Abstract
Recruitment of the homologous recombination machinery to sites of double-strand breaks is a cell cycle-regulated event requiring entry into S phase and CDK1 activity. Here, we demonstrate that the central recombination protein, Rad52, forms foci independent of DNA replication, and its recruitment requires B-type cyclin/CDK1 activity. Induction of the intra-S-phase checkpoint by hydroxyurea (HU) inhibits Rad52 focus formation in response to ionizing radiation. This inhibition is dependent upon Mec1/Tel1 kinase activity, as HU-treated cells form Rad52 foci in the presence of the PI3 kinase inhibitor caffeine. These Rad52 foci colocalize with foci formed by the replication clamp PCNA. These results indicate that Mec1 activity inhibits the recruitment of Rad52 to both sites of DNA damage and stalled replication forks during the intra-S-phase checkpoint. We propose that B-type cyclins promote the recruitment of Rad52 to sites of DNA damage, whereas Mec1 inhibits spurious recombination at stalled replication forks.
Keywords: checkpoints, DNA damage, recombination, replication
Introduction
The cell follows a discrete programme as it undergoes growth and cell division. The cell cycle delineates four main stages for the eukaryotic cell: it first undergoes a period of growth (G1), replicates its genetic material (S), enters a second phase of growth (G2), and then divides (M). All stages are important for cell viability, and the cell has evolved pathways for coping with DNA damage during each stage. S phase is of particular interest, as it is during this stage that the cell copies the entirety of its nuclear-encoded genetic material.
The G1 to S transition is coupled to the appearance of a bud in Saccharomyces cerevisiae, representing polar growth of the future daughter cell. CDC4 encodes the F-box component of the SCF—Skp1, cullin, and F-box—complex comprised of Skp1, Cdc34, and Cdc53. The SCF complex targets proteins for degradation by the 26S proteasome at the G1 to S transition (Skowyra et al, 1997; Lyapina et al, 1998; Patton et al, 1998; Seol et al, 1999; Deshaies and Ferrell, 2001). One such target is the B-type cyclin inhibitor Sic1, which blocks the initiation of DNA replication (Verma et al, 1997; Deshaies and Ferrell, 2001; Nash et al, 2001). Once the cell enters S phase, DNA replication proceeds by the coordinated initiation of DNA replication at multiple origins of replication assembled during G1.
Replication requires that multiple protein complexes associate with origins of DNA in a highly coordinated manner. The complexes bind sequentially to the origin in preparation for replication, termed origin licensing (Blow, 1993; Coverley et al, 1993; Diffley et al, 1994; Chevalier and Blow, 1996). The origin recognition complex (ORC) composed of Orc1–Orc6, stably associates with origins of DNA (Rao and Stillman, 1995; Newlon, 1997), and in late M/G1 phase, Cdc6 associates with ORC and is required for loading the replicative helicase, Mcm2-7, onto DNA (Liang et al, 1995; Donovan et al, 1997; Tanaka et al, 1997; Bowers et al, 2004; Randell et al, 2006). The S-phase kinase Cdc7 and its activation partner Dbf4 are required to recruit another essential protein, Cdc45, onto origins (Zou and Stillman, 2000). Once origin licensing has occurred, Clb-CDK1 activity initiates early origin firing, and DNA replication proceeds (Nougarede et al, 2000).
DNA damage can arise due to mechanical stresses that occur during DNA replication or as a consequence of DNA protein adducts resulting from faulty DNA metabolism. The repair of induced DNA damage also presents difficulties for cells in S phase. Repair or bypass of damage along a single strand of duplex DNA—such as abasic sites or a thymidine dimer induced by UV irradiation—can occur during G1 or G2 phase as the complementary strand remains intact (Swanson et al, 1999; Hubscher et al, 2002). Damage along a single strand of the DNA presents a serious obstacle for the cell if it stalls replication forks or interferes with origin firing (Sun et al, 1996; Myung and Kolodner, 2002; Cobb et al, 2003; Katou et al, 2003; Chin et al, 2006; Cordon-Preciado et al, 2006). Stalled DNA replication forks must necessarily undergo replication restart if the replisome dissociates. Collapsed forks lead to the appearance of both single-stranded DNA (ssDNA) and exposed DNA ends, two major signals that induce the DNA damage checkpoint response and recruit the homologous recombination (HR) machinery (Melo et al, 2001; Usui et al, 2001; Petrini and Stracker, 2003; Zou and Elledge, 2003; Lisby et al, 2004).
To cope with damage sensed during DNA replication, the S. cerevisiae homologue of ATR, Mec1, and its downstream target kinase Rad53 activate the intra-S-phase checkpoint (Lopes et al, 2001; Tercero and Diffley, 2001; Cha and Kleckner, 2002). In response to DNA damage affecting replication, the checkpoint stabilizes stalled replication forks. In the absence of Mec1 or Rad53 kinase activity, the intra-S-phase checkpoint is absent, and replication forks collapse upon encountering DNA damage (Lopes et al, 2001; Tercero and Diffley, 2001). Although checkpoint events leading to replication fork stabilization have been identified, the steps necessary to restart stalled or collapsed replication forks are not well understood.
Recruiting the DNA damage checkpoint and repair machinery to DSBs is a highly regulated process and the method of repair depends upon both cell cycle phase and the type of DNA damage induced. In G1, cells repair single-strand DNA breaks prior to the onset of DNA replication using mismatch repair and base-excision repair processes (Taylor and Lehmann, 1998; Harfe and Jinks-Robertson, 2000; Plosky et al, 2002). Furthermore, DNA DSBs are preferentially repaired by NHEJ in G1, unless the DNA ends require processing before repair can proceed (Karathanasis and Wilson, 2002; Limbo et al, 2007; Sartori et al, 2007; Takeda et al, 2007). In S. cerevisiae, recruitment of the HR machinery, initiated by the Rad52 protein, is restricted to cells that have entered S phase (Lisby et al, 2001).
To understand how the cell copes with DNA damage during S phase, we examined the cellular response to both spontaneous and induced DNA damage in vivo by monitoring Rad52 focus formation. We treated various mutants involved in DNA replication initiation and progression with DNA-damaging agents such as hydroxyurea (HU) to arrest cells in S phase and study the response to DNA damage. We find that inhibiting B-type cyclin activity hinders Rad52 focus formation, whereas abrogating DNA replication does not. Furthermore, HU arrest actively inhibits Rad52 focus formation at both stalled replication forks and at DSBs in an Mec1-dependent manner. These results show that HR is activated by Clb-CDK1 in a replication-independent manner, whereas HR is inhibited by the checkpoint kinase Mec1 at stalled replication forks.
Results
Rad52 focus formation requires Clb-CDK1 activity
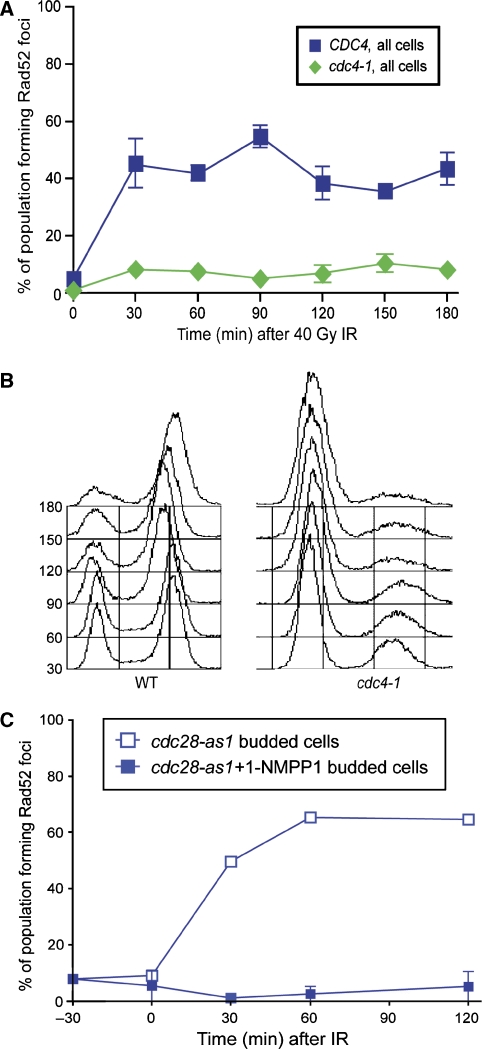
The cyclin-dependent kinase CDK1 regulates resection and recruitment of the ssDNA-binding protein RPA to DNA following DSB induction (Aylon et al, 2004; Ira et al, 2004). Recruitment of the homologous recombination protein Rad52 is also regulated as Rad52 focus formation occurs only in budded cells (Lisby et al, 2001). To determine how HR is cell cycle regulated, we examined mutants that uncouple cell cycle stage from morphology, allowing budding and axial growth in the absence of entry into S phase (Goh and Surana, 1999; Nash et al, 2001). A temperature-sensitive allele of the essential gene CDC4, cdc4-1, arrests cells in G1 upon shift to the non-permissive temperature (Nash et al, 2001). These cdc4-1 cells show an elongated budded morphology; however, they do not initiate DNA replication and have low Clb-CDK1 activity due the high level of the B-type cyclin inhibitor SIC1 (Schwob et al, 1994, Piatti et al, 1996). In response to ionizing radiation (IR), WT cells form Rad52 foci within 30 min, whereas cdc4-1 cells form foci in fewer than 10% of cells (Figure 1A). Furthermore, in cdc4-1 mutant cells, >85% of the population has not undergone DNA replication, as shown by FACS analysis, while WT cells accumulate in a G2/M checkpoint arrest (Figure 1B). The majority of Rad52 focus-forming cells in the cdc4-1 population can be attributed to the non-arrested fraction.
Figure 1.
Rad52 foci do not form in the absence of Cdc28–B-type cyclin activity. (A) cdc4-1 cells are first arrested by shifting to non-permissive temperature for 2 h. cdc4-1 cells do not form Rad52 foci in response to 40 Gy IR, in contrast to WT cells. (B) FACS profile of DNA content of WT and cdc4-1 cells upon shift to 37°C upon exposure to IR. (C) Cells containing an analogue-sensitive allele of Cdc28, cdc28-as1, are exposed to 40 Gy IR in the presence and absence of the specific inhibitor 1-NMPP1.
To determine whether the inhibition of B-type cyclins can also prevent Rad52 focus formation, we overexpressed a stabilized version of Sic1, the B-type cyclin inhibitor present during G1 (Tanaka and Diffley, 2002). Rad52 focus formation in response to IR is delayed in sic1-C70td cells as compared with WT cells (Supplementary Figure S1). However, overexpression of Sic1 alone is not sufficient to maintain a G1-arrested cell population in the presence of DNA damage, likely accounting for the Rad52 foci that are observed later (Supplementary Figure S1). To ascertain whether Cdc28 activity is responsible for Rad52 focus formation, we used an analogue-sensitive allele of Cdc28, cdc28-as1, to block kinase activity in the presence of the inhibitor 1-NMPP1 (Bishop et al, 2000; Ubersax et al, 2003). Indeed, abrogation of Cdc28 kinase activity inhibits Rad52 focus formation in response to IR, whereas cdc28-as1 cells readily form Rad52 foci in response to IR in the absence of inhibitor (Figure 1C). Altogether, these results suggest that B-type cyclin activation of Cdc28 is required for the recruitment of Rad52 into foci following IR-induced DNA damage.
Delaying DNA replication affects Rad52 focus formation
Clb5 and Clb6 are the S-phase B-type cyclins expressed at the G1/S transition, and their activation of CDK1 promotes DNA replication (Schwob and Nasmyth, 1993; Dahmann et al, 1995). Although both Clb5 and Clb6 are expressed in mitotically growing cells, deletion of Clb6 has little to no observed phenotype. However, cells lacking Clb5 are sensitive to both HU and camptothecin and experience a delay in DNA replication, indicating that the other four B-type cyclins, Clb1-4, are redundant for the role of Clb5 in promoting entry into S phase and DNA replication (Epstein and Cross, 1992; Schwob and Nasmyth, 1993). To see whether the S-phase cyclins have a function in recruiting the HR machinery to DSBs, we examined Rad52 focus formation in cells lacking Clb5 and Clb6. Rad52 foci form rapidly following DNA damage in clb6Δ cells, similar to WT (Supplementary Figure S2). On the other hand, clb5Δ cells exhibit elevated levels of spontaneous Rad52 foci (30% of budded cells, compared with 5% in WT cells). Furthermore, Rad52 foci are induced in response to IR in both clb5Δ and clb5Δ clb6Δ cells. However, in clb5Δ clb6Δ cells, focus formation is delayed compared with WT (Supplementary Figure S2). This defect in Rad52 focus formation may contribute to the DNA damage sensitivity of clb5Δ cells. Thus, CDK1 activation by the B-type cyclins represents a critical transition point after which the HR machinery can be recruited to both spontaneous and induced DNA double-strand breaks.
Rad52 focus formation occurs in the absence of DNA replication
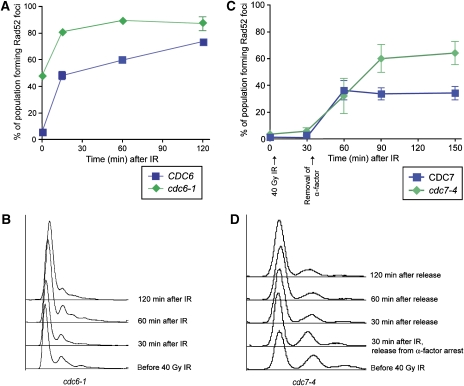
B-type cyclin activity governs entry into S phase and the initiation of DNA replication (Verma et al, 1997). To determine whether DNA replication is required for recruiting the HR machinery, temperature-sensitive mutants of both Cdc6 and Cdc7 were examined for their response to IR. Shifting the cells to the non-permissive temperature prevents the initiation of DNA synthesis, even though B-type cyclins are present in the cells. After IR, cdc6-1 cells form Rad52 foci near WT levels, indicating that efficient origin firing is not necessary for the recruitment of Rad52 to the sites of DNA damage (Figure 2A and B). Additionally, cdc7-4 cells at the non-permissive temperature, which are blocked for origin firing and DNA elongation (Bousset and Diffley, 1998; Pasero et al, 1999), also form Rad52 foci in response to IR with similar kinetics to WT cells (Figure 2C and D). Cdc7-deficient cells at non-permissive temperature have B-type cyclin-CDK1 activity (Piatti et al, 1996; Duncker et al, 1999). We find Rad52 foci do not form in response to IR in cdc7-4 cells where Cdc28 kinase activity is also inactivated (Supplementary Figure S3). Taken together, these results indicate that although Clb-CDK1 activity is required for Rad52 focus formation, ongoing DNA replication is not.
Figure 2.
Rad52 foci form in the absence of bulk DNA replication. (A) cdc6-1 cells are first held at the non-permissive temperature for 120 min, exposed to 40 Gy IR, and samples are taken for microscopy. cdc6-1 mutant cells show elevated levels of spontaneous Rad52 foci (time 0), and also form foci in response to IR. (B) FACS analysis showing DNA content of the cells shown in (A). (C) cdc7-4 cells are first arrested in G1 with α-factor, then shifted to the non-permissive temperature for 2 h. Cells were then exposed to 40 Gy IR while in α-factor arrest and released 30 min after exposure. (D) Representative FACS samples of cdc7-4 cells shown in (C).
Interestingly, both cdc6-1 and cdc7-4 cells exhibit elevated levels of spontaneous Rad52 foci (48 and 30%, respectively versus 5–10% in WT cells; Figure 2A and Supplementary Figure S3, time 0). These observations indicate that defects in origin firing and DNA elongation lead to genomic instability requiring the HR machinery. DNA replication initiation must be highly regulated, not only to replicate the DNA once in each cell cycle but also to inhibit DNA damage arising at improperly regulated origins.
HU inhibits the recruitment of Rad52 to DSBs
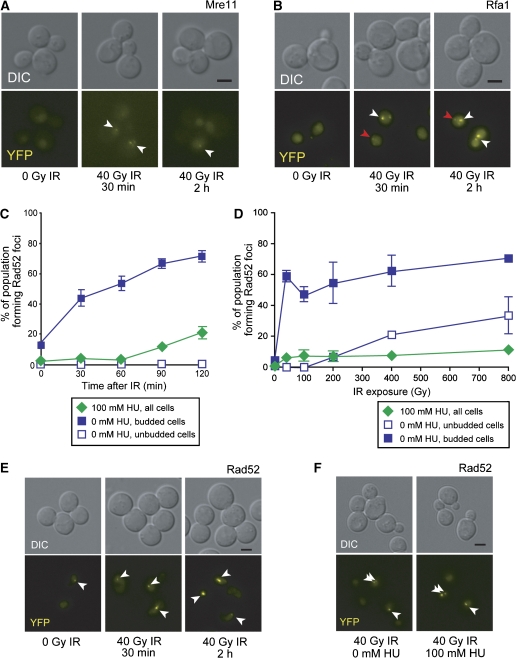
In response to DNA damage sensed during replication, the cell activates the intra-S-phase checkpoint. HU stalls DNA replication by depleting dNTP pools through inhibition of ribonucleotide reductase (RNR) (Slater, 1973; Nordlund and Reichard, 2006). To determine how the cell responds to DSBs after activation of the intra-S-phase checkpoint, we first arrested cells in HU and then exposed them to IR. After HU treatment, Mre11 focus levels remain unchanged (7%), suggesting that the intra-S-phase checkpoint does not recruit the MRX complex to stalled forks (Supplementary Figure S4) (Lisby et al, 2004). In HU-arrested cells, both Mre11 and Rfa1 form foci in response to IR (Figure 3A and B; Supplementary Figure S4). These results suggest that IR-induced DSBs are recognized by the MRX complex and the ends are processed into ssDNA bound by RPA. We also find that Rfa1 foci colocalize with a marked I-SceI DSB site in cells released from G1 into HU-containing media (data not shown), further supporting the view that RPA is recruited to DSBs during the intra-S-phase checkpoint.
Figure 3.
HU suppresses Rad52 but not Mre11 or Rfa1 focus formation. (A) Mre11 foci form in response to IR in HU-arrested cells. (B) Rfa1 foci form in response to IR during HU arrest. White arrowheads indicate bright IR-induced foci, whereas red arrowheads point to fainter foci characteristic of those observed during DNA replication. (C) Rad52 focus formation in HU-treated cells over time. HU-arrested cells do not form appreciable levels of Rad52 foci in response to 40 Gy IR in comparison to untreated cells. (D) Rad52 focus formation in HU-treated cells at high doses of IR. HU-arrested cells are exposed to a range of IR doses, and then assayed for Rad52 focus formation after 60 min. WT budded cells form Rad52 foci at 40 Gy IR, whereas unbudded G1 cells form Rad52 foci at IR doses of 400 Gy and higher. HU-arrested cells, on the other hand, do not form Rad52 foci at any level of IR dose tested. (E) HU does not inhibit Rad52 focus formation in G2-arrested cells. Cells first arrested in G2/M with nocodazole are treated with 100 mM HU. Rad52 forms foci in response to IR in G2/M-arrested cells, even in the presence of HU. (F) The addition of HU does not dissociate Rad52 foci formed in response to IR. Cells first exposed to IR are treated with HU 90 min after exposure. The addition of HU does not lead to the disappearance of the Rad52 foci that form in response to IR.
We next examined the HU-arrested cell population for its ability to recruit Rad52 for both spontaneous and IR-induced damage. As we reported previously, spontaneous Rad52 focus levels drop from the ∼10% seen in the budded cell population down to ∼0%, suggesting that HU treatment inhibits spontaneous Rad52 focus formation (Figure 3C; Lisby et al, 2004). To determine whether Rad52 recruitment to DSBs is inhibited during HU arrest, we exposed HU-arrested cells to IR. When cells are first arrested in HU, we find that Rad52 foci do not form after IR, even up to 120 min after exposure (Figure 3C). It is also possible that Rad52 may form foci in response to DNA damage at high doses, similar to that observed in G1 cells (400–800 Gy; Lisby et al, 2001). As shown in Figure 3D, WT budded cells form Rad52 foci at doses of 40 Gy and higher, and WT unbudded cells form foci at levels of 400 Gy or higher. However, HU-treated cells do not form Rad52 foci even in response to 800 Gy IR (Figure 3D). To determine whether HU alone inhibits Rad52 focus formation, we arrested cells in G2 using the microtubule poison, nocodazole. Indeed, Rad52 foci form in G2-arrested cells in response to IR in the presence of HU (Figure 3E). Furthermore, Rad52 foci formed in response to IR do not dissociate upon addition of HU (Figure 3F). Thus, either blocking ongoing DNA replication or activation of the intra-S-phase checkpoint inhibits Rad52 from being recruited to the sites of DNA damage in the presence of stalled replication forks.
Mec1-dependent intra-S-phase checkpoint activation inhibits Rad52 focus formation
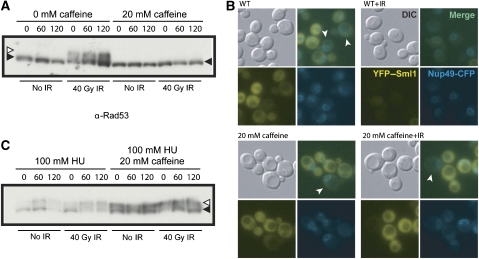
Mec1 is a key checkpoint protein involved in the intra-S-phase checkpoint response and is required, along with its downstream effector kinase Rad53, for stabilization of stalled replication forks in the presence of HU (Lopes et al, 2001; Tercero and Diffley, 2001; Zhao et al, 2001). Deletion of either MEC1 or RAD53 is lethal; however, this lethality can be rescued by deletion of SML1, which encodes a small protein inhibitor of RNR (Zhao et al, 1998). Indeed, Sml1 degradation is an extremely sensitive indicator of checkpoint activation (Torres-Rosell et al, 2007; Barlow et al, 2008). To determine whether Mec1-dependent intra-S-phase checkpoint activation inhibits Rad52 focus formation in the presence of HU, we used the small molecule caffeine, a potent inhibitor of the PI3 checkpoint kinases Mec1 and Tel1, the homologues of vertebrate ATR and ATM, respectively (Gentner and Werner, 1975; Hall-Jackson et al, 1999; Heffernan et al, 2002). In the presence of caffeine, both Mec1 and Tel1 kinase activities are inhibited, effectively blocking further checkpoint activation. We monitored Rad53 phosphorylation by protein blotting and Sml1 degradation in vivo by fluorescence of YFP–Sml1 to assess checkpoint activation. In WT cells, Rad53 is hyperphosphorylated in response to IR, whereas in caffeine-treated cells it is not, demonstrating that kinase activity has been effectively inhibited (Figure 4A). Furthermore, we also find that caffeine inhibits the degradation of Sml1 in response to IR (Figure 4B).
Figure 4.
Caffeine inhibits the Rad53-mediated DNA damage checkpoint response. (A) Caffeine inhibits phosphorylation of Rad53 in response to 40 Gy of IR. Asynchronously growing cells are incubated in 20 mM caffeine, and half the culture is then exposed to IR. Extracts were taken from cells and blotted for Rad53 (filled-in arrowhead). Cells exposed to 40 Gy IR exhibit Rad53 hyperphosphorylation (open arrowhead), whereas cells treated with caffeine do not. (B) Caffeine-induced inhibition of Mec1/Tel1 kinase activity blocks damage-induced, but not S phase, degradation of Sml1. The levels of Sml1 protein in vivo were assessed by monitoring YFP–Sml1 fluorescent protein levels. WT cells normally have high levels of cytoplasmic Sml1 and rapidly degrade Sml1 in response to exposure to IR. S-phase cells (arrowheads) degrade Sml1 in an Mec1-independent manner. Caffeine-treated cells degrade Sml1 in S-phase cells (arrowheads), similar to WT, but do not degrade Sml1 in response to damage, indicating that the IR-induced damage requires Mec1/Tel1 activity, whereas the S-phase degradation does not. (C) HU treatment induces Rad53 phosphorylation; however, hyperphosphorylated Rad53 (open arrowhead) does not disappear with the addition of caffeine to HU-treated cells but does abrogate cell cycle arrest (FACS analysis, data not shown), indicating that persistent Mec1 activity is required for maintenance of the intra-S checkpoint.
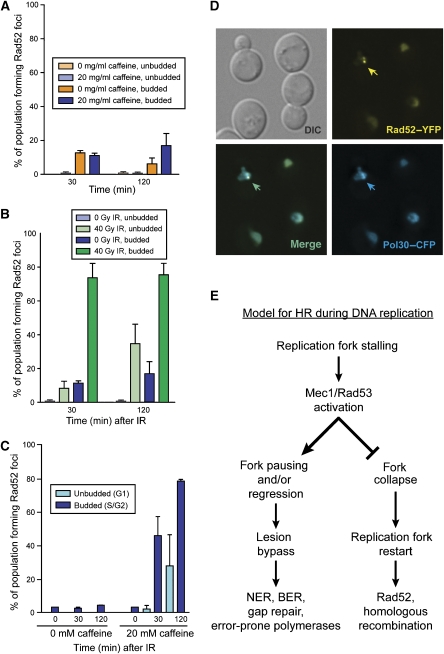
Previously, we found that spontaneous Rad52 focus levels are elevated in mec1Δ sml1Δ cells (Lisby et al, 2004). By 120 min, caffeine-treated cells also exhibit marginally higher levels of spontaneous Rad52 foci compared with untreated cells, suggesting that blocking the DNA damage checkpoint elicits a response from the HR machinery (Figure 5A). In response to IR, caffeine-treated budded cells form Rad52 foci similar to WT cells. However, unlike the WT unbudded G1 cells where Rad52 foci rarely form, 10% of caffeine-treated G1 cells form Rad52 foci 30 min after IR, and these levels increase to 40% by 120 min (Figure 5B). This increase in Rad52 focus formation is likely due to defective G2/M checkpoint arrest allowing cells to divide before the lesion can be repaired.
Figure 5.
Rad52 recruitment during S phase. (A) Rad52 focus formation after caffeine treatment. The addition of 20 mM caffeine to asynchronously growing cells induces a small increase in Rad52 focus formation by 120 min, yet this increase is not statistically significant, suggesting that the Rad52 foci observed in mec1Δ cells accumulate over multiple cell divisions in response to spontaneous DNA damage. (B) Caffeine treatment causes increased Rad52 foci in G1 cells in response to IR. The addition of caffeine does not change Rad52 focus formation in response to IR in budded cells. However, the foci in the unbudded cell population increases, consistent with loss of the Mec1-dependent G2/M checkpoint. (C) HU-arrested cells treated with caffeine form Rad52 foci in response to IR. Cells are first treated with 100 mM HU, then with 20 mM caffeine for 30 min, and finally exposed to IR. (D) Rad52 foci colocalize with PCNA in HU-arrested cells treated with caffeine. Cells treated first with 100 mM HU for 90 min and then with 20 mM caffeine for 30 min form Rad52–YFP foci and these foci colocalize with CFP–Pol30 foci present in S-phase cells. (E) Model for Rad52 recruitment to damage during S phase. After DNA damage, Mec1/Rad53 activity prevents replication fork collapse at lesions by stabilizing stalled replication forks, channelling lesion bypass through BER, NER, and so on. In the absence of Mec1/Rad53 activity, replication forks collapse, requiring recombination for replication restart, manifested as increased Rad52 foci.
As WT cells do not form Rad52 foci during HU arrest even after exposure to IR (Figures 3C, D and 5C), we also treated these cells with caffeine. Rad52 foci readily form in response to IR in HU-arrested cells treated with caffeine (Figure 5C). Furthermore, when HU treatment induces Rad52 foci in the presence of caffeine, these foci colocalize with the DNA replication clamp PCNA—encoded by POL30 in S. cerevisiae—suggesting that they form at collapsed replication forks (Figure 5D). Interestingly, we also find that the phosphorylated Rad53 present in HU-arrested cells remains after the addition of caffeine (Figure 4C). From these observations, we conclude that continued Mec1/Tel1 kinase activity is required to inhibit the recruitment of Rad52 during the intra-S-phase checkpoint at both stalled replication forks and DSBs.
Discussion
DNA repair is a highly regulated event, incorporating multiple inputs to elicit an effective DNA damage response. To prevent HR at inappropriate times, the cell has coupled the recruitment of the HR machinery to the cell cycle. The central HR protein Rad52 is loaded onto the sites of DNA damage when the B-type cyclins activate CDK1 and the cell has entered into S phase. Indeed, it would be detrimental to recruit the HR machinery to ssDNA lesions and base mismatches, as it may increase genome instability and more efficient pathways for DNA repair in G1 cells exist such as NER, BER, and UV photoproduct repair. Furthermore, ssDNA lesions can be repaired faithfully in G1 cells without the aid of a homologous template. In addition, double-stranded DNA lesions can be repaired through NHEJ during G1, whereas those requiring HR for repair must wait for the cell cycle to proceed to undergo further steps of DNA repair. Rad52 is not recruited until the B-type cyclin inhibitor Sic1 is degraded by the 26S proteasome in a Cdc4/SCF-dependent manner and CDK1 is activated by pairing with the B-type cyclins (Figure 1). Interestingly, CDK1 also activates the Sae2 nuclease, responsible for cleaving hairpin ends as well as other DNA end structures (Lengsfeld et al, 2007; Sartori et al, 2007; Huertas et al, 2008). However, the regulation of Sae2 activity alone cannot account for the lack of foci observed in the absence of CDK1 kinase activity, as sae2Δ cells show only a slight delay in Rad52 focus formation in response to IR (Lisby et al, 2004). Thus, CDK1 kinase activity promotes HR by both enhancing the processing of DNA ends and by promoting the recruitment of Rad52.
To explore this regulation in more detail, we examined cells lacking either Cdc6 or Cdc7 for their ability to form Rad52 foci. Rad52 forms foci in response to IR even in the absence of DNA replication in cdc6-1 and cdc7-4 cells (Figure 2A and C). Furthermore, both cdc6-1 and cdc7-4 cells show elevated levels of spontaneous Rad52 foci (Figure 2A; Supplementary Figure S3), indicating that the lack of origin firing in cdc6-1 cells or DNA replication elongation in cdc7-4 cells leads to increased genomic instability. Perhaps origin firing in the absence of Cdc6 or Cdc7 results in the pre-RC remaining bound to the DNA, thereby blocking subsequent replication fork progression through the unfired origin. Another possibility is that an initiated origin results in a replication bubble, which, without the replication machinery, contains ssDNA that recruits the DSBR machinery. Rad52 is recruited, in the presence or absence of ongoing DNA replication once the B-type cyclins activate CDK1. On the other hand, we find that in the event of perturbed replication, Mec1 and Rad53 activate the intra-S-phase checkpoint, both stabilizing the DNA replication machinery at stalled forks and inhibiting Rad52 focus formation (Figure 3C and D). Furthermore, when Mec1 is inactivated by the addition of caffeine, Rad52 forms foci in response to HU as well as IR (Figure 5B and C). Indeed, in an mec1Δ sml1Δ background, virtually all cells form Rad52 foci in response to HU, most likely due to replication fork collapse. Taken together, these results support a model where the recruitment of homologous recombination proteins is promoted by Clb-CDK1 kinase activity, and inhibited at stalled replication forks by the checkpoint kinase Mec1 (Figure 5E).
The coupling of Mec1-dependent replication fork stabilization and the inhibition of Rad52 focus formation provides S-phase cells with a window of opportunity to restart replication without initiating homologous recombination. This additional time may allow replication to restart simply by fork pausing followed by lesion excision and repair or recruitment of lesion bypass proteins. Although HR is active in S and G2 phases of the cell cycle (Takata et al, 1998; Aylon et al, 2004; Ira et al, 2004), in S phase, recruitment of HR proteins such as Rad52 may actually be detrimental to the cell, particularly in regions with active DNA replication forks. Instead, it is likely that replication fork restart without the initiation of HR is preferred, so that replication can be re-established quickly. By delaying the recruitment of Rad52, the cell limits delays in replication restart and also mutagenic DSBR processes such as HR from a heterologous template, heteroallelic recombination from a homologue rather than a sister chromatid, or break-induced replication, leading to gene conversion or loss of heterozygosity. Alternatively, inhibiting HR by blocking the recruitment of Rad52 to stalled forks may allow cells to restart replication by alternative mechanisms. A recent report suggests that stalled replication forks induced by the replisome running into RNA polymerase can restart using mRNA as a template (Pomerantz and O'Donnell, 2008). However, HR is also important in S-phase cells as replication restart after fork collapse requires the HR machinery, and the failure to repair a collapsed fork will lead to significant loss of genetic material and possibly death. Thus, Mec1 activation carefully regulates the decision to enter into recombination during S phase.
The Mec1/Rad53-dependent intra-S-phase checkpoint actively inhibits the recruitment of the recombination protein Rad52 to the sites of DNA damage, both at stalled replication forks and at IR-induced DSBs. These results indicate that the intra-S-phase checkpoint elicits a global, diffusible signal inhibiting recombination throughout the nucleus and not simply at the stalled forks themselves. Both RPA and Rad52 are phosphorylated upon entry into S phase and in response to DNA damage (Din et al, 1990; Brush et al, 1996; Antunez de Mayolo et al, 2006). Rad52 is not directly phosphorylated by Mec1 (Antunez de Mayolo et al, 2006), but the ssDNA-binding protein RPA that recruits Rad52 to the sites of DNA damage is a target of Mec1 phosphorylation (Din et al, 1990; Brush et al, 1996; Kim and Brill, 2003; Bartrand et al, 2004), which may control Rad52 recruitment. Alternatively, a checkpoint kinase downstream of Mec1, such as Dun1 or Rad53 itself may control the recruitment of Rad52 in S phase. The fact that phosphorylated Rad53 is stable in the presence of caffeine implies that Rad53 activity does not inhibit Rad52 focus formation, even in the presence of HU. Thus, Mec1 itself—or an alternative kinase for example Chk1—may block the recruitment of the HR machinery during the intra-S-phase checkpoint.
Here, we show that the B-type cyclins and the Mec1/Tel1 checkpoint kinases govern differential DNA checkpoint responses to DNA damage sensed in the G1 and S phases of the cell cycle. During G1, Tel1 and Mec1 cooperatively activate the DNA damage checkpoint, whereas the B-type cyclins and Mec1/Rad53 regulate the DNA damage checkpoint response in S phase. We hypothesize that G1 and S/G2 cells initiate a different checkpoint response very early in the signal kinase cascade to determine subsequent repair events. We propose that progressing replication forks function as potent sensors of DNA damage, tightly regulated by the Clb-CDK1 and Mec1/Rad53 kinases. These results indicate that the recruitment of a key recombination protein, Rad52, is modulated to maintain genome integrity without impairing DNA replication and cell cycle progression.
Materials and methods
Yeast strains and media
Yeast strains used in this study are listed in Table I. Fluorescently tagged proteins and chromosomal sites were described previously (Lisby et al, 2001, 2004). Microscopy experiments were carried out at 23°C in synthetic complete (SC) medium supplemented with 2% glucose, raffinose, or galactose as noted. Expression of the I-SceI enzyme from the Gal1-10 promoter was induced by the addition of galactose to a final concentration of 2% in cultures growing in 2% raffinose.
Table 1.
Strains used in this study
| Strains | Relevant genotypea |
|---|---|
| W3749-14C | MATa ADE2 bar1∷LEU2 RAD52-YFP |
| W4466-15B | MATa ADE2 bar1∷LEU2 RAD52-YFP cdc4-1 |
| W5712-7b | MATa ADE2 bar1∷LEU2 RAD52-YFP ura3∷Gal-SIC1C70td∷URA3 |
| W5288-16C | MATa ADE2 RAD52-YFP cdc6-1 |
| W5640-10A | MATa ADE2 bar1∷LEU2 RAD52-YFP cdc7-4 |
| W4470-7B | MATa ADE2 RAD52-YFP clb6∷LEU2 |
| W4470-6D | MATa ADE2 RAD52-YFP clb5∷URA3 |
| W4470-15C | MATa ADE2 RAD52-YFP clb5∷URA3 clb6∷LEU2 |
| W3483-10A | MATa ADE2 bar1∷LEU2 MRE11-YFP |
| W3775-12C | MATa ADE2 bar1∷LEU2 RFA1-YFP |
| W6032-19A | MATa ADE2 bar1∷LEU2 RAD52-YFP CFP-POL30 |
| W4622-14B | MATa ADE2 bar1∷LEU2 YFP-SML1+ pWJ1323 CFP-NUP49 |
| W7780-16A | MATa ADE2 bar1∷LEU2 YFP-SML1 RFA1-CFP |
| W8375-3C | MATa ADE2 lys2Δ cdc28-as1∷URA3 CDC7 Rad52-YFP |
| W8375-3D | MATa ADE2 TRP1 cdc28-as1∷URA3 cdc7-4 Rad52-YFP |
| aAll strains are isogenic to W1588-4C, unless otherwise noted (Zhao et al, 1998). | |
Cell synchronizations and drug treatment
Temperature-sensitive mutant strains—cdc6-1, cdc7-4 and cdc4-1—were held at the non-permissive temperature, 37°C, for 120 min, by which time the bulk of the population was arrested with a 1C content of DNA. The culture was then exposed to 40 Gy IR and then samples were taken for microscopy and FACS analysis. For HU arrest in S phase, cells were placed in 100 mM HU for 90 min prior to subsequent treatments. For HU arrest in G2 phase, cells were placed in 5 μg/ml nocodazole for 2 h prior to exposure to IR or treatment with HU. Caffeine-treated cells were grown in fresh media for 2 h and then 20 mM caffeine was added to the media for 30 min prior to exposure to γ-radiation. Cells were arrested in G2 by adding 5 μg/ml nocodazole for 2 h prior to further treatment.
FACS analysis
Cells were suspended in 70% ethanol and then fixed overnight at 25°C. Fixed samples were prepared for FACS by first incubating in 50 mM sodium citrate/0.25 μg/μl RNase to remove RNA for 1 h, followed by a 1-h incubation in 1 μg/μl proteinase K at 50°C. DNA was then dyed with propidium iodide (8 μg/ml) for 30 min and then sorted for cell size and DNA content on a Beckman Coulter FACS Calibur.
γ-Irradiation
The γ-ray sensitivity of strains was determined by growing cultures in YPD to mid-log phase at 23°C. An appropriate number of cells were plated on YPD plates and exposed to different doses of γ-rays using a Gammacell-220 60Co irradiator (Atomic Energy of Canada). Cells analysed by microscopy were pre-grown in SC at 23°C until OD600 reached 0.2. At this point, the liquid cultures were exposed to defined doses of irradiation and aliquots of the cultures were processed immediately for imaging.
Live cell imaging and fluorescent microscopy
Cells were prepared for fluorescent microscopy as described previously (Lisby et al, 2001). Live cell images were captured with a cooled Orca-ER CCD camera (Hamamatsu, Japan) mounted on a Zeiss Axioplan II microscope (Carl Zeiss, Thornwood, NY). All images were captured at 100-fold magnification using a Plan-Apochromat × 100, 1.4 NA objective lens. The illumination source was a 100 W mercury arc lamp (Osram, Munich, Germany). For each field of cells, 11 fluorescent images at each of the relevant wavelengths were obtained at 0.3 μm intervals along the z-axis to allow inspection of all focal planes of each cell. Images were acquired and pseudo-coloured using OpenLab software (Improvision, Lexington, MA) and prepared for publication in Adobe Photoshop (Adobe, San Jose, CA). Fluorophores were visualized using band-pass CFP (31044 v2), YFP (41028), and RFP (41002c) filter sets from Chroma (Brattleboro, VT). 3D reconstruction and measurement of fluorescent intensity were carried out using Volocity software (Improvision). Image acquisition times for fluorophore-tagged proteins are as follows: Rad52–YFP (1500 ms), Rfa1–YFP (2500 ms), Ddc1–YFP (2000 ms), Ddc2–YFP (2000 ms), Mre11–YFP (3000 ms) and TetI–RFP (1000 ms), and were taken with a 10% neutral density filter in place to reduce photobleaching. Fluorophores used in this study were the red- and blue-shifted enhanced variants YFP (10C) (Ormo et al, 1996) and CFP (W7) (Heim and Tsien, 1996) of the GFP gene and the monomeric version of DsRed (mRFP1) (Campbell et al, 2002).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure Legends
Acknowledgments
We thank the members of the Rothstein lab for helpful discussions and advice. We thank Michael Lisby for experimental input and both Michael Lisby and Peter Thorpe for comments on the paper. We also thank John Diffley, Fred Cross, and Orna Cohen-Fix for yeast strains and advice. We also thank Lorraine Symington and Jean Gautier for helpful discussions. This study was supported by GM50237 (RR), GM67055 (RR), GM70088 (JHB), and GM73568 (JHB).
References
- Antunez de Mayolo A, Lisby M, Erdeniz N, Thybo T, Mortensen UH, Rothstein R (2006) Multiple start codons and phosphorylation result in discrete Rad52 protein species. Nucleic Acids Res 34: 2587–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Liefshitz B, Kupiec M (2004) The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J 23: 4868–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JH, Lisby M, Rothstein R (2008) Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol Cell 30: 73–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartrand AJ, Iyasu D, Brush GS (2004) DNA stimulates Mec1-mediated phosphorylation of replication protein A. J Biol Chem 279: 26762–26767 [DOI] [PubMed] [Google Scholar]
- Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, Wood JL, Morgan DO, Shokat KM (2000) A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407: 395–401 [DOI] [PubMed] [Google Scholar]
- Blow JJ (1993) Preventing re-replication of DNA in a single cell cycle: evidence for a replication licensing factor. J Cell Biol 122: 993–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousset K, Diffley JF (1998) The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev 12: 480–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JL, Randell JC, Chen S, Bell SP (2004) ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol Cell 16: 967–978 [DOI] [PubMed] [Google Scholar]
- Brush GS, Morrow DM, Hieter P, Kelly TJ (1996) The ATM homologue MEC1 is required for phosphorylation of replication protein A in yeast. Proc Natl Acad Sci USA 93: 15075–15080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY (2002) A monomeric red fluorescent protein. Proc Natl Acad Sci USA 99: 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha RS, Kleckner N (2002) ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297: 602–606 [DOI] [PubMed] [Google Scholar]
- Chevalier S, Blow JJ (1996) Cell cycle control of replication initiation in eukaryotes. Curr Opin Cell Biol 8: 815–821 [DOI] [PubMed] [Google Scholar]
- Chin JK, Bashkirov VI, Heyer WD, Romesberg FE (2006) Esc4/Rtt107 and the control of recombination during replication. DNA Repair (Amst) 5: 618–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM (2003) DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J 22: 4325–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordon-Preciado V, Ufano S, Bueno A (2006) Limiting amounts of budding yeast Rad53 S-phase checkpoint activity results in increased resistance to DNA alkylation damage. Nucleic Acids Res 34: 5852–5862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coverley D, Downes CS, Romanowski P, Laskey RA (1993) Reversible effects of nuclear membrane permeabilization on DNA replication: evidence for a positive licensing factor. J Cell Biol 122: 985–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmann C, Diffley JF, Nasmyth KA (1995) S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr Biol 5: 1257–1269 [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Ferrell JE Jr (2001) Multisite phosphorylation and the countdown to S phase. Cell 107: 819–822 [DOI] [PubMed] [Google Scholar]
- Diffley JF, Cocker JH, Dowell SJ, Rowley A (1994) Two steps in the assembly of complexes at yeast replication origins in vivo. Cell 78: 303–316 [DOI] [PubMed] [Google Scholar]
- Din S, Brill SJ, Fairman MP, Stillman B (1990) Cell-cycle-regulated phosphorylation of DNA replication factor A from human and yeast cells. Genes Dev 4: 968–977 [DOI] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JF (1997) Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci USA 94: 5611–5616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncker BP, Pasero P, Braguglia D, Heun P, Weinreich M, Gasser SM (1999) Cyclin B-CDK1 kinase stimulates ORC- and CDC6-independent steps of semiconservative plasmid replication in yeast nuclear extracts. Mol Cell Biol 19: 1226–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein CB, Cross FR (1992) CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev 6: 1695–1706 [DOI] [PubMed] [Google Scholar]
- Gentner NE, Werner MM (1975) Repair in Schizosaccharomyces pombe as measured by recovery from caffeine enhancement of radiation-induced lethality. Mol Gen Genet 142: 171–183 [DOI] [PubMed] [Google Scholar]
- Goh PY, Surana U (1999) Cdc4, a protein required for the onset of S phase, serves an essential function during G(2)/M transition in Saccharomyces cerevisiae. Mol Cell Biol 19: 5512–5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Jackson CA, Cross DA, Morrice N, Smythe C (1999) ATR is a caffeine-sensitive, DNA-activated protein kinase with a substrate specificity distinct from DNA-PK. Oncogene 18: 6707–6713 [DOI] [PubMed] [Google Scholar]
- Harfe BD, Jinks-Robertson S (2000) Mismatch repair proteins and mitotic genome stability. Mutat Res 451: 151–167 [DOI] [PubMed] [Google Scholar]
- Heffernan TP, Simpson DA, Frank AR, Heinloth AN, Paules RS, Cordeiro-Stone M, Kaufmann WK (2002) An ATR- and Chk1-dependent S checkpoint inhibits replicon initiation following UVC-induced DNA damage. Mol Cell Biol 22: 8552–8561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim R, Tsien RY (1996) Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol 6: 178–182 [DOI] [PubMed] [Google Scholar]
- Hubscher U, Maga G, Spadari S (2002) Eukaryotic DNA polymerases. Annu Rev Biochem 71: 133–163 [DOI] [PubMed] [Google Scholar]
- Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP (2008) CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455: 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, Haber JE, Foiani M (2004) DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431: 1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karathanasis E, Wilson TE (2002) Enhancement of Saccharomyces cerevisiae end-joining efficiency by cell growth stage but not by impairment of recombination. Genetics 161: 1015–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K (2003) S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424: 1078–1083 [DOI] [PubMed] [Google Scholar]
- Kim HS, Brill SJ (2003) MEC1-dependent phosphorylation of yeast RPA1 in vitro. DNA Repair (Amst) 2: 1321–1335 [DOI] [PubMed] [Google Scholar]
- Lengsfeld B, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT (2007) Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell 28: 638–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Weinreich M, Stillman B (1995) ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell 81: 667–676 [DOI] [PubMed] [Google Scholar]
- Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P (2007) Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell 28: 134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R (2004) Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713 [DOI] [PubMed] [Google Scholar]
- Lisby M, Rothstein R, Mortensen UH (2001) Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci USA 98: 8276–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M (2001) The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412: 557–561 [DOI] [PubMed] [Google Scholar]
- Lyapina SA, Correll CC, Kipreos ET, Deshaies RJ (1998) Human CUL1 forms an evolutionarily conserved ubiquitin ligase complex (SCF) with SKP1 and an F-box protein. Proc Natl Acad Sci USA 95: 7451–7456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo JA, Cohen J, Toczyski DP (2001) Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev 15: 2809–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Kolodner RD (2002) Suppression of genome instability by redundant S-phase checkpoint pathways in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 99: 4500–4507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M (2001) Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414: 514–521 [DOI] [PubMed] [Google Scholar]
- Newlon CS (1997) Putting it all together: building a prereplicative complex. Cell 91: 717–720 [DOI] [PubMed] [Google Scholar]
- Nordlund P, Reichard P (2006) Ribonucleotide reductases. Annu Rev Biochem 75: 681–706 [DOI] [PubMed] [Google Scholar]
- Nougarede R, Della Seta F, Zarzov P, Schwob E (2000) Hierarchy of S-phase-promoting factors: yeast Dbf4–Cdc7 kinase requires prior S-phase cyclin-dependent kinase activation. Mol Cell Biol 20: 3795–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ (1996) Crystal structure of the Aequorea victoria green fluorescent protein. Science 273: 1392–1395 [DOI] [PubMed] [Google Scholar]
- Pasero P, Duncker BP, Schwob E, Gasser SM (1999) A role for the Cdc7 kinase regulatory subunit Dbf4p in the formation of initiation-competent origins of replication. Genes Dev 13: 2159–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Willems AR, Sa D, Kuras L, Thomas D, Craig KL, Tyers M (1998) Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev 12: 692–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrini JH, Stracker TH (2003) The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol 13: 458–462 [DOI] [PubMed] [Google Scholar]
- Piatti S, Bohm T, Cocker JH, Diffley JFX, Nasmyth K (1996) Activation of S-phase-promoting CDKs in late G1 defines a ‘point of no return' after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev 10: 1516–1531 [DOI] [PubMed] [Google Scholar]
- Plosky B, Samson L, Engelward BP, Gold B, Schlaen B, Millas T, Magnotti M, Schor J, Scicchitano DA (2002) Base excision repair and nucleotide excision repair contribute to the removal of N-methylpurines from active genes. DNA Repair (Amst) 1: 683–696 [DOI] [PubMed] [Google Scholar]
- Pomerantz RT, O'Donnell M (2008) The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature 456: 762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randell JC, Bowers JL, Rodriguez HK, Bell SP (2006) Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase. Mol Cell 21: 29–39 [DOI] [PubMed] [Google Scholar]
- Rao H, Stillman B (1995) The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc Natl Acad Sci USA 92: 2224–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP (2007) Human CtIP promotes DNA end resection. Nature 450: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob E, Böhm T, Mendenhall MD, Nasmyth K (1994) The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79: 233–244 [DOI] [PubMed] [Google Scholar]
- Schwob E, Nasmyth K (1993) CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev 7: 1160–1175 [DOI] [PubMed] [Google Scholar]
- Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Deshaies RJ, Shevchenko A, Deshaies RJ (1999) Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev 13: 1614–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW (1997) F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin–ligase complex. Cell 91: 209–219 [DOI] [PubMed] [Google Scholar]
- Slater ML (1973) Effect of reversible inhibition of deoxyribonucleic acid synthesis on the yeast cell cycle. J Bacteriol 113: 263–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Fay DS, Marini F, Foiani M, Stern DF (1996) Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev 10: 395–406 [DOI] [PubMed] [Google Scholar]
- Swanson RL, Morey NJ, Doetsch PW, Jinks-Robertson S (1999) Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol Cell Biol 19: 2929–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S (1998) Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J 17: 5497–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Nakamura K, Taniguchi Y, Paull TT (2007) Ctp1/CtIP and the MRN complex collaborate in the initial steps of homologous recombination. Mol Cell 28: 351–352 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Diffley JF (2002) Deregulated G1-cyclin expression induces genomic instability by preventing efficient pre-RC formation. Genes Dev 16: 2639–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Knapp D, Nasmyth K (1997) Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell 90: 649–660 [DOI] [PubMed] [Google Scholar]
- Taylor EM, Lehmann AR (1998) Conservation of eukaryotic DNA repair mechanisms. Int J Radiat Biol 74: 277–286 [DOI] [PubMed] [Google Scholar]
- Tercero JA, Diffley JF (2001) Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412: 553–557 [DOI] [PubMed] [Google Scholar]
- Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, Reid R, Jentsch S, Rothstein R, Aragon L, Lisby M (2007) The Smc5–Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol 9: 923–931 [DOI] [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO (2003) Targets of the cyclin-dependent kinase CDK1. Nature. 425: 859–864 [DOI] [PubMed] [Google Scholar]
- Usui T, Ogawa H, Petrini JH (2001) A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol Cell 7: 1255–1266 [DOI] [PubMed] [Google Scholar]
- Verma R, Annan RS, Huddleston MJ, Carr SA, Reynard G, Deshaies RJ (1997) Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 278: 455–460 [DOI] [PubMed] [Google Scholar]
- Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R (2001) The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J 20: 3544–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Muller EG, Rothstein R (1998) A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell 2: 329–340 [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
- Zou L, Stillman B (2000) Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p–Dbf4p kinase. Mol Cell Biol 20: 3086–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure Legends