Abstract
Variations in tryptophan fluorescence intensities confirm that copper(II) interacts with α-synuclein, a protein implicated in Parkinson’s disease. Trp4 fluorescence decay kinetics measured for the F4W protein show that Cu(II) binds tightly (Kd ∼ 100 nM) near the N-terminus at pH 7. Work on a F4W/H50S mutant indicates that a histidine imidazole is not a ligand in this high-affinity site.
Parkinson’s disease is characterized by the accumulation of neuronal deposits comprising largely the protein α-synuclein (α-syn).1,2 Although rare heritable forms of the disease have been documented,3–6 the sporadic form is far more common and possibly connected to environmental factors that promote oxidative stress and aberrant metal-mediated redox chemistry.7–12 Indeed, metal-enhanced oligomerization has been observed for α-syn in vitro,13,14 and there is evidence that specific metal−protein interactions are critical in neurodegenerative diseases involving other amyloidogenic polypeptides such as amyloid β peptide (Alzheimer’s disease),15–17 prion protein (spongiform encephalopathies),18,19 and superoxide dismutase (amyotrophic lateral sclerosis).20,21 A difficult issue to resolve is whether metal ions perturb protein structures and thereby alter function, whether metal−protein complexes directly participate in the production of reactive oxygen species, or whether both mechanisms are at work.
We have employed tryptophan fluorescence measurements to probe Cu(II)−protein binding in four mutants containing single W substitutions (F4W, Y39W, F94W, and Y125W: Figure 1).(22) Upon addition of Cu(II) (0.3−60 μΜ), the tryptophan fluorescence is quenched site-specifically (Figures 1S and 2S in Supporting Information). The fluorophore at position 4 is the most strongly quenched (>80% at [Cu(II)] = [protein] = 5 µM, vide infra), while W39 exhibits minimal quenching until more than 1 equiv of Cu(II) has been added (∼20% fluorescence decrease at 10 equiv of Cu(II), Figure 3S in Supporting Information). Even though the participation of the N-terminal amino group(23) and residues in the vicinity of 3−9(24) have been implicated in Cu(II) binding, our observation was somewhat unexpected because H50 was identified previously as the Cu(II) ligand with the strongest affinity.23,24 Furthermore, neither W94 nor W125 reporter in the highly acidic (15 carboxylate groups) C-terminal region(25) is sensitive to the presence of Cu(II) under these solution conditions, and fluorescence energy transfer kinetics measured for W39→Y(NO2) pairs near H50 (Y19−W39 and W39−Y55) show no detectable changes upon the addition of Cu(II). The data confirm that Cu(II) binding to α-syn does not induce a significant conformational change between residues 19 and 55 (data not shown).
Figure 1.

Primary amino acid sequence of human α-synuclein. Potential side-chain copper(II) ligands (N-terminus, Lys, His, Asp, Glu, and Met) are colored. Underlined residues indicate the location of Trp probes.
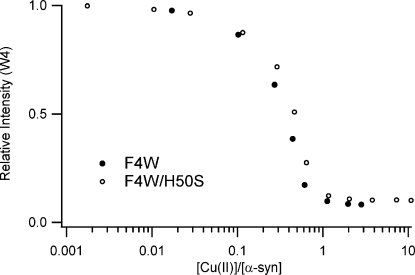
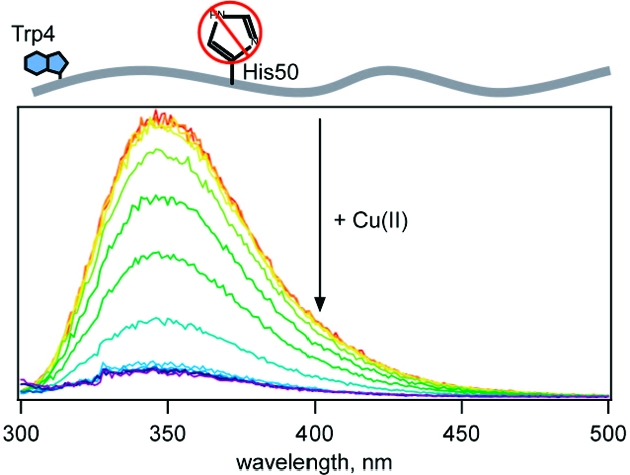
We examined the F4W protein in greater depth to probe binding in the N-terminal region because W4 is most sensitive to the presence of Cu(II). In addition, we replaced H50 with a serine residue (F4W/H50S variant) to assess the role, if any, of imidazole ligation to Cu(II). Upon copper(II) addition (25 nM to 50 µM CuSO4 in deoxygenated 20 mM MOPS, 100 mM NaCl pH 7, 25 °C), tryptophan fluorescence is dramatically quenched, owing to direct Cu(II)−protein interactions (Figure 2).(26) Titration curves for the two proteins (F4W and F4W/H50S) clearly demonstrate that one copper ion binds per protein, with a dissociation constant of <5 µM.
Figure 2.
Intensity of α-synuclein fluorescence as a function of added Cu(II); [α-syn] = 5 µM in deoxygenated 20 mM MOPS, 100 mM NaCl, pH 7 buffer.
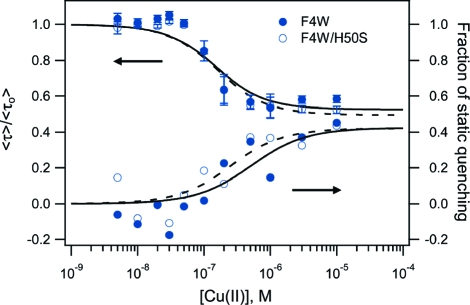
To obtain reliable values for Cu−syn dissociation constants, we measured tryptophan excited-state decay kinetics at submicromolar protein concentrations ([α-syn] = 100 nM, Figure 4S in Supporting Information). The kinetics revealed two fluorescence quenching modes. Although 1 equiv of Cu(II) reduces steady-state W fluorescence to ∼10% of its initial value (Figure 2), acceleration of W4 fluorescence decay (Supporting Information, Figures 5S and 6S) only accounts at most for a ∼40% reduction in W4 fluorescence intensity (Figure 3, top). The remainder of the observed steady-state quenching must arise from formation of a nonluminescent ground-state complex between W4 and Cu(II) (static quenching) or a dynamic process that proceeds on a time scale substantially shorter than the resolution of our decay measurements (<100 ps).
Figure 3.
Average W4 fluorescence lifetimes and fraction of static quenching extracted from NNLS fits (Supporting Information) of F4W (●) and F4W/H50S (○) α-synuclein variants ([protein]= 100 nM) as functions of added Cu(II) in deoxygenated 20 mM MOPS, 100 mM NaCl, pH 7 buffer. Fits to a single-site binding model are shown as solid (F4W) and dashed (F4W/H50S) lines.
We can fit the titration curves derived from the average lifetime data using a single-site binding model (Figure 3, Supporting Information: F4W, Kd = 100(50) nM and F4W/H50S, Kd = 110(50) nM). The similarity in binding constants for the F4W and F4W/H50S proteins clearly indicates that the high-affinity site does not involve the H50 imidazole group,(27) an observation that is consistent with recent NMR measurements on an H50A mutant.(23) Difficulties associated with determining the degree of fluorescence quenching (Figure 3) lead to greater error in the estimated dissociation constant.
W4 fluorescence quenching by Cu(II) likely involves an electron-transfer process involving contact between the metal ion and the singlet-excited indole group. The dual quenching modes observed in W4 α-syn could arise from two distinct Cu(II)−protein sites with similar binding affinities: one in which W4 and Cu(II) are within static quenching distance (contact), and the other where W4 is sufficiently close to Cu(II) to make contact during the ∼3 ns excited-singlet lifetime. Alternatively, the complex fluorescence quenching behavior may reflect a broad distribution of W4−Cu(II) distances in a structurally heterogeneous polypeptide that allows the indole side chain to sample different conformations. Those structures with short W4−Cu(II) distances would give rise to static quenching, whereas those with longer distances would be associated with dynamic processes.
The high-affinity N-terminal Cu(II)-binding site revealed by W4 fluorescence quenching does not involve H50 nor global polypeptide rearrangements that bring W39, W94, or W125 into proximity. We suggest that the tight binding involves the α-amino group of M1 (human protein contains a free amino terminus(28)) and the carboxylate group of D2, along with deprotonated amides and carbonyl groups from the peptide backbone, consistent with previous work on synthetic N-terminal fragments29,30 and similar to the structurally characterized Cu(II) site in the octarepeat domain of the prion protein.(31) Indeed, the outer-sphere association of a tryptophan indole side chain with the Cu(II)−peptide complex would likely produce static fluorescence quenching analogous to that observed in W4 α-syn. The details of intracellular copper ion homeostasis are not well understood,(32) so it is difficult to assess the importance of Cu(II)−α-syn binding in the natural function of the protein or the disease state with which it is associated.
Our experiments demonstrate that tryptophan fluorescence is a sensitive indicator of copper(II) binding to α-synuclein. Because we can make measurements at extremely low protein concentrations, we are able to extract reliable dissociation constants for high-affinity binding sites. With this approach, we can determine how different α-syn subpopulations interact with other biomolecules and compounds that are implicated in Parkinson’s disease.
Acknowledgments
Supported by the Intramural Research Program of the National Institutes of Health, the National Heart, Lung, and Blood Institute (JCL), and grants from the Ellison Medical Foundation (Senior Scholar Award in Aging to H.B.G.) and NIH (GM068461 to J.R.W.; DK19038 to H.B.G.). Initial work was supported by a Beckman Senior Research Fellowship (J.C.L.).
Supporting Information Available
Figures 1S−6S. Experimental details and data analyses of fluorescence decay curves. Complete refs (4) and (5). This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Cookson M. R. Annu. Rev. Biochem. 2005, 74, 29. [DOI] [PubMed] [Google Scholar]
- Dawson T. M.; Dawson V. L. Science 2003, 302, 819. [DOI] [PubMed] [Google Scholar]
- Kruger R.; Kuhn W.; Muller T.; Woitalla D.; Graeber M.; Kosel S.; Przuntek H.; Epplen J. T.; Schols L.; Riess O. Nat. Genet. 1998, 18, 106. [DOI] [PubMed] [Google Scholar]
- Polymeropoulos M. H.; et al. Science 1997, 276, 2045. [DOI] [PubMed] [Google Scholar]
- Singleton A. B.; et al. Science 2003, 302, 841. [DOI] [PubMed] [Google Scholar]
- Zarranz J. J.; Alegre J.; Gomez-Esteban J. C.; Lezcano E.; Ros R.; Ampuero I.; Vidal L.; Hoenicka J.; Rodriguez O.; Atares B.; Llorens V.; Tortosa E. G.; del Ser T.; Munoz D. G.; de Yebenes J. G. Ann. Neurol. 2004, 55, 164. [DOI] [PubMed] [Google Scholar]
- Tabner B. J.; El-Agna O. M. A.; Fullwood N. J.; Allsop D. Biochem. Soc. Trans. 2005, 33, 1082. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Dawson V. L.; Dawson T. M. Neurobiol. Dis. 2000, 7, 240. [DOI] [PubMed] [Google Scholar]
- Smeyne R. J.; Jackson-Lewis V. Mol. Brain Res. 2005, 134, 57. [DOI] [PubMed] [Google Scholar]
- Brown D. FEBS J. 2007, 274, 3766. [DOI] [PubMed] [Google Scholar]
- Conway K. A.; Rochet J. C.; Bieganski R. M.; Lansbury P. T. Science 2001, 294, 1346. [DOI] [PubMed] [Google Scholar]
- Gotz M. E.; Double K.; Gerlach M.; Youdim M. B. H.; Riederer P. Ann. N.Y. Acad. Sci. 2004, 1012, 193. [DOI] [PubMed] [Google Scholar]
- Uversky V. N.; Li J.; Fink A. L. J. Biol. Chem. 2001, 276, 44284. [DOI] [PubMed] [Google Scholar]
- Yamin G.; Glaser C. B.; Uversky V. N.; Fink A. L. J. Biol. Chem. 2003, 278, 27630. [DOI] [PubMed] [Google Scholar]
- Karr J. W.; Kaupp L. J.; Szalai V. A. J. Am. Chem. Soc. 2004, 126, 13534. [DOI] [PubMed] [Google Scholar]
- Atwood C. S.; Moir R. D.; Huang X.; Scarpa R. C.; Bacarra N. M.; Romano D. M.; Hartshorn M. A.; Tanzi R. E.; Bush A. I. J. Biol. Chem. 1998, 273, 12817. [DOI] [PubMed] [Google Scholar]
- Donnelly P.; Xiao Z.; Wedd A. Curr. Opin. Chem. Biol. 2007, 11, 128. [DOI] [PubMed] [Google Scholar]
- Millhauser G. Annu. Rev. Phys. Chem. 2007, 58, 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klewpatinond M.; Davies P.; Bowen S.; Brown D.; Viles J. J. Biol. Chem. 2008, 283, 1870. [DOI] [PubMed] [Google Scholar]
- Valentine J. S.; Hart P. J. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaggelli E.; Kozlowski H.; Valensin D.; Valensin G. Chem. Rev. 2006, 106, 1995. [DOI] [PubMed] [Google Scholar]
- Prior fluorescence and energy-transfer kinetics studies revealed that Trp excited-singlet lifetimes at positions 4, 39, and 94 exhibit different protein concentration dependences, indicating preferred regions of association. Moreover, donor−acceptor distance distributions extracted from tryptophan to 3-nitrotyrosine [W → Y(NO2)] energy-transfer kinetics measurements reveal heterogeneous protein conformations:; Lee J. C.; Langen R.; Hummel P. A.; Gray H. B.; Winkler J. R. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 16466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung Y.-H.; Rospigliosi C.; Eliezer D. Biochim. Biophys. Acta 2006, 1764, 5. [DOI] [PubMed] [Google Scholar]
- Rasia R. M.; Bertoncini C. W.; Marsh D.; Hoyer W.; Cherny D.; Zweckstetter M.; Griesinger C.; Jovin T. M.; Fernández C. O. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik S. R.; Shin H.-J.; Lee J.-H.; Chang C.-S.; Kim J. Biochem. J. 1999, 340, 821. [PMC free article] [PubMed] [Google Scholar]
- The fluorescence intensity is nearly recoverable (≥95%) by the addition of Cu(II) chelator EDTA. A model fluorophore (N-acetyltryptophanamide) is not quenched appreciably (<5%) by Cu(II)
- Our experiments do not preclude the possibility of H50 binding to Cu(II) ions under different solution conditions, but rather that it is not part of the N-terminal site we have identified. Similar results were obtained at [α-syn] = 25 µM and between pH 6 and 7 using either MOPS or MES buffer. The data indicate that there are multiple Cu(II) binding sites
- Li W.; West N.; Colla E.; Pletnikova O.; Troncoso J. C.; Marsh L.; Dawson T. M.; Jäkälä P.; Hartmann T.; Price D. L.; Lee M. K. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalik-Jankowska T.; Rajewska A.; Wiśniewska K.; Grzonka Z.; Jezierska J. J. Inorg. Biochem. 2005, 99, 2282. [DOI] [PubMed] [Google Scholar]
- Kowalik-Jankowska T.; Rajewska A.; Jankowska E.; Grzonka Z. Dalton Trans. 2006, 2006, 5068. [DOI] [PubMed] [Google Scholar]
- Burns C.; Aronoff-Spencer E.; Dunham C.; Lario P.; Avdievich N.; Antholine W.; Olmstead M.; Vrielink A.; Gerfen G.; Peisach J.; Scott W.; Millhauser G. Biochemistry 2002, 41, 3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney L. A.; O’Halloran T. V. Science 2003, 300, 931. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.