Abstract
We propose here KineticDB, a systematically compiled database of protein folding kinetics, which contains about 90 unique proteins. The main goal of the KineticDB is to provide users with a diverse set of protein folding rates determined experimentally. The search for determinants of protein folding is still in progress, aimed at obtaining a new understanding of the folding process. Comparison with experimental protein folding rates has been the main tool for validation of both theoretical models and empirical relationships during the last 10 years. It is, therefore, necessary to provide a researcher with as much data as possible in a simple and easy-to-use way. At present, the KineticDB contains the results of folding kinetics measurements of single-domain proteins and separate protein domains as well as short peptides without disulfide bonds. It includes data on about 90 unique proteins and many mutants that have been systematically accumulated over the last 10 years and is the largest collection of protein folding kinetic data presented as a database. The KineticDB is available at http://kineticdb.protres.ru/db/index.pl.
INTRODUCTION
The problem of protein folding is one of the most fundamental in molecular biology. The progress in understanding the protein folding helps predicting protein 3D structures (1), resulting recently in the designing of principally novel proteins (2,3). The ever-increasing computer potential gives an opportunity to perform molecular dynamics simulations for the folding of small proteins (4,5). Also, in the last decade, the understanding of protein folding processes has resulted in the development of first crude models of protein folding provided the protein 3D structure is known (6–11). The relevance of protein folding models is often tested as the ability to predict protein folding rates (8–11), although reproducing other features of protein folding such as the ‘all-or-none’ transition or the folding nucleus is also important. Simultaneously, a number of empirical and bioinformational methods has been developed, which provide additional information on protein folding determinants as well as allowed predicting protein folding rates from tertiary, secondary or primary protein structure (12–15). Prediction of protein folding rates is of special value because aggregation directly depends on the rate of protein folding.
The validation of predictions using experimental data was first undertaken in the empirical study of Plaxco and coworkers (12). At the same time, Jackson published her seminal review (16) that reports folding kinetics data of all proteins studied by that moment. Since then the test for correlation of predicted values with experimental results has become widely used in theoretical studies of protein folding (8–11,17). However, updating the initial dataset collected by Jackson was a rather hard job since experimental papers most often described one experiment per paper and there was no protocol for presenting folding kinetic results. Such a protocol was suggested only in 2005 by Maxwell and coworkers (18), where the folding kinetics data for 30 proteins having no evident folding intermediates were collected at standard conditions. Simultaneously, the Protein Folding Database (PFD) was developed (19,20). It has a well-developed interface and systematically collected experimental data on protein folding kinetic studies. Also, it has a form for depositing researcher's own folding kinetics data. At the moment, the PFD contains folding kinetics data of about 40 unique proteins and many mutants.
In this article, we present our KineticDB with folding kinetics data of about 90 unique proteins, which is available at http://kineticdb.protres.ru/db/index.pl. The current version of the KineticDB contains single-domain proteins, separate protein domains and short peptides without disulfide bonds in their native structure. The KineticDB is the result of our 10-year manual collection of protein folding kinetic data from literature used in our theoretical research. The dataset underlying the KineticDB database has proved to be useful for a number of theoretical, empirical and bioinformational studies of protein folding (15,21–24). The KineticDB is a valuable additional resource alternative to the PFD.
DESCRIPTION OF KINETICDB
The KineticDB is a relational database realized using MySQL and a number of Perl scripts. Each record of the KineticDB relates to a single protein folding kinetics measurement extracted from the original paper and gives details of the experimentally studied protein, its best available tertiary structure, experimental conditions, reference to the original paper and experimental results.
Details of the experimentally studied protein include the full name of the protein, its acronym, its source organism (‘synthetic’ for de novo designed proteins), the protein sequence and its length, the initial and end positions related to the whole sequence if a fragment of the protein was used for experimental studies.
Details of the best available structure corresponding to an experimentally studied protein include the code of the file with the structure according to the Protein Data Bank (26), the corresponding chain identifier inside the file, the identifiers of the start and end residues of the fragment corresponding to the experimentally studied protein, the sequence of the fragment, its length and mutation with respect to the wild-type sequence, and the identifier of the fragment according to the Structural Classification of Proteins (27). In addition, the method of structure resolution with the resolution value (in the case of X-ray structure) and with the number of models (in the case of structure determined by the method of nuclear magnetic resonance) is also included. For some proteins, there is no exact match in the Protein Data Bank to the protein studied experimentally. In this case, the structure of the closest homolog is given. Though, in the case when there is no structure of a close homolog, nothing is given at all. We understand that the choice of the best available structure corresponding to the experimentally studied protein is ambiguous. In order to take this into account, there is a possibility to change the Protein Data Bank identifier of the best structure or to have even several structures for a protein at the organizational level of the database. It should be noted that for proteins studied by Maxwell et al. (18), we took Protein Data Bank structure identifiers recommended in their paper, while for other proteins we took PDB structures that were selected during the theoretical and empirical investigations on the prediction of protein folding rates (11,13,15,25).
Details of experimental conditions include pH, temperature, denaturant concentration, buffer and type of denaturing agent. The field ‘Other’ contains all other relevant information. All conditions refer to the point where logarithms of folding and unfolding rate in water are obtained. Thus, in the case when the denaturing agent is a chemical denaturant, the denaturant concentration in this section is given as zero. Other conditions are suggested to be kept constant at all denaturant concentrations studied. However, we do not focus very much on the experimental conditions; the main goal of this section is to show to what extent conditions differ from the standard ones (18).
Experimental results include natural logarithms of protein folding and unfolding rates extrapolated to water, the natural logarithm of the mid-transition rate of folding (which is equal to the mid-transition rate of unfolding), transition state coordinate, free energy of unfolding in water, type of protein folding kinetics behavior: two-state (single-exponential throughout all experimental conditions studied) or multi-state (if multi-exponential kinetics was observed at least at some range of denaturant concentration). Also, there are slopes of changing the free energy values of unfolding and natural logarithms of protein folding and unfolding rates with denaturant (the so-called ‘m-values’) that are given only if a chemical denaturant is used. And finally, the temperature and denaturant concentration of the mid-transition are given. It should be noted that if a chemical denaturant is used, the temperature of mid-transition is the same as the temperature corresponding to in-water folding/unfolding rates, while in the case of an experiment with temperature denaturation the denaturant concentration is the same as in the case of in-water protein folding/unfolding rates.
It should be noted that in the current design the database reflects our theoretical and empirical studies of protein folding rates prediction (9,13,15). That is, if a protein was studied in several different conditions, we selected the measurement done at conditions closest to the standard ones: 25°C, pH 7.0 and the absence of a denaturant. This is also in agreement with the paper of Maxwell et al. (18). However, in the future we may include also additional experiments with the same protein.
USE OF KINETICDB
The KineticDB has a simple interface consisting of a few pages.
The home page offers an opportunity either to go to the database summary table or to search in the database for particular protein(s). In the menu there is a link to the ‘Help’ option that describes the meaning of all fields of the database. The main page contains links to the related resources as well.
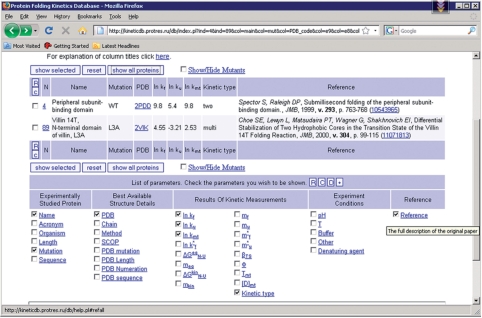
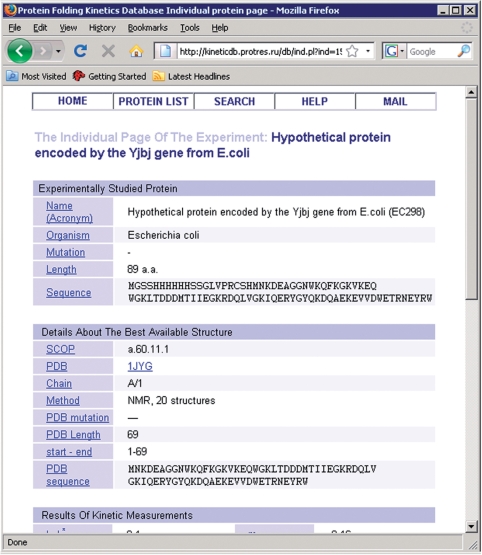
The page with the list of proteins (Figure 1) initially contains only a small part of the database records, for which several fields are shown. Using controls on the page one can choose to display all database records. By checking appropriate boxes one can choose any set of database records with any parameters to be shown. The protein list can be sorted by any parameter, ascending or descending. Each parameter name is supplied with a pop-up hint with the meaning of the parameter (Figure 1). Each protein has links to the Protein Data Bank (26), Structural Classification of Proteins (27) and PubMed databases. The database identifier in the first column is linked to the individual page of the experiment (Figure 2).
Figure 1.
Screenshot of the central part of the page with the list of proteins. There is the menu for displaying different parameters as well as the table with protein folding kinetics measurements.
Figure 2.
Screenshot of part of an individual page with an example of protein folding kinetic measurement.
An advanced search page allows searching in the database by keywords and filter the results by some parameters.
Our database is made for researchers who would like to test model relationships both for all proteins experimentally studied by now and for different groups of proteins. Analytical tools are being developed to make use of the accumulated data to support the selected set of the already developed different methods of protein folding rate prediction.
CONCLUSIONS AND FUTURE DIRECTIONS
We have proposed here the basic design of KineticDB, a systematically compiled database of protein folding kinetics. The main goal of the KineticDB is to provide users with regularly updated information about diverse data on protein folding kinetics in a well-documented manner. At the moment the search for determinants of protein folding kinetics is still in progress with the goal of obtaining a new understanding of the folding process. It is, therefore, necessary to keep as much data as possible in a simple and easy-to-use way to facilitate testing new models and theories of protein folding against experimental data. Also, the KineticDB can be used as a unified dataset to compare performance of different methods of prediction of protein folding rates.
At present the KineticDB contains the results of protein folding kinetics measurements of single-domain proteins or separate protein domains as well as short peptides without disulfide bonds. It includes about 90 unique proteins and many mutants that have been systematically accumulated over the last 10 years, and is the widest collection of protein folding kinetics data compiled as a database. Moreover, it is possible to add the measurements of new proteins and/or mutants as new information becomes available; the impending work is to include in the database protein folding kinetics measurements of proteins with disulfide bonds as well as the measurements in conditions other than standard. Also, we are going to incorporate the results of using multiple variants of protein structure. In order to make the database as wide and up-to-date as possible, we are addressing research community with a request to send us references containing new protein folding kinetics data. We will be grateful for any contribution to the database concerning both bug reports and new protein folding kinetics data.
FUNDING
Russian Foundation for Basic Research; program ‘Molecular and cellular biology’; INTAS (05-1000004-7747); Howard Hughes Medical Institute (55005607). Open Access charges were waived by Oxford University Press.
ACKNOWLEDGEMENTS
We are grateful to Sergiy Garbuzinskiy, Oxana Galzitskaya, Alexei Finkelstein and everybody who participated in the collection of the protein folding kinetics data.
REFERENCES
- 1.Qian B, Raman S, Das R, Bradley P, McCoy AJ, Read RJ, Baker D. High-resolution structure prediction and the crystallographic phase problem. Nature. 2007;450:259–264. doi: 10.1038/nature06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang L, Althoff EA, Clemente FR, Doyle L, Rothlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas C.F., III, et al. De novo computational design of retro-aldol enzymes. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, et al. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 4.Snow CD, Nguyen H, Pande VS, Gruebele M. Absolute comparison of simulated and experimental protein-folding dynamics. Nature. 2002;420:102–106. doi: 10.1038/nature01160. [DOI] [PubMed] [Google Scholar]
- 5.Mayor U, Guydosh NR, Johnson CM, Grossmann JG, Sato S, Jas GS, Freund SM, Alonso DO, Daggett V, Fersht AR. The complete folding pathway of a protein from nanoseconds to microseconds. Nature. 2003;421:863–867. doi: 10.1038/nature01428. [DOI] [PubMed] [Google Scholar]
- 6.Galzitskaya OV, Finkelstein AV. A theoretical search for folding/unfolding nuclei in three-dimensional protein structures. Proc. Natl Acad. Sci. USA. 1999;96:11299–11304. doi: 10.1073/pnas.96.20.11299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alm E, Baker D. Prediction of protein-folding mechanisms from free-energy landscapes derived from native structures. Proc. Natl Acad. Sci. USA. 1999;96:11305–11310. doi: 10.1073/pnas.96.20.11305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munoz V, Eaton WA. A simple model for calculating the kinetics of protein folding from three-dimensional structures. Proc. Natl Acad. Sci. USA. 1999;96:11311–11316. doi: 10.1073/pnas.96.20.11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivankov DN, Finkelstein AV. Theoretical study of a landscape of protein folding-unfolding pathways. Folding rates at midtransition. Biochemistry. 2001;40:9957–9961. doi: 10.1021/bi010625p. [DOI] [PubMed] [Google Scholar]
- 10.Alm E, Morozov AV, Kortemme T, Baker D. Simple physical models connect theory and experiment in protein folding kinetics. J. Mol. Biol. 2002;322:463–476. doi: 10.1016/s0022-2836(02)00706-4. [DOI] [PubMed] [Google Scholar]
- 11.Garbuzynskiy SO, Finkelstein AV, Galzitskaya OV. Outlining folding nuclei in globular proteins. J. Mol. Biol. 2004;336:509–525. doi: 10.1016/j.jmb.2003.12.018. [DOI] [PubMed] [Google Scholar]
- 12.Plaxco KW, Simons KT, Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. J. Mol. Biol. 1998;277:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 13.Ivankov DN, Garbuzynskiy SO, Alm E, Plaxco KW, Baker D, Finkelstein AV. Contact order revisited: influence of protein size on the folding rate. Protein Sci. 2003;12:2057–2062. doi: 10.1110/ps.0302503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong H, Isom DG, Srinivasan R, Rose GD. Local secondary structure content predicts folding rates for simple, two-state proteins. J. Mol. Biol. 2003;327:1149–1154. doi: 10.1016/s0022-2836(03)00211-0. [DOI] [PubMed] [Google Scholar]
- 15.Ivankov DN, Finkelstein AV. Prediction of protein folding rates from the amino acid sequence-predicted secondary structure. Proc. Natl Acad. Sci. USA. 2004;101:8942–8944. doi: 10.1073/pnas.0402659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson SE. How do small single-domain proteins fold? Fold Des. 1998;3:R81–R91. doi: 10.1016/S1359-0278(98)00033-9. [DOI] [PubMed] [Google Scholar]
- 17.Makarov DE, Keller CA, Plaxco KW, Metiu H. How the folding rate constant of simple, single-domain proteins depends on the number of native contacts. Proc. Natl Acad. Sci. USA. 2002;99:3535–3539. doi: 10.1073/pnas.052713599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maxwell KL, Wildes D, Zarrine-Afsar A, De Los Rios MA, Brown AG, Friel CT, Hedberg L, Horng JC, Bona D, Miller EJ, et al. Protein folding: defining a “standard” set of experimental conditions and a preliminary kinetic data set of two-state proteins. Protein Sci. 2005;14:602–616. doi: 10.1110/ps.041205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fulton KF, Devlin GL, Jodun RA, Silvestri L, Bottomley SP, Fersht AR, Buckle AM. PFD: a database for the investigation of protein folding kinetics and stability. Nucleic Acids Res. 2005;33:283. doi: 10.1093/nar/gki016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fulton KF, Bate MA, Faux NG, Mahmood K, Betts C, Buckle AM. Protein Folding Database (PFD 2.0): an online environment for the International Foldeomics Consortium. Nucleic Acids Res. 2007;35:D304–D307. doi: 10.1093/nar/gkl1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma BG, Guo JX, Zhang HY. Direct correlation between proteins' folding rates and their amino acid compositions: an ab initio folding rate prediction. Proteins. 2006;65:362–372. doi: 10.1002/prot.21140. [DOI] [PubMed] [Google Scholar]
- 22.Galzitskaya OV, Garbuzynskiy SO. Entropy capacity determines protein folding. Proteins. 2006;63:144–154. doi: 10.1002/prot.20851. [DOI] [PubMed] [Google Scholar]
- 23.Gromiha MM, Thangakani AM, Selvaraj S. FOLD-RATE: prediction of protein folding rates from amino acid sequence. Nucleic Acids Res. 2006;34:W70–W74. doi: 10.1093/nar/gkl043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naganathan AN, Munoz V. Scaling of folding times with protein size. J. Am. Chem. Soc. 2005;127:480–481. doi: 10.1021/ja044449u. [DOI] [PubMed] [Google Scholar]
- 25.Galzitskaya OV, Reifsnyder DC, Bogatyreva NS, Ivankov DN, Garbuzynskiy SO. More compact protein globules exhibit slower folding rates. Proteins. 2008;70:329–332. doi: 10.1002/prot.21619. [DOI] [PubMed] [Google Scholar]
- 26.Berman HM, Bhat TN, Bourne PE, Feng Z, Gilliland G, Weissig H, Westbrook J. The Protein Data Bank and the challenge of structural genomics. Nat. Struct. Biol. 2000;7(Suppl):957–959. doi: 10.1038/80734. [DOI] [PubMed] [Google Scholar]
- 27.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]