Abstract
The evolutionary mechanisms maintaining genetic variation in life span, particularly post-reproductive life span, are poorly understood. We characterized the effects of spontaneous mutations on life span in the rhabditid nematodes Caenorhabditis elegans and C. briggsae and standing genetic variance for life span and correlation of life span with fitness in C. briggsae. Mutations decreased mean life span, a signature of directional selection. Mutational correlations between life span and fitness were consistently positive. The average selection coefficient against new mutations in C. briggsae was approximately 2% when homozygous. The pattern of phylogeographic variation in life span is inconsistent with global mutation–selection balance (MSB), but MSB appears to hold at the local level. Standing genetic correlations in C. briggsae reflect mutational correlations at a local scale but not at a broad phylogeographic level. At the local scale, results are broadly consistent with predictions of the “mutation accumulation” hypothesis for the evolution of aging.
Keywords: Mutation, genetic variation, nematode, life span, selection
HOW genetic variation is maintained in nature is a long-standing question in evolutionary biology (e.g., 1,2). Variation in life history traits has attracted particular attention because life history traits (defined sensu 3, i.e., all else equal, an increase in trait value results in an increase in fitness) are inherent components of fitness and directional selection removes additive genetic variance for fitness (4), so the presence of additive genetic variance for fitness in an equilibrium population represents something of a paradox. However, life history traits typically exhibit more additive genetic variance than do morphological or behavioral traits, not less (5).
Mutation plays a key role as the ultimate source of genetic variation. Three mechanisms link mutation with variation. These range from positive selection (balancing or diversifying), in which mutation plays a minor role, to mutation-drift equilibrium (MDE) and mutation–selection balance (MSB), in which variation is maintained by a balance between mutational input and elimination by either drift or purifying selection (5,6). Of the three mechanisms, the potential role of MSB has received much attention. It has been argued that (at least for Drosophila melanogaster, the only taxon for which substantial data exist) there is too much additive genetic variance for MSB to completely explain the variation in most life history traits (3,7), but different people can (and do) reach opposing conclusions from the same data (e.g., 5).
One life history trait that has been extensively studied is life span. Genetic variation for life span is often observed both within species and between closely related species (for review in fishes, see 8). In particular, post-reproductive life span has attracted a great deal of theoretical and experimental attention (9–13) because post-reproductive life would seem to provide no fitness benefit, except under special circumstances, like those proposed for humans (14–16), in which parental care may be important. Investigations into variation in life span generally take one of two approaches: (a) explicitly investigate age-specific patterns in mortality (e.g., 17–19) or (b) estimating genetic or mutational variance in mean life span in laboratory or wild populations (e.g., 20,21–25). However, results from taxa that do not exhibit reproductive senescence have limited applicability to organisms with extensive post-reproductive life span, such as humans.
Additive genetic variance for life span shows that it can evolve; differences between groups in life span prove that it does. There are two general classes of evolutionary scenarios invoked to explain the evolution of aging (i.e., senescence), both of which stem from the diminishing efficiency of natural selection with age (4,26). Under the “antagonistic pleiotropy” (AP) hypothesis, mutations that increase early reproduction but reduce life span will be beneficial because early reproduction contributes more to fitness than late reproduction, so senescence is due to the deleterious pleiotropic effects expressed in late life of alleles that increase reproduction in early life (27). Conversely, the “mutation accumulation” (MA*; the “*” is used to distinguish the hypothesis from the protocol of mutation accumulation) hypothesis predicts that at some point in life natural selection becomes so inefficient that deleterious alleles with effects past that age are effectively neutral and can accumulate due to genetic drift (28).
Distinguishing between the two hypotheses has proven extraordinarily difficult foremost because the two are not mutually exclusive, and supporting evidence exists for both (for review, see 21,29). A necessary component to understanding the evolution of aging is an accurate characterization of the effects of new mutations allowed to accumulate under relaxed selection. Two attributes of new mutations— their effect on mean life span and their correlation with (some measure of) fitness—are key. If mean life span changes with MA, it provides prima facie evidence that life span is in fact under directional selection and is thus a proper life history trait. Second, if the standing genetic covariance of life span with fitness differs from the mutational covariance, it strongly suggests that the standing covariance has been shaped by natural selection in some way.
Here, we report a study in which we characterize the effects of spontaneous mutation on life span and related demographic parameters in two species of self-fertilizing nematodes in the genus Caenorhabditis, C. briggsae and C. elegans. Further, we characterize the standing genetic (co)variance for life span in a worldwide collection of natural isolates of C. briggsae and compare the standing (co)variance with the mutational input. The study is motivated by two primary factors. First, and foremost, we are interested in providing a comprehensive characterization of the effects of spontaneous mutations on these traits in a taxon whose post-reproductive life span is at least qualitatively similar to that of humans; such data are notably lacking. There are considerable data from large-effect mutants, which we believe may be atypical and misleading (see the Discussion section).
Second, we address the possibility that MSB is a sufficient explanation for the patterns of genetic (co)variance observed in C. briggsae. In a large population at MSB, the standing genetic variance is related to the mutational variance by the relationship VG ≈ VM/S (5), where VG represents the standing genetic variance, VM the per-generation input of genetic variance by mutation, and S the average strength of selection against a new mutant allele. VG and VM can be calculated directly from the data; if the inferred value of S is unrealistically large or small, it suggests that MSB is probably not a sufficient explanation for the observed standing variance. Finally, we interpret the results in the context of the evolution of aging.
METHODS
MA Lines and Life-Span Assay
We characterized life span in two strains (= genotypes) each of C. briggsae (HK104 and PB800) and C. elegans (N2 and PB306) that had undergone 250 generations of MA as described elsewhere (30). Briefly, replicate lines of a highly inbred stock population were allowed to evolve at very small effective population size (∼1), allowing all but the most highly deleterious mutations to accumulate at the neutral rate. Relative to the ancestral control stocks, MA lines evolved significantly decreased mean fitness (30) and body size (31) and the genetic (among-line) component of variance in those traits increased due to the input of new genetic variation by mutation. All stocks (ancestral control and generation 250 MA lines) were cryopreserved at −80°C using standard methods (32). All stocks were maintained at 20°C on nematode growth medium (NGM) plates seeded with Escherichia coli (OP50 strain).
Prior to the life-span assay, ancestral controls were thawed and replicated 10 times; each of these replicates constitutes a (pseudo) line, analogous to the MA lines. For each of the four strains, we assayed life span in 10 ancestral control lines and in 25–41 (of the initial 100) generation 250 MA lines. Life-span measurements were conducted in four blocks, with each strain represented in two blocks and lines randomly assigned to each block. At the beginning of a block, three replicates of all lines were established and age synchronized by standard bleaching technique (33). Two days after age synchronization, 10 worms (approximately L2 to L3 stage) from each replicate were randomly selected and transferred to each of three plates (30 worms total per MA line, 10 worms total per ancestral control pseudoline). Each plate was assigned a random number and was handled in numerical order. Worms were transferred daily to new plates during their reproductive period. Life span was measured daily by scoring each worm by touch response. Worms confirmed dead were removed from the plates; the date of removal was noted as an uncensored data point. Worms that disappeared from the plates (no carcass found) were counted as censored data, with the date of disappearance recorded.
Life-Span Assay in Wild Worm Isolates
To characterize standing genetic variation for life span in C. briggsae, we employed a similar life-span assay protocol for 43 wild isolates of C. briggsae, generously provided by Asher Cutter, Marie-Anne Félix, and The Caenorhabditis Genetics Center (see Supplementary Table 1 for list of isolates). Each natural isolate (strain) was divided into three replicate sublines and inbred for six generations by transferring a single L4 hermaphrodite to a new NGM plate. At generation six, inbred sublines were allowed to expand to a large population size and cryopreserved. Frozen stocks were thawed and allowed to expand back to a large population size. Each subline was replicated three times and maintained by single-worm descent for three generations after thawing to remove maternal and grandmaternal effects. To assay life span, five individuals from each replicate were randomly selected (at the L4 stage) and placed individually on NGM plates (15 worms total per strain). Subsequent to cessation of reproduction, life span was measured daily by scoring each worm by touch response. Worms were transferred daily to new plates during their reproductive period. Censored and uncensored data were recorded as aforementioned except that notation of censored data did not start until after the cessation of reproduction (approximately day 5).
Data Analysis
Characterization of life span and demographic models: Because we could not clearly differentiate between a missing (but alive) worm and a worm that had died and whose carcass we could not find, we took a conservative approach and used only uncensored data (confirmed deaths) for calculations of life span, mortality models, and mutational (co)variances in life span. However, because censored data can influence mortality models and mortality parameter estimates, we also present the results of the mortality models as calculated with censored and uncensored data (complete data set).
Descriptive statistics of the life-span data, demographic statistics, best-fit models, and parameter estimates of mortality models were obtained by maximum likelihood methods using WinModest (34). Data were fitted to one of four models, Gompertz (G, μx = aebx), Logistic (L, μx = aebx + f(s)), Makeham (M, μx = c + aebx), or Logistic–Makeham (LM, μx = c + aebx + f(s)), where μx represents age-specific mortality rate at age x, a is the initial adult mortality rate (intercept), b is the rate of exponential increase in mortality with age (slope), c is the age-independent mortality risk, and s is the degree of deceleration in mortality at advanced ages (34). To make statistical comparisons between mortality parameters for strains that were best fit to different models, data from all strains were fit to the Logistic–Makeham model, which incorporates all four model parameters. Pairwise comparisons of parameters were made by the log-likelihood ratio test of WinModest. This test compares a model with independently estimated parameters with models that constrain individual parameters to be equal. Twice the difference between maximum likelihood estimates for the constrained model and the unconstrained models fit a chi-square distribution with one degree of freedom. A Bonferroni correction was used to compensate for multiple pairwise comparisons. Significant differences in life span between ancestral controls and MA lines, modeled as survival probability to adult worm age (in days), were determined using an extension of the nonparametric Gehan's generalized Wilcoxon test, which assigns a score to each individual's survival time and then calculates and analyzes a chi-square value for each treatment group (Statistica v. 7.1; StatSoft, Inc., Tulsa, OK). Pairwise comparisons were made, when appropriate, using the nonparametric Gehan's generalized Wilcoxon test (Statistica). Graphical representations of mortality rates (as ln(μx), uncensored data only; in Supplementary Figures 1 and 2) include symbols representing mean ± standard error calculated across lines (MA data) or strains (wild worm data). Lines were generated using the model type (aforementioned equations) and model parameters calculated by WinModest; note that the goodness of fit is constrained by small sample size and the low mortality rate at early time points (35).
To evaluate patterns in censored data, we first assessed the timing of the censored data points with respect to the experimental design (i.e., when the worms were handled). We next scaled the latest date that a censored data point occurred on any given plate (set of 10 worms) to the mean life span of the uncensored worms from that plate and then used t tests to test whether these scaled data tended to be different in the MA lines in comparison with the ancestral controls. A similar analysis was conducted on the censored data points for the wild worm isolates, except that the latest date of a censored data point was scaled to the strain mean life span because worms were assayed individually and differences among clade were analyzed by analysis of variance (ANOVA).
Per-generation change in the mean and mutational variance of life span: There are two fundamental observable quantities of interest in an MA experiment—the per-generation change in the trait mean and the per-generation increase in the among-line variance (≈ the mutational variance VM). The per-generation change in the mean can be considered either on the raw scale (Rm, the slope of the regression of the mean trait value against time, measured in generations of MA) or scaled as a fraction of the generation 0 mean ( , where
, where 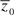 is the trait mean at generation 0). ΔM is typically the more meaningful of the two because the average mutational effect is meaningful only relative to the starting phenotype. Similarly, the per-generation increase in genetic variation can be considered either on the raw scale (VM = [VL,MA − VL,0]/2t, where VM is the mutational variance, VL,MA the among-line component of variance of the MA lines, VL,0 the among-line variance of the ancestral controls, and t the number of generations of MA), scaled by the trait mean (the mutational coefficient of variation or the “opportunity for selection”; see following), or scaled by the environmental component of variance (the mutational heritability , where VE is the within-line [environmental] component of variance). The mutational coefficient of variation is
is the trait mean at generation 0). ΔM is typically the more meaningful of the two because the average mutational effect is meaningful only relative to the starting phenotype. Similarly, the per-generation increase in genetic variation can be considered either on the raw scale (VM = [VL,MA − VL,0]/2t, where VM is the mutational variance, VL,MA the among-line component of variance of the MA lines, VL,0 the among-line variance of the ancestral controls, and t the number of generations of MA), scaled by the trait mean (the mutational coefficient of variation or the “opportunity for selection”; see following), or scaled by the environmental component of variance (the mutational heritability , where VE is the within-line [environmental] component of variance). The mutational coefficient of variation is  , where
, where  is the trait mean; and the opportunity for selection is
is the trait mean; and the opportunity for selection is 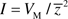 . CVM and I are perfectly correlated, but I has a specific biologic interpretation: for a trait under directional selection, I establishes the upper bound on the rate of evolution (36–38). Because mean life span decreases with MA (see following), I is the most meaningful measure of mutational variance in this case.
. CVM and I are perfectly correlated, but I has a specific biologic interpretation: for a trait under directional selection, I establishes the upper bound on the rate of evolution (36–38). Because mean life span decreases with MA (see following), I is the most meaningful measure of mutational variance in this case.
Trait means and variance components were determined from the following general linear mixed model as implemented by the MIXED procedure in SAS v. 9.1, performed separately for each strain. The model is as follows: life span = treatment + block + line + residual. Treatment (MA vs control) is a fixed effect; the other effects are random. The within-replicate term was omitted because including it greatly increased the run time of some analyses (see following). Variance components for block, line, and residual were estimated separately for MA and control treatments (PROC MIXED option GROUP=<Treatment>). Trait means were estimated by least squares, given the random effects; variance components were estimated by restricted maximum likelihood.
In our comparative context, the hypotheses of interest can be stated formally as follows: Does statistic A (i.e., ΔM or I) differ significantly between groups X and Y (e.g., between species)? The statistics of interest, ΔM and I, are composite variables and are not amenable to standard methods of hypothesis testing (e.g., F or likelihood ratio tests). We therefore employ a bootstrap method to construct approximate empirical confidence intervals for the statistics of interest and consider groups for whom the 95% confidence intervals do not overlap as significantly different. The bootstrap method is as follows: A pseudo-data set is constructed by resampling the data with replacement at the level of line (i.e., including all replicates within a line), maintaining the same number of control and MA lines as in the original data set, and analyzed using the general linear mixed model given previously. This procedure is repeated 1,000 times; the standard deviation of the pseudo-estimates is an approximate standard error of the mean, and the upper and lower 2.5% of pseudo-estimates establish approximate 95% confidence limits on the statistics of interest (39).
Our bootstrap protocol treats the within-line variance as a fixed property of a line and thus potentially underestimates the total variance (resampling at multiple hierarchical levels can result in biased estimates of variance components). To account for this possibility, we also conducted randomization tests of the hypothesis that the statistic of interest—ΔM or IM—differs significantly from zero within a strain. Data (i.e., life span) were randomly permuted within the given treatment/line/replicate structure and the statistic of interest calculated from the randomized data. This procedure was repeated 5,000 times and the observed statistic compared with the null distribution. If the absolute value of the observed statistic falls in the upper 2.5% of the null distribution, it is considered as significantly different from zero in a two-tailed test. The bootstrap confidence intervals appear somewhat conservative compared with the randomization tests (e.g., the results for IM in the HK104 strain; Table 2) and we rely on the bootstrap confidence intervals for among-group comparisons. Results of the randomization tests are presented in Tables 1 and 2.
Table 2.
Summary Statistics for Mutational (Co)Variance in Life Span
| Strain (species) | HK104 (Caenorhabditis briggsae) | PB800 (C. briggsae) | N2 (C. elegans) | PB306 (C. elegans) |
| VM × 103 | 6.663 ± 5.3 | 17.10 ± 5.1 | 6.247 ± 2.0 | 9.73 ± 3.1 |
| CVM | 0.401 ± 0.2 | 0.813 ± 0.1 | 0.474 ± 0.1 | 0.649 ± 0.1 |
| IM × 104 | 0.341 ± 0.2 (0–0.8), PRAND < .0065 | 0.694 ± 0.2 (0.3–1.1), PRAND < .0002 | 0.231 ± 0.1 (0.1–0.4), PRAND < .0002 | 0.490 ± 0.1 (0.2–0.8), PRAND < .0002 |
| × 103 | 0.740 ± 0.6 | 2.81 ± 1.2 | 1.221 ± 0.4 | 2.358 ± 0.9 |
| rM(W,LS) | 0.33 ± 0.30 | 0.30 ± 0.21 | 0.99 ± 0.14 | 0.47 ± 0.26 |
Note: Statistics are presented as mean ± standard error of mean (confidence interval where appropriate). CVM = mutational coefficient of variation; = mutational heritability; IM = opportunity for selection; PRAND = frequency of estimates of IM from randomized data more extreme than the observed mean; rM(W,LS) = mutational correlation between life span and lifetime reproductive success; VM mutational variance at generation 250. See text for details of calculations.
Table 1.
Life Span and Mortality Models in Mutation Accumulation (MA) Lines
| Strain (species) |
HK104 (Caenorhabditis briggsae) |
PB800 (C. briggsae) |
N2 (C. elegans) |
PB306 (C. elegans) |
||||
| MA Generation | 0 | 250 | 0 | 250 | 0 | 250 | 0 | 250 |
| n lines | 10 | 25 | 10 | 30 | 10 | 41 | 10 | 35 |
| Life span ± SEM, (min–max) | 19.4 ± 0.2 (12–24) | 14.7 ± 0.3* (6–26) | 17.7 ± 0.6 (13–24) | 15.1 ± 0.4 (7–23) | 17.6 ± 0.4 (11–22) | 16.1 ± 0.2 (9–22) | 16.1 ± 0.5 (12–21) | 13.8 ± 0.3* (7–23) |
| ΔM × 103 (95% CI) | −0.887 ± 0.13 (−1.1 to −0.6), PRAND < .0002 | −0.497 ± 0.13 (−0.7 to −0.2), PRAND < .001 | −0.331 ± 0.09 (−0.5 to −0.2), PRAND < .004 | −0.597 ± 0.12 (−0.8 to −0.4), PRAND < .0004 | ||||
| Mortality model | ||||||||
| Uncensored | Gompertz | Logistic | Logistic | Logistic | Gompertz | Logistic | Gompertz | Logistic |
| Complete | Gompertz | Logistic | Logistic | Logistic | Gompertz | Logistic | Logistic | Logistic |
| a × 104 (95% CI) | ||||||||
| Uncensored | 0.7 (0.1 to 6.7) | 8.1† (2 to 34) | 0‡ (0 to 1) | 9.0§ (2 to 41) | 0.05 (0.1 to 5) | 0.6 (0.1 to 7) | 2.7 (0.4 to 19) | 0.7 (0.1 to 8) |
| Complete | 2.4 (0.5 to 13.2) | 3.1 (0.6 to 15.8) | 0 (0 to 13.8) | 1.6 (0.2 to 9.9) | 0.3 (0.03 to 3.1) | 0.2 (0.02 to 2.7) | 0.03 (0 to 48) | 0.1 (0.005 to 3) |
| b (95% CI) | ||||||||
| Uncensored | 0.42 (0.3 to 0.6) | 0.43 (0.3 to 0.6) | 1.34 (0.7 to 2.5) | 0.39§ (0.3 to 0.6) | 0.49 (0.4 to 0.6) | 0.55† (0.4 to 0.8) | 0.42 (0.3 to 0.6) | 0.72§ (0.5 to 1) |
| Complete | 0.31 (0.2 to 0.4) | 0.49 (0.3 to 0.7) | 1.33 (0.6 to 2.9) | 0.52 (0.4 to 0.7) | 0.51 (0.4 to 0.6) | 0.61 (0.4 to 0.8) | 0.79 (0.4 to 1.7) | 0.89 (0.6 to 1.3) |
| s (95% CI) | ||||||||
| Uncensored | — | 1.05 (0.5 to 2.3) | 5.3 (2.3 to 12.5) | 0.61† (0.2 to 2.2) | — | 0.74 (0.3 to 2.1) | n.d. | 1.85 (1 to 3.5) |
| Complete | — | 2.51 (1.2 to 5.2) | 5.4 (1.9 to 15.3) | 1.9 (0.8 to 4.6) | — | 1.3 (0.6 to 2.9) | 1.9 (0.4 to 9.6) | 4.5 (2.5 to 8) |
| VM (a) × 108 | 8.2 | 6.8 | 2.8 | 5.8 | ||||
| VM (b) × 106 | 7.8 | 1.9 | 3.2 | 23 | ||||
| (a) × 104 | 4.1 | 7.6 | 5.5 | 6.3 | ||||
| (b) × 104 | 24 | 0.83 | 2.3 | 9.9 | ||||
| ra,b | 0.293 | −0.548‖ | −0.853‖ | −0.399 | ||||
Notes: Life span was measured in 30 worms per MA line and 10 worms per ancestral control line. Mortality model fitting was conducted with the uncensored data alone and with the complete data set. All other calculations were conducted with uncensored data alone. ΔM × 103 = per-generation change in the mean scaled as a fraction of the control mean; a = initial adult mortality rate; b = slope of mortality rate function, rate of exponential increase in mortality with age; CI = confidence interval; = mutational heritability at generation 250; PRAND = frequency of estimates of ΔM from randomized data more extreme than the observed mean; ra,b = correlation coefficient of relationship between a and b; s = rate of senescence, decline in mortality rate at advanced ages; SEM = standard error of mean; VM = mutational variance at generation 250. See text for details of calculations.
Significant decrease in mean life span between generations 0 and 250, determined by an extension of the nonparametric Gehan's generalized Wilcoxon test, p ≤ .0075.
Significant change in mortality parameter for generation = 0 vs. generation = 250, significant after Bonferroni correction, p < .01.
Actual value, 2.1 × 10−9, indistinguishable from 0.
Significant change in mortality parameter for generation = 0 vs. generation = 250, significant before Bonferroni correction, p < .05.
Significant correlation between a and b, p < .05.
Mutational covariance between life span and fitness: Components of (co)variance were determined for life span and fitness (measured at generation 200 at 20°C and/or generation 220 at 25°C; see (40) for details) from the linear model y = trait + block + line + error where the fixed effect trait represents log(life span) and log(1+fitness). “Fitness” is lifetime fecundity weighted by probability of survivorship, which is highly correlated with demographic measures of fitness (r ≈ .9, C.F.B., unpublished data, 2007). The among-line component of covariance between traits is the genetic covariance, which in this case is the mutational covariance. Genetic correlations rM and their associated standard errors were calculated from the among-line components of (co)variance. Because fitness was measured at generation 200 or 220 and life span at generation 250, the last 30–50 generations of MA in the life span data do not contribute to the covariance with fitness. To account for the difference in numbers of generations, we multiplied the variance in life span by the average difference, 210/250, so  . The analysis was implemented in SAS v. 9.2 PROC MIXED with the unstructured covariance (“TYPE=UNR”) option. Some analyses using untransformed data failed to converge.
. The analysis was implemented in SAS v. 9.2 PROC MIXED with the unstructured covariance (“TYPE=UNR”) option. Some analyses using untransformed data failed to converge.
Standing genetic variance in life span in C. briggsae: Variance in life span among wild isolates of C. briggsae was partitioned taking several historical factors into account. Molecular evidence indicates that there are three major clades of C. briggsae, a tropical clade, an equatorial clade, and a temperate clade (41), and that the clades differ in the level of standing molecular variation. Preliminary analysis revealed that life span differs among clades; worms from the temperate clade live longer in our experimental conditions than do those from the other clades. Thus, we include clade as a fixed effect in the subsequent analysis. Moreover, samples (strains) were not collected in a uniform manner (unsurprisingly), and some strains are likely descended very recently from a common ancestor (e.g., five of the six “equatorial” lines had the exact same multilocus nuclear haplotype; 41). To account for the possibility that multiple strains from one collecting site are actually the same strain, we partitioned the variance within a clade into sampling location, strain within sampling location, and inbred line within strains. The full model is as follows: life span = clade + location + strain(location) + inbred line(strain(location)) + residual. Clade is a fixed effect, the other effects are random; the residual term is the environmental component of variance. Clade mean life spans were determined by least squares, given the random effects. Half the sum of the three random effects constitutes the genetic component of variance; we tested for significant genetic variance by likelihood ratio test of the comparison of the full model against a model with the three random effects removed; twice the difference in log likelihoods between the two models is chi square distributed with three degrees of freedom. Note that the total genetic variance includes a between-location component, which leads to some complications in assessing MSB; those complications are considered explicitly in the Discussion section. The genetic coefficient of variation, CVG, and opportunity for selection, IG, were calculated using the unweighted mean of the three clade mean life spans. The standard error of the joint parameter VM/VG (≈ S) was calculated by the Delta method (equation A1.19b, 42) with the standard errors of the individual parameters VM and VG calculated as described previously. We report the asymptotic 95% confidence interval.
Standing covariance between life span and fitness in C. briggsae: We first attempted to calculate the genetic correlation between life span and fitness in the wild isolates (Salomon et al., unpublished data, 2009) using the same hierarchical model as for the standing variance, but the analysis did not converge. Instead, we determined the trait averages for each inbred line and calculated the correlations of inbred line means from the raw (untransformed) data using the model y = trait + location + line(location) + inbred line(line(location)) + error; the analysis failed to converge when clade was included in the model. The analysis was implemented in SAS v. 9.2 PROC MIXED with the unstructured covariance (“TYPE=UNR”) option. Significance was assessed using likelihood ratio tests of the model with the unconstrained correlation (“TYPE=UNR”) versus the model with the correlation constrained to be 0 (“TYPE=UN(1)”). In this case, the correlation of line means is an upper bound on the absolute value of the genetic correlation because when traits are measured on the same individual the correlation of line means is confounded by 1/n times the environmental correlation, where n is the number of replicates of each line (43).
RESULTS
Change in the Mean and Variance
Mean life span (uncensored data only) decreased for 250 generations of MA in all four strains (Table 1 and Supplementary Figure 1). The per-generation change in mean life span (ΔM) declined significantly in all four strains. The rank order of decline was HK104 > PB306 > PB800 > N2; HK104 declined significantly faster than N2. Mean life spans of the ancestral controls of the four strains differed significantly (Kruskal–Wallis ANOVA, p = .0003), with HK104 ancestral controls having significantly longer life span than PB306 (p = .00009) and PB800 (p = .043), but not N2 (p = .10).
For a trait under directional selection (see following), I, the opportunity for selection, represents the upper bound for adaptive evolution and is the most meaningful measure of variance (36–38). Although there were no significant differences in IM among the four strains, N2 had the smallest IM and PB800 had the largest (Table 2). Mutational heritabilities were of the order 10−3 and mutational CVs were of the order 0.5–1, both typical for life history traits (5).
Mortality Models for MA Lines
We determined the best-fit model for uncensored data from the MA and ancestral control lines (Table 1) using the maximum likelihood methods implemented in WinModest (34). For three of the strains (N2, PB306, HK104), the ancestral control lines were best described by the Gompertz model, which includes terms for initial adult mortality rate (a, intercept) and the rate of exponential increase in mortality with age (b, slope). Although PB306 had a substantially higher mortality intercept than N2 and HK104, the three strains had very similar mortality slopes. Ancestral control lines of PB800 were best fit to the logistic model, which also includes a term for the degree of deceleration in mortality at advanced ages (s). PB800 ancestral control lines had the smallest intercept of any controls, but the highest slope. The MA lines of all four strains were best described by the Logistic model.
To compare mortality parameters between lines fit to different models, we fit all lines to the Logistic–Makeham model, which has four mortality parameters, and made pairwise comparisons using the log-likelihood ratio test of WinModest and with a Bonferroni correction for multiple tests. The N2 MA lines had a significantly higher slope than the ancestral control lines of N2 (p < .01). This increase in slope in the MA lines also was detected for PB306 and PB800 but was not significant after the Bonferroni correction. Caenorhabditis briggsae had more mutational variance for the mortality intercept than did C. elegans, specifically with the largest variance in HK104. Interestingly, this pattern did not hold for the rate of mortality slope, given the large variance in PB306. We found significant negative correlations between the mortality intercepts and slopes in PB800 and N2.
The percentages of censored data points (missing worms) in the MA lines of each strain were slightly but not significantly higher than in the respective ancestral control lines (data not shown; Mann–Whitney U test, p > .161 for all strains). For nearly all lines, 43%–51% of the censored data points occurred on days 3–6 (data not shown), which were the days that the worms were transferred during their reproductive period. The PB800 ancestral control lines differed from this pattern, with only 28% of the censored data points occurring on days 3–6. In contrast, fewer than 7% (on average) of the nearly 5,000 uncensored data points (confirmed deaths) occurred during the reproductive period (data not shown). On average, the last days that censored data points were recorded (scaled to the mean life span of the uncensored worms on each individual plate) were slightly but not significantly lower in the ancestral control lines of HK104, N2, and PB306 than in the respective MA lines (p ≥ .359); this trend was reversed for PB800 (p = .156). When the complete data set was analyzed (censored and uncensored), determination of the best-fit mortality model was altered only for the PB306 ancestral control (Gompertz → Logistic; Table 1). Confidence limits of the estimates of the mortality parameters in the models of the complete data sets overlapped with the confidence limits of the models of the uncensored data. Because these results suggest that the main contribution to the censored data set was handling error during worm transfers, all other analyses outside of determination of mortality model were conducted on the uncensored data sets only.
Mutational Correlation Between Life Span and Fitness
In three of the four strains, the mutational correlation between life span and fitness rM(W,LS) did not differ significantly from zero, although it is positive in all cases (Table 2). Point estimates from the two C. briggsae strains are very similar to each other (≈0.3); the PB306 strain of C. elegans is slightly larger (≈0.47). In N2, however, rM(W,LS) ≈ 1. Previous estimates of mutational correlations in the N2 strain range from 0.2 (44) to about 0.5 (45); estimates from two TE mutator strains derived from N2 are also around 0.5 (44). It is unclear why the value for N2 is so much greater than those of the other strains or from other experiments with N2, although sampling variance is always a possible explanation.
Life Span in Natural Isolates of Caenorhabditis briggsae
Worms from the temperate clade lived longer than worms from the equatorial or tropical clades (p < .05; Table 3 and Supplementary Figure 2; uncensored data only). The mean life span of worms from wild isolates was longer than those of the ancestral controls of the four strains used in the MA experiment, presumably due to consistent differences in the protocols of the two experiments. We detected significant genetic variation for life span (VG) in the wild worm isolates (averaged over the three clades, p < .01; likelihood ratio test (LRT) χ2 = 11.8, df = 3), although the broad sense heritability for life span was quite low (H2 = 0.054).
Table 3.
Life Span, Mortality Models, and Estimates of (Co)Variance for Wild Caenorhabditis briggsae Isolates
| Clade | Equatorial | Temperate | Tropical |
| Life span ± SEM (min–max) | 20.8 ± 1.2 (14–28) | 24.8 ± 1.5* (16–32) | 19.7 ± 1.5 (13–28) |
| n (locations, strains, no. of worms/strain) | (1, 6, 8–13) | (8, 31, 6–17) | (2, 6, 9–12) |
| Mortality model | |||
| Uncensored | Gompertz | Gompertz | Gompertz |
| Complete | Logistic | Logistic | Logistic |
| a (×104) (95% CI) | |||
| Uncensored | 5.6 (0.5–73) | 4.97 (0.5–64) | 10.1 (0.9–140) |
| Complete | 0.6 (0.04–10.8) | 3.2 (1.4–7) | 0.06 (0†–5.9) |
| b (95% CI) | |||
| Uncensored | 0.29 (0.2–0.5) | 0.26 (0.2–0.4) | 0.28 (0.2–0.5) |
| Complete | 0.42 (0.3–0.6) | 0.25 (0.2–0.3) | 0.64 (0.4–1.1) |
| s (95% CI) | |||
| Complete | 0.89 (0.3–2.8) | 0.26 (0.1–0.7) | 2.7 (1.2–5.9) |
| VG (a) × 107 | 1.618 | ||
| VG (b) × 103 | 2.166 | ||
| ra,b | −0.7018‡ | ||
| VG (LS) all clades | 1.27§ (0.72) | ||
| VE (LS) all clades | 23.54 (1.62) | ||
| IG (LS) all clades | 0.0026 (0–0.0058)‖ | ||
| H2 (LS) all clades | 0.054 (0–0.13)‖ | ||
| S (=VM/VG) | 0.020 (0–0.057)‖ | ||
| r(W.LS, inbred line) | 0.3¶ ± 0.12 | ||
| r(W.LS, strain) | 0.25 ± 0.70 | ||
| r(W.LS, location) | −0.66 ± 0.33 | ||
Notes: Life span was measured in 15 worms per strain. Mortality model fitting was conducted with the uncensored data alone and with the complete data set. All other calculations were conducted with uncensored data alone. a = initial adult mortality rate; b = slope of mortality rate function, rate of exponential increase in mortality with age; CI = confidence interval; H2 = broad-sense heritability; IG = opportunity for selection; ra,b = correlation coefficient of relationship between a and b; s = selection coefficient as calculated from VG/VM; VE = environmental variance; VG = standing genetic variance. See text for details of calculation.
Life span of temperate worms significantly longer than equatorial and tropical, determined by an extension of the nonparametric Gehan's generalized Wilcoxon test, p < .05.
Actual value, 6.1 × 10−8, indistinguishable from 0.
Significant correlation of line means between a and b, p < .05.
p < .01 by likelihood ratio test of the full model vs. the null hypothesis of no genetic variance (χ2 = 11.8, df = 3).
Asymptotic 95% CI.
p < .02 by likelihood ratio test of the full model vs. the null hypothesis of no genetic variance (χ2 = 5.6, df = 1).
All clades were best described by the Gompertz model (uncensored data only), with no significant differences among clades in the slope or intercept. As found in some of the laboratory strains, the intercepts and slopes were significantly negatively correlated in the wild worm isolates (calculated from data pooled across clades). There were no significant differences in the percentages of censored data points (missing worms) across the three clades (p = .181). The averaged last days that censored data points were recorded (scaled to the mean life span of the uncensored worms for each strain) did not differ among clades (p = .662). When the complete data set was analyzed (censored and uncensored), determination of the best-fit mortality model was altered for all three clades (Gompertz → Logistic; Table 3). To what extent this alteration is influenced by the lack of censored data points prior to day 5 (data not recorded) is unknown. Confidence limits of the estimates of the mortality parameters in the models of the complete data sets overlapped with the confidence limits of the models of the uncensored data.
Approximate genetic correlations between life span and fitness for the different hierarchical levels are reported in Table 3. Point estimates of correlations at the level of inbred line (r = .30, LRT χ2 = 5.6, df = 1, p < .02) and strain (r = .25, p > .7) closely reflect the mutational correlation for C. briggsae; in contrast, the correlation at the level of location (r = −.65, p > .14; we were unable to include clade in the model) was large and negative. We consider the potential meaning of the qualitative difference at the different hierarchical levels in the Discussion section.
DISCUSSION
Spontaneous mutations consistently decrease post-reproductive life span in C. elegans and C. briggsae. These results are broadly consistent with our characterizations of mutational effects on fitness and body size in these strains (30,31,40) and on life span in fruit flies (46–48). A consistent change in mean phenotype with MA under relaxed selection implies that the trait is under directional (not necessarily direct) selection. Previous studies with the N2 strain of C. elegans have produced conflicting results (23,45,49), and our data show that life span in N2 does appear to decline more slowly than in the other strains, in particular the HK104 strain of C. briggsae. Mutational variance is inherently more difficult to measure than the change in the trait mean (50), but the patterns in mutational variance are quite similar to the change in the mean. The exception to the generality of these results is that in all previous studies, the PB800 strain of C. briggsae has also consistently declined faster and accumulated mutational variance faster than the two strains of C. elegans. There is no obvious reason why life span should be different, although sampling variance is an obvious possibility. Interestingly, the HK104 ancestral controls lived significantly longer than the other strains, suggesting an underlying difference in some aspect of the genetic architecture of life span between HK104 and the other strains. Moreover, the ancestral HK104 has consistently lower reproductive output than the other strains (30). One possibility is that the ancestral HK104 harbors an allele of large effect that increases life span and simultaneously decreases reproduction, such alleles are known from the literature (e.g., 51). If so, it implies that the allele of large effect acts epistatically in such a way to exacerbate the cumulative effects of mutations of small effect at other loci, that is, epistasis is synergistic.
In laboratory populations of rhabditid nematodes, as in humans, the post-reproductive period is lengthy. Whether this feature of their life history occurs in nature or is an artifact of the laboratory environment is unclear (52). It seems extremely unlikely that post-reproductive life span is the direct target of selection in nematodes; the most likely explanation is that mutant alleles that affect life span have deleterious pleiotropic effects on other traits that directly underlie fitness. The mutational correlation of life span with lifetime reproductive output (i.e., fitness) is consistently positive, more so in C. elegans than in C. briggsae. This result is consistent with a pleiotropic relationship between life span and fitness, although the alternative explanation of a genetic correlation resulting from different numbers of (nonpleiotropic) mutations in different lines cannot be ruled out (53; see following). Previous studies with the N2 strain of C. elegans and its derivatives have also found a positive correlation between life span and fecundity (23,45). Given that many alleles of known large effect tend to simultaneously increase life span and reduce fecundity, this pattern suggests that alleles of small to modest effect, such as are likely to accumulate in an MA experiment, have qualitatively different pleiotropic effects than do (many) alleles of large effect.
Mortality Models
The strength of interpretation of mortality patterns are tightly linked to the number of individuals within a group (within each MA line or within each inbred wild worm strain) (18,54), and the sample sizes in this study are on the low end (30 worms per MA line, 10 worms per ancestral control pseudoline, 15 worms per wild worm strain). Nonetheless, we found some interesting patterns in mortality models, both within and across species. For three of the four strains (HK104, N2, PB306), mortality patterns of the ancestral control lines were best fit by the Gompertz model and MA lines were best fit by the Logistic model. These two models differ by the inclusion of an extra term in the Logistic model that describes the rate of deceleration in mortality at advanced ages. Most characterizations of mortality curves in C. elegans include an early period of exponential increase in mortality (e.g., 55), and a period of deceleration in mortality at advanced ages (e.g., 56,57). The shift from Gompertz to Logistic in three of the strains is intriguing and suggests that the mutations have age-specific effects that slow mortality at advanced ages, which could be confirmed by an analysis of age-specific mutational variances for life span (18,46). The initial mortality rates (a, intercept) for the ancestral controls of HK104, N2, and PB306 were in the range previously reported for C. elegans (56), and in three of the four strains were higher in the MA lines than in ancestral controls. Similar increases in initial mortality rates following MA have been documented in D. melanogaster (47,48). The mortality slopes detected in the worm MA lines are substantially higher than comparable studies of fruit flies (18,46–48,58), in which age-specific mortality rates appear to be under direct selection, particularly in young flies (18,46). Across strains we found a mixed pattern of changes in mortality slope with MA, as has been documented previously (48). The most unusual strain was PB800, in which both the ancestral controls and MA lines fit the Logistic model, the mortality intercept in the ancestral controls was extremely low, and MA lines had decreased slope in comparison with ancestral controls. It is unclear why the PB800 strain behaved differently from the others, a trend we did not detect in previous studies of these MA lines (30,31,40).
When these analyses were repeated with the censored data (missing worms) included, the overall results were fairly similar to those discussed previously. In most cases (but not the PB800 ancestral control), the majority of the censored data points occurred during the days that the worms were transferred to new plates to avoid confusing parents and offspring. We cannot determine to what degree this pattern of accumulation of early-censored data points is due to mishandling of worms versus other causes of worm disappearance. Nonetheless, analysis of the complete data set did result in several interesting changes to the mortality model results, particularly the shift from Gompertz to Logistic dynamics for the PB306 ancestral control and the increase in the rates of deceleration of mortality in all strains.
Very little is known about mutational variances of mortality models parameters because MA studies typically measure variance in life span without fitting life-span data to mortality models (e.g., 23,25,49), or assess mortality models but not mutational variances in model parameters (e.g., 47,48). In one notable exception (46), 19 generations of MA in female D. melanogaster resulted in significant increases in mutational variance for all mortality parameters (Gompertz and Logistic models). However, because Pletcher and coworkers (46) did not report environmental variances for the mortality model parameters, we are unable to directly compare our mortality model results. Mutational heritabilities for the mortality parameters in our MA lines were slightly lower than those of other life history traits in nematodes (25,31,40,59,60) and lower than those for life span as detected in this study. The relatively lower mutational heritability likely is due to increased environmental variance (within-line component of variance) in our estimates of the mortality models.
In three strains (not HK104), we detected a negative correlation between the mortality slope and intercept (significant in PB800 and N2). This is consistent with previous studies (e.g., 61,62), and suggests that low baseline mortality may be associated with accelerating mortality rates at advanced ages. Interestingly, we detected the opposite (but nonsignificant) pattern in HK104. HK104 has the somewhat mysterious property of having consistently large main effects (ΔM, CVM) and low mutational correlations between traits (31,40) (except perhaps life span; Table 2). HK104 is also particularly susceptible to random environmental variation (63). We suspect that the generally weak genetic correlation between traits in HK104 is due to the greater influence of random environmental effects in that strain than in the others.
The patterns of mortality in the three clades of wild worms were best described by the Gompertz model, characterized by high initial mortality rates but relatively low mortality slopes. The mortality rates and slopes in the wild worm isolates are comparable with those of wild-caught D. simulans (19,64) and bean beetles (54). Analysis of the complete data set shifted the best-fit model type from Gompertz to Logistic, but we cannot evaluate whether this actually reflects deceleration in mortality at advanced ages when the censored data are included since censored data were not recorded during the reproductive period (worm transferring period) in this experiment. The standing genetic variances for the mortality parameters, particularly for the slope, are lower than estimates of additive genetic variance for these parameters in fruit flies (18,61). We detected greater genetic variance for the intercept than the slope (when scaled to the trait mean), consistent with patterns of additive genetic variance in some (18,61), but not all (20), studies of fruit flies. As seen in three of the four strains used for MA and documented previously (19), the mortality slopes and intercepts were negatively correlated.
Standing Genetic Variance in Life Span
The significant relationship of life span with clade membership in the wild isolates of C. briggsae suggests that global MSB cannot completely explain the standing genetic variance for life span in this species. This result is at odds with our findings for fitness and body size, in which there was no significant relationship of trait value with clade (43). Cutter and coworkers (65) have argued that the temperate clade of C. briggsae was recently derived from a tropical ancestor, and it is possible that life span diverged during the founding. Genetic drift associated with small founding population sizes and genetic draft (ie, hitchhiking; 66) associated with adaptive divergence are plausible explanations.
On a more local (and recent) scale, the pattern of genetic variation is quite consistent with a local MSB scenario. Summing over the three hierarchical levels of genetic variation (among locations, among populations, and among inbred lines) and employing the large population MSB formulation (VG ≈ VM/S), the point estimate of the average selection coefficient, S, against mutations affecting life span is about 2%. This value is very similar to the point estimate of the strength of selection against mutations affecting fitness in C. briggsae (also about 2%; 43), although the confidence limits on both estimates are large. The observed decline in mean life span with MA (∼0.05%–0.1% per generation; see Table 1) provides strong evidence that mutations affecting life span are deleterious. If we take the point estimate of the mutational correlation of life span with fitness in C. briggsae (∼0.3) at face value, we conclude that the estimated selection coefficient of 2% against mutations affecting life span is perhaps somewhat of an overestimate but not grossly so. It is important to note that divergence between populations (adaptive or neutral) will cause the VM/VG formulation to underestimate the average selection coefficient (by inflating the denominator), so if the inferred strength of selection against mutations affecting life span is an overestimate, it is not a result of pooling the components of genetic variance as we have done in this calculation.
The standing genetic correlation between life span and fitness in the wild isolates of C. briggsae reveals an interesting pattern. The genetic component of covariance (and hence correlation) can be partitioned into among inbred lines within a strain (i.e., due to the shared effects of alleles within an ancestral individual), among strains within a location (i.e., due to the shared effects of alleles segregating in the population), and among locations within a clade (due to the shared effects of alleles fixed or at different frequencies in the different locations). The point estimates of the among-inbred line correlation (r = .30) and the among-strain correlation (r = .25) are very close to the average mutational correlation of 0.3. That result strongly suggests that the pattern of genetic correlation observed within and between strains within a location faithfully reflects the mutational correlation and reinforces the conclusion that MSB is a sufficient explanation for the observed genetic variation in life span at that hierarchical level. In contrast, the genetic correlation among locations is large and negative (r = −.66). We were unable to include clade in the model due to failure to converge, so the among-location correlation includes both within- and among-clade effects. Although there are only three clades, the rank order (from highest to lowest) of fitness and life span is tropical > equatorial > temperate and temperate > equatorial > tropical, respectively, that is, there is a qualitative negative correlation between fitness and life span at the clade level. Qualitatively, the only major difference is that temperate clade worms have a substantially longer life span than the others. A tempting hypothesis is that in the course of diverging from the tropical ancestor, an allele of large effect that increases life span but decreases fecundity became fixed in the temperate populations.
It is important to recognize that genetic correlations among MA lines (what we have called mutational correlations) can result both from pleiotropy and from linkage disequilibrium (LD) due to different MA lines harboring different numbers of mutations (53,67). This result derives from the explicit assumption that the number of mutations is Poisson distributed among lines and is independent of the expected number of mutations accumulated. The details depend on the distribution of mutational effects, but even in the absence of any pleiotropy, the mutational correlation will always be nonzero due to this LD as long as some lines harbor at least one mutation affecting each trait and the distribution of mutational effects is not completely symmetrical around zero. Estes and Phillips (67) demonstrated that for mutations with fairly large phenotypic effect (E(a) ∼ 0.05–0.2) these nonpleiotropic correlations are not terribly large, although we assume they contribute to the observed correlation. This phenomenon obviously extends to natural populations, although we are unaware of any theoretical treatment in this regard.
It is interesting to consider these results in the context of the evolution of “aging” (i.e., senescence) (21,26–29). Under the AP hypothesis, mutations that increase early reproduction but reduce life span will be beneficial because early reproduction contributes more to fitness than late reproduction. If variation in life span is solely due to alleles that exhibit AP, there is no reason to expect a decrease in life span with relaxation of selection, and in fact, mean life span might plausibly increase because deleterious alleles that reduce early reproduction might cause a pleiotropic increase in life span. Conversely, if variation in life span is solely due to alleles with deleterious effects on both life span and fitness, as predicted under the MA* hypothesis, we predict that mean life span will decrease with relaxed selection, as we observe here. Viewed in the context of genetic correlations, AP predicts a negative mutational correlation between fitness (as defined here) and life span because longer-lived worms have lower fitness. Conversely, MA* predicts a positive mutational correlation between fitness and life span because deleterious alleles that reduce both life span and fitness can now accumulate.
The variation in the direction of the standing genetic correlations in C. briggsae with hierarchical scale has implications for the causes of variation in aging. Within locations (i.e., clades), the positive genetic correlation between fitness and life span suggests a primary role for MA* and, more importantly, provides no evidence for any role of AP. That is, MSB is a sufficient explanation for the observed variation in life span, and for the relationship between life span and fitness. Between locations, the negative genetic correlation suggests that alleles with antagonistic effects on life span and fitness as defined here may have differently fixed in the different clades (or at least the allele frequencies differ) and that deterministic MSB* is not a sufficient explanation for the variation in life span.
Finally, it should be noted that although post-reproductive life span as measured in the lab is unlikely to be the direct target of selection, there may be different, and relevant, correlations that we have not measured. Most obvious is the possible correlation of post-reproductive adult life span as we have measured it with survival during the dauer stage, which is quite plausibly a target of direct selection, such that longer-lived dauers have higher (68) fitness (but see 69). Mutant alleles of large effect that increase post-reproductive adult life span often also influence the dauer stage (for review, see 70–72). Because fitness is inversely correlated with life span in these large-effect mutants (opposite of the MA trend we have documented), there is some reason to expect the MA trend in dauer life span would also differ.
SUPPLEMENTARY MATERIAL
Supplementary material can be found at: http://biomed.gerontologyjournals.org/.
FUNDING
National Institutes of Health (NIH)National Research Service Award postdoctoral fellowship to C.F.B.; NIH award to C.F.B. and D. R. Denver (1 R01GM072639-01A2); University of Florida Claude D. Pepper Older Americans Independence Center pilot study grant to J.J.-M.; and University of Florida University Scholars Award to A.U.
Supplementary Material
Acknowledgments
This work had its origins when C.F.B. was a postdoc in Mike Lynch's laboratory at the Indiana University. We thank the Indiana worm crew for 200 generations of hard labor. We are indebted to the Florida worm crew: Veronica Chik, Smokin’ Joe Hong, Khyzer Izhar, Chikako Matsuba, Laurence Sylvestre, Rebecca Theobald, and Judit Ungvari-Martin. We thank D. R. Denver, M. Lynch, R. D. Holt, L. F. Matos, and M. L. Wayne for their helpful advice and suggestions. We thank the two anonymous reviewers for their many helpful comments, especially the second reviewer for catching a potentially embarrassing lapse in (C.F.B.’s) logic.
References
- 1.Lewontin RC. The Genetic Basis of Evolutionary Change. New York: Columbia University Press; 1974. [Google Scholar]
- 2.Singh RS, Krimbas CB. Evolutionary Genetics: From Molecules to Morphology. Cambridge, UK: Cambridge University Press; 2000. [Google Scholar]
- 3.Charlesworth B, Hughes KA. The maintenance of genetic variation in life-history traits. In: Singh RS, Krimbas CB, editors. Evolutionary Genetics: From Molecules to Morphology. Cambridge, UK: Cambridge University Press; 1999. pp. 369–392. [Google Scholar]
- 4.Fisher RA. The Genetical Theory of Natural Selection. Oxford, UK: Oxford University Press; 1930. [Google Scholar]
- 5.Houle D, Morikawa B, Lynch M. Comparing mutational variabilities. Genetics. 1996;143:1467–1483. doi: 10.1093/genetics/143.3.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barton NH. Pleiotropic models of quantitative variation. Genetics. 1990;124:773–782. doi: 10.1093/genetics/124.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukai T, Nagano S. The genetic structure of natural populations of Drosophila melanogaster. XVI. Excess of additive genetic variance of viability. Genetics. 1983;105:115–134. doi: 10.1093/genetics/105.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reznick D, Ghalambor C, Nunney L. The evolution of senescence in fish. Mech Ageing Dev. 2002;123:773–789. doi: 10.1016/s0047-6374(01)00423-7. [DOI] [PubMed] [Google Scholar]
- 9.Packer C, Tatar M, Collins A. Reproductive cessation in female mammals. Nature. 1998;392:807–811. doi: 10.1038/33910. [DOI] [PubMed] [Google Scholar]
- 10.Reznick D, Bryant M, Holmes D. The evolution of senescence and post-reproductive lifespan in guppies (Poecilia reticulata) PLoS Biol. 2006;4:e7. doi: 10.1371/journal.pbio.0040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Headrick DH, Goeden RD. The biology of nonfrugivorous tephritid fruit flies. Annu Rev Entomol. 1998;43:217–241. doi: 10.1146/annurev.ento.43.1.217. [DOI] [PubMed] [Google Scholar]
- 12.Carey JR, Liedo P, Müller H-G, et al. Biodemography of a long-lived tephritid: reproduction and longevity in a large cohort of female Mexican fruit flies, Anastrepha ludens. Exp Gerontol. 2005;40:793–800. doi: 10.1016/j.exger.2005.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bateman MA. The ecology of fruit flies. Annu Rev Entomol. 1972;17:496–518. [Google Scholar]
- 14.Hawkes K, O’Connell JF, Blurton-Jones NG, Alvarez H, Charnov EL. Grandmothering, menopause, and the evolution of human life histories. Proc Natl Acad Sci U S A. 1998;95:1336–1339. doi: 10.1073/pnas.95.3.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shanley DP, Kirkwood TBL. Evolution of the human menopause. Bioessays. 2001;23:282–287. doi: 10.1002/1521-1878(200103)23:3<282::AID-BIES1038>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 16.Lahdenpera M, Lummaa V, Helle S, Tremblay M, Russell AF. Fitness benefits of prolonged post-reproductive lifespan in women. Nature. 2004;428:178–181. doi: 10.1038/nature02367. [DOI] [PubMed] [Google Scholar]
- 17.Pletcher SD, Khazaeli AA, Curtsinger JW. Why do life spans differ? Partitioning mean longevity differences in terms of age-specific mortality parameters. J Gerontol A Biol Sci Med Sci. 2000;55:B381–B389. doi: 10.1093/gerona/55.8.b381. [DOI] [PubMed] [Google Scholar]
- 18.Promislow DEL, Tatar M, Khazaeli AA, Curtsinger JW. Age-specific patterns of genetic variance in Drosophila melanogaster. I. Mortality. Genetics. 1996;143:839–848. doi: 10.1093/genetics/143.2.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spencer CC, Promislow DEL. Age-specific changes in epistatic effects on mortality rate in Drosophila melanogaster. J Hered. 2005;96:513–521. doi: 10.1093/jhered/esi071. [DOI] [PubMed] [Google Scholar]
- 20.Hughes KA, Charlesworth B. A genetic analysis of senescence in Drosophila. Nature. 1994;367:64–66. doi: 10.1038/367064a0. [DOI] [PubMed] [Google Scholar]
- 21.Curtsinger JW, Fukui HH, Khazaeli AA, et al. Genetic variation and aging. Annu Rev Genet. 1995;29:553–575. doi: 10.1146/annurev.ge.29.120195.003005. [DOI] [PubMed] [Google Scholar]
- 22.Wilson AJ, Nussey DH, Pemberton JM, et al. Evidence for a genetic basis of aging in two wild vertebrate populations. Curr Biol. 2007;17:2136–2142. doi: 10.1016/j.cub.2007.11.043. [DOI] [PubMed] [Google Scholar]
- 23.Bégin M, Schoen DJ. Low impact of germline transposition on the rate of mildly deleterious mutation in Caenorhabditis elegans. Genetics. 2006;174:2129–2136. doi: 10.1534/genetics.106.065508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Houle D, Hughes KA, Hoffmaster DK, et al. The effects of spontaneous mutation on quantitative traits. I. Variances and covariances of life history traits. Genetics. 1994;138:773–785. doi: 10.1093/genetics/138.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vassilieva LL, Lynch M. The rate of spontaneous mutation for life-history traits in Caenorhabditis elegans. Genetics. 1999;151:119–129. doi: 10.1093/genetics/151.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamilton WD. The moulding of senescence by natural selection. J Theor Biol. 1966;12:12–45. doi: 10.1016/0022-5193(66)90184-6. [DOI] [PubMed] [Google Scholar]
- 27.Williams GC. Pleiotropy, natural selection, and the evolution of senescence. Evolution. 1957;11:398–411. [Google Scholar]
- 28.Partridge L, Barton NH. Optimality, mutation and the evolution of ageing. Nature. 1993;362:305–311. doi: 10.1038/362305a0. [DOI] [PubMed] [Google Scholar]
- 29.Hughes KA, Reynolds RM. Evolutionary and mechanistic theories of aging. Annu Rev Entomol. 2005;50:421–445. doi: 10.1146/annurev.ento.50.071803.130409. [DOI] [PubMed] [Google Scholar]
- 30.Baer CF, Shaw F, Steding C, et al. Comparative evolutionary genetics of spontaneous mutations affecting fitness in rhabditid nematodes. Proc Natl Acad Sci U S A. 2005;102:5785–5790. doi: 10.1073/pnas.0406056102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ostrow D, Phillips N, Avalos A, et al. Mutational bias for body size in rhabditid nematodes. Genetics. 2007;176:1653–1661. doi: 10.1534/genetics.107.074666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wood WB. The Nematode Caenorhabditis elegans. Plainview, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 33.Stiernagle T. Maintenance of C. elegans. In: Hope IA, editor. C. elegans: A Practical Approach. London: Oxford University Press; 1999. pp. 51–67. [Google Scholar]
- 34.Pletcher SD. Model fitting and hypothesis testing for age-specific mortality data. J Evol Biol. 1999;12:430–439. [Google Scholar]
- 35.Promislow DEL, Tatar M, Pletcher SD, Carey JR. Below-threshold mortality: implications for studies in evolution, ecology and demography. J Evol Biol. 1999;12:314–328. [Google Scholar]
- 36.Houle D. Comparing evolvability and variability of quantitative traits. Genetics. 1992;130:195–204. doi: 10.1093/genetics/130.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crow JF. Some possibilities for measuring selection intensities in man. Hum Biol. 1958;30:1–13. [PubMed] [Google Scholar]
- 38.Wade MJ. Natural selection. In: Fox CW, Wolf JB, editors. Evolutionary Genetics. New York: Oxford University Press; 2006. pp. 49–64. [Google Scholar]
- 39.Efron B, Tibshirani R. An Introduction to the Boostrap. New York: Chapman and Hall; 1993. [Google Scholar]
- 40.Baer CF, Phillips N, Ostrow D, et al. Cumulative effects of spontaneous mutations for fitness in Caenorhabditis: role of genotype, environment, and stress. Genetics. 2006;174:1387–1395. doi: 10.1534/genetics.106.061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dolgin ES, Félix MA, Cutter AD. Hakuna Nematoda: genetic and phenotypic diversity in African isolates of Caenorhabditis elegans and C. briggsae. Heredity. 2008;100:304–315. doi: 10.1038/sj.hdy.6801079. [DOI] [PubMed] [Google Scholar]
- 42.Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 43.Via S. The quantitative genetics of polyphagy in an insect herbivore. 2. Genetic correlations in larval performance within and among host plants. Evolution. 1984;38:896–905. doi: 10.1111/j.1558-5646.1984.tb00360.x. [DOI] [PubMed] [Google Scholar]
- 44.Bégin M, Schoen DJ. Transposable elements, mutational correlations, and population divergence in Caenorhabditis elegans. Evolution. 2007;61:1062–1070. doi: 10.1111/j.1558-5646.2007.00097.x. [DOI] [PubMed] [Google Scholar]
- 45.Vassilieva LL, Hook AM, Lynch M. The fitness effects of spontaneous mutations in Caenorhabditis elegans. Evolution. 2000;54:1234–1246. doi: 10.1111/j.0014-3820.2000.tb00557.x. [DOI] [PubMed] [Google Scholar]
- 46.Pletcher SD, Houle D, Curtsinger JW. Age-specific properties of spontaneous mutations affecting mortality in Drosophila melanogaster. Genetics. 1998;148:287–303. doi: 10.1093/genetics/148.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yampolsky LY, Pearse LE, Promislow DEL. Age-specific effects of novel mutations in Drosophila melanogaster. Genetica. 2001;110:11–29. doi: 10.1023/a:1017582625191. [DOI] [PubMed] [Google Scholar]
- 48.Gong Y, Thompson JN, Woodruff RC. Effect of deleterious mutations on life span in Drosophila melanogaster. J Gerontol A Biol Sci Med Sci. 2006;61:1246–1252. doi: 10.1093/gerona/61.12.1246. [DOI] [PubMed] [Google Scholar]
- 49.Keightley PD, Caballero A. Genomic mutation rates for lifetime reproductive output and lifespan in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1997;94:3823–3827. doi: 10.1073/pnas.94.8.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shabalina SA, Yapolsky LY, Kondrashov AS. Rapid decline of fitness in panmictic populations of Drosophila melanogaster maintained under relaxed natural selection. Proc Nat Acad Sci U S A. 1997;94:13034–13039. doi: 10.1073/pnas.94.24.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jenkins NL, McColl G, Lithgow GJ. Fitness cost of extended lifespan in Caenorhabditis elegans. Proc Biol Sci. 2004;271:2523–2526. doi: 10.1098/rspb.2004.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Voorhies WA, Fuchs J, Thomas S. The longevity of Caenorhabditis elegans in soil. Biol Lett. 2005;1:247–249. doi: 10.1098/rsbl.2004.0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keightley PD, Davies EK, Peters AD, Shaw RG. Properties of ethylmethane sulfonate-induced mutations affecting life-history traits in Caenorhabditis elegans and inferences about bivariate distributions of mutation effects. Genetics. 2000;156:143–154. doi: 10.1093/genetics/156.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tatar M, Carey JR. Genetics of mortality in the bean beetle Callosobruchus maculatus. Evolution. 1994;48:1371–1376. doi: 10.1111/j.1558-5646.1994.tb05320.x. [DOI] [PubMed] [Google Scholar]
- 55.Brooks A, Lithgow GJ, Johnson TE. Mortality rates in a genetically heterogeneous population of Caenorhabditis elegans. Science. 1994;263:668. doi: 10.1126/science.8303273. [DOI] [PubMed] [Google Scholar]
- 56.Johnson TE, Wu D, Tedesco P, Dames S, Vaupel JW. Age-specific demographic profiles of longevity mutants in Caenorhabditis elegans show segmental effects. J Gerontol A Biol Sci Med Sci. 2001;56:B331–B339. doi: 10.1093/gerona/56.8.b331. [DOI] [PubMed] [Google Scholar]
- 57.Vanfleteren JR, de Vreese A, Braeckman BP. Two-parameter logistic and Weibull equations provide better fits to survival data from isogenic populations of Caenorhabditis elegans in axenic culture than does the Gompertz model. J Gerontol A Biol Sci Med Sci. 1998;53:B393–B403. doi: 10.1093/gerona/53a.6.b393. [DOI] [PubMed] [Google Scholar]
- 58.Curtsinger JW, Fukui HH, Townsend DR, Vaupel JW. Demography of genotypes: failure of the limited life-span paradigm in Drosophila melanogaster. Science. 1992;258:461–463. doi: 10.1126/science.1411541. [DOI] [PubMed] [Google Scholar]
- 59.Ajie BC, Estes S, Lynch M, Phillips PC. Behavioral degradation under mutation accumulation in Caenorhabditis elegans. Genetics. 2005;170:655–660. doi: 10.1534/genetics.104.040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Azevedo RBR, Keightley PD, Lauren-Maatta C, et al. Spontaneous mutational variation for body size in Caenorhabditis elegans. Genetics. 2002;162:755–765. doi: 10.1093/genetics/162.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hughes KA. The evolutionary genetics of male life-history characters in Drosophila melanogaster. Evolution. 1995;49:521–537. doi: 10.1111/j.1558-5646.1995.tb02284.x. [DOI] [PubMed] [Google Scholar]
- 62.Strehler BL, Mildvan AS. General theory of mortality and aging. Science. 1960;132:14–21. doi: 10.1126/science.132.3418.14. [DOI] [PubMed] [Google Scholar]
- 63.Baer CF. Quantifying the decanalizing effects of spontaneous mutations in rhabditid nematodes. Am Nat. 2008;172:272–281. doi: 10.1086/589455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Melvin RG, Ballard JWO. Intraspecific variation in survival and mitochondrial oxidative phosphorylation in wild-caught Drosophila simulans. Aging Cell. 2006;5:225–233. doi: 10.1111/j.1474-9726.2006.00211.x. [DOI] [PubMed] [Google Scholar]
- 65.Cutter AD, Félix M-A, Barrière A, Charlesworth D. Patterns of nucleotide polymorphism distinguish temperate and tropical wild isolates of Caenorhabditis briggsae. Genetics. 2006;173:2021–2031. doi: 10.1534/genetics.106.058651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gillespie JH. Genetic drift in an infinite population. The pseudohitchhiking model. Genetics. 2000;155:909–919. doi: 10.1093/genetics/155.2.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Estes S, Phillips PC. Variation in pleiotropy and the mutational underpinnings of the G-matrix. Evolution. 2006;60:2655–2660. [PubMed] [Google Scholar]
- 68.Klass M, Hirsh D. Non-ageing developmental variant of Caenorhabditis elegans. Nature. 1976;260:523–525. doi: 10.1038/260523a0. [DOI] [PubMed] [Google Scholar]
- 69.Kim S, Paik Y-K. Developmental and reproductive consequences of prolonged non-aging dauer in Caenorhabditis elegans. Biochem Biophys Res. 2008;368:588–592. doi: 10.1016/j.bbrc.2008.01.131. [DOI] [PubMed] [Google Scholar]
- 70.Johnson TE, Henderson S, Murakami S, et al. Longevity genes in the nematode Caenorhabditis elegans also mediate increased resistance to stress and prevent disease. J Inherit Metab Dis. 2002;25:197–206. doi: 10.1023/a:1015677828407. [DOI] [PubMed] [Google Scholar]
- 71.Gershon H, Gershon D. Caenorhabditis elegans—a paradigm for aging research: advantages and limitations. Mech Ageing Dev. 2002;123:261–274. doi: 10.1016/s0047-6374(01)00401-8. [DOI] [PubMed] [Google Scholar]
- 72.Braeckman BP, Houthoofd K, Vanfleteren JR. Insulin-like signaling, metabolism, stress resistance and aging in Caenorhabditis elegans. Mech Ageing Dev. 2001;122:673–693. doi: 10.1016/s0047-6374(01)00222-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


