Abstract
White collar-1 (WC-1) and white collar-2 (WC-2) are essential for light-mediated responses in Neurospora crassa, but the molecular mechanisms underlying gene induction and the roles of other real and putative photoreceptors remain poorly characterized. Unsupervised hierarchical clustering of genome-wide microarrays reveals 5.6% of detectable transcripts, including several novel mediators, that are either early or late light responsive. Evidence is shown for photoreception in the absence of the dominant, and here confirmed, white collar complex (WCC) that regulates both types of light responses. VVD primarily modulates late responses, whereas light-responsive submerged protoperithecia-1 (SUB-1), a GATA family transcription factor, is essential for most late light gene expression. After a 15-min light stimulus, the WCC directly binds the sub-1 promoter. Bioinformatics analysis detects many early light response elements (ELREs), as well as identifying a late light response element (LLRE) required for wild-type activity of late light response promoters. The data provide a global picture of transcriptional response to light, as well as illuminating the cis- and trans-acting elements comprising the regulatory signalling cascade that governs the photobiological response.
Keywords: light, microarray, Neurospora, photoreceptors, wc-1
Introduction
Neurospora crassa has served as a model organism to study light responses in eukaryotic cells for several decades (Dunlap and Loros, 2004; Purschwitz et al, 2006; Bahn et al, 2007; Corrochano, 2007; Heintzen and Liu, 2007; Herrera-Estrella and Horwitz, 2007). Two GATA family zinc finger transcription factors (TFs), white collar-1 (WC-1) and white collar-2 (WC-2), have been shown to play an essential role in diverse UV/blue light regulated physiological processes, including maintenance and resetting of the circadian clock, carotenoid biosynthesis, asexual conidiation and, in the sexual cycle, the formation of protoperithecia and the direction of ascospores release. Molecularly, WC-1 is both a flavin-adenine dinucleotide-binding photoreceptor and a TF. WC-1 interacts with WC-2 through its Per-Arnt-Sim domain and, after sensing light, forms a large white collar complex (L-WCC) that activates downstream target genes, presumably through the recognition of a consensus sequence, the light responsive element (LRE) (Froehlich et al, 2002; He et al, 2002; Cheng et al, 2003). Notably, despite extensive efforts to identify blind strains in several labs, only wc-1 and wc-2 mutants have been isolated (Linden et al, 1997), strongly suggesting their unique and dominant role in mediating light signals. In addition to WC-1, VVD is the other blue light photoreceptor that has been implicated in light sensing. In vvd loss-of-function strains, the induction of light responses is largely normal, but there subsequently appears a defect in photoadaptation, the ability of the organism to sense incremental changes in light intensity (Heintzen et al, 2001; Schwerdtfeger and Linden, 2001, 2003; Shrode et al, 2001).
In the Neurospora genome sequence, several putative photoreceptors have been identified based on sequence similarities, including a cryptochrome and two phytochrome homologues (PHY-1 and PHY-2) (Galagan et al, 2003). The presence of phytochromes was a surprise, as all known light responses in Neurospora are restricted to blue light (Dunlap and Loros, 2006). Studies further show that PHY-2 not only can covalently bind either biliverdin or phycocyanobilin but also is capable of undergoing a photocycle in vitro. However, due to the absence of a detectable phenotype in the knockout strains (Froehlich et al, 2005), the biological function of PHY-1 and PHY-2 remains to be discovered. Another potential photoreceptor, NOP-1, is structurally related to archaeal rhodopsins and is also capable of absorbing light and undergoing a photocycle. However, like other putative photoreceptor mutants in Neurospora, the knockout stain has only a weak phenotype at best (Bieszke et al, 1999a, 1999b; Furutani et al, 2004). Undoubtedly, a more comprehensive and systematic approach is needed to elucidate functions of these light-sensing proteins.
Here, using microarrays with full-genome coverage (Dunlap et al, 2007; Tian et al, 2007) and large-scale quantitative analysis, we provide a catalogue of the 314 genes showing strong early or late light responses and describe functional and sequence analysis, suggesting a clear correspondence between the timing of induction and gene function. Data confirm the dominant role of WC-1 and WC-2 in initiating the light responses and of VVD in modulating the responses but show that residual light responsivity remains in a wc-1 loss-of-function mutant. Identification of SUB-1 as a novel light-responsive TF and of novel cis-acting early light response elements (ELREs) and late light response elements (LLREs) allows a more global understanding of the regulatory cascade governing the response of the organism to light.
Results
Unsupervised hierarchical clustering of light-inducible responses identifies ELRGs and LLRGs
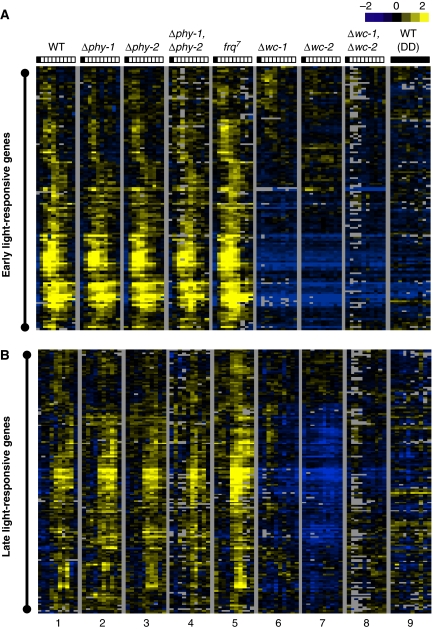
We took a systematic approach using long-oligomer constructed microarrays with full-genome coverage (Tian et al, 2007) to characterize white light-inducible transcriptional changes in wild-type (WT) and various photoreceptor knockout strains. Earlier studies have shown that Neurospora can display dramatic transcriptional changes within 5 min or less (Crosthwaite et al, 1995; Linden et al, 1997). Therefore, we sampled most intensively within the first 30 min, namely at 0, 5, 10, 15, 30, 60, 90, 120, 180 and 240 min after lights-on. To eliminate intrinsic systemic variation between microarrays and have a direct comparison between experiments, we adopted an unsupervised hierarchical clustering approach (Eisen et al, 1998) to identify bona fide light-responsive genes (see Materials and methods for details). An assumption made is that genes that are truly responsive to light should behave more similar to each other across sequential time points and different strains. After clustering all of the data across 90 microarrays, a subcluster containing 314 genes (Supplementary Table I), equal to 5.6% of the total detectable transcripts, became evident with a robust and coherent light-responsive pattern. This ratio is slightly higher but close to a prior estimate (3%) from spotted cDNA microarrays covering 1343 genes (Lewis et al, 2002). Within this subcluster of genes, several divisions were merged and re-grouped as either early or late light-responsive genes based on visual inspection (ELRGs and LLRGs cover 92% of the total identified light-responsive genes, Supplementary Tables II and III). As shown in Figure 1A and B, lanes 1–4, light-induced transcriptional changes are extremely consistent in strains reported earlier to have normal light responses, including single- and double-knockout strains of phy-1 and phy-2 (lanes 2–4) (Froehlich et al, 2005) (The few genes showing altered expression in phytochrome gene knockouts are the subject of a separate study). In the frq7 strain, a long-period clock mutant, we observed a generally higher amplitude of induction for both types of light responses (lane 5, Supplementary Figure 1), presumably due to the positive feedback loop between FRQ and WC-1 level (Lee et al, 2000).
Figure 1.
Unsupervised hierarchical clustering of light-inducible responses identifies early and late light-responsive genes. (A) Clustering of early light-responsive genes. (B) Clustering of late light-responsive genes. Experimental strains: Lane 1, 74A (WT); Lane 2, Δphy-1; Lane 3, Δphy-2; Lane 4, Δphy-1, Δphy-2; Lane 5, frq7; Lane 6, Δwc-1; Lane 7, Δwc-2; Lane 8, Δwc-1, Δwc-2; Lane 9, 74A (DD). In each lane, from left to right, the individual columns correspond to light treatment lasting for 0, 5, 10, 15, 30, 45, 60, 90, 120, 240 min, respectively. For each row, the data were centred so that each measurement reflects transcript abundance relative to the mean expression across all 90 microarrays (a common reference RNA design). Yellow squares indicate transcripts with increased expression; Blue squares indicate transcripts with decreased expression; Black squares indicate transcripts with expression levels close to mean value; Gray squares represent missing data.
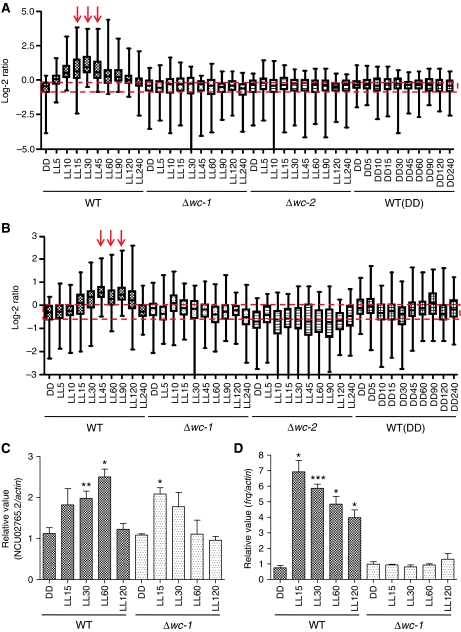
As predicted, both early and late light responses are severely impaired in wc-1/wc-2 single- and double-knockout strains (lanes 6, 7 and 8). To enhance the robustness of clustering and to serve as a reference dataset for visual comparison, we combined microarray data from a WT strain in constant darkness (DD, lane 9). Overall, although quantitatively expanding the number of light-regulated genes by about eight-fold, these data are qualitatively consistent with what is known about Neurospora photobiology in general, with waves of ELRGs followed by LLRGs (Sommer et al, 1989; Linden et al, 1997; Lewis et al, 2002). To facilitate a direct comparison between WT and wc knockout strains, we plotted the microarray readouts of WT, Δwc-1, Δwc-2 and WT (DD) side by side in another dimension. As shown in Figure 2A, the ELRGs peak between 15 and 45 min after onset of light, whereas LLRGs peak later between 45 and 90 min (Figure 2B). Reflecting photoadaptation, both types of light responses return to their basal level of expression after 4 h of constant light (Figure 2A and B, DD versus LL240 time point in the WT strain). The microarray data from Δwc-1, Δwc-2 and the WT strain in constant darkness (Figure 2A and B) show no induction, suggesting that there is no increase in gene expression due to development or other changes over the 4-h time course. The distinct kinetics of induction that distinguishes the early and late light responses suggested to us that they are separately regulated, and that there might be common molecular mechanisms underlying the timing control of each response.
Figure 2.
Most but not all light-induced transcriptional responses are absent in strains lacking WC-1. (A) Relative microarray readouts of early light-responsive genes. (B) Relative microarray readouts of late light-responsive genes in WT, Δwc-1, Δwc-2 and WT (DD). Median value is shown as a horizontal bar with the 25–75% range are indicated as a box and extreme values are indicated as an extended line. Red arrows indicate where peaks of induction are. Red dashed lines enclose the 25–75% range of the WT readout in the first DD time point to facilitate a visual comparison for light induction. (C, D) RT–QPCR analysis of light induction of NCU02765.2 and frq in WT and Δwc-1. The results obtained by three independent replicate experiments are shown. Columns represent mean values±standard error; asterisks indicate statistical significance when compared with the DD time point, as determined by paired t-test, *P<0.05, **P<0.01, ***P<0.001. A full-colour version of this figure is available at The EMBO Journal Online.
In addition to the visual inspection, the same coherent light-responsive clusters, one early and one late, were identified as statistically significant by an unbiased, scale-free bootstrapping analysis covering 53 genes (Shimodaira, 2004) (Supplementary Figure 2, see Supplementary data for details), supporting the clustering outcome as a valid estimate of regulatory similarity. To add further statistical verification to our list of light-responsive genes, we re-analysed our data with the SAM (significance analysis of microarrays) package (Tusher et al, 2001). Using 10% false discovery rate (FDR) as a cutoff, SAM identified 152 genes (Supplementary Table VI, see Materials and methods for details) that show at least a 2.2-fold increase after light treatment, whereas none of the light-repressed genes could be identified with the same FDR cutoff. Interestingly, the clustering approach identified twice as many target genes (314 versus 152) while covering 93% of the statistically significant genes and additionally found no evidence for a light-repressed subcluster. If a split in a cluster produces a nearly homogeneous pattern with regard to a unique property (i.e. light responsive) and encompasses most known targets (Supplementary Table IV, e.g. see Bell-Pedersen et al (1996); Linden et al (1997); Lewis et al (2002)) and statistically significant genes (Supplementary Table VI), such a subcluster of candidate light-induced genes should be considered interesting and valid (Murray et al, 2004).
Most but not all light-induced transcriptional responses are absent in strains lacking WC-1
Although most if not all light responses are impaired in the wc knockout strains (as shown in Figure 2A and B), residual light responses revealed in the Δwc-1 background (Figure 1B, lane 6) are interesting, suggesting that Neurospora might not be completely blind in the absence of WC-1. To validate these residual light responses, we performed quantitative RT–QPCR analysis with RNA samples from three independent biological replicates. After testing the induction of six candidate genes identified by the microarray (NCU01555.2, NCU02765.2, NCU04415.2, NCU04510.2, NCU04605.2 and NCU05490.2), we could detect a consistent, low increase of NCU02765.2 in the Δwc-1 background (Figures 2C, P=0.02, as determined by paired t-test). NCU02765.2 encodes a small (116 aa) conserved, nonessential protein of unknown functions (data not shown). However, other genes examined showed variable responses to light stimulus (data not shown). Induction of frq is shown in Figure 2D as an internal control for our sample preparation. These residual light responses appear to be relatively weak and transient in contrast to the conventional light responses but are surprising nonetheless given the central role of the WC-1 photoreceptor in Neurospora light responses.
Light-sensing machinery in Neurospora co-regulates genes with similar biological functions
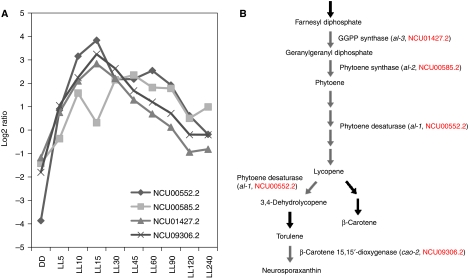
Co-regulation of transcript levels as shown by many microarray analyses is often seen among genes in a metabolic or developmental pathway and can be used to predict the functions of earlier unstudied genes (Eisen et al, 1998; Harmer et al, 2000; Kim et al, 2001; Kasuga et al, 2005). In this context, it is informative that enrichment of genes in specific developmental pathways is seen among the light-responsive genes of Neurospora. For instance, four genes having almost identical kinetics of induction (Figure 3A) encode enzymes that execute consecutive steps in the carotenoid biosynthesis pathway (Figure 3B). Particularly, NCU09306.2, previously unknown but predicted to be a gene related to β-carotene 15,15′-dioxygenase, has been shown recently to have a role in torulene cleavage, the last step of carotenoid biosynthesis in Neurospora, and named cao-2 (carotenoid oxygenase-2) (Saelices et al, 2007). This is consistent with the notion that information from co-regulated light responses can be used to predict the roles for genes of unknown function. The induction kinetics of each light-responsive gene in WT is included in Supplementary Table VII.
Figure 3.
Light-sensing machinery in Neurospora co-regulates genes with similar biological functions. (A) Microarray readouts of four early light-responsive genes involved in the carotenoid biosynthesis pathway. Representative data from the WT strain are used for plotting. (B) The enzyme pathway of carotenoid biosynthesis in Neurospora crassa. The figure is modified from The Neurospora Compendium (Perkins et al, 2001). NCU numbers of light-regulated genes are shown in red. A full-colour version of this figure is available at The EMBO Journal Online.
Approximately half of the genes identified as light responsive remain unclassified as to function. Large-scale analysis of putative functions using FunCatDB (http://mips.gsf.de/proj/funcatDB/search_main_frame.html) indicates that there is a clear correspondence between the timing of induction and the underlying nature of the biological processes (Table I), such that some functional groups of genes are highly enriched. For instance, ELRGs involved in the synthesis of photoprotective pigments (the lipid, fatty acid and isoprenoid metabolism category) comprise 7.1% of the early light-responsive genes but only 2.7% of genes in the genome. This enrichment has a probability of only 8e−03 of occurring by chance, calculated based on hypergeometric distribution (Ruepp et al, 2004). Similarly, genes involved in the synthesis of vitamins, cofactors and prosthetic groups (4.7%), secondary metabolism (4.7%), DNA processing (such as DNA repair genes, 6.3%), cellular signalling (5.5%) and several key pathways implicated in cellular sensing and response to external stimuli (such as os-2, wc-1, vvd and frq) are over-represented relative to their percentage in the genome as well. Genes highly enriched among the late light-response group are those involved in carbohydrate metabolism (20%), oxidation of fatty acids (1.9%) and components involved in oxygen/radical detoxification reaction (2.5%, including catalase-1, catalase-2, catalase-4 and superoxide dismutase). In summary, gene within distinct functional categories shows specific time-dependent regulation, illustrating a temporal sequence of important cellular events for Neurospora in response to the daily light stimulus.
Table 1.
Functional analysis of ELRGs and LLRGs using FunCatDB
| Functional categorya | This study gene matches | Genome matches (%) | P-valueb |
|---|---|---|---|
| Early light-responsive genes (ELRGs) | |||
| Lipid, fatty acid and isoprenoid metabolism | 7.1% (9/127) | 2.7 | 8e−03 |
| Biosynthesis of vitamins, cofactors, and prosthetic groups | 4.7% (6/127) | 0.8 | 4.6e−04 |
| Secondary metabolism | 4.7% (6/127) | 1.3 | 5.9e−04 |
| DNA processing | 6.3% (8/127) | 1.9 | 3e−03 |
| Cellular signalling | 5.5% (7/127) | 1.8 | 7.9e−03 |
| Osmotic and salt stress response | 1.6 (2/127) | 0.1 | 4.6e−03 |
| Photoperception and response | 1.6% (2/127) | 0.04 | 1e−03 |
| Circadian rhythm | 1.6% (2/127) | 0.1 | 6e−03 |
| Unclassified protein | 36% (49/127) | 57 | 1 |
| Late light-responsive genes (LLRGs) | |||
| C-compound and carbohydrate metabolism | 20% (31/157) | 5.4 | 1.9e−10 |
| Oxidation of fatty acids | 1.9% (3/157) | 0.2 | 1.7e−03 |
| Oxygen and radical detoxification | 2.5% (4/157) | 0.2 | 1.4e−04 |
| Unclassified protein | 50% (78/157) | 57 | 1 |
| aMIPS Functional Catalogue Database (FunCatDB, http://mips.gsf.de/proj/funcatDB/search_main_frame.html). | |||
| bFunctional categories selected to present here were based on P-value (<1e−02). | |||
SUB-1, an early light-responsive TF, is involved in regulating some early and most late light responses
Given the finding that the light-responsive TF WC-1 (Froehlich et al, 2002; He et al, 2002) is the primary component in mediating light signalling and acts with kinetics consistent with the early light responses, we predicted that it might initiate a regulatory cascade leading to the activation of late responses. Although such a cascade could take many forms, the simplest would be through a transcriptional hierarchy. Our analysis identified six light-responsive TFs, including WC-1 (Table II). We verified their light induction and WC-1 dependence by RT–QPCR analysis, using RNA samples from three independent biological replicates (Supplementary Figure 3). To investigate the function of these TFs in response to light, we used additional microarray analysis to follow the light-induced transcriptional changes in the strains bearing a gene-replacement knockout of each TF (Colot et al, 2006) (Figure 4A and B, lanes 2–6). Given that the light-responsive targets were known from the initial microarray analysis, only five sequential time points were followed for each knockout strain ranging from 0, 15, 30, 60 to 120 min after onset of light. Lane 1 in Figure 4A and B represents an independent biological replicate of WT, serving as an internal control for the microarray measurements.
Table 2.
Light-responsive transcription factors in Neurospora crassa
| NCU no. | Class | Comment | Ratioa |
|---|---|---|---|
| NCU 02356.2 | GATA zinc finger TF | wc-1 | 2/6 |
| NCU 01154.2 | GATA zinc finger TF | sub-1 | |
| NCU 02713.2 | C2H2 zinc finger TF | csp-1b | 2/43 |
| NCU 04179.2 | C2H2 zinc finger TF | sah-1 | |
| NCU 06407.2 | Zn(II)2Cys6 fungal binuclear TF | vad-3 | 2/77 |
| NCU 03643.2 | Zn(II)2Cys6 fungal binuclear TF | Cutinase TF-1β | |
| aThe ratio indicates the number of light-responsive transcription factors versus total number of transcription factors within the same family ([10]). | |||
| bThe identity of csp-1 was recently reported (Lambreghts et al, 2009). | |||
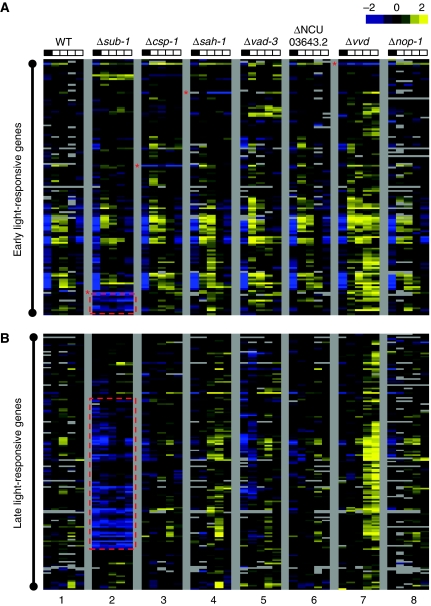
Figure 4.
SUB-1, an early light-responsive TF, is involved in regulating some early and most late light responses. (A) Comparison of early light-responsive genes. (B) Comparison of late light-responsive genes. Lane 1, 74A (WT); Lane 2, Δsub-1 (NCU01154.2); Lane 3, Δcsp-1 (NCU02713.2); Lane 4, Δsah-1 (NCU04179.2); Lane 5, Δvad-3 (NCU06407.2); Lane 6, ΔNCU03643.2; Lane7, Δvvd; Lane 8, Δnop-1. For each lane, from left to right, the individual columns correspond to light treatment for 0, 15, 30, 60, 120 min, respectively. For each row, the data were centred across different columns before clustering. Square colours are as described in Figure 1. Red asterisk to the left indicates the row corresponding to the knocked out gene in the respective knockout strain. Vertical red dashes identify clusters of genes whose light induction is abrogated in the Δsub-1 strains.
Interestingly, as shown in Figure 4A and B, the regulation of most ELRGs and LLRGs appears to be more or less unchanged in the different TF knockout strains with the exception of Δsub-1 (lane 2). In the Δsub-1 strain, some early and most late light responses are severely impaired across sequential time points, suggesting that light-responsive SUB-1 might be involved in regulating light responses as WC-1. For other TF knockout strains, the variations in their late light responses (Figure 4B, lanes 3–6) did not signal a consistent pattern in the same manner as Δsub-1 as showed by SAM analysis (Supplementary Figure 6).
Identification of a full set of bona fide light-responsive genes also provides a unique opportunity to re-explore the function of another putative photoreceptor, NOP-1 (Bieszke et al, 2007), at the transcriptional level. As shown in Figure 4A and B (lanes 8), there appeared to be no systematic changes in the regulation of ELRGs and LLRGs, suggesting that the biological function of NOP-1 might not be in the modulation of light-induced transcriptional changes, and thus remains to be determined. Interestingly but not surprisingly, we confirmed the photoadaptation defects in the Δvvd strain (Heintzen et al, 2001; Schwerdtfeger and Linden, 2001, 2003; Shrode et al, 2001) for both ELRGs and LLRGs (Figure 4A and B, lane 7). Clearly, the timing of induction is not altered for either early or late light responses in Δvvd, suggesting VVD is not directly involved in the initial regulation of light responses. Instead, once transcription is turned on by the light-activated WCC, VVD can serve as a universal brake to repress both types of light responses under prolonged light exposure. The data from the Δvvd strain is not only consistent with earlier observations on a small number of light-responsive targets but also provides a completely independent validation of the genes we identified as light responsive in Figure 1.
SUB-1 is required for efficient transduction of light signals to some ELRGs and most LLRGs under WCC control
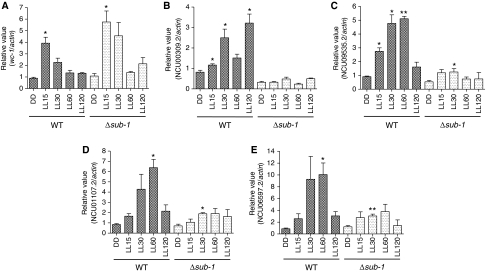
To gain further insight into the light function of SUB-1, we choose two ELRGs, NCU00309.2 and NCU009535.2, and two LLRGs, NCU01107.2 and NCU06597.2, to validate the impaired light responses in the Δsub-1 strain. The early light-responsive gene wc-1, whose light induction has no obvious difference between WT and Δsub-1 as predicted by our microarray analysis, was monitored as quality control for our light treatment and RT–QPCR analysis. As shown in Figure 5A, wc-1 is properly induced by 4–5-fold in both strains, indicating that sub-1 is not required for light activation of wc-1 transcript. In contrast, four candidate genes revealed by microarray analysis as either early or late light responsive in WT but defective in Δsub-1, all reproducibly show a marked impairment in their light responses in the absence of sub-1 (Figure 5B–E), confirming the microarray measurements. However, with the exception of NCU00309.2, a slight but significant increase of transcript in response to light could still be observed in Δsub-1 for genes NCU09535.2, NCU01107.2 and NCU06597.2 (Figure 5C–E), indicating that sub-1 is not the sole regulator of these genes, and/or may work to modulate the action of other regulator(s). Also worth mentioning, the Δsub-1 strain, unlike Δwc-1, appeared to have a normal accumulation of carotenoid in constant light (data not shown).
Figure 5.
SUB-1 is required for efficient transduction of light signals to some ELRGs and most LLRGs under WCC control. (A) RT–QPCR analysis of light induction of wc-1 in WT and Δsub-1. (B, C) RT–QPCR analysis of light induction of two early light-responsive genes, NCU00309.2 and NCU09535.2 in WT and Δsub-1. (D, E) RT–QPCR analysis of light induction of two late light-responsive genes, NCU01107.2 and NCU06597.2 in WT and Δsub-1. The results obtained by three independent biological replicate experiments are shown here. Columns represent mean values±standard error; asterisks indicate statistical significance when compared with DD time point, as determined by paired t-test, *P<0.05, **P<0.01.
The circadian clock is not altered in the absence of sub-1
For many genes, light responses and circadian regulation or function are linked. To explore possible clock functions of these light-responsive TFs, we introduced the dominant RAS allele, ras-1bd (Belden et al, 2007a), into the individual TF knockout strains by genetic crossing. As shown in Supplementary Figure 4, the only arrhythmic strain is the knockout of wc-1, in agreement with earlier data (Crosthwaite et al, 1997). Other TF knockout strains all display a largely normal clock phenotype (n⩾5), indicating that these TFs are not required for the circadian clock, although there may be some small perturbations in clock period and phase.
An LLRE contributes to mediating late light responses in vivo
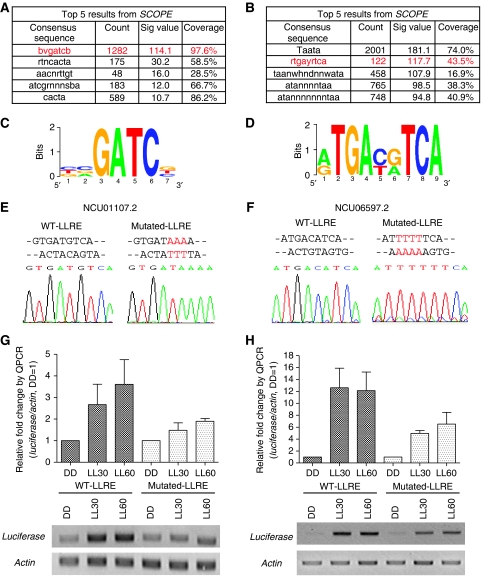
To search for distinct cis-acting regulatory motifs important for the induction of light responses, we used SCOPE (Carlson et al, 2007) to screen the entire intergenic regions for both early and late light-responsive genes. Briefly, SCOPE uses three algorithms to identify sequence motifs: BEAM (Carlson et al, 2006a) finds nondegenerate motifs (e.g. ACGCGT), PRISM (Carlson et al, 2006b) finds degenerate motifs (e.g. ACSCGW) and SPACER (Chakravarty et al, 2007) finds bipartite motifs (e.g. ACGnnnnnnnCGW). The results from all three algorithms are merged and the best scoring motifs are presented in rank order. Sig value in SCOPE is a measure of how likely it is that the consensus sequence in question could have been over-represented as observed by chance alone. For a detailed discussion of the Sig value see van Helden et al (1998) and Chakravarty et al (2007). For both types of light responses, the top five motifs identified and ranked by SCOPE are shown in Figure 6A and B. The number one motif identified from early light responses (Figure 6C, total 1282 counts for 123 genes with 97.6% coverage, Sig value=114.1) is related to the core consensus sequence of the LRE that is bound by the WCC and has been shown to be necessary and sufficient for mediating light induction of the clock gene frq in vivo (Froehlich et al, 2002). Although SCOPE identified the core sequence, it is likely that this core is paired with another in close proximity to be effective, as seen in the frq promoter. This result supports the accuracy of motif prediction by SCOPE, although it is clear that additional context or sequence information, not detected by SCOPE, is needed to govern ELRGs. The distribution of this ELRE core motif among all early light-responsive genes is shown in Supplementary Figure 5A. In terms of regulating the late light responses, one of the top putative LLREs seemed particularly interesting (Figure 6D, ranked as number two in Figure 6B). Although this element is associated with only 154 genes (43.5% coverage), its relatively constant position (mostly within 500 bp upstream from the promoter of late light-responsive genes, Supplementary Figure 5B) and infrequent occurrence (∼1.8 counts per gene) suggested functionality. For other over-represented putative ELREs and LLREs (Figure 6A and B), the detailed information can be obtained, along with the list of early and late light-responsive genes (Supplementary Tables II and III) at SCOPE website: http://genie.dartmouth.edu/scope/.
Figure 6.
An LLRE contributes to mediating late light responses in vivo. (A, B) Top 5 ELREs and LLREs as predicted by SCOPE. (C, D) Sequence logo of a selected ELRE and LLRE. (E, F) Sequencing data of the WT LLRE versus a mutated LLRE. The mutated nucleotides are shown in red. (G, H) RT–QPCR analysis of light induction from a luciferase-containing transcript using the promoter sequence from NCU01107.2 and NCU06597.2 with either WT or mutated LLRE. Upper panel: data from RT–QPCR. Lower panel: data from semiquantitative RT–PCR. The results obtained by three independent replicate experiments are shown here. Columns represent mean values±standard error.
The promoter sequences from two verified late light-responsive genes, NCU01107.2 (Figure 5D) and NCU06597.2 (Figure 5E), were used to test the light function of this putative LLRE in vivo. As revealed by SCOPE, there are two overlapping LLRE motifs in each of the promoter sequences, each being 1 bp apart on opposite strands (Figure 6E and F). The intergenic regions upstream of the NCU01107.2 (622 bp) and NCU06597.2 (1140 bp) translation start site were fused to a luciferase reporter (Gooch et al, 2008) and integrated into the his-3 locus. The reporter precisely recapitulates the endogenous late light-responsive pattern of expression as shown in Figure 6G and H (compared with their endogenous light induction in Figure 5D and E), with a peak at LL60. We used the expression of actin, which is not altered by light, as an internal reference for comparison (bottom panel, Figure 6G and H). Site-directed mutagenesis (Figure 6E and F) was used to change the sequences of the LLRE driving luciferase, and this construct was examined for function after transformation and integration at his-3. In these transformants, the amplitude of light induction driven by the promoters with mutated LLRE is decreased by about 50% compared with the promoters with WT LLRE. The data suggest that this LLRE is indeed a functional motif and is required for full induction of late light responses in vivo. However, a consistent light response was still observed from the mutated promoters, and this coupled with the fact that only 43.5% of late light-responsive genes contain this LLRE, which suggests that additional cis-regulatory motifs remain to be discovered for regulating late light responses.
Light enhances the direct binding of the WCC to the sub-1 promoter
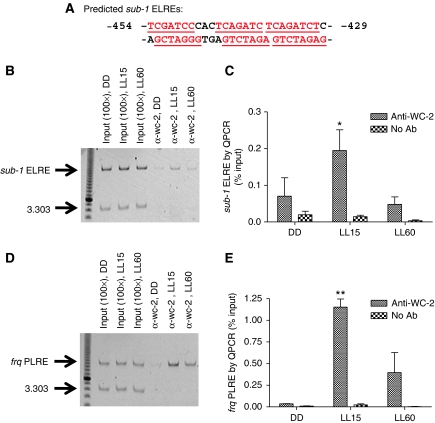
To establish a direct molecular connection between WCC and the induction of sub-1, we asked whether the WCC can directly bind to the sub-1 promoter in response to a light stimulus. As shown by an earlier study (Froehlich et al, 2002), a promoter sequence with adjacent repeats of the LRE might serve as a good indicator for where the WCC binds. As revealed by SCOPE, there are 11 putative core ELREs within 3000 bp upstream of the sub-1 translation start site, with six appearing within a 26-bp region between −454 and −429 bp upstream of the sub-1 start codon (Figure 7A). Therefore, we chose this region as a candidate for a direct binding site of the WCC.
Figure 7.
Light enhances the direct binding of the WCC to the sub-1 promoter. (A) Six adjacent ELREs on the sub-1 promoter (shown in red). (B, C) ChIP analysis of sub-1 ELRE and (D, E) frq PLRE was performed with antibodies against WC-2. 3.303 indicates a primer pair targeting a noncoding region in the Neuropsora genome (Belden et al, 2007a). No Ab indicates no-antibody control. QPCR data were obtained for three independent replicate experiments. Columns represent mean values±standard error. Asterisks indicate statistical significance when compared with the DD time point, as determined by paired t-test, *P<0.05, **P<0.01. A full-colour version of this figure is available at The EMBO Journal Online.
Chromatin immunoprecipitation (ChIP) analysis using antibodies against WC-2 has been successfully applied to study the binding of the WCC to the promoter of several ELRGs in vivo (He and Liu, 2005; Belden et al, 2007b), so ChIP was performed to examine the direct binding of WCC to the sub-1 promoter. As shown in Figure 7B (semiquantitative PCR) and Figure 7C (QPCR analysis), 15 min of light significantly enhances the direct binding of WCC to the sub-1 promoter compared with DD time point. The interaction is eliminated after 60 min in constant light, presumably reflecting the ongoing photoadaptation process. As a positive control for the light treatment and ChIP sample preparation, we confirmed the direct interaction between the WCC and the proximal LRE (PLRE) on the frq promoter in response to light (Belden et al, 2007b) (Figure 7D and E).
Discussion
The transcriptional response of Neurospora to light has been described here on a genome-wide scale. Consistent with data from many labs (Dunlap and Loros, 2004, 2006; Corrochano, 2007; Herrera-Estrella and Horwitz, 2007), the majority of light responses depend on the WCC, which initiates a cascade of subsequent activities (Figure 8), including WC-1-dependent acetylation of histone H3 associated with the early light-responsive gene, al-3 (Grimaldi et al, 2006). Once activated by light, WC-1 undergoes a light-induced conformational change and forms an L-WCC with WC-2 that activates early light responses by a direct binding to the ELRE of ELRGs, including vvd (He and Liu, 2005), wc-1 (Froehlich et al, 2002; He et al, 2002) and, as shown here, sub-1. This happens rapidly after lights-on, certainly within minutes, at least for some genes (Crosthwaite et al, 1995). Late light responses, which peak about 30 min later or more, are shown here to be regulated in part by SUB-1, an early light-responsive TF. Without SUB-1, the amplitude of light induction appears to be severely impaired for some ELRGs and most LLRGs, supporting its important role in the Neurospora light signalling cascades. Interestingly, we found that the amplitude of light responses is tightly linked to the level of FRQ as suggested in earlier study (Merrow et al, 2001). In the frq7 strain, the amplitude of both early and late light responses was noticeably boosted. FRQ has been shown to positively regulate the amount of WC-1 posttranscriptionally (Lee et al, 2000; Schafmeier et al, 2006), and these data provide additional molecular evidence that the light function of WC-1 is indeed under the control of the clock, possibly through direct interaction with FRQ. In addition, we observed photoadaptation defects in the Δvvd strain for both early and late light responses, suggesting that there might be a common underlying mechanism used by VVD to serve as a universal brake on light effects. One possible explanation is that VVD, after light activation, might temporarily inhibit the function of WCC through some posttranslational modification, presumably phosphorylation (Heintzen et al, 2001; Schwerdtfeger and Linden, 2001; He and Liu, 2005). Therefore, when VVD is absent, WCC constitutively activates ELRGs, including sub-1. In turn, increased SUB-1 together with other, unknown, activators continuously activates LLRGs, resulting in photoadaptation defects on a genome-wide scale.
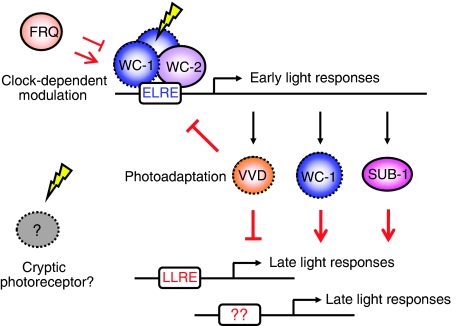
Figure 8.
Model of a hierarchical light-sensing cascade in Neurospora crassa. Our results reveal an ordered sequence of molecular events resulting in light-regulated gene expression in Neurospora. The induction of early light responses is controlled primarily by WCC through direct binding to the ELRE on the promoter of ELRGs, including vvd, wc-1 and sub-1. In turn, SUB-1 is required for efficient transduction of light signals to some ELRGs and most LLRGs. Meanwhile, in contrast to the ELRE, a novel LLRE is shown to be required for the full induction of late light responses in vivo. In addition to the modulation of induction amplitude by the clock protein FRQ, both ELRGs and LLRGs in constant light are subject to the repression mediated by VVD. Intriguingly, some residual light responses can still be observed in the absence of WC-1. Components capable of sensing light directly through a chromophore are marked with dashed lines in the figure.
These data also confirm the existence of residual, albeit low amplitude, light responses in the absence of WC-1 in Neurospora. This has been a topic of some confusion with an initial result later linked to the use of wc strains that were still partially functional mutants (Dragovic et al, 2002; Heintzen and Liu, 2007). Studies using gene replacements of each wc gene (Collett et al, 2002; Lee et al, 2003) showed complete loss of light-induced gene expression for specific individual genes. This genome-wide microarray analysis has confirmed the total loss of light responsivity in earlier studied genes but did identify a few novel genes of unknown function whose expression responded to light in strains lacking WC-1 but not in WC-2. We are pursuing the nature of this response, including the determination of whether it is actually mediated by a photoreceptor as distinct from a physical process. But, notably, in the absence of a WC-1 homologue, residual light responses have been reported in Trichoderma (Rosales-Saavedra et al, 2006) and are a possible explanation, although not the only one, for light responses in Phycomyces (Idnurm et al, 2006). Consistent with the findings here, these residual light responses appear to be comparatively weak in most cases. Therefore, the physiological significance of these residual light responses remains to be determined. It can be noted that, even without a specified photoreceptor, endogenous reactive oxygen species generated by a photon stimulus might serve as an alternative input to pass on light signals to downstream targets (Schroeder and Johnson, 1995; Bouvier et al, 1998; Yoshida and Hasunuma, 2004; Iigusa et al, 2005). Such an ancillary pathway might become more influential when the primary light-sensing machinery is absent.
A novel and important factor in light responses, SUB-1, was identified in this study. Preliminary data (not shown) suggested that simple expression of SUB-1 by itself in the dark was not sufficient to induce late light responses, suggesting that SUB-1 was activated or modified in a light-dependent manner, or was interacting with a light-regulated factor(s), leading to downstream activation of late light responses. As shown here, even without SUB-1, other factors can still activate several LLRGs although with a much lower amplitude of induction. Given the fact that late light responses share many regulatory features with early light responses (e.g. modulation by both FRQ and VVD; complete dependence upon functional WCC), WCC becomes a likely candidate for the missing factor. However, attempts to show a physical interaction between SUB-1 and WCC by co-immunoprecipitation were not successful (data not shown), indicating their interaction is weak or transient or direct physical interaction is not required for mediating late light responses. Both the substantial reduction in gene activation in response to light when the LLRE is mutated and severe loss of the late light response in Δsub-1 strains mutually support SUB-1 as a light-responsive TF interacting with this LLRE. Nevertheless, attempts to show a direct interaction in vivo using a ChIP assay with V5-tagged SUB-1 were inconclusive. Clearly, both SUB-1 and the LLRE contribute to the late light response, although the entire relationship between SUB-1 and the LLRE is not yet fully understood.
It should be noted that the culture conditions and experimental design may have altered the set of light-responsive genes identified. For instance, we failed to identify two known light-inducible genes, ccg-1 (Arpaia et al, 1995) and fluffy (Belden et al, 2007a), as well as a few light-responsive targets reported in a separate microarray study that used 2% sucrose as the major carbon source (Lewis et al, 2002). The function of CCG-1 is still unknown, but its expression is strongly repressed in the presence of glucose (McNally and Free, 1988; Xie et al, 2004). Therefore, it is not surprising that the light induction of ccg-1 was impaired as well under our high glucose condition (2% glucose). However, because most light responsive genes actually have a higher amplitude of induction under high glucose condition (2% compared with 0.1% glucose medium, data not shown), medium with 2% glucose was used for this study to enhance the sensitivity of the microarray detection. Meanwhile, several light-responsive genes have also been shown to have altered induction in the presence of the ras-1bd allele that has commonly been present in strains analysed for photoresponses (Dunlap and Loros, 2005; Liu and Bell-Pedersen, 2006). For instance, light induction of the TF fluffy is significantly reduced in the WT strain 74A (Belden et al, 2007a) when compared with strains carrying the dominant RAS allele ras-1bd. To reflect the native light responses in Neurospora, we used strains without the ras-1bd mutation in this study. A recent study focusing on genetic network models of the circadian clock in Neurospora (Dong et al, 2008) report light-responsive genes from microarray analysis using a ras-1bd strain grown in galactose/Fries-based medium. Response to light was followed between 20 min and 24 h in constant light although total growth time was not controlled. After 24 h of light treatment, 60% of their total detectable transcripts were reported to be potentially light responsive. The involvement of RAS signalling pathways in both light input and clock output further highlights the complexity underlying the regulatory networks of the Neurospora light-sensing cascade.
SUB-1 is a GATA type zinc finger TF, first identified as a mutant resulting in the development of submerged protoperithecia (Colot et al, 2006). There are only six GATA zinc finger family members (out of 176 TFs) in the Neurospora genome (Borkovich et al, 2004). In addition to SUB-1, WC-1 and WC-2 are members in this family and are both essential components in controlling Neurospora light responses. Therefore, although some biological functions have been associated with the other three GATA members (Fu and Marzluf, 1990; Zhou and Marzluf, 1999; Feng et al, 2000) and none were light induced, their involvement with light responses might be worth of re-evaluating. Notably, unlike other zinc finger family members, GATA factors are found exclusively in eukaryotic organisms, which might provide insight into the evolution and conservation of light signalling components in the fungal and possibly other kingdoms.
Are there additional photoreceptors or light-inducible TFs beyond WC-1, VVD and SUB-1 involved in controlling transcriptional responses to light in Neurospora? On the basis of our profiling data, the influence from additional TFs and putative photoreceptors appears to be minor at most. Their biological function in response to the light stimulus might either not relate to the modulation of transcription or could become evident at specific developmental stages or under different growth conditions. In a recent study, the nop-1 gene was reported not to be light responsive, as additionally found here, but to have a regulatory function late in development (Bieszke et al, 2007). Thus, in the case of nop-1, sample collection time was critical in uncovering function. This principle may hold true for the light responsive TFs identified in this study.
Materials and methods
Strains
The WT strain used here is OR74A. All single-knockout strains came from the Neurospora knockout project (Colot et al, 2006) and have been deposited in the Fungal Genetics Stock Center, Kansas City (www.fgsc.net). Double-knockout strains, 559-4 (wc-1∷hph; wc-2∷hph) and 560-8 (phy-1∷hph; phy-2∷hph), were generated by crossing single-knockout strains and confirmed by Southern blot. 521-2 (WT, frq7) was generated by crossing 585-70 (bd, frq7) with 74A to remove the band (ras-1bd) mutation (Belden et al, 2007a). Strain 324-8 (ras-1bd) was used for the ChIP assay. NCU strain numbers are from Neurospora annotation (http://www.broad.mit.edu/annotation/genome/neurospora/Home.html).
Culture conditions and light treatment
Frozen conidia were inoculated onto a minimal slant (Davis and De Serres, 1970) one week in advance to generate fresh conidia. On day 0, conidia were suspended in sterile water for quantification. To form a mycelial layer, 1 × 107 of conidia or similar amount of mass (e.g. no asexual sporulation in Δcsp-1) were inoculated into a 10-cm Petri dish with 20 ml Bird medium (Metzenberg, 2004) containing 2% glucose. After 24 h of incubation in darkness at 25°C, a mycelia plug was cut with a No. 4 cork borer (8 mm diameter) and transferred into a 125-ml flask with 50 ml Bird medium containing 2% glucose. All procedures were performed under a low red light environment to avoid any possible light-stimulating effects (Aronson et al, 1994).
After another 24 h of culture with constant shaking (125 r.p.m.) in darkness (DD) at 25°C, the flasks were moved to a shaker at 25°C with a continuous white light stimulus (LL), covering a wide range of the spectrum from 400 to 700 nm (cool white fluorescent light bulb, General Electric F20T12-CW, 40–50 μmole photons/m2/s), and then harvested before (DD) and after 5 (LL5), 10 (LL10), 15 (LL15), 30 (LL30), 45 (LL45), 60 (LL60), 90 (LL90), 120 (LL120) or 240 (LL240) minutes of white light treatment. Using vacuum filtration, mycelia were harvested and immediately frozen in liquid nitrogen and stored at −80°C until RNA extraction.
Microarray probe sequence and layout format
Detailed information can be found online at Home of the Filamentous Fungal Microarray Database (http://www.yale.edu/townsend/Links/ffdatabase/downloads.html).
Image and data analysis
The slides were scanned with a GMS 418 microarray scanner (Affymetrix). After griding with ScanAlyze 2.51, the raw data were uploaded and normalized with GeneTraffic software (Stratagene). Spots of insufficient quality were flagged by visual inspection and signal to background noise ratio. Because of the fact that some light-inducible genes have almost no transcription in the dark, only spots with a fluorescent intensity more than one-fold (at least over 200) in the sample channel and more than two-fold (at least over 400) in the reference channel over local/average background were selected for further analysis. Genes missing more than 25% of their measurements from 90 microarrays were all excluded. Under the culture conditions used, we obtained reliable measurements for 5588 spots equivalent to about 54% of the genes in Neurospora. Genes with absolute values of a log-2 ratio greater than 0.5 on at least two arrays while having more than four-fold changes (max–min) across 90 microarrays were all included for final clustering and SAM analysis. In total, 2864 out of 10 372 spots passed these criteria. Unsupervised hierarchical clustering was done using Cluster 3.0 (Eisen et al, 1998) with the method of average linkage after centering with mean value. Graphical representations of clustering results were generated using TreeView 1.0.13. Complete datasets of all 135 arrays are available on Gene Expression Omnibus (reference number for access: GSE8932, http://www.ncbi.nlm.nih.gov/projects/geo).
Sample preparation for microarray, microarray hybridization, quantitative RT–PCR, semiquantitative RT–PCR, site-directed mutagenesis, ChIP assay, bootstrapping and SAM analysis
These methods are described in Supplementary data available at The EMBO Journal Online (http://www.embojournal.org).
Supplementary Material
Supplementary Figures 1–6
Supplementary Figure Legends
Supplementary Tables
Supplementary data
Acknowledgments
We gratefully acknowledge Michael Whitfield, Lacy George and David A Jewell for microarray and statistical analysis, Luis Larrondo for kindly providing plasmid pLL07, Bill Belden for technical assistance with the ChIP assays, Randy Lambreghts for critical reading of the manuscript and the Fungal Genetics Stock Center, Kansas City. This work was supported by grants from the National Institutes of Health to JJL and JCD (RO1 GM08336), to JCD (RO1 GM34985 and PO1GM68087), and by the core grant to the Norris Cotton Cancer Center at Dartmouth.
References
- Aronson B, Johnson K, Loros JJ, Dunlap JC (1994) Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science 263: 1578–1584 [DOI] [PubMed] [Google Scholar]
- Arpaia G, Loros JJ, Dunlap JC, Morelli G, Macino G (1995) Light induction of the clock-controlled gene ccg-1 is not transduced through the circadian clock in Neurospora crassa. Mol Gen Genet 247: 157–163 [DOI] [PubMed] [Google Scholar]
- Bahn YS, Xue C, Idnurm A, Rutherford JC, Heitman J, Cardenas ME (2007) Sensing the environment: lessons from fungi. Nat Rev Microbiol 5: 57–69 [DOI] [PubMed] [Google Scholar]
- Belden WJ, Larrondo LF, Froehlich AC, Shi M, Chen CH, Loros JJ, Dunlap JC (2007a) The band mutation in Neurospora crassa is a dominant allele of ras-1 implicating RAS signaling in circadian output. Genes Dev 21: 1494–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belden WJ, Loros JJ, Dunlap JC (2007b) Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol Cell 25: 587–600 [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D, Shinohara ML, Loros JJ, Dunlap JC (1996) Circadian clock-controlled genes isolated from Neurospora crassa are late night- to early morning-specific. Proc Natl Acad Sci USA 93: 13096–13101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieszke JA, Braun EL, Bean LE, Kang S, Natvig DO, Borkovich KA (1999a) The nop-1 gene of Neurospora crassa encodes a seven transmembrane helix retinal-binding protein homologous to archaeal rhodopsins. Proc Natl Acad Sci USA 96: 8034–8039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieszke JA, Li L, Borkovich KA (2007) The fungal opsin gene nop-1 is negatively-regulated by a component of the blue light sensing pathway and influences conidiation-specific gene expression in Neurospora crassa. Curr Genet 52: 149–157 [DOI] [PubMed] [Google Scholar]
- Bieszke JA, Spudich EN, Scott KL, Borkovich KA, Spudich JL (1999b) A eukaryotic protein, NOP-1, binds retinal to form an archaeal rhodopsin-like photochemically reactive pigment. Biochemistry 38: 14138–14145 [DOI] [PubMed] [Google Scholar]
- Borkovich KA, Alex LA, Yarden O, Freitag M, Turner GE, Read ND, Seiler S, Bell-Pedersen D, Paietta J, Plesofsky N, Plamann M, Goodrich-Tanrikulu M, Schulte U, Mannhaupt G, Nargang FE, Radford A, Selitrennikoff C, Galagan JE, Dunlap JC, Loros JJ et al. (2004) Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol Mol Biol Rev 68: 1–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier F, Backhaus RA, Camara B (1998) Induction and control of chromoplast-specific carotenoid genes by oxidative stress. J Biol Chem 273: 30651–30659 [DOI] [PubMed] [Google Scholar]
- Carlson JM, Chakravarty A, Deziel CE, Gross RH (2007) SCOPE: a web server for practical de novo motif discovery. Nucleic Acids Res 35 (Web Server issue): W259–W264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson JM, Chakravarty A, Gross RH (2006a) BEAM: a beam search algorithm for the identification of cis-regulatory elements in groups of genes. J Comput Biol 13: 686–701 [DOI] [PubMed] [Google Scholar]
- Carlson JM, Chakravarty A, Khetani RS, Gross RH (2006b) Bounded search for de novo identification of degenerate cis-regulatory elements. BMC Bioinformatics 7: 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty A, Carlson JM, Khetani RS, DeZiel CE, Gross RH (2007) SPACER: identification of cis-regulatory elements with non-contiguous critical residues. Bioinformatics 23: 1029–1031 [DOI] [PubMed] [Google Scholar]
- Cheng P, Yang Y, Wang L, He Q, Liu Y (2003) WHITE COLLAR-1, a multifunctional Neurospora protein involved in the circadian feedback loops, light sensing, and transcription repression of wc-2. J Biol Chem 278: 3801–3808 [DOI] [PubMed] [Google Scholar]
- Collett MA, Garceau N, Dunlap JC, Loros JJ (2002) Light and clock expression of the Neurospora clock gene frequency is differentially driven by but dependent on WHITE COLLAR-2. Genetics 160: 149–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA 103: 10352–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrochano LM (2007) Fungal photoreceptors: sensory molecules for fungal development and behaviour. Photochem Photobiol Sci 6: 725–736 [DOI] [PubMed] [Google Scholar]
- Crosthwaite SK, Dunlap JC, Loros JJ (1997) Neurospora wc-1 and wc-2: transcription, photoresponses, and the origins of circadian rhythmicity. Science 276: 763–769 [DOI] [PubMed] [Google Scholar]
- Crosthwaite SK, Loros JJ, Dunlap JC (1995) Light-induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript. Cell 81: 1003–1012 [DOI] [PubMed] [Google Scholar]
- Davis RH, De Serres JF (1970) Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol 17: 79–143 [Google Scholar]
- Dong W, Tang X, Yu Y, Nilsen R, Kim R, Griffith J, Arnold J, Schuttler HB (2008) Systems biology of the clock in Neurospora crassa. PLoS ONE 3: e3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragovic Z, Tan Y, Gorl M, Roenneberg T, Merrow M (2002) Light reception and circadian behavior in ‘blind' and ‘clock-less' mutants of Neurospora crassa. EMBO J 21: 3643–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC, Borkovich KA, Henn MR, Turner GE, Sachs MS, Glass NL, McCluskey K, Plamann M, Galagan JE, Birren BW, Weiss RL, Townsend JP, Loros JJ, Nelson MA, Lambreghts R, Colot HV, Park G, Collopy P, Ringelberg C, Crew C et al. (2007) Enabling a community to dissect an organism: overview of the Neurospora functional genomics project. Adv Genet 57: 49–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ (2004) The Neurospora circadian system. J Biol Rhythms 19: 414–424 [DOI] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ (2005) Analysis of circadian rhythms in Neurospora: overview of assays and genetic and molecular biological manipulation. Methods Enzymol 393: 3–22 [DOI] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ (2006) How fungi keep time: circadian system in Neurospora and other fungi. Curr Opin Microbiol 9: 579–587 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Haas H, Marzluf GA (2000) ASD4, a new GATA factor of Neurospora crassa, displays sequence-specific DNA binding and functions in ascus and ascospore development. Biochemistry 39: 11065–11073 [DOI] [PubMed] [Google Scholar]
- Froehlich AC, Liu Y, Loros JJ, Dunlap JC (2002) White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science 297: 815–819 [DOI] [PubMed] [Google Scholar]
- Froehlich AC, Noh B, Vierstra RD, Loros J, Dunlap JC (2005) Genetic and molecular analysis of phytochromes from the filamentous fungus Neurospora crassa. Eukaryot Cell 4: 2140–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YH, Marzluf GA (1990) nit-2, the major nitrogen regulatory gene of Neurospora crassa, encodes a protein with a putative zinc finger DNA-binding domain. Mol Cell Biol 10: 1056–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani Y, Bezerra AG Jr, Waschuk S, Sumii M, Brown LS, Kandori H (2004) FTIR spectroscopy of the K photointermediate of Neurospora rhodopsin: structural changes of the retinal, protein, and water molecules after photoisomerization. Biochemistry 43: 9636–9646 [DOI] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, FitzHugh W, Ma LJ, Smirnov S, Purcell S, Rehman B, Elkins T, Engels R, Wang S, Nielsen CB, Butler J, Endrizzi M, Qui D, Ianakiev P, Bell-Pedersen D et al. (2003) The genome sequence of the filamentous fungus Neurospora crassa. Nature 422: 859–868 [DOI] [PubMed] [Google Scholar]
- Gooch VD, Mehra A, Larrondo LF, Fox J, Touroutoutoudis M, Loros JJ, Dunlap JC (2008) Fully codon-optimized luciferase uncovers novel temperature characteristics of the Neurospora clock. Eukaryot Cell 7: 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi B, Coiro P, Filetici P, Berge E, Dobosy JR, Freitag M, Selker EU, Ballario P (2006) The Neurospora crassa White Collar-1 dependent blue light response requires acetylation of histone H3 lysine 14 by NGF-1. Mol Biol Cell 17: 4576–4583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- He Q, Cheng P, Yang Y, Wang L, Gardner KH, Liu Y (2002) White collar-1, a DNA binding transcription factor and a light sensor. Science 297: 840–843 [DOI] [PubMed] [Google Scholar]
- He Q, Liu Y (2005) Molecular mechanism of light responses in Neurospora: from light-induced transcription to photoadaptation. Genes Dev 19: 2888–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzen C, Liu Y (2007) The Neurospora crassa circadian clock. Adv Genet 58: 25–66 [DOI] [PubMed] [Google Scholar]
- Heintzen C, Loros JJ, Dunlap JC (2001) The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell 104: 453–464 [DOI] [PubMed] [Google Scholar]
- Herrera-Estrella A, Horwitz BA (2007) Looking through the eyes of fungi: molecular genetics of photoreception. Mol Microbiol 64: 5–15 [DOI] [PubMed] [Google Scholar]
- Idnurm A, Rodriguez-Romero J, Corrochano LM, Sanz C, Iturriaga EA, Eslava AP, Heitman J (2006) The Phycomyces madA gene encodes a blue-light photoreceptor for phototropism and other light responses. Proc Natl Acad Sci USA 103: 4546–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iigusa H, Yoshida Y, Hasunuma K (2005) Oxygen and hydrogen peroxide enhance light-induced carotenoid synthesis in Neurospora crassa. FEBS Lett 579: 4012–4016 [DOI] [PubMed] [Google Scholar]
- Kasuga T, Townsend JP, Tian C, Gilbert LB, Mannhaupt G, Taylor JW, Glass NL (2005) Long-oligomer microarray profiling in Neurospora crassa reveals the transcriptional program underlying biochemical and physiological events of conidial germination. Nucleic Acids Res 33: 6469–6485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Lund J, Kiraly M, Duke K, Jiang M, Stuart JM, Eizinger A, Wylie BN, Davidson GS (2001) A gene expression map for Caenorhabditis elegans. Science 293: 2087–2092 [DOI] [PubMed] [Google Scholar]
- Lambreghts R, Shi M, Belden WJ, deCaprio D, Park D, Henn MR, Galagan JE, Basturkmen M, Birren B, Sachs MS, Dunlap JC, Loros JJ (2009) A high-density SNP map for Neurospora crassa. Genetics 181: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Dunlap JC, Loros JJ (2003) Roles for WHITE COLLAR-1 in circadian and general photoperception in Neurospora crassa. Genetics 163: 103–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Loros JJ, Dunlap JC (2000) Interconnected feedback loops in the Neurospora circadian system. Science 289: 107–110 [DOI] [PubMed] [Google Scholar]
- Lewis ZA, Correa A, Schwerdtfeger C, Link KL, Xie X, Gomer RH, Thomas T, Ebbole DJ, Bell-Pedersen D (2002) Overexpression of White Collar-1 (WC-1) activates circadian clock-associated genes, but is not sufficient to induce most light-regulated gene expression in Neurospora crassa. Mol Microbiol 45: 917–931 [DOI] [PubMed] [Google Scholar]
- Linden H, Ballario P, Macino G (1997) Blue light regulation in Neurospora crassa. Fungal Genet Biol 22: 141–150 [DOI] [PubMed] [Google Scholar]
- Liu Y, Bell-Pedersen D (2006) Circadian rhythms in Neurospora crassa and other filamentous fungi. Eukaryot Cell 5: 1184–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally MT, Free SJ (1988) Isolation and characterization of a Neurospora glucose-repressible gene. Curr Genet 14: 545–551 [DOI] [PubMed] [Google Scholar]
- Merrow M, Franchi L, Dragovic Z, Gorl M, Johnson J, Brunner M, Macino G, Roenneberg T (2001) Circadian regulation of the light input pathway in Neurospora crassa. EMBO J 20: 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzenberg RL (2004) Bird medium: an alternative to Vogel Medium. Fungal Genet Newsl 51: 19–20 [Google Scholar]
- Murray JI, Whitfield ML, Trinklein ND, Myers RM, Brown PO, Botstein D (2004) Diverse and specific gene expression responses to stresses in cultured human cells. Mol Biol Cell 15: 2361–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D, Radford A, Sachs M (2001) The Neurospora Compendium Chromosomal Loci. San Diego:Academic Press [Google Scholar]
- Purschwitz J, Muller S, Kastner C, Fischer R (2006) Seeing the rainbow: light sensing in fungi. Curr Opin Microbiol 9: 566–571 [DOI] [PubMed] [Google Scholar]
- Rosales-Saavedra T, Esquivel-Naranjo EU, Casas-Flores S, Martinez-Hernandez P, Ibarra-Laclette E, Cortes-Penagos C, Herrera-Estrella A (2006) Novel light-regulated genes in Trichoderma atroviride: a dissection by cDNA microarrays. Microbiology 152: 3305–3317 [DOI] [PubMed] [Google Scholar]
- Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, Tetko I, Guldener U, Mannhaupt G, Munsterkotter M, Mewes HW (2004) The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res 32: 5539–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saelices L, Youssar L, Holdermann I, Al-Babili S, Avalos J (2007) Identification of the gene responsible for torulene cleavage in the Neurospora carotenoid pathway. Mol Genet Genomics 278: 527–537 [DOI] [PubMed] [Google Scholar]
- Schafmeier T, Kaldi K, Diernfellner A, Mohr C, Brunner M (2006) Phosphorylation-dependent maturation of Neurospora circadian clock protein from a nuclear repressor toward a cytoplasmic activator. Genes Dev 20: 297–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder WA, Johnson EA (1995) Singlet oxygen and peroxyl radicals regulate carotenoid biosynthesis in Phaffia rhodozyma. J Biol Chem 270: 18374–18379 [DOI] [PubMed] [Google Scholar]
- Schwerdtfeger C, Linden H (2001) Blue light adaptation and desensitization of light signal transduction in Neurospora crassa. Mol Microbiol 39: 1080–1087 [DOI] [PubMed] [Google Scholar]
- Schwerdtfeger C, Linden H (2003) VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J 22: 4846–4855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H (2004) Approximately unbiased tests of regions using multistep-multiscale bootstrap resampling. Ann Stat 32: 2616–2641 [Google Scholar]
- Shrode LB, Lewis ZA, White LD, Bell-Pedersen D, Ebbole DJ (2001) vvd is required for light adaptation of conidiation-specific genes of Neurospora crassa, but not circadian conidiation. Fungal Genet Biol 32: 169–181 [DOI] [PubMed] [Google Scholar]
- Sommer T, Chambers JA, Eberle J, Lauter FR, Russo VE (1989) Fast light-regulated genes of Neurospora crassa. Nucleic Acids Res 17: 5713–5723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Kasuga T, Sachs MS, Glass NL (2007) Transcriptional profiling of cross pathway control in Neurospora crassa and comparative analysis of the Gcn4 and CPC1 regulons. Eukaryot Cell 6: 1018–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden J, Andre B, Collado-Vides J (1998) Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J Mol Biol 281: 827–842 [DOI] [PubMed] [Google Scholar]
- Xie X, Wilkinson HH, Correa A, Lewis ZA, Bell-Pedersen D, Ebbole DJ (2004) Transcriptional response to glucose starvation and functional analysis of a glucose transporter of Neurospora crassa. Fungal Genet Biol 41: 1104–1119 [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Hasunuma K (2004) Reactive oxygen species affect photomorphogenesis in Neurospora crassa. J Biol Chem 279: 6986–6993 [DOI] [PubMed] [Google Scholar]
- Zhou L, Marzluf GA (1999) Functional analysis of the two zinc fingers of SRE, a GATA-type factor that negatively regulates siderophore synthesis in Neurospora crassa. Biochemistry 38: 4335–4341 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1–6
Supplementary Figure Legends
Supplementary Tables
Supplementary data