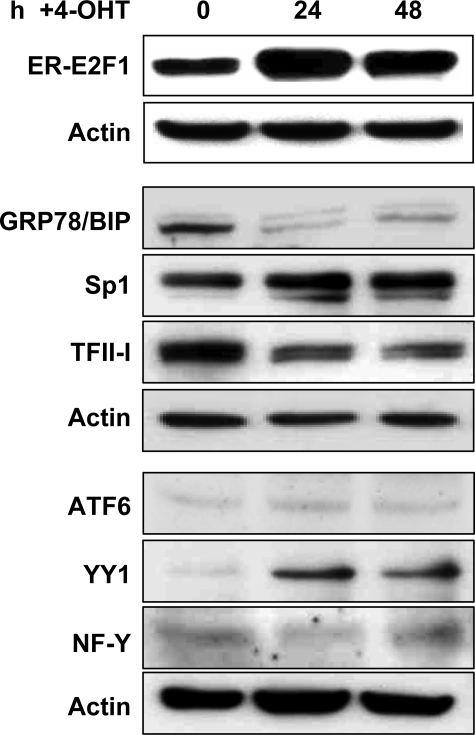
Influence of E2F1 on Protein Expression of ERSE-binding Transcription Factors—Stress-induced genes are activated through the assembly of transcription factors on ERSEs in target gene promoters. The ER stress response element-binding factor (ERSF) complex contains NF-Y/CBF, YY1, TFII-I, and the nuclear form of ATF6 (17, 19, 33). Activation of the stress response is further attenuated by Sp family proteins (16). In addition to the observed dissociation of DNA-Sp1·TFII-I transcription factor complexes by binding of E2F1 to the ERSE sequence, we explored the impact of activated endogenous E2F1 on protein expression of key molecules involved in the regulation of GRP78/BIP transcriptional activity in serum-starved Saos-2ERE2F1 cells. Expression levels of the ERSF complex components were analyzed by Western blot 24 and 48 h after 4-OHT treatment (Fig. 6). In accordance with a down-regulation of GRP78/BIP mRNA by E2F1, enhanced nuclear translocation of the transcription factor significantly reduced GRP78/BIP on the protein level. The results also show that the enhanced E2F1 activity is associated with decreased TFII-I expression and increasing amounts of YY1, whereas the expression levels of Sp1, NF-Y, and ATF6 remain unaffected.
FIGURE 6.
E2F1 modulates ERSE-binding transcription factors on protein level. Serum-starved Saos-2 cells stably transfected with ER-E2F1 were grown in the presence of 4-OHT at the indicated times, followed by Western blot analysis. Equivalent amounts of total cell proteins were fractionated by SDS-PAGE and probed with antibodies specific for E2F1, GRP78/BIP, Sp1, TFII-I, ATF6, YY1, and NF-Y. Equal protein loading was confirmed by reprobing the stripped blots with anti-actin antibody. Detection was achieved by chemiluminescence.
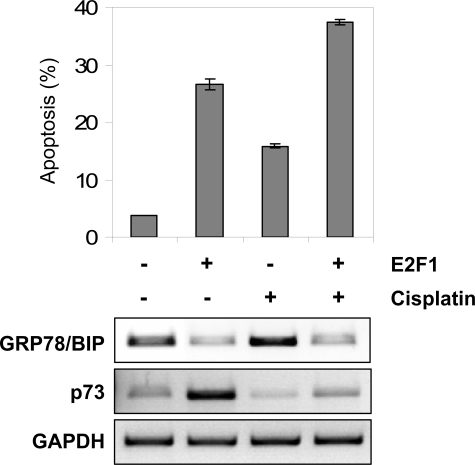
Repression of GRP78/BIP Expression by E2F1 Contributes to E2F1-mediated Sensitization of Cancer Cells to Drug Treatment—p53-deficient cancer cells are often resistant to conventional chemotherapy. E2F1 has been shown to trigger apoptosis, and its overexpression can effectively sensitize various cancer cells to chemotherapeutic agents. However, the molecular mechanisms that underlie the cooperation between deregulated E2F1 and DNA damage induced by genotoxic drugs in human cancers are not fully understood. In this regard, it has been shown that cisplatin induces expression of antiapoptotic GRP78/BIP (34), which might contribute to resistance of tumor cells against this anticancer drug. To assess whether E2F1-mediated repression of GRP78/BIP promotes drug sensitivity, Hep3B cells were infected with AdERE2F1 and subsequently treated with cisplatin (30 μm). At 24 h, the population of cells with activated E2F1 has lower GRP78/BIP and a higher rate of apoptotic cells (Fig. 7). In accordance with earlier reports, cisplatin increased GRP78/BIP expression (Fig. 7, bottom). At 24 h post-treatment, the percentage of apoptotic cells measured by flow cytometry had significantly increased when E2F1 was combined with cisplatin, and enhanced cell death occurred in the population showing reduced GRP78/BIP levels. Taken together, our data demonstrate that enhanced apoptosis induction by combined treatment with E2F1 and cisplatin correlates with decreased expression of cytoprotective GRP78/BIP, indicating that repression of GRP78/BIP by E2F1 is critical for E2F1-mediated chemosensitization of neoplastic cells.
FIGURE 7.
Repression of GRP78/BIP by E2F1 accounts for E2F1-associated chemosensitization of cancer cells. Apoptosis induction after AdERE2F1 infection, 30 μm cisplatin, or combination treatment was analyzed in Hep3B cells by flow cytometry. The percentage of apoptotic cells (as determined by cells with sub-G1 DNA content) 24 h after treatment is as indicated. Semiquantitative reverse transcription-PCR analysis revealed differences in GRP78/BIP transcript levels for each treatment. p73 was used as a positive control. Data were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) values.
DISCUSSION
Development of resistance to chemotherapy is a major obstacle to successful cancer treatment. Poor oxygenation (hypoxia) and glucose deprivation due to the poor vascularization in the majority of human solid tumors leads to the activation of unfolded protein response as a basic survival mechanism of the eukaryotic cell. From the known ER chaperones GRP78/BIP seems to play a major role in this prosurvival machinery, which results in therapy resistance, cancer progression, and possibly metastasis (3, 35). The level of GRP78/BIP is strongly elevated in a variety of cancer cell lines, solid tumors, and human cancer biopsies, correlating with higher pathologic grade, recurrence, and poor patient survival in several cancers (36, 37). In support of the notion that GRP78/BIP is more critically needed for the survival of stressed cells, such as cancer, tumor progression was significantly impeded in heterozygous GRP78 mice as indicated by a longer latency period, reduced tumor size, and increased tumor apoptosis (38). GRP78/BIP exhibits its antiapoptotic function by blocking the executor caspase-7 activated in response to ER stress and genotoxic drugs (39), the proapoptotic protein BIK, and its downstream target BAX and by prevention of cytochrome c release from the mitochondria (7, 40). Given the importance of GRP78/BIP in cancer cell survival, its inhibition is a prime task for the development of successful tumor treatment.
Stimulation of cell death by E2F1 occurs via multiple pathways, some of which involve the tumor suppressor p53 and are autonomous of p53 and Rb. Currently, the best characterized mechanism for E2F1-induced apoptosis is by activating proapoptotic genes (29, 41, 42). In these studies, the DNA binding activity of E2F1 rather than transactivation was proven to be essential for its apoptotic function. Our data support a second concept of E2F1-mediated cell death through the inhibition of antiapoptotic target genes (22, 23). We identified E2F1 as major inhibitor of the prosurvival factor GRP78/BIP in p53-deficient cancer cells. Both overexpression and activation of endogenous E2F1 lead to a significant decrease in GRP78/BIP mRNA and protein expression levels by means of repressing the GRP78/BIP promoter. In agreement with previous studies regarding the requirements for E2F1 to induce p53-independent cell death, we found that repression of GRP78/BIP promoter activity by E2F1 requires DNA binding but not transactivation and occurs independent from Rb. Our data indicate that two regions of the proximal core promoter between -371 and -304 and between -159 and -109 containing the distal GC-box and the ER stress response element 1, which mediate maximum promoter activation, are essential for the full inhibitory effect of E2F1. Using several promoter constructs in chromatin immunoprecipitation and gel mobility shift assays, we show that E2F1 does indeed bind to the GRP78/BIP promoter both in vitro and in vivo via noncanonical E2F1-like sequences, one that partially overlaps the distal GC-box and a second within the Sp/TFII-I sites of the GC-rich N9 motif in ERSE 1. Direct binding of E2F1 to this site is accompanied by the displacement of the nuclear transcription factors Sp1 and TFII-I from DNA, as evident by reduced complex formation.
This mechanism appears to be different from regulation of the two other currently known antiapoptotic targets of E2F1, Bcl-2 and its family member Mcl-1, where transcriptional repression by E2F1 occurs via a classical E2F1 regulatory element (22). An Sp1 site has generally been shown to convey regulation by E2F1 (31, 32), and the E2F1 protein is capable of binding Sp1 (31, 32, 43). This interaction takes place in the N-terminal region of E2F1, involves zinc finger 1 and part of finger 2 of the Sp1 DNA binding domain, and is in line with the hypothesis that E2F1 functions as a growth- and cell cycle-regulated tethering factor between Sp1 and the basic transcription machinery (32, 44-46). Interestingly, cooperative binding of these two proteins also appears to be the causative mechanism for E2F1-mediated repression of the cyclin D1 promoter (47). In this case, the E2F1 amino terminus and DNA binding domain are both required for regulation through Sp1 binding sites, suggesting that E2F1 exhibits negative regulation of genes involved in cell cycle progression through formation of an E2F1-Sp1 complex. Another example supporting the involvement of Sp sites in transcriptional repression by E2F1 in human tumor cells is the hTERT promoter (48). E2F1 has been reported to interact with Sp1 through the C-terminal region of Sp1 (31), which is also crucial for the interactions with other Sp1 proteins to form a homomultimeric complex that synergistically activates transcription (49). The hTERT promoter belongs to the kind of promoters bearing multiple GC-boxes that have been demonstrated to be activated by Sp1 with a high degree of synergism. Based on their data, repression of Sp1-mediated activation of the hTERT promoter by E2F1 is therefore likely due to an inhibition of the formation of the Sp1 multimeric complex, whereas noncanonical E2F1 sites in the promoter reported by Crowe et al. (50) had almost no repressive effect (48). E2F1-mediated inhibition of antiapoptotic GRP78/BIP thus differs from both described repression models, since the repressor function of E2F1 on the human GRP78/BIP promoter may occur through direct DNA binding of E2F1 to GC-rich motifs, thereby establishing a new essential mechanism for apoptosis induction by E2F1. Our results are in accordance with a recent location analysis of E2F1 binding sites in the human genome, indicating that E2F1 is recruited to the promoter regions of 25-35% of genes, but only a small fraction of identified E2F1 sites possess the consensus binding motif (51).
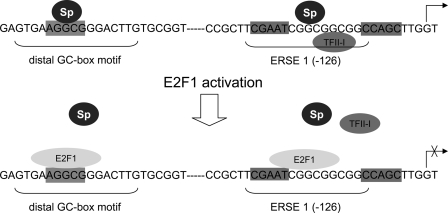
Specifically, our findings support the concept that in cancer cells, E2F1 induces apoptosis independently of p53 by suppressing GRP78/BIP expression through interfering with the binding of positive regulatory transcription factors, such as Sp1 and TFII-I, to GC motifs. In this regard, the 9-bp sequence strikingly rich in GC in the tripartite structure of the consensus mammalian ER stress response element has recently been shown to be essentially needed for the interaction with ERSF and maximal stress-dependent transactivation (12). Together with NF-Y and YY1 that bind to the distal motifs of the tripartite ERSE, the TFII-I and Sp transcription factors (Sp1, Sp3, and Sp4) are integral parts of the ERSF complex that are important for stress-induced responses through their binding to the GC-rich sequence in tumor cells (16). Since binding of E2F1 to the distal ERSE would also abolish the interaction of NF-Y with the promoter, repression of GRP78/BIP by E2F1 through a direct mechanism may therefore be mediated through competition for critical positive regulators of transcription, such as Sp1, TFII-I, and NF-Y (Fig. 8), which may interrupt transmission of an activation signal to the basic transcription machinery. In support of this model, van Ginkel et al. (45) previously showed that in complex promoters the main role of E2F is to keep transcription low. In this scenario, the decision of whether E2F sites regulate activation or repression is made by the promoter context. E2F requires cooperation with other transcription factors for stable binding to promoter DNA to activate transcription, whereas the E2F1 transactivation domain only in an E2F1 minimal promoter is sufficient to induce transactivation.
FIGURE 8.
Model of E2F1-inducible changes in transcription factor occupancy of the GRP78/BIP promoter. In stressed cells, Sp proteins are in contact with the distal GC-box and GC motifs of the ERSE-N9 region in the GRP78/BIP promoter. Upon E2F1 activation, E2F1 binds to its putative sites within the GC sequence motifs, resulting in the displacement of transcription factor binding to these regions, thereby acting as a transcriptional repressor.
Interestingly, our findings suggest that E2F1, apart from its direct inhibitory effect on the GRP78/BIP promoter, also regulates GRP78/BIP expression by modulating the protein levels of key components of the ERSF complex. In our studies, the transcription factor TFII-I was clearly reduced, whereas YY1 increased in response to E2F1 activation. The constitutively expressed Yin Yang 1 (YY1) protein plays a multifunctional role in various biological processes as initiator, activator, and repressor of transcription of many genes and also activates the GRP78/BIP promoter under ER stress conditions (52). Analysis of the YY1 promoter revealed at least six potential binding sites for E2F1 (53). Moreover, synergistic cooperation between YY1 and E2F1 in the regulation of the p73 promoter (54) emphasizes the necessity of increased YY1 protein levels during activation of E2F1-mediated apoptosis. A potentially direct up-regulation of YY1 by E2F1, however, did not affect the repressive activity of E2F1 on the GRP78/BIP promoter. Consistently, E2F1-mediated down-regulation of TFII-I may play an important role for induction of cell cycle arrest and apoptosis. TFII-I is tyrosine-phosphorylated in response to extracellular growth signals and transcriptionally activates growth-promoting genes. Most strikingly, upon cell cycle arrest resulting from genotoxic stress, TFII-I is ubiquitinated and targeted for proteasomal degradation (55). However, it has been shown that this mechanism is p53- and ATM-dependent. Since our experiments have been performed in p53-null cells, this event has to be further analyzed.
Several studies demonstrated that deregulated E2F1 expression leads to rapid and widespread cell death by apoptosis in a variety of human cancer cells (56-60). There is increasing evidence that execution of the apoptotic program is achieved by an intense cross-talk between the mitochondria and the ER (61), suggesting that the ER might regulate apoptosis by sensitizing mitochondria to extrinsic and intrinsic stimuli (62). In this context, E2F1 mainly acts by integrating diverse apoptotic stimuli into a common death pathway that eventually includes the release of cytochrome c from the mitochondria. This occurs regardless of p53 through both direct induction of proapoptotic and repression of antiapoptotic Bcl-2 family members in tumor cells (22, 23, 63). Therefore, the direct down-regulation of GRP78/BIP that prevents cytochrome c release is another efficient mechanism for apoptosis progression, indicating that E2F1 is essentially important for promoting mitochondrial apoptosis. Notably, E2F1 has been shown capable of sensitizing various cancer cells to genotoxic treatment (64-66), although the mechanism of this action is not yet fully understood. Conversely, in vitro studies suggest that GRP78/BIP confers chemoresistance in malignant cells (9, 67). Thus, we have characterized the repression of GRP78/BIP expression by E2F1 as a biologically relevant event in the progression of cells toward apoptosis. In fact, we can show that the E2F1-induced decrease in chemoprotective GRP78/BIP directly correlates with enhanced apoptosis of cisplatin-treated Hep3B hepatoma cells. Together, our results suggest that GRP78/BIP is a critical target of E2F1, whose suppression may potentiate its capacity to induce apoptosis and to improve responsiveness of tumor cells to chemotherapy.
Supplementary Material
Acknowledgments
We thank Anja Stoll for technical assistance.
This work was supported by Forschungsförderung der Medizinischen Fakultät der Rostocker Universität Grant 889007. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
Footnotes
The abbreviations used are: ER, endoplasmic reticulum; ERSE, ER stress response element; 4-OHT, 4-hydroxytamoxifen; TA, transactivation; Rb, retinoblastoma protein.
References
- 1.Brown, J. M., and Giaccia, A. J. (1998) Cancer Res. 58 1408-1416 [PubMed] [Google Scholar]
- 2.Feldman, D. E., Chauhan, V., and Koong, A. C. (2005) Mol. Cancer Res. 3 597-605 [DOI] [PubMed] [Google Scholar]
- 3.Ni, M., and Lee, A. S. (2007) FEBS Lett. 581 3641-3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haze, K., Okada, T., Yoshida, H., Yanagi, H., Yura, T., Negishi, M., and Mori, K. (2001) Biochem. J. 355 19-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang, Y., Shen, J., Arenzana, N., Tirasophon, W., Kaufman, R. J., and Prywes, R. (2000) J. Biol. Chem. 275 27013-27020 [DOI] [PubMed] [Google Scholar]
- 6.Okada, T., Yoshida, H., Akazawa, R., Negishi, M., and Mori, K. (2002) Biochem. J. 366 585-594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu, Y., Li, J., and Lee, A. S. (2007) Cancer Res. 67 3734-3740 [DOI] [PubMed] [Google Scholar]
- 8.Daneshmand, S., Quek, M. L., Lin, E., Lee, C., Cote, R. J., Hawes, D., Cai, J., Groshen, S., Lieskovsky, G., Skinner, D. G., Lee, A. S., and Pinski, J. (2007) Hum. Pathol. 38 1547-1552 [DOI] [PubMed] [Google Scholar]
- 9.Lee, E., Nichols, P., Spicer, D., Groshen, S., Yu, M. C., and Lee, A. S. (2006) Cancer Res. 66 7849-7853 [DOI] [PubMed] [Google Scholar]
- 10.Dong, D., Ko, B., Baumeister, P., Swenson, S., Costa, F., Markland, F., Stiles, C., Patterson, J. B., Bates, S. E., and Lee, A. S. (2005) Cancer Res. 65 5785-5791 [DOI] [PubMed] [Google Scholar]
- 11.Sugawara, S., Takeda, K., Lee, A., and Dennert, G. (1993) Cancer Res. 53 6001-6005 [PubMed] [Google Scholar]
- 12.Roy, B., and Lee, A. S. (1999) Nucleic Acids Res. 27 1437-1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, W. W., Hsiung, Y., Zhou, Y., Roy, B., and Lee, A. S. (1997) Mol. Cell Biol. 17 54-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mori, K. (2000) Cell 101 451-454 [DOI] [PubMed] [Google Scholar]
- 15.Parker, R., Phan, T., Baumeister, P., Roy, B., Cheriyath, V., Roy, A. L., and Lee, A. S. (2001) Mol. Cell Biol. 21 3220-3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdelrahim, M., Liu, S., and Safe, S. (2005) J. Biol. Chem. 280 16508-16513 [DOI] [PubMed] [Google Scholar]
- 17.Yoshida, H., Okada, T., Haze, K., Yanagi, H., Yura, T., Negishi, M., and Mori, K. (2000) Mol. Cell Biol. 20 6755-6767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshida, H., Okada, T., Haze, K., Yanagi, H., Yura, T., Negishi, M., and Mori, K. (2001) Mol. Cell Biol. 21 1239-1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong, M., Lin, M. Y., Huang, J. M., Baumeister, P., Hakre, S., Roy, A. L., and Lee, A. S. (2005) J. Biol. Chem. 280 16821-16828 [DOI] [PubMed] [Google Scholar]
- 20.Dornan, D., Shimizu, H., Perkins, N. D., and Hupp, T. R. (2003) J. Biol. Chem. 278 13431-13441 [DOI] [PubMed] [Google Scholar]
- 21.Ianari, A., Gallo, R., Palma, M., Alesse, E., and Gulino, A. (2004) J. Biol. Chem. 279 30830-30835 [DOI] [PubMed] [Google Scholar]
- 22.Croxton, R., Ma, Y., Song, L., Haura, E. B., and Cress, W. D. (2002) Oncogene 21 1359-1369 [DOI] [PubMed] [Google Scholar]
- 23.Eischen, C. M., Packham, G., Nip, J., Fee, B. E., Hiebert, S. W., Zambetti, G. P., and Cleveland, J. L. (2001) Oncogene 20 6983-6993 [DOI] [PubMed] [Google Scholar]
- 24.Chen, M., Capps, C., Willerson, J. T., and Zoldhelyi, P. (2002) Circulation 106 2707-2713 [DOI] [PubMed] [Google Scholar]
- 25.Li, Z., Kreutzer, M., Mikkat, S., Mise, N., Glocker, M. O., and Putzer, B. M. (2006) Proteomics 6 5735-5745 [DOI] [PubMed] [Google Scholar]
- 26.Stiewe, T., and Putzer, B. M. (2000) Nat. Genet. 26 464-469 [DOI] [PubMed] [Google Scholar]
- 27.Furukawa, Y., Nishimura, N., Satoh, M., Endo, H., Iwase, S., Yamada, H., Matsuda, M., Kano, Y., and Nakamura, M. (2002) J. Biol. Chem. 277 39760-39768 [DOI] [PubMed] [Google Scholar]
- 28.Stanelle, J., Stiewe, T., Rodicker, F., Kohler, K., Theseling, C., and Putzer, B. M. (2003) Cardiovasc. Res. 59 512-519 [DOI] [PubMed] [Google Scholar]
- 29.Hsieh, J. K., Fredersdorf, S., Kouzarides, T., Martin, K., and Lu, X. (1997) Genes Dev. 11 1840-1852 [DOI] [PubMed] [Google Scholar]
- 30.Stanelle, J., Stiewe, T., Theseling, C. C., Peter, M., and Putzer, B. M. (2002) Nucleic Acids Res. 30 1859-1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, S. Y., Black, A. R., Kostic, D., Pajovic, S., Hoover, C. N., and Azizkhan, J. C. (1996) Mol. Cell Biol. 16 1668-1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karlseder, J., Rotheneder, H., and Wintersberger, E. (1996) Mol. Cell Biol. 16 1659-1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, M., Baumeister, P., Roy, B., Phan, T., Foti, D., Luo, S., and Lee, A. S. (2000) Mol. Cell Biol. 20 5096-5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mandic, A., Hansson, J., Linder, S., and Shoshan, M. C. (2003) J. Biol. Chem. 278 9100-9106 [DOI] [PubMed] [Google Scholar]
- 35.Gazit, G., Lu, J., and Lee, A. S. (1999) Breast Cancer Res. Treat. 54 135-146 [DOI] [PubMed] [Google Scholar]
- 36.Dong, D., Dubeau, L., Bading, J., Nguyen, K., Luna, M., Yu, H., Gazit-Bornstein, G., Gordon, E. M., Gomer, C., Hall, F. L., Gambhir, S. S., and Lee, A. S. (2004) Hum. Gene Ther. 15 553-561 [DOI] [PubMed] [Google Scholar]
- 37.Zhang, J., Jiang, Y., Jia, Z., Li, Q., Gong, W., Wang, L., Wei, D., Yao, J., Fang, S., and Xie, K. (2006) Clin. Exp. Metastasis 23 401-410 [DOI] [PubMed] [Google Scholar]
- 38.Lee, A. S. (2001) Trends Biochem. Sci. 26 504-510 [DOI] [PubMed] [Google Scholar]
- 39.Reddy, R. K., Mao, C., Baumeister, P., Austin, R. C., Kaufman, R. J., and Lee, A. S. (2003) J. Biol. Chem. 278 20915-20924 [DOI] [PubMed] [Google Scholar]
- 40.Ranganathan, A. C., Zhang, L., Adam, A. P., and Aguirre-Ghiso, J. A. (2006) Cancer Res. 66 1702-1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bates, S., Phillips, A. C., Clark, P. A., Stott, F., Peters, G., Ludwig, R. L., and Vousden, K. H. (1998) Nature 395 124-125 [DOI] [PubMed] [Google Scholar]
- 42.Phillips, A. C., Bates, S., Ryan, K. M., Helin, K., and Vousden, K. H. (1997) Genes Dev. 11 1853-1863 [DOI] [PubMed] [Google Scholar]
- 43.Rotheneder, H., Geymayer, S., and Haidweger, E. (1999) J. Mol. Biol. 293 1005-1015 [DOI] [PubMed] [Google Scholar]
- 44.Shin, E. K., Tevosian, S. G., and Yee, A. S. (1996) J. Biol. Chem. 271 12261-12268 [DOI] [PubMed] [Google Scholar]
- 45.van Ginkel, P. R., Hsiao, K. M., Schjerven, H., and Farnham, P. J. (1997) J. Biol. Chem. 272 18367-18374 [DOI] [PubMed] [Google Scholar]
- 46.Blais, A., Monte, D., Pouliot, F., and Labrie, C. (2002) J. Biol. Chem. 277 31679-31693 [DOI] [PubMed] [Google Scholar]
- 47.Watanabe, G., Albanese, C., Lee, R. J., Reutens, A., Vairo, G., Henglein, B., and Pestell, R. G. (1998) Mol. Cell Biol. 18 3212-3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Won, J., Yim, J., and Kim, T. K. (2002) FASEB J. 16 1943-1945 [DOI] [PubMed] [Google Scholar]
- 49.Pascal, E., and Tjian, R. (1991) Genes Dev. 5 1646-1656 [DOI] [PubMed] [Google Scholar]
- 50.Crowe, D. L., Nguyen, D. C., Tsang, K. J., and Kyo, S. (2001) Nucleic Acids Res. 29 2789-2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bieda, M., Xu, X., Singer, M. A., Green, R., and Farnham, P. J. (2006) Genome Res. 16 595-605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baumeister, P., Luo, S., Skarnes, W. C., Sui, G., Seto, E., Shi, Y., and Lee, A. S. (2005) Mol. Cell Biol. 25 4529-4540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joshi, B., Rastogi, S., Morris, M., Carastro, L. M., DeCook, C., Seto, E., and Chellappan, S. P. (2007) Biochem. J. 401 155-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu, S., Murai, S., Kataoka, K., and Miyagishi, M. (2008) Biochem. Biophys. Res. Commun. 365 75-81 [DOI] [PubMed] [Google Scholar]
- 55.Desgranges, Z. P., Ahn, J., Lazebnik, M. B., Ashworth, T., Lee, C., Pestell, R. C., Rosenberg, N., Prives, C., and Roy, A. L. (2005) Mol. Cell Biol. 25 10940-10952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hunt, K. K., Deng, J., Liu, T. J., Wilson-Heiner, M., Swisher, S. G., Clayman, G., and Hung, M. C. (1997) Cancer Res. 57 4722-4726 [PubMed] [Google Scholar]
- 57.Fueyo, J., Gomez-Manzano, C., Yung, W. K., Liu, T. J., Alemany, R., Mc-Donnell, T. J., Shi, X., Rao, J. S., Levin, V. A., and Kyritsis, A. P. (1998) Nat. Med. 4 685-690 [DOI] [PubMed] [Google Scholar]
- 58.Liu, T. J., Wang, M., Breau, R. L., Henderson, Y., El-Naggar, A. K., Steck, K. D., Sicard, M. W., and Clayman, G. L. (1999) Cancer Gene Ther. 6 163-171 [DOI] [PubMed] [Google Scholar]
- 59.Rodicker, F., Stiewe, T., Zimmermann, S., and Putzer, B. M. (2001) Cancer Res. 61 7052-7055 [PubMed] [Google Scholar]
- 60.Yang, H. L., Dong, Y. B., Elliott, M. J., Liu, T. J., Atienza, C., Jr., Stilwell, A., and McMasters, K. M. (1999) Clin. Cancer Res. 5 2242-2250 [PubMed] [Google Scholar]
- 61.Kim, R., Emi, M., Tanabe, K., and Murakami, S. (2006) Apoptosis 11 5-13 [DOI] [PubMed] [Google Scholar]
- 62.Rao, R. V., Castro-Obregon, S., Frankowski, H., Schuler, M., Stoka, V., del Rio, G., Bredesen, D. E., and Ellerby, H. M. (2002) J. Biol. Chem. 277 21836-21842 [DOI] [PubMed] [Google Scholar]
- 63.Putzer, B. M. (2007) J. Cell Mol. Med. 11 239-251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meng, R. D., Phillips, P., and El-Deiry, W. S. (1999) Int. J. Oncol. 14 5-14 [PubMed] [Google Scholar]
- 65.Dong, Y. B., Yang, H. L., Elliott, M. J., and McMasters, K. M. (2002) Cancer Res. 62 1776-1783 [PubMed] [Google Scholar]
- 66.Elliott, M. J., Farmer, M. R., Atienza, C., Jr., Stilwell, A., Dong, Y. B., Yang, H. L., Wong, S. L., and McMasters, K. M. (2002) Tumour Biol. 23 76-86 [DOI] [PubMed] [Google Scholar]
- 67.Pyrko, P., Schonthal, A. H., Hofman, F. M., Chen, T. C., and Lee, A. S. (2007) Cancer Res. 67 9809-9816 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.