Abstract
The evolutionary relationships of 46 Shigella strains representing each of the serotypes belonging to the four traditional Shigella species (subgroups), Dysenteriae, Flexneri, Boydii, and Sonnei, were determined by sequencing of eight housekeeping genes in four regions of the chromosome. Analysis revealed a very similar evolutionary pattern for each region. Three clusters of strains were identified, each including strains from different subgroups. Cluster 1 contains the majority of Boydii and Dysenteriae strains (B1–4, B6, B8, B10, B14, and B18; and D3–7, D9, and D11–13) plus Flexneri 6 and 6A. Cluster 2 contains seven Boydii strains (B5, B7, B9, B11, B15, B16, and B17) and Dysenteriae 2. Cluster 3 contains one Boydii strain (B12) and the Flexneri serotypes 1–5 strains. Sonnei and three Dysenteriae strains (D1, D8, and D10) are outside of the three main clusters but, nonetheless, are clearly within Escherichia coli. Boydii 13 was found to be distantly related to E. coli. Shigella strains, like the other pathogenic forms of E. coli, do not have a single evolutionary origin, indicating convergent evolution of Shigella phenotypic properties. We estimate the three main Shigella clusters to have evolved within the last 35,000 to 270,000 years, suggesting that shigellosis was one of the early infectious diseases of humans.
Shigella is a well known human pathogen that is prevalent in less developed countries where conditions of poor sanitation and personal hygiene increase the incidence of disease. The low infectious dose (10 cells) (1), allows the disease to be spread effectively by infected food or water, and also by person-to-person contact.
Historically Shigella was first described as Bacillus dysenteriae, the cause of bacilliary dysentery. It was clearly related to Bacillus (now Escherichia) coli but was given a different name because B. coli was known as a commensal organism and, at that time, there was no concept of a species including strains with such diverse characteristics. In the 1940s, four species of the new genus Shigella were recognized. The traditional classification of Shigella is a product of this history, which has been well documented by Ewing (2) and Bensted (3) and follows the recommendations of Ewing (4) with four species: S. boydii, S. dysenteriae, S. flexneri, and S. sonnei, also known as Shigella subgroups A, B, C, and D respectively. Nevertheless, Shigella and E. coli have always been considered to be very closely related, and for some time it has been clear that they are sufficiently similar to be placed in the same species (2, 5).
However, it was necessary to find characteristics that would enable Shigella strains to be distinguished from E. coli to confirm a diagnosis, and the major characteristics used were inability to ferment lactose and nonmotility. The matter was complicated in the 1940s when pathogenic strains were found that had some of the sugar fermentation and other characteristics of E. coli rather than Shigella, and these were treated as pathogenic forms of E. coli.
The two genera have been retained largely because of the serious nature of the disease shigellosis and its association with the name of the genus. However it now seems clear that Shigella strains are not a discrete group within E. coli and so do not even constitute a subspecies. For this reason, we refer to all Shigella strains as forms of E. coli (6). We consider it more accurate and less confusing to use a nomenclature that reflects well established natural relationships.
Subdivision into serotypes is based on the O antigen only, because Shigella strains lack the flagella H antigen and capsular K antigen also used in typing other E. coli. Forty-six Shigella serotypes are recognized. Flexneri serotypes 1 through 5 (12 including “subtypes”) are closely related and have a common basic structure (7) encoded by a common O antigen gene cluster (8). The differences are conferred by addition to the basic structure of glucosyl and/or O-acetyl residues (9). Flexneri serotypes 6 and 6A are not closely related genetically to the other Flexneri forms but crossreact with them serologically because of similarity in part of the O antigen structure (8).
Boydii and Dysenteriae have 18 and 13 well differentiated serotypes, respectively, with only occasional serological crossreaction between serotypes (2). Their repeat unit structures, where known, are unique among Shigella, although many Boydii and Dysenteriae O antigens are either identical to or related to a conventional E. coli O antigen (2). Sonnei has only one serotype, which has been shown to comprise a single clone (10). The Sonnei and Flexneri O antigens are not found otherwise in E. coli.
Earlier population genetic studies using multilocus enzyme electrophoresis (MLEE) showed that Shigella strains fall within E. coli (11). We used MLEE and sequencing of the mdh gene to demonstrate that Shigella strains do not form a single discrete set of strains within E. coli (6). In this study, we look at the relationships of all Shigella serotypes by sequencing four regions of the chromosome.
Materials and Methods
We used 46 Shigella strains to represent the known serotypes. Details are given in supplementary Table 1, which is published as supplemental data on the PNAS web site, www.pnas.org. For Dysenteriae, Flexneri, and Boydii, for which there are multiple serotypes, we refer to strains as D1, F1A, and B1 for Dysenteriae 1, Flexneri 1A, and Boydii 1, respectively, and so forth. For Sonnei, we use the abbreviation SS in tables and figures. We also used ECOR set strains ECOR7, ECOR28, ECOR30, ECOR33, ECOR37, ECOR50, ECOR54, and ECOR59, selected to represent nonpathogenic E. coli strains, and Salmonella enterica LT2, which was used as an outgroup in the analysis.
Four segments of the chromosome were sequenced directly from PCR product. Primers for PCR and sequencing (supplementary Table 2) were based on the E. coli K-12 sequence and were selected taking into account conservation in sequence from related species. The primers were chosen to give two overlapping PCR amplicons for sequencing each of the four segments.
DNA sequences were assembled and edited by using programs phred, phrap, and consed (12). Further analysis was undertaken by using programs available from the Australian National Genomic Information Service (ANGIS) at The University of Sydney. Sequence comparisons were analyzed by using the multicomp package (13), which gives pairwise comparisons of DNA and amino acid sequences. multicomp calculates nucleotide diversity (π) by the method described by Nei and Miller (14) and average pairwise percentage difference. Calculation of synonymous and nonsynonymous substitution rates was done by using the program kindly provided by W.-H. Li (15). Molecular evolutionary relationships among each of the genes studied were examined by the neighbor-joining method of tree construction (16, 17), based on distance estimated by using the two-parameter method of Kimura (18). Phylogenetic trees and bootstrap analysis to determine the statistical stability of each node were done by using phylip (version 3.4 written by Joseph Felsenstein, Department of Genetics, University of Washington, Seattle, WA).
Results
A total of 7,160 bp from four regions was sequenced for each strain, except for D8 and B13, where one or two segments (600–950 bp), respectively, could not be amplified (see supplementary Table 3). The four regions are approximately equidistant on the chromosome and cover eight housekeeping genes. The 7,160 bp comprises 2,032 bp of thrB-thrC region; 1,486 bp of trpB-trpC region; 2,101 bp of purM-purN region; and 1,541 of mdh-argR region. Details of the genes are presented in supplementary Table 3.
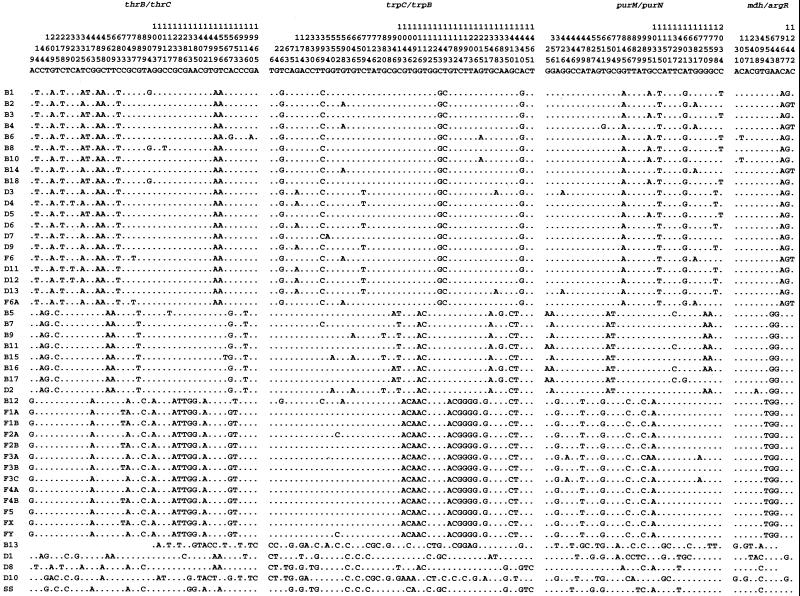
The sequence alignment for informative sites is shown in Fig. 1. It is immediately clear that all but five of the Shigella strains fall into one of three major clusters, and that the same strains are in each cluster for all four regions of the chromosome. It is also clear that the strains within each cluster are very similar in sequence, but that there is little in common among the three clusters. Most of the variation involves base substitutions, but there are exceptions. B11 has a 52-bp deletion in the intergenic region between mdh and argR. B13 is particularly divergent in the purM-purN region, having a codon insertion (cysteine) at position 623 of the purN gene, and deletions of bases 13–14, 28–33, and 45–46 in the intragenic region downstream of the purN gene, resulting in a total of 2,094 bp for this strain.
Figure 1.
Alignment of the informative sites for the four regions sequenced. Note that for B13, D1, D8, D10, and Sonnei, which have very divergent sequences, the use of informative sites leads only to omission of many polymorphic sites.
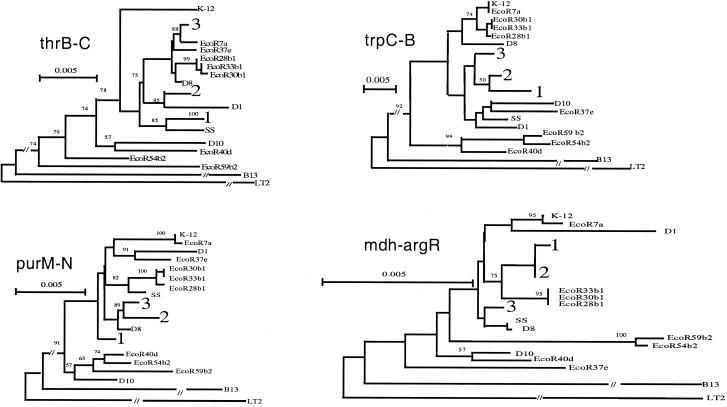
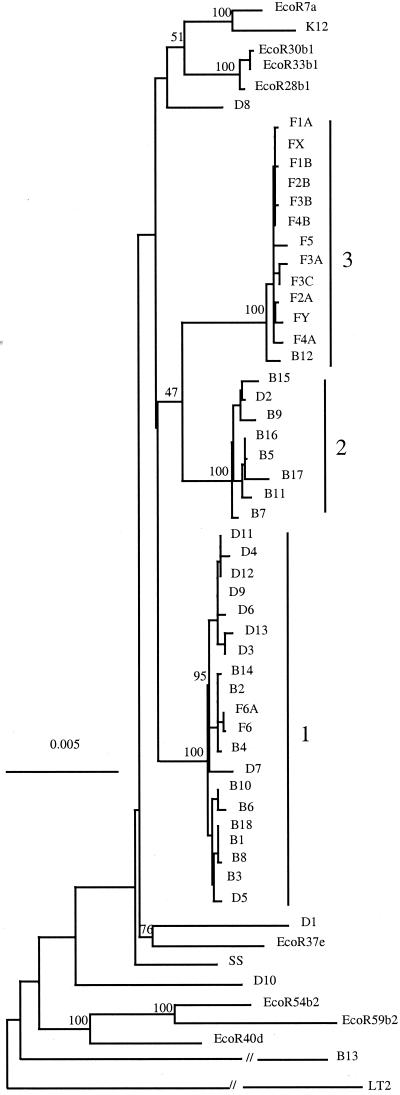
Trees for the four regions are shown in Fig. 2, and a tree for the combined data is shown in Fig. 3. Cluster 1 contains the majority of Boydii and Dysenteriae strains, plus F6 and F6A. Cluster 2 consists of seven Boydii strains (B5, B7, B9, B11, B15, B16, and B17) and D2. Cluster 3 consists of the Flexneri strains except F 6 and F6A, and B12. Sonnei D1, D8, and D10 are outside of the three main Shigella clusters but, nonetheless, are clearly within E. coli. B13 is only distantly related to other E. coli strains (Figs. 1 and 2, and supplementary Table 4). We applied bootstrap analysis using 1,000 replicas, and the high bootstrap values (100% for all three Clusters) confirm that the three clusters identified are robust.
Figure 2.
Phylogenetic trees generated by the neighbor-joining method for the four regions sequenced, showing different placement of the three clusters among ECOR set strains. The three major clusters are identified by numbers 1, 2, and 3 respectively. For detailed branching within each cluster, see Supplementary Fig. 4. Bootstrap values are percentages of 1000 replications and are indicated at the nodes. LT2 is used as the outgroup.
Figure 3.
Combined phylogenetic tree generated by the neighbor-joining method. Includes the sequences from the four regions sequenced to give a total of 7160 bp. F, Flexneri; B, Boydii; D, Dysenteriae; SS, Sonnei, followed by the serotype number. Bootstrap values are percentages of 1000 replications and are indicated at the nodes. LT2 is used as the outgroup.
Discussion
Historically, what is now Dysenteriae 1 (D1), the type form of Shigella, was discovered soon after E. coli and, because of its human pathogenesis, put in a separate genus. As more pathogenic strains were found, the absence of motility and lactose fermentation were found to be useful diagnostic criteria for dysentery bacteria placed in Shigella, although their relationship to E. coli was always evident. In our previous work (6), we showed by MLEE and sequence of the mdh gene that some Shigella strains at least fall into discrete clusters within E. coli. The sequence data we now present from four regions around the chromosome consistently give three clusters of strains, each including strains from more than one traditional Shigella species. In addition, Sonnei falls outside of these clusters, as do D1, D8, D10, and B13, the last being very divergent. The consistency of the data from four regions gives great confidence in the new groupings, showing that the four existing species are not valid taxa even as groups within E. coli, raising questions as to their origins and convergent evolution.
Multiple Origins of Shigella Strains.
The presence of three major clusters and five forms not closely related to any other suggests that the Shigella phenotype has arisen eight times, or seven if one disregards B13 for this purpose, it being so divergent (supplementary Table 4) that it does not fall into E. coli, and is best treated as the only known representative of an unnamed species and left out of current discussion of the origins of the Shigella phenotype. We note that Manolov (19) considered “Shigella 13” strains not to be members of the genus Shigella, and that the classification of B13 as a Shigella or E. coli strain has also been observed to be incorrect by Ewing (20) and Brenner (21, 22).
It is interesting to note that D1, the type form for Shigella and one of the prominent strains in outbreaks, and Sonnei, the most commonly isolated in industrialized countries, are both outside of the three main clusters. D1 is also unusual in production of Shiga toxin.
The current study included eight ECOR set E. coli strains, and, for three of the four regions, some fall between major Shigella clusters (Fig. 2), indicating that the three clusters are not derived from a single ancestral Shigella form. The distribution of Shigella strains among the ECOR set of 72 strains is more clearly seen in the earlier work using MLEE (6). There were cluster 1 and cluster 3 strains in that study, and it can be seen they are not closely related. Unfortunately, the limited number of Shigella strains used did not include any in cluster 2. The relationships among the ECOR set strains vary, as do the relationships of the three Shigella clusters and five nonclustered Shigella strains among themselves and with the ECOR set strains. This variation is not unexpected because E. coli has a quite high level of recombination (23). The mdh gene has been used in several studies, and a tree was constructed of all available E. coli sequences, including 31 ECOR set strains (6, 24), 32 pathogenic strains (6), and 21 isolates sampled from native rats in Australia (unpublished data) and all strains in this study. No non-Shigella strains fell within any of the three Shigella clusters, confirming that each is a cluster of closely related Shigella-only strains presumably derived from a single parental Shigella form.
Convergent Evolution of the Shigella Phenotype.
Shigella strains have a characteristic form of pathogenesis involving invasion of mucosal epithelium cells of the large intestine. The genes for the invasive property reside on a plasmid present in all Shigella strains tested thus far (25). However, Shigella strains have traditionally been defined and identified by characteristics either known or thought to be determined by chromosomal genes. In particular, Shigella strains lack catabolic pathways otherwise widely present in E. coli (2). With few or no exceptions, lactose and mucate are not used, and lysine is not decarboxylated. Shigella strains are also nonmotile. Other E. coli strains are commonly positive for these properties. The independent evolution of the Shigella phenotype many times raises questions on the loss of motility and catabolic pathways by convergent evolution.
The studies by Al Mamun et al. (26) show that the basis for loss of motility in Shigella strains varies and can be due to deletion in the fliF operon (flagellar coding region) or an ISI insertion mutation in the flhD operon. The lack of lactose fermentation was studied in several Shigella strains by Ito (27). Southern hybridization of lacY, lacA, and lacZ genes showed that Flexneri 1 and 3 (cluster 3 in this study) and Bodyii 2 (cluster 2) and 4 (cluster 1) do not contain any of the lac genes and that Dysenteriae 1 has lacY and lacA but not lacZ, whereas Sonnei has all three genes. It was further shown that, in Sonnei, the defect lies in the permase activity. These observations support the concept of multiple origins of the Shigella phenotype by convergent evolution.
It seems most likely that acquisition of the plasmid is a prerequisite for adoption of Shigella properties, and that the other characteristics, originally used together with symptoms of dysentery to define Shigella strains, are acquired by mutation later.
Convergent evolution of phenotypes is also seen for characters not universally present in Shigella strains. Shigella strains commonly lack ability to metabolize substrates otherwise widely utilized by E. coli. For example, mannitol utilization is present in 98% of typical E. coli, and its absence in many Shigella strains is presumably due to loss of function. There appear to have been seven independent events: three in outlier strains D1, D8, and D10, D2 in cluster 2, D5 and D7 in cluster 1, and the ancestor of the seven Dysenteriae strains (D3, D4, D6, D9, D11, D12, and D13) grouped together within cluster 1. Likewise, 96% of E. coli are positive for indole production, and loss of function is presumed to have occurred in Shigella cluster 3 and also in three of the outlier strains. A similar situation applies for utilization of xylose, rhamnose, and glycerol, present in 82%, 83%, and 89% of E. coli isolates, respectively [biochemical data from Ewing (2)].
The Shigella subgroups (2) are generally distinguished by differences in their biochemical profiles (Supplementary Table 5) although not unambiguously and often only in combination with serotyping. It is interesting that, when the strains are rearranged according to their respective sequence-based clusters, the profile of biochemical reactions is no less consistent than that of strains arranged by subgroup (compare supplementary Tables 5 and 6). In the case of indole production, there is a much better fit with the sequence-based clusters than the traditional subgroups. Mannitol fermentation, used to differentiate Boydii and Dysenteriae, naturally gives a very clear picture when strains are arranged in traditional subgroups. However, the far better correlation of indole production with genetic relatedness suggests that indole production would have been a better criterion for subdivision of Shigella strains. Note that the Dysenteriae strains in cluster 1 are grouped together, except D5 and D7, effectively allowing the use of mannitol to subdivide cluster 1.
It seems that Shigella strains have a general tendency to lose catabolic pathways, and this tendency gives rise to convergent evolution. Some of these characteristics have been used to define Shigella strains whereas others are not so widespread. One can speculate that Shigella strains, which live inside epithelial cells, do not need the range of catabolic pathways that generally characterize E. coli as a species. In the case of lysine decarboxylase, it has been shown that this property is deleterious for Shigella such that there would be strong selection for its loss (28).
The situation with enteroinvasive E. coli (EIEC) strains is also interesting. Many have Shigella-like characteristics: they may be lac−, nonmotile, low level indole producing, and/or may not produce gas during fermentation. Their invasion properties are due to presence of a plasmid similar to those of Shigella strains. However, the characteristics of these strains are not well studied in comparison to those of Shigella strains, and we have not included them in this study.
The EIEC strains do not have the full set of characters that define Shigella strains, but they may be strains developing the full Shigella phenotype. None of the five EIEC strains included in an earlier study (6) was in any of the three clusters of Shigella strains in the mdh tree referred to above, suggesting that the distinction between Shigella and EIEC strains is not entirely arbitrary.
Age of the Three Main Shigella Clusters.
The mean distance between alleles at synonymous sites (Ks) (supplementary Table 7) was used to estimate the time since descent from a common ancestor for each of the three main clusters of Shigella strains. We used the molecular clock rates of Whittam (29) and of Gutman and Dykhuizen (23) as done by Achtman et al. (30) to obtain a range of estimates. We calculated the Ks values for each gene and used the average of the eight genes as the distance for calculations. The estimated time is between 35,000 to 270,000 years for the three clusters. Clusters 1 and 2 have similar estimated times of origin 50,000–270,000 year ago, whereas cluster 3 apparently arose later, at 35,000 to 170,000 years ago. This is a very short time in comparison with that for diversification of E. coli in general, as illustrated by divergence of group A, one of the major ECOR set groups, estimated to have diverged from other major groups 8–22 million years ago (31).
These conclusions on the origins of Shigella can be compared with those for other human-specific pathogens for which similar data are available.
Yersinia pestis, the cause of plague, lacks variation in any of six gene segments sequenced and is proposed to be a clone that evolved from closely related Y. pseudotuberculosis 1,500 to 20,000 years ago (30). Plasmodium falciparum, a major cause of malaria, likewise lacks variation in five housekeeping genes and is estimated to have an origin as a clone after a selective sweep about 6,000 years ago (32).
The Mycobacterium tuberculosis complex, hich includes M. bovis, M. africanum,and M. microtis as well as M. tuberculosis, has virtually no silent substitution even between “species. There are many substitutions associated with drug resistance, but these are likely to have arisen by selection for drug resistance, and it is silent substitutions that give an estimate for age of the clone. On this basis, M. tuberculosis is proposed to have undergone an evolutionary bottleneck about 15,000 to 20,000 years ago (30) (this time frame presumably applies to the complex although there are significant host specificity differences between M. bovis, M. microtis, and M. tuberculosis, with M. africanum resembling M. tuberculosis in high degree of specificity for humans).
The Shigella strains of E. coli are essentially human specific as are M. tuberculosis and P. falciparum.
The Shigella strains and Y. pestis are in effect clones of more diverse species found in a range of animal hosts. However, the three clusters of Shigella strains carry much more neutral variation than the Y. pseudotuberculosis clone known as Y. pestis, or M. tuberculosis or Plasmodium falciparum. The dates for origin of Y. pestis and the sweep/bottlenecks of M. tuberculosis and P. falciparum are within or close to the start of the neolithic, with a change to settlement based population structure and rapid expansion of human population, and for Y. pestis a change in the ecology of the rat. The sequence variation within each of the three major Shigella clusters suggests that they arose much earlier, which is very interesting when looked at in relation to evolution of humans, the host species.
It has been proposed that that organisms like Shigella, which cause short-term infections with no carrier state and no alternative host, could not have survived in the paleolithic because early man lived in small bands with infrequent contact with other closed bands (33, 34). It is thought that such organisms could only survive after the development of agriculture with large settlements, a situation thought to have arisen in the neolithic about 10,000 years ago.
The estimated time of origin of Shigella forms of E. coli correlates with the origin and expansion of Homo sapiens rather than with the development of agriculture. It is not clear what this correlation means. It may be that Shigella strains had a greater capacity to survive in small hunter gatherer bands in the paleolithic than currently assumed. Perhaps we should also consider the possibility that we have underestimated the complexity of human populations 50,000 to 200,000 years ago during the rise to dominance of H. sapiens and the paleolithic expansion.
Antigenic Diversity in Shigella Strains.
There is considerable diversity of O antigen in the three clusters of Shigella strains. Those of cluster 3 are mostly flexneri strains with O antigens that differ only in phage-encoded properties, but each of the 28 strains in clusters 1 and 2 has a quite different O antigen, with the exception of F6 and F6A, which have variants of the same O antigen. Of the 27 unique O antigens, 9 are reported to be identical to antigens in other E. coli strains (serological data from Ewing, ref. 2). That leaves 18 O antigens found only in one or other of these clusters. This diversity must have arisen by transfer of O antigen gene clusters to strains of these clusters during the 50,000 to 270,000 years since the last common ancestor of the current members of each cluster. The fact that 18 of the 27 O antigens have not been reported in other E. coli strains suggests that a high proportion of them may have come directly from outside of E. coli. D1 has one of the genes for O antigen synthesis on a plasmid (35), and the whole of the O antigen gene cluster of Sonnei is on a plasmid (36). The plasmid location is a good indication of recent lateral transfer and there is direct evidence for recent transfer of the Sonnei O antigen gene cluster from Plesiomonas shigelloides (J. Sheperd, L. Wang, and P.R.R., unpublished data). In any case, this expansion of O antigen diversity over a very short period relative to the time frame for diversification of the extant E. coli (see above) indicates that O antigen specificity is very important for Shigella strains. This study provides the first data that allow us to give a time frame for the generation of antigenic diversity in a closely related group of strains and an indication of the frequency of lateral transfer of such gene clusters.
The Classification of Shigella Strains.
As discussed above with the exception of Sonnei, which is a clone (10), the four generally recognized subgroups of Shigella do not represent natural groupings. Some of the relationships evident in Fig. 2 have been recognized for some time: e.g., that F6 is distinct from the other Flexneri strains, and that B13 is very distinct. Indeed Brenner (37) suggested the transfer of F6 to the Boydii subgroup but was rejected for historical reasons.
It is interesting to compare the results of this study with those of Dodd and Jones (38). They used 102 Shigella strains to determine relationships by using the methods of numerical taxonomy. In that study, Shigella strains formed a taxon not only distinct from E. coli but more closely related to Proteus/Providencia, Citrobacter, and Salmonella than to E. coli. We suggest that the reason for the discrepancy is that, in numerical taxonomy, the characters used are in general those developed by taxonomists because of their use in identification of generally recognized taxa. In a group like Shigella, which has been difficult to distinguish from E. coli, these characters will be strongly biased in favor of those that confirm the preferred taxonomy. This example illustrates the value of sequence data for determining true relationships in bacteria.
A good understanding of the origins of Shigella will be most important as more detail of the molecular basis for pathogenicity emerges. It will also help us to better understand clinical variation among Shigella forms.
Supplementary Material
Acknowledgments
We are grateful to Drs. Johanne Lefebvre and Pierre Harbec for providing strains shown in supplementary Table 1, and to Christine Dodd for providing additional information on the work reported in ref. 38. We thank David Ryan for technical assistance. We thank the anonymous referees for suggestions on improvement of the manuscript. The research was supported by a grant from the National Health and Medical Research Council (Australia).
Abbreviations
- MLEE
multilocus enzyme electrophoresis
- EIEC
enteroinvasive E. coli
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF293105–AF293320).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.180094797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.180094797
References
- 1.Dupont H L, Levine M M, Hornick R B, Formal S B. J Infect Dis. 1989;159:1126–1128. doi: 10.1093/infdis/159.6.1126. [DOI] [PubMed] [Google Scholar]
- 2.Ewing W H. Edwards and Ewing's Identification of the Enterobacteriaceae. Amsterdam: Elsevier Science; 1986. [Google Scholar]
- 3.Bensted H. Can J Microbiol. 1956;2:163–174. doi: 10.1139/m56-022. [DOI] [PubMed] [Google Scholar]
- 4.Ewing W H. J Bacteriol. 1949;57:633–638. doi: 10.1128/jb.57.6.633-638.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenner D J. In: Bergey's Manual of Systematic Bacteriology. Krieg N R, Holt J G, editors. Vol. 1. Baltimore: Williams and Wilkins; 1984. pp. 408–420. [Google Scholar]
- 6.Pupo G M, Karaolis D K R, Lan R, Reeves P R. Infect Immun. 1997;65:2685–2692. doi: 10.1128/iai.65.7.2685-2692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simmons D A R, Romanowska E. J Med Microbiol. 1987;23:289–302. doi: 10.1099/00222615-23-4-289. [DOI] [PubMed] [Google Scholar]
- 8.Cheah K-Y, Beger D W, Manning P A. FEMS Microbiol Lett. 1991;83:213–218. doi: 10.1016/0378-1097(91)90356-f. [DOI] [PubMed] [Google Scholar]
- 9.Huan T P, Bastin D A, Whittle B L, Lindberg A A, Verma N. Gene. 1997;195:217–227. doi: 10.1016/s0378-1119(97)00143-1. [DOI] [PubMed] [Google Scholar]
- 10.Karaolis D K R, Lan R, Reeves P R. J, Clin Microbiol. 1994;32:796–802. doi: 10.1128/jcm.32.3.796-802.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ochman H, Whittam T S, Caugant D A, Selander R K. J Gen Microbiol. 1983;129:2715–2726. doi: 10.1099/00221287-129-9-2715. [DOI] [PubMed] [Google Scholar]
- 12.Gordon D, Abajian C, Green P. PCR Methods Appl. 1998;8:195–202. [Google Scholar]
- 13.Reeves P R, Farnell L, Lan R. Comput Appl Biol Sci. 1994;10:281–284. doi: 10.1093/bioinformatics/10.3.281. [DOI] [PubMed] [Google Scholar]
- 14.Nei M, Miller J C. Genetics. 1990;125:873–879. doi: 10.1093/genetics/125.4.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W-H. J Mol Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- 16.Nei M. Molecular Evolutionary Genetics. New York: Columbia Univ. Press; 1987. [Google Scholar]
- 17.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 18.Kimura M. J Mol Evol. 1980;116:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 19.Manolov D G. J Hyg Epidemiol Microbiol Immunol. 1959;3:84. [PubMed] [Google Scholar]
- 20.Ewing W H, Reavis R W, Davis B R. Can J Microbiol. 1958;4:89–107. doi: 10.1139/m58-012. [DOI] [PubMed] [Google Scholar]
- 21.Brenner D J, Fanning G R, Miklos G V, Steigerwalt A G. Int J Syst Bacteriol. 1973;23:1–7. [Google Scholar]
- 22.Brenner D J, Steigerwalt A G, Wathen H G, Gross R J, Rowe B. J Clin Microbiol. 1982;16:432–436. doi: 10.1128/jcm.16.3.432-436.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guttman D S, Dykhuizen D E. Science. 1994;266:1380–1383. doi: 10.1126/science.7973728. [DOI] [PubMed] [Google Scholar]
- 24.Boyd E F, Nelson K, Wang F-S, Whittam T S, Selander R K. Proc Natl Acad Sci USA. 1994;91:1280–1284. doi: 10.1073/pnas.91.4.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hale T L. Microbiol Rev. 1991;55:206–224. doi: 10.1128/mr.55.2.206-224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al Mamun A A M, Tominaga A, Enomoto M. J Bacteriol. 1997;179:4493–4500. doi: 10.1128/jb.179.14.4493-4500.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito H, Kido N, Arakawa Y, Ohta M, Sugiyama T, Kato N. Appl Environ Microbiol. 1991;57:2912–2917. doi: 10.1128/aem.57.10.2912-2917.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maurelli A T, Fernandez R E, Bloch C A, Rode C K, Fasano A. Proc Natl Acad Sci USA. 1998;95:3943–3948. doi: 10.1073/pnas.95.7.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whittam T S. In: Escherichia and Salmonella Cellular and Molecular Biology. Neidhardt F C, Curtiss R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Vol. 2. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 2708–2720. [Google Scholar]
- 30.Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. Proc Natl Acad Sci USA. 1999;96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bergthorsson U, Ochman H. Mol Biol Evol. 1998;15:6–16. doi: 10.1093/oxfordjournals.molbev.a025847. [DOI] [PubMed] [Google Scholar]
- 32.Rich S M, Light M C, Hudson R R, Ayala F J. Proc Natl Acad Sci USA. 1998;95:4425–4430. doi: 10.1073/pnas.95.8.4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fenner F. In: The Impact of Civilization on the Biology of Man. Boyden S W, editor. Toronto: Univ. Toronto Press; 1970. pp. 48–68. [Google Scholar]
- 34.McKeown T. The Origins of Human Disease. Oxford: Blackwell; 1988. [Google Scholar]
- 35.Klena J D, Asford R S, II, Schnaitman C A. J Bacteriol. 1992;174:7297–7307. doi: 10.1128/jb.174.22.7297-7307.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kopecko D J, Washington O, Formal S B. Infect Immun. 1980;29:207–214. doi: 10.1128/iai.29.1.207-214.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brenner D J. Int J Syst Bacteriol. 1984;34:87–88. [Google Scholar]
- 38.Dodd C E R, Jones D. J Gen Microbiol. 1982;128:1933–1957. doi: 10.1099/00221287-128-9-1933. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.