Abstract
Recent studies have demonstrated that non-coding RNAs (ncRNAs) play important roles during development and evolution. Chicken, the first genome-sequenced non-mammalian amniote, possesses unique features for developmental and evolutionary studies. However, apart from microRNAs, information on chicken ncRNAs has mainly been obtained from computational predictions without experimental validation. In the present study, we performed a systematic identification of intermediate size ncRNAs (50–500 nt) by ncRNA library construction and identified 125 chicken ncRNAs. Importantly, through the bioinformatics and expression analysis, we found the chicken ncRNAs has several novel features: (i) comparative genomic analysis against 18 sequenced vertebrate genomes revealed that the majority of the newly identified ncRNA candidates is not conserved and most are potentially bird/chicken specific, suggesting that ncRNAs play roles in lineage/species specification during evolution. (ii) The expression pattern analysis of intronic snoRNAs and their host genes suggested the coordinated expression between snoRNAs and their host genes. (iii) Several spatio-temporal specific expression patterns suggest involvement of ncRNAs in tissue development. Together, these findings provide new clues for future functional study of ncRNAs during development and evolution.
INTRODUCTION
The completion of an increasing number of genome sequencing projects have revealed that sequences corresponding to protein-coding genes only comprise around 1.5–2% of vertebrate genomes (1–4). Accumulating evidence suggests that considerable proportions of eukaryotic genomes are transcribed as non-coding RNAs (ncRNAs) (5–8). A number of different types of ncRNAs have been identified (9–14), ranging in size from around 20 nt, e.g. miRNAs and piRNAs, to over 10 000 nt. Importantly, a growing body of evidence has shown that ncRNAs can act as key regulators of development and other biological processes, supporting the notion that ncRNAs are indispensable players in the control of eukaryotic life/biology (15–20).
The chicken (Gallus gallus) possesses unique features for the study of developmental processes of organogenesis and has for several decades been used as a model organism for vertebrate developmental biology. The domesticated chicken originates from the red jungle fowl (G. gallus), and shares a common ancestor with mammals ∼310 million years ago (Mya). The chicken was the first non-mammalian amniote to have its genome sequenced; the genome size is about 40% of the average mammal genome and contains 20 000–23 000 predicted protein-coding genes. As a representative of the birds, chicken occupies an important evolutionary position which bridges the evolutionary gap between mammals and other vertebrates (4), and some genomic syntenic regions are more conserved between human and chicken than between human and mouse (21). Therefore, comparative genomic analyses of the chicken genome could provide important information for functional and evolutionary studies of vertebrate protein coding and non-protein-coding genes.
Genome-wide identification of chicken ncRNAs has mainly relied on computational predictions by searching for sequences with homology to ncRNAs identified in other species. The first draft of the chicken genome sequencing project only predicted 571 RNA genes, mainly including highly conserved tRNAs, rRNAs, snRNAs, RNase P, Y RNA, SRP, 7SK and a few conserved miRNAs and snoRNAs (4). The updated data from ENSEMBL (Release 52) annotated 1026 chicken ncRNA genes due to an increased number of predicted snoRNAs and miRNAs based on searching for homologs of human and/or mouse RNAs (560 miRNAs and 148 snoRNAs) (22). Very recently, Qu’s group reported 155 chicken snoRNAs predicted by snoSeeker (23). Thus far, 5732 human and 3287 mouse RNA genes, respectively, have been annotated by the ENSEMBL Release 52, and based on these numbers it is reasonable to expect that there exist at least 3000–4000 ncRNA genes in the chicken genome that have not yet been identified. The identification of ncRNAs escaping bioinformatical/computational prediction will rely on experimental strategies and systematic screening. Screenings for chicken microRNAs have been performed by several groups (24–26), but genome-wide identification of other types of chicken ncRNAs by experimental RNomics has yet not been reported and this prompted us to perform a profiling of the chicken non-miRNA ncRNAs. In the present study, we have focused on the systematic identification of intermediate size ncRNAs with lengths in the size range 50–500 nt.
A majority of known ncRNAs in the size range 50–500 nt are snoRNAs, snRNAs, tRNAs, 5SrRNAs and 5.8SrRNAs. During the past few years, a number of intermediate size ncRNAs have been identified in a variety of organisms spanning from Escherichia coli to Homo sapiens by experimental approaches or computational prediction or combination of the two strategies (9,27–31). snoRNAs, which mainly function in modification of ribosomal RNAs (rRNAs), small nuclear RNAs (snRNAs) and tRNAs (32,33), represent one of the largest groups of functional ncRNAs currently known in eukaryotic cells. Based on their sequence and structural features, snoRNAs can be classified into two families, the C/D box snoRNAs that generally guide 2′-O-methylation and the H/ACA box snoRNAs that guide rRNA pseudouridylation, both by pairing with targets via sequence complementarity (17,34). Although it has been reported that some human C/D box snoRNAs are independently transcribed as evidenced by the presence of methylated guanosine caps at their 5′-ends (35), most vertebrate snoRNAs are encoded in the intronic regions of protein-coding genes with ribosomal or other translation-related functions (36). In general, the biogenesis of intron-encoded snoRNAs is dependent on the transcription of their host genes (37). snoRNAs were originally assumed to guide rRNA modification, however, recent findings have demonstrated that transcripts with snoRNA characteristics can also target other types of RNAs such as mRNAs and thereby regulate their post-transcriptional expression or splicing patterns. The C/D box snoRNA HBII-52 regulated alternative splice site usage in a reporter construct carrying exons IV, Va, Vb and VI of the serotonin receptor 5-HT2CR when transfected into immortalized neuronal cells (19). In another report it was shown that an artificial C/D box snoRNA could induce pre-mRNA splicing impairment (38). In addition, it has been demonstrated that a subset of H/ACA box snoRNAs located in the Cajal bodies are involved in the regulation of telomerase RNA localization (36,39). Moreover, tissue-specific and developmentally regulated expression indicates that some snoRNAs have regulatory functions during development (40,41). In summary, previous findings suggest the existence of numerous snoRNAs in eukaryotic genomes with functional roles extending beyond those merely related to ribosomal RNA modification. Therefore, identification and characterization of broad sets of snoRNAs and other intermediate size regulatory ncRNAs in various organisms is crucial for a better understanding of ncRNA functions in development and evolution.
In this study, we carried out a genome-wide systematic identification of chicken ncRNAs by constructing chicken ncRNAs libraries (50–500 nt). A total of 125 unique ncRNAs were cloned, including 68 recently predicted ncRNA candidates (23). We examined the sequence conservation of the ncRNAs among 18 vertebrate species and found that most of the newly identified ncRNAs are potentially chicken- or bird-specific. The expression pattern analysis of intronic snoRNAs and their host genes suggested the coordinated expression between snoRNAs and their host genes. Finally, several ncRNAs exhibited specific tissue and temporal expression patterns, indicating functional roles in tissue development.
MATERIALS AND METHODS
Identification and classification of chicken ncRNAs
Total RNA was isolated from chicken tissues at embryonic Day 14, post-hatch 1 day and post-hatch 4 weeks. At each time-point, five randomly selected male individuals were sacrificed for tissue sampling. Full-length ncRNA specific libraries of both capped and uncapped transcripts were generated according to a previously described method (30) with modifications. Total RNA was fractionated on Qiagen-tips with a 0.6–1.0 M NaCl gradient elution of QRW2 buffer (Qiagen RNA/DNA handbook). Highly abundant rRNAs (5.8S rRNAs and 5S rRNAs) and snRNAs (U1 snRNA, U2 snRNA, U4 snRNA and U5 snRNA) were removed from the small RNA fraction (50–500 nt) with the Ambion MicrobExpress kit. The remaining RNAs were dephosphorylated with calf intestine alkaline phophatase (Fermentas), and ligated to the 3′ adaptor with T4 RNA ligase (Fermentas). After removal of excessive 3′ adaptor, the ligation products were split into two aliquots, of which one was treated with PolyNucleotide Kinase (PNK, Fermentas) to phosphorylate non-capped RNA, and the other was treated with Tobacco Acid Pyrophosphatase (TAP, Epicentre) to remove 5′-end methyl-guanosine caps from capped RNA. Thereafter, both samples were ligated to the 5′ adaptor and reverse transcribed with Thermoscript reverse transcriptase (Invitrogen) using oligo 3RT as the RT primer. The cDNA was amplified by PCR for 13 cycles using Platinum Taq (Invitrogen) with 3RT and 5CD primers, cloned into pGEM-T vector (Promega) and sequenced. All oligonucleotide sequences used in this study are shown in Supplementary Data file 4.
After removing redundant sequences, the obtained unique ncRNA sequences were first used as queries in a BlastN search against the NCBI nucleotide database and the Ensemble annotated chicken genome sequences to remove matches to rRNAs, tRNAs and mRNAs. The remaining sequences were used for ncRNA prediction and classification using the INFERNAL (42) and SnoReport software (43). The genomic loci of snoRNAs were obtained with the BLAT program (44), and comparisons of cloned snoRNAs with snoRNAs recorded in NCBI, Ensemble and Rfam databases was performed according to the genomic loci of snoRNAs. Known snoRNAs from other species were downloaded from the Rfam database. Transcripts that could not be annotated by any former databases or tools were regarded as unclassified ncRNAs.
For the experimentally identified snoRNA candidates, their potential targets were computationally predicted based on their base complementary information by using pairwise alignment. We searched against several known chicken non-coding transcripts including four rRNAs (18S, 28S, 5.8S, 5S) and eight snRNAs (U1, U2, U4a, U4b, U4b1, U4c, U4x and U5a).
Northern blot hybridization
Total RNA extracted from eight chicken tissues, including heart, liver, brain, lung, kidney, intestine, spleen and skeletal muscle at different development stages, were separated by 8% PAGE (7 M urea) and transferred to Nylon membrane (N+, Amersham). Antisense RNAs against the ncRNAs were labeled with Digoxigenin-11-UTP by in vitro transcription with T7 and SP6 RNA polymerase. The RNA blots were hybridized in ULTRAhyb (Ambion) at 68°C overnight, washed with 2× SSC/0.1% SDS washing buffer at 68°C for 2 × 5 min, following stringent washing with 0.1× SSC/0.1% SDS washing buffer at 68°C for 2 × 30 min. Thereafter, the RNA blots were blocked with blocking buffer for 30–60 min at room temperature and incubated for 30 min with anti-DIG-AP (1 : 10 000 diluted in blocking buffer). Hybridization signals were detected by CDP-star (Roche). Chemiluminescent signals were exposed to X-ray film. For detection of snoRNA expression in different tissues, RNA samples from chicken at 2 weeks post hatching were used.
Analysis of snoRNA host genes
Intronic ncRNAs were identified according to the ENSEMBL release 52 Gallus gallus genome annotation version WASHUC2. The Gene Ontology analysis of host genes of intronic snoRNAs was carried out using the GOEAST software (45). The microarray data used in this study covered expression profiles of chicken skeletal muscle tissues at post-hatch 1 day, 2, 4, 6 and 8 weeks. Three biological replicates were performed for each time point, and the average signal for each gene over the three biological replicates were used for further analysis. The expression changes shown in Figure 4B were calculated as the expression of a gene at a time point minus its expression at the immediate preceding time point.
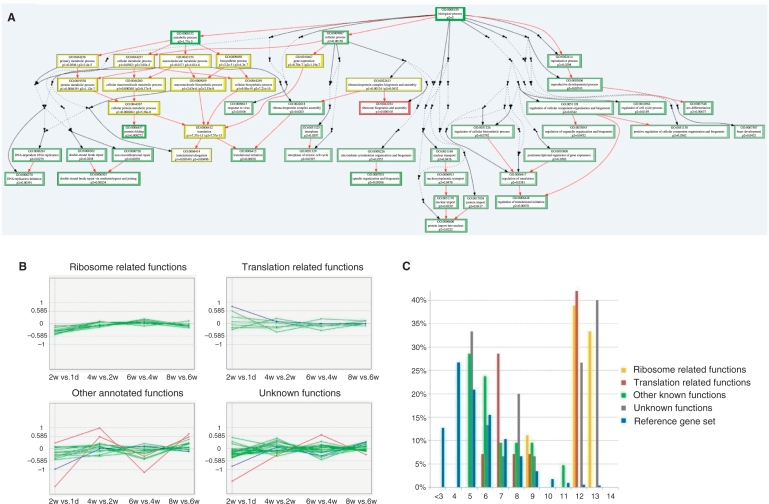
Figure 4.
Analysis of snoRNA host genes. (A) Functional analysis of snoRNA host genes according to the Biological Process category of Gene Ontology. GO terms enriched among host genes of C/D box snoRNAs and H/ACA box snoRNAs are shown as red and green boxes, respectively, and GO terms enriched among host genes of both C/D box and H/ACA box snoRNAs are shown as yellow boxes. The P-values for the enrichment of each GO term are shown within the boxes. Non-enriched GO terms in the hierarchical tree of GO terms are shown as dots to reduce the figure size. (B) Expressional changes of snoRNA host gene in chicken skeletal muscle tissue across different developmental stages. The log2 value of the expression difference between post-hatch time points 1 day, 2, 4, 6 and 8 weeks are shown. Genes with expression variance less than 1.5-fold between any two continues time points are shown as green lines, genes with expression variance larger than 1.5-fold but less than 2-fold between any two continues time points are shown as blue lines, and genes with expression variance larger than 2-fold between any two continues time points are shown as red lines. (C) Expression of snoRNA host genes in chicken skeletal muscle tissues. The log2 values of the average expression level of each snoRNA host gene across all examined time points are shown on the X-axis. The proportions of snoRNA host genes within each expression level range are shown on the Y-axis. The expression of all genes detected on the microarray is used as the reference gene set.
Conservation analysis of ncRNAs
The genomic sequences of chicken and 18 vertebrate genomes were downloaded from the UCSC genome browser. The conservation of chicken ncRNAs in the 18 vertebrate genomes was determined by using chicken ncRNAs as queries in BLAT searches of other genomes (44). Conservation scores were calculated based on the maximal alignment length and the identity of the alignment of the BLAT hits on each vertebrate genome. The conservation status of all cloned ncRNAs was clustered hierarchically according to their conservation scores in the 18 genomes using the hclust package in R.
Preparation and transfection of chicken fetal myoblasts
Chicken fetal myoblasts (CFMs) were prepared from pectoralis muscle tissue from 11th day White Leghorn chicken embryos according to a modified protocol described by Yang et al. (46). Briefly, the pectoralis muscle tissue was collected and washed with phosphate buffered saline (PBS). The tissue was minced and digested with 0.1% collagenase type I in PBS buffer at 37°C for 30 min with constantly shaking. The digested tissue was collected by centrifuging, resuspended in PBS buffer by repeated pipetting and filtrated to remove aggregated cells and myotubes. The collected cells were resuspended in α-MEM medium, seeded into a 100-mm dish, and cultured for 1 h at 37°C. Rapidly adhering cells were mainly composed of fibroblasts and were discarded. Myoblasts remaining in suspension were collected, transferred to a six-well gelatin-coated plate at 1 × 105 cells/cm2 with α-MEM containing 10% Horse Serum, 100 IU/ml penicillin and 100 μg/ml streptomycin (Invitrogen), and cultured for 24 h at 37°C in humidified 95% air–5% CO2. The myoblasts were transfected with the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. The transfected cells were lysed after 48 h of transfection, and total RNA was extracted using the TRIzol reagent (Invitrogen). The primers for pfkm minigene generation are shown in Supplementary Data file 4.
RESULTS
Identification and Classification of Chicken ncRNAs
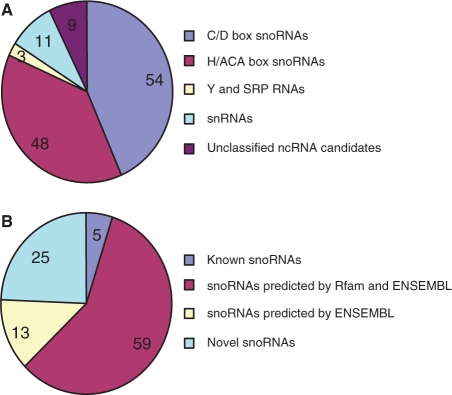
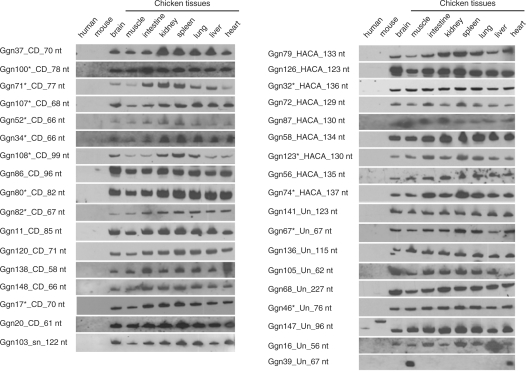
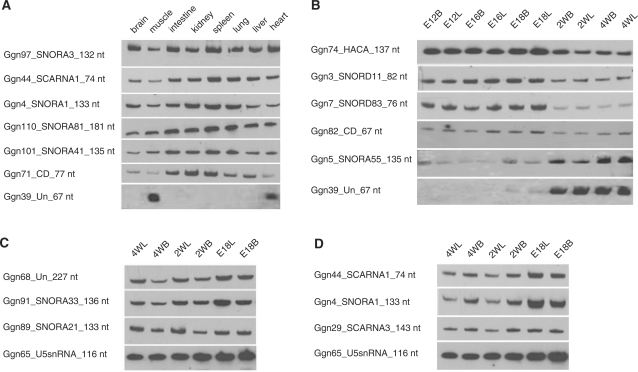
Full-length intermediate size ncRNAs (50–500 nt) enriched libraries were constructed according to a previously described method (30) with some modifications. This procedure ensured that the library contained a substantial fraction of full-length ncRNA clones with defined 5′ and 3′ termini. The total RNA used for library construction was extracted from a mix of 10 chicken tissues (heart, liver, brain, lung, kidney, intestine, glandular stomach, gizzard, spleen, skeletal muscle) harvested at three developmental stages (14 days old embryo, 1 day and 4 weeks chicken after hatching). Altogether, 3468 clones from two full-length cDNA libraries were sequenced. After removing matches to tRNAs and rRNAs, the remaining 201 sequences were taken as ncRNA candidates. Among these, 33 clones with a match to EST or RefSeq RNA sequences were excluded in this report because they can not be detected the same size band as obtained from library sequencing, suggesting they could be mRNA degradation products. The 43 other ncRNA candidates were also excluded because they did not yield detectable expression signals by northern blot. The remaining 125 ncRNAs candidates validated by northern blot included 90 ncRNA loci predicted by the ENSEMBL or Rfam (22,47) and 35 novel candidates (Figure 1, Supplementary Data files 1 and 2). The previously predicted 90 ncRNAs included 77 snoRNAs, 10 snRNA, two Y RNAs and one SRP RNA. The 35 novel ncRNA candidates were comprised of 25 transcripts with possible snoRNA characteristics, one transcript predicted to be a snRNA, and nine transcripts that could not be assigned to any known class of ncRNAs (Table 1). Among the 25 novel snoRNA candidates, three C/D box snoRNA candidates could target chicken rRNAs, whereas 10 of the novel snoRNA candidates might be orphan snoRNAs with no apparent target in chicken rRNAs or snRNAs. The other 12 novel snoRNA candidates were also independently predicted by Qu’s group (23) (Table 1 and Supplementary Data file 3). The expression of the 35 novel ncRNA candidates was shown in Figure 2. They are ubiquitously expressed in the eight tested chicken tissues, the single exception being Ggn39, which is specifically expressed in skeletal muscle and heart tissue. The expression patterns of the other ncRNAs were shown in Supplementary Figure S1.
Figure 1.
Classification of cloned chicken ncRNAs. (A) Classification of unique ncRNA sequences. (B) Distribution of cloned snoRNAs.
Table 1.
General information for 35 novel ncRNA candidates identified in this study
| GGN_ID | GenBank_ID | Reads_no | Length | ncRNA_ type | Target | Genomic _location in_chick | chr_location_ in_zebra finch | Seq_identity_ with_zebra finch |
|---|---|---|---|---|---|---|---|---|
| GGN11 | EU240230 | 2 | 85 | CD | 28S rRNA | Intergenic | No hit | No hit |
| GGN20 | EU240238 | 1 | 61 | CD | 28S rRNA | Intergenic | No hit | No hit |
| GGN37 | EU240254 | 1 | 70 | CD | 28S rRNA | Intergenic | chr4 13645396 13645464 + | 85.30% |
| GGN86 | EU240302 | 9 | 96 | CD | Orphan | Intergenic | chr22_random 663757 663840 + | 92.50% |
| GGN120 | EU240333 | 1 | 71 | CD | Orphan | Intronic | No hit | No hit |
| GGN138 | EU240346 | 1 | 58 | CD | Orphan | Intergenic | No hit | No hit |
| GGN148 | EU240352 | 2 | 66 | CD | Orphan | No_genomic_loci | No hit | No hit |
| GGN100a | EU240315 | 3 | 78 | CD | 18S rRNA | Intronic | chr7 21747676 21747750 − | 85.40% |
| GGN71a | EU240287 | 1 | 77 | CD | 18S rRNA | Intronic | chr5 61606460 61606536 − | 85.80% |
| GGN107a | EU240321 | 6 | 68 | CD | 28S rRNA | Intronic | chr8 3321788 3321851 − | 87.50% |
| GGN52a | EU240268 | 1 | 66 | CD | 18S rRNA | Intronic | chr8 3323611 3323676 − | 87.90% |
| GGN34a | EU240252 | 1 | 66 | CD | 28S rRNA | Intronic | chr2 121527086 121527152 − | 88.20% |
| GGN108a | EU240322 | 2 | 99 | CD | 28S rRNA | Intronic | chr8 17728445 17728543 + | 91.00% |
| GGN80a | EU240296 | 4 | 82 | CD | 18S rRNA | Intronic | chr17 6882327 6882410+ | 95.90% |
| GGN82a | EU240298 | 2 | 67 | CD | 18S rRNA | Intronic | chr19 7185695 7185736 + | 97.50% |
| GGN17a | EU240236 | 1 | 70 | CD | 28S rRNA | Intronic | No hit | No hit |
| GGN79 | EU240295 | 3 | 133 | HACA | Orphan | Intronic | chr15 7584209 7584339 − | 86.30% |
| GGN126 | EU240339 | 1 | 123 | HACA | Orphan | Intergenic | chr5 9310745 9310867 + | 87.90% |
| GGN72 | EU240288 | 5 | 129 | HACA | Orphan | Intronic | chr1 80099319 80099434 + | 90.60% |
| GGN87 | EU240303 | 3 | 130 | HACA | Orphan | Intergenic | chr23 92846 92969 − | 92.40% |
| GGN58 | EU240274 | 11 | 134 | HACA | Orphan | Intergenic | No hit | No hit |
| GGN56 | EU240272 | 2 | 135 | HACA | Orphan | Intronic | No hit | No hit |
| GGN32a | EU240250 | 1 | 136 | HACA | 28S rRNA | Intergenic | chr1 57671975 57672107 + | 90.00% |
| GGN123a | EU240336 | 5 | 130 | HACA | 18S rRNA | Intronic | No hit | No hit |
| GGN74a | EU240290 | 2 | 137 | HACA | 28S rRNA | Intronic | No hit | No hit |
| GGN103 | EU240318 | 4 | 122 | snRNA | Intergenic | No hit | No hit | |
| GGN141 | EU240348 | 1 | 123 | Unclassified | Intergenic | chr1 80104054 80104169 + | 83.70% | |
| GGN67a | EU240283 | 2 | 69 | Unclassified | Intronic | chr1 90202832 90202890+ | 88.20% | |
| GGN136 | EU240344 | 1 | 115 | Unclassified | Intergenic | chr20 6905097 6905198− | 90.90% | |
| GGN105 | EU240320 | 3 | 62 | Unclassified | Intronic | chr8 3324079 3324144− | 91.70% | |
| GGN68 | EU240284 | 1 | 227 | Unclassified | Intronic | LGE22 223377 223624− | 92.40% | |
| GGN46a | EU240262 | 1 | 76 | Unclassified | Intronic | chr1A 50331437 50331502+ | 100% | |
| GGN46a | EU240262 | 1 | 76 | Unclassified | Intronic | chr1A 50337731 50337796+ | 100% | |
| GGN147 | EU240351 | 1 | 96 | Unclassified | Intronic | No hit | No hit | |
| GGN16 | EU240235 | 1 | 56 | Unclassified | Intronic | No hit | No hit | |
| GGN39 | EU240256 | 1 | 67 | Unclassified | Intronic | No hit | No hit |
ancRNAs independently predicted by Qu’s group (23).
Figure 2.
Tissue expression pattern of 35 novel ncRNA candidates. The expression pattern of 35 novel ncRNA candidates was examined by northern blot analysis of total RNA from chicken heart, liver, lung, spleen, kidney, intestine, skeletal muscles and brain. Total RNA from human and mouse skeletal muscle were included in each blot in order to test for possible expression of the ncRNA candidates in different species. The ncRNA candidates are labeled as GgnX_class_size on the left of each northern blot. (For example: Ggn37_CD_70 nt means Ggn37 is a predicted C/D box snoRNA with length 70 nt. ‘Un’ denotes ‘unclassified’.) Asterisks indicate that ncRNAs were independently predicted by Qu’s group (23).
Potentially bird-specific ncRNAs
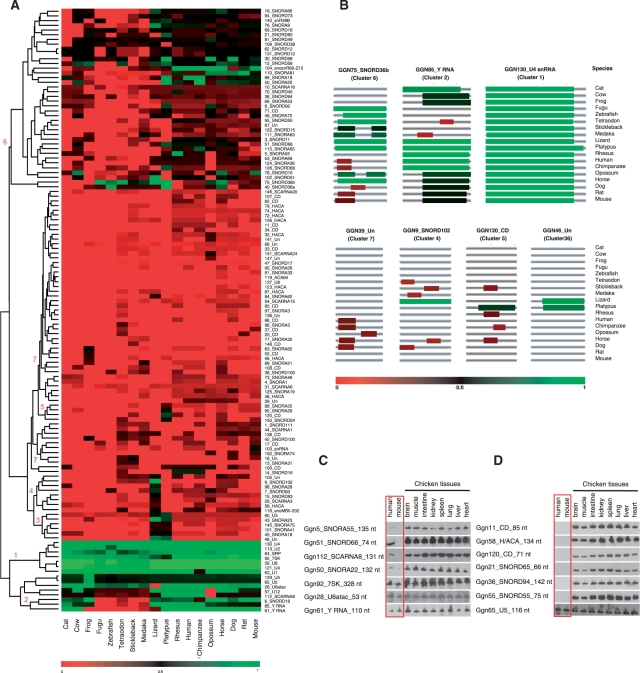
To analyze the evolutionary conservation of the ncRNAs, we searched for homologs of all the 125 chicken ncRNAs in the genomes of the lizard and 6 fish and 11 mammalian species, and hierarchically clustered the ncRNAs according to their conservation scores. The 125 chicken ncRNAs could be grouped into seven clusters according to their sequence conservation scores in each of the 18 queried genomes (Figure 3A), and one ncRNA from each of the seven clusters was selected as a representative and shown in Figure 3B. As shown in Figure 3A, compared to the highly conserved snRNAs, previously predicted snoRNAs are less conserved, whereas the newly identified snoRNAs and unclassified ncRNAs are poorly or not conserved. Ten snRNAs, two Y RNA and SRP RNA are highly conserved and grouped into clusters 1 and 2 (e.g. Ggn130_U4 and Ggn85_Y RNA in Figure 3B), and only the predicted snRNA (Ggn103) shows relatively low conservation (cluster 7). Unlike snRNAs, snoRNAs are generally less conserved. Of the 77 previously predicted snoRNAs, none are grouped into cluster 1 and only two are grouped into cluster 2 (Ggn112_SCARNA8 and Ggn6_SNORD18), whereas 36 (46.7%) belong to cluster 6, which is composed of transcripts with very limited sequence conservation (e.g. Ggn75_SNORD36 in Figure 3B). Interestingly, 16 snoRNAs, fall into clusters 3–5 (e.g. Ggn46, Ggn9 and Ggn120 in Figure 3B) and are only conserved in the lizard and/or platypus genomes but not in other mammals, suggesting the existence of an ancient group of snoRNAs that have been preserved in the reptile–bird lineage.
Figure 3.
Comparative genomics analysis of 125 chicken ncRNAs. (A) Heatmap of the conservations scores of 125 chicken ncRNAs in 18 vertebrate genomes. The conservation degree decreases as color changes from green to red. The ncRNAs were hierarchically clustered based on the conservation scores in the 18 genomes. (B) Representatives of ncRNAs from each of the seven clusters. (C) Representatives of conserved ncRNAs. The identity of each ncRNA is shown on the left of each blot. The tested chicken tissues are indicated on the upper of the figure. Human and mouse total RNA samples used in the experiments are as described in Figure 2. (D) Representatives of less conserved ncRNAs. The U5 snRNA was included as a loading control.
The 35 newly identified snoRNAs and unclassified ncRNA candidates are all not well conserved. In total, 22 of 25 novel snoRNA candidates (88%) are grouped into cluster 7 which show no conservation in any other examined genome/organism (Figure 3A). Six of the nine unclassified ncRNA candidates (67%) are also unconserved and are grouped into cluster 7 (e.g. Ggn39 in Figure 3B). Consistent with their low conservation score in mammalian genomes, only one ncRNA out of 35 novel ncRNAs can be detected in human and mouse tissues by northern blot (Figure 2). Interestingly, three snoRNAs (Ggn5, Ggn51 and Ggn112) can be detected in human but not in mouse tissues (Figure 3C and Supplementary Figure 2). In support of this observation, genomic sequence analysis showed these three snoRNAs have higher sequences similarity between chicken and human than between chicken and mouse, indicating that these chicken snoRNAs are shared by chicken and a limited number of mammalian lineages.
We further searched for homologs of 35 novel ncRNA candidates in the recently released zebra finch (Taeniopygia guttata) genome. Twenty one of the novel transcripts have homologs in the zebra finch with sequence identity above 85%, whereas the remaining 14 ncRNAs had no hits in the current version of the zebra finch genome (Table 1). Although this may be due to the incomplete genomic sequence available for the zebra finch it is also possible that some of these ncRNAs are specific to chicken.
Host gene analysis of intron-encoded snoRNAs
Of the 102 snoRNAs, 89 were encoded by intronic regions of protein-coding genes and 11 were located in intergenic regions (Table 2). For two of the snoRNA transcripts no genomic loci could be found in the current version of the chicken genome sequence. The 11 non-intronic snoRNAs appear all to be unique transcriptional units. For the intronic snoRNAs, we found 12 protein-coding genes each hosting two intronic snoRNAs, two genes hosting three snoRNAs and two genes hosting four snoRNAs (Supplementary Data file 2). Only in two cases, a single intron encodes the two ncRNA loci, and in both cases the two loci within the same intron encoded identical ncRNA transcripts (Supplementary Figure S3).
Table 2.
Eleven snoRNAs with intergenic loci
| Ggn_ID | GenBank_ID | Annotation | Reads_no | Length | Sequences |
|---|---|---|---|---|---|
| Ggn29 | EU240247 | ENSGALT00000042330 :SCARNA3 | 1 | 143 | CTGGAAGCTGTTGGGACCAGTTGGGACCGGTGGTGGGCAA AACAAGTGTTTTGTCCACTCTTGCAGTCTTCCTAGAGT AAAGTGGAGGTCTCTGTCTGCTTTAGGAGAGCCAACT AAACTGGATCGTTCCCCTCCATACATGT |
| Ggn119a | EU240332 | RF01225:ACA64 | 1 | 134 | TTGGCACAGTCTGAAAATCACCTGCGGTCTCCCGTGGCCCC CGGTGAGATAACCGTGCCGACAGCATAGGGAACATCA AAGCTTATTGCCCACGGTGACAGTGTGTGGGGAGTGA AACCCGTGTTCCCATAACC |
| Ggn38 | EU240255 | ENSGALT00000042195 :SNORD100 | 1 | 72 | ATGAAAATTTGGCTCCCTCTACTGAAACAAATGAGGAGAAC TGCCTTAATCGTTACCCAAATCTTCTGAGAT |
| Ggn11 | EU240230 | Predicted CD box snoRNA | 2 | 85 | TGCTCAATGATGATCATCCTCCTTTGATTACCTGGTGAGGT AATATGAGAGGACATGGAATAATTTCACCGGCGAAC TGAGAGCA |
| Ggn138 | EU240346 | Predicted CD box snoRNA | 1 | 58 | AGTGCAGTGATCATAAACCAAAGCTGAACATGGAGCTCAGT GTGGTTTATTGTAGAGG |
| Ggn20 | EU240238 | Predicted CD box snoRNA | 1 | 61 | GCTGGTGATGAGATAGTTATCCCTGTCCGAAACGTTCCTCTG TGGAAGCGTGACTCTGAGG |
| Ggn37 | EU240254 | Predicted CD box snoRNA | 1 | 70 | CGTCCGATGATGAACCTCAATGCTGTTCACATCCTGACACGC CGTGACGAGCGCTGTCGAGCTGAGGACG |
| Ggn86 | EU240302 | Predicted CD box snoRNA | 9 | 96 | CGATCCTTCCGGTTCATAGCAAATGATGAATGGGAGTTGCAC GCGGCTGCGTGACGTGTGCGCCCTTTGTTACGACGTGC ACAGCCCCTTCTGAGC |
| Ggn58 | EU240274 | Predicted HACA box snoRNA | 11 | 134 | CATGCCCCAGTCGTGTTGCAGATATGGCTGTAGTGCCATGTT TGTGTCATTAGGTGGCAGAAAGGAAAAGGCTGTGTCTT TGCTAATGCTCTGAAACCGGTGAGCACTCAGGAATGAC TAGCAACCTGACAAAT |
| Ggn87 | EU240303 | Predicted HACA box snoRNA | 3 | 130 | CTGCATGTTAATCCAAGAGCTGTGGCTCTGACGTAGCTGC AGGTCTCCAACAACATGCAAGAGCAACGGGAAGGTCT TTGACTGCTCGGCCTCTTCTGCCTGTTGCTGTCACTCAC CCCTCCTATATATT |
| Ggn126 | EU240339 | Predicted HACA box snoRNA | 1 | 123 | GGCTCGCGCAATTCCAAACCTGACAGTGGTTCTGGTTTGCT GTCAGCCTCATAGAGCAAAAGCGAGGGTTTATTCACT GAAAAGGTGAAGCCGTTTCCTTTTTTCCTTGCCTCCC TCACACTC |
aIndependently predicted by Qu’s group (23).
We performed functional analysis of the snoRNA host genes based on Gene Ontology annotations. Host genes of both C/D box and H/ACA box snoRNAs were enriched with functional annotation relating to ribosome biogenesis, translation and cellular protein metabolic processes. We also found that host genes of both C/D box and H/ACA box snoRNAs were enriched for some other functions, including initiation of DNA duplication, protein transportation, regulation of translation initiation, regulation of cell cycle and other processes (Figure 4A and Supplementary Figure S4).
To further explore the possible coordinated expression pattern between intronic snoRNAs and their host genes, we examined the expression of snoRNA host genes using our recently published Affymetrix microarray data (48). RNA samples from chicken skeletal muscle tissues at 1 day, or 2, 4, 6 and 8 weeks after hatching were used for the array analysis. The results show that in general, the expression of snoRNA host genes did not vary much across the examined developmental stages, and that genes with ribosome related functions had least variation in expression (Figure 4B). Host genes with functions related to cell cycle and apoptosis showed the most variable expression across developmental stages (Figure 4B). Correspondingly, snoRNAs encoded in intronic regions of ribosome related genes also exhibited constant expression across different developmental stages, whereas snoRNAs hosted by non-ribosome related genes showed variable expression levels during postnatal development (Supplementary Figure S5). It is worth noting that snoRNA host genes with ribosome and translation related functions had much higher expression levels than the average coding gene expression, whereas the expression levels of host genes with other known functions were relatively low (Figure 4C). Similarly, such expression discrepancy also observed in the expression pattern of snoRNAs during chicken muscle development (Supplementary Figure S5).
Tissue and developmental stage-specific expression of chicken ncRNAs
The ncRNA expression patterns may yield important clues to their functions. Thus, we investigated the expression of ncRNAs in various chicken tissues, including heart, liver, lung, spleen, kidney, intestine, brain and skeletal muscle. All examined ncRNAs were detected by northern blot analysis (Supplementary Figure S1). Unlike snRNAs and Y-RNAs, which showed constant expression levels across all examined tissues, some of the snoRNAs tended to have lower expression in brain, muscle and heart than in other tissues (Figure 5A). Interestingly, we found that one of the unclassified ncRNA (Ggn39) was only expressed in the chicken heart and skeletal muscle (Figure 5A). To look at the possible functional roles of the ncRNAs during skeletal muscle development, we examined the expression pattern of these ncRNAs in chicken skeletal muscle tissues at different developmental stages by northern blot analysis. Our results showed that several of the snoRNAs had developmentally regulated expression patterns (Figure 5B and Supplementary Figure S5). For example, snoRNAs Ggn74, Ggn3 and Ggn7 were highly expressed during embryonic development, whereas snoRNA Ggn5 was more abundantly expressed after hatch (Figure 5B). Ggn39, which is specifically expressed in skeletal muscle and heart, was predominantly expressed at the post-hatching stages (Figure 5B), suggesting that this unclassified ncRNA might have specific functions in myogenesis during postnatal development.
Figure 5.
Spatio-temporal ncRNA expression. (A) Tissue-specific ncRNA expression. (B) ncRNAs with developmentally regulated expression in chicken muscle tissues. The tested samples were from Pectoralis major muscles at different developmental stages of broiler (B) and layer (L) chickens, including embryonic Days 12, 16, 18 (E12B, E12L, E16B, E16L, E18B, E18L) and 2 and 4 weeks after hatch (2WB, 2WL, 4WB, 4WL). (C) Three ncRNAs with higher expression level in skeletal muscle of layer (L) than of broiler (B). (D) Three ncRNAs with higher expression level in skeletal muscle of broiler (B) than of layer (L).
To further investigate the possible involvement of ncRNAs in chicken myogenesis, we analyzed the expression pattern of the 105 ncRNAs in the skeletal muscle tissues of two chicken lines with divergent muscle growth rates (fast growing ‘broiler’ chicken for meat production and slow growing ‘layer’ chicken for egg production). We found that several ncRNAs were differentially expressed in skeletal muscle tissues between broilers and layers. The expression levels of Ggn68, Ggn91 and Ggn89 were significantly higher in layers than in broilers at post-hatching 2 and 4 weeks (Figure 5C), whereas Ggn44, Ggn4 and Ggn29 showed higher expression in broilers than in layers (Figure 5D). The differential expression patterns of these ncRNAs suggest that they may be important regulators for the divergent muscle growth phenotype of broilers and layers.
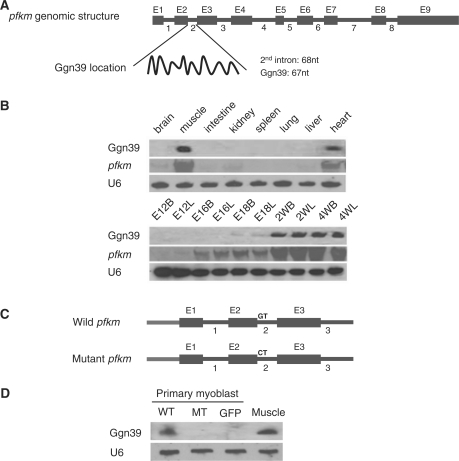
Ggn39 is generated from the pfkm gene in a splicing-dependent manner
Mapping the novel transcripts to the chicken genome showed that Ggn39 is located in the second intron of the phosphofructokinase M (pfkm) gene (Figure 6A). The size of this intron is 68 nt, whereas the cloned Ggn39 transcript is 67 nt long corresponding to 1–67 bp of intron sequence. Northern blot analysis showed that Ggn39 and its host gene pfkm were specifically detected in skeletal and heart muscle tissues, and are most abundantly expressed during postnatal myogenesis (Figure 6B). The similarity in spatio-temporal expression-patterns of the ncRNA and host gene loci suggests that Ggn39 may be generated by intron splicing from the pfkm pre-mRNA. We tested this possibility by transfecting the chicken myoblasts with a wild-type pfkm minigene (pcDNA3-pfkm) comprising exons 1, 2, 3 and introns 1, 2 and the first 37 bp of intron 3, and a mutant pfkm minigene containing a mutation in the 5′ splicing site (GT to CT) of the second intron (pcDNA3-pfkm-mutant) (Figure 6C). Northern blot analysis demonstrated that the Ggn39 ncRNA was only detected in the myoblasts transfected with the wild-type minigene but not in myoblast transfected with the mutant construct (Figure 6D), suggesting that Ggn39 is generated in a splicing-dependent manner.
Figure 6.
Ggn39 is generated in a splicing-dependent manner. (A) The genomic organization of Ggn39. Boxes and lines represent exons (E1–E9) and introns (1–8), respectively, of the pfkm gene. (B) The spatio-temporal expression pattern of Ggn39 and its host gene pfkm. The expression patterns of Ggn39 and pfkm were examined by northern blot analysis of total RNA from chicken tissues. The corresponding antisense RNA labeled with DIG-11-UTP were used as probes. U6 snRNA was used as loading control. (C) Schematic structure of the pfkm minigene and the second intron 5′ splicing site mutant. (D) The biogenesis of Ggn39 is dependent on splicing of the second intron of the pfkm gene. Chicken primary myoblasts were transfected with wild-type (WT) and mutant (MT) pfkm minigenes. GFP denotes pcDNA3-EGFP that served as transfection control. Northern blot analysis was performed on total RNAs prepared from the transfected cells. As the Ggn39 is highly expressed in skeletal muscle, chicken skeletal muscle RNA was used as positive control for the northern blot experiments.
DISCUSSION
Increasing numbers of ncRNAs are being identified from various model organisms by experimental RNomics combined with bioinformatics analyses (49,50). Herein, we have performed the first systematic profiling of the chicken intermediate size ncRNA transcriptome by experimental RNomics, and cloned 125 chicken ncRNAs. Comparative genomic analysis with blast against 18 sequenced vertebrate genomes showed that only a few (5 out of 35) of the novel ncRNAs have homologs in non-avian vertebrates, and the remaining novel ncRNAs are likely to be specific to the bird or chicken lineages. Our findings are in line with recent studies on other species. Computer analysis of genomic tiling array data revealed 300 putative candidates for primate specific ncRNAs (51). Unclassified novel ncRNAs often show little sequence conservation and low cellular concentrations (30). Less conserved ncRNAs might be important factors in animal development (8), and a remaining challenge for a better understanding of ncRNA function is therefore the detection of less conserved, low abundance ncRNAs in different organisms. Together with previous reports, the identification of chicken specific ncRNAs indicated that there may exist numerous types of ncRNAs that are specific to a lineage or even a single species.
It has been reported that a majority of the identified vertebrate snoRNAs are encoded in introns of protein-coding genes whose functions are mostly related to synthesis, structure or function of the translational apparatus (36). This mode of gene organization and expression provides a molecular basis for functional prediction for recently discovered ncRNAs. Similar analyses have been proved invaluable for predicting the functional roles of miRNAs host genes (52). In this study, we have sought to combine GO annotation on host gene function and cellular localizations with experimentally validated expression profiles of snoRNA host genes to predict possible functions of the intronic snoRNAs. We have found that snoRNA host genes encode proteins with a broad spectrum of biological roles ranging from DNA duplication initiation, protein transportation, translation initiation, regulation of cell cycle and other developmental processes. Our results indicate that the host genes of H/ACA have a broader spectrum of cellular locations than do host genes of C/D box snoRNAs, suggesting that H/ACA and C/D box snoRNAs may function in different cellular processes. This observation was further supported by the microarray data on host gene expression during skeletal muscle development. snoRNA host genes with functions related to ribosome biogenesis and translation were much more abundantly expressed during myogenesis than host genes with other functions (such as cell cycle and apoptosis), which showed lower and more variable expression across the developmental stages. Our findings could provide information for further studies of the functions of snoRNAs from host genes with different biological functions, as well as for the study of the functional and regulatory coordination between snoRNAs and their host genes during development.
The spatio-temporally regulated expression patterns of the novel ncRNAs provide further functional information about the role of these ncRNAs during development. It is well accepted that some ncRNAs, especially some microRNAs and piRNAs, have specific spatio-temporal expression patterns and play functional roles in cell differentiation and organogenesis during development. For instance, miRNAs with muscle-specific expression are involved in development and disease of the skeletal and heart muscle (53,54). In general, the spatio-temporally regulated expression of other classes of ncRNAs, particularly the unclassified ncRNAs, has been less well explored. We investigated this question by northern blot analysis of a substantial number of the ncRNAs identified in the present work. Our data showed that the majority of the novel ncRNAs exhibited ubiquitous tissue distribution; some were abundantly expressed only in a few tissues. Most interestingly, we found that one of the unclassified ncRNAs (Ggn39) was generated from the second intron of pfkm gene in a splicing-dependent manner, and that Ggn39 ncRNA, like its host gene pfkm, showed specific expression in the chicken heart and skeletal muscle cells, especially during postnatal development. As a key enzyme in the glucose metabolism, PFKM plays important roles in the metabolism and development of skeletal muscle (55). The coordinated expression pattern of Ggn39 and its host gene suggests a functional correlation between Ggn39 and PFKM which may provides useful information for further studies of the metabolic and developmental functions of Ggn39 and its host gene pfkm.
Recently, brain-specific snoRNAs have been implicated in brain development and neurological disease (41,56,57). In chicken we identified an H/ACA-box snoRNA (Ggn146) encoded in the intron of the chicken ortholog of the Williams Beuren syndrome chromosome region 22 gene. Since Williams Beuren syndrome is caused by microdeletion within the syntenic region containing this gene in human (58), it is an intriguing possibility that an ortholog of Ggn146 may also be involved in the pathogenesis of the Williams Beuren syndrome in human. The broiler and layer chickens are an excellent model systems for the study of molecular mechanisms involved in skeletal muscle development because of their significant muscle growth differences during myogenesis. Up to now, the molecular genetics underlying the divergent muscle growth rate between broilers and layers have remained unclear. Similar to muscle-specific miRNAs (e.g. miRNA-1), ncRNAs that are differentially expressed in skeletal muscle tissues between broiler and layer chickens might function as muscle growth regulators. Identification of these ncRNAs could facilitate ncRNA functional studies and provide essential information for a better understanding of the functional roles of ncRNAs during development.
ACCESSION NUMBERS
EU240220, EU240222–EU240239, EU240246–EU240250, EU240252–EU240263, EU240265–EU240328, EU240331–EU240352, EU240354–EU240355, EU240357.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Basic Research Program of China (2007CB946903, 2007CB946901, 2005CB522405, 2005CB522505, 2009CB941602, 2009CB825403); National Natural Science Foundation of China (30721063, 30871248); Chinese National Programs for High Technology Research and Development (2006AA10A121, 2007AA02Z109).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 3.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 4.Hillier LW, Miller W, Birney E, Warren W, Hardison RC, Ponting CP, Bork P, Burt DW, Groenen MA, Delany ME, et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 5.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 6.Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SP, Gingeras TR. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- 7.Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 8.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huttenhofer A, Kiefmann M, Meier-Ewert S, O'B;rien J, Lehrach H, Bachellerie JP, Brosius J. RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J. 2001;20:2943–2953. doi: 10.1093/emboj/20.11.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Numata K, Kanai A, Saito R, Kondo S, Adachi J, Wilming LG, Hume DA, Hayashizaki Y, Tomita M. Identification of putative noncoding RNAs among the RIKEN mouse full-length cDNA collection. Genome Res. 2003;13:1301–1306. doi: 10.1101/gr.1011603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl Acad. Sci. USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 13.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 14.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 15.Storz G, Altuvia S, Wassarman KM. An abundance of RNA regulators. Annu. Rev. Biochem. 2005;74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- 16.Plasterk RH. Micro RNAs in animal development. Cell. 2006;124:877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 17.Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the ‘genome complexity’ conundrum. Genes Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- 19.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 20.Couzin J. MicroRNAs make big impression in disease after disease. Science. 2008;319:1782–1784. doi: 10.1126/science.319.5871.1782. [DOI] [PubMed] [Google Scholar]
- 21.Burt DW, Bruley C, Dunn IC, Jones CT, Ramage A, Law AS, Morrice DR, Paton IR, Smith J, Windsor D, et al. The dynamics of chromosome evolution in birds and mammals. Nature. 1999;402:411–413. doi: 10.1038/46555. [DOI] [PubMed] [Google Scholar]
- 22.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shao P, Yang JH, Zhou H, Guan DG, Qu LH. Genome-wide analysis of chicken snoRNAs provides unique implications for the evolution of vertebrate snoRNAs. BMC Genomics. 2009;10:86. doi: 10.1186/1471-2164-10-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shao P, Zhou H, Xiao ZD, He JH, Huang MB, Chen YQ, Qu LH. Identification of novel chicken microRNAs and analysis of their genomic organization. Gene. 2008;418:34–40. doi: 10.1016/j.gene.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Burnside J, Ouyang M, Anderson A, Bernberg E, Lu C, Meyers BC, Green PJ, Markis M, Isaacs G, Huang E, et al. Deep sequencing of chicken microRNAs. BMC Genomics. 2008;9:185. doi: 10.1186/1471-2164-9-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glazov EA, Cottee PA, Barris WC, Moore RJ, Dalrymple BP, Tizard ML. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Res. 2008;18:957–964. doi: 10.1101/gr.074740.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan G, Klambt C, Bachellerie JP, Brosius J, Huttenhofer A. RNomics in Drosophila melanogaster: identification of 66 candidates for novel non-messenger RNAs. Nucleic Acids Res. 2003;31:2495–2507. doi: 10.1093/nar/gkg361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eddy SR. Computational genomics of noncoding RNA genes. Cell. 2002;109:137–140. doi: 10.1016/s0092-8674(02)00727-4. [DOI] [PubMed] [Google Scholar]
- 29.Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager JG, Huttenhofer A, Wagner EG. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng W, Zhu X, Skogerbo G, Zhao Y, Fu Z, Wang Y, He H, Cai L, Sun H, Liu C, et al. Organization of the Caenorhabditis elegans small non-coding transcriptome: genomic features, biogenesis, and expression. Genome Res. 2006;16:20–29. doi: 10.1101/gr.4139206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vitali P, Royo H, Seitz H, Bachellerie JP, Huttenhofer A, Cavaille J. Identification of 13 novel human modification guide RNAs. Nucleic Acids Res. 2003;31:6543–6551. doi: 10.1093/nar/gkg849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu. Rev. Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 33.Zemann A, op de Bekke A, Kiefmann M, Brosius J, Schmitz J. Evolution of small nucleolar RNAs in nematodes. Nucleic Acids Res. 2006;34:2676–2685. doi: 10.1093/nar/gkl359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balakin AG, Smith L, Fournier MJ. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 35.Tycowski KT, Aab A, Steitz JA. Guide RNAs with 5′ caps and novel box C/D snoRNA-like domains for modification of snRNAs in metazoa. Curr. Biol. 2004;14:1985–1995. doi: 10.1016/j.cub.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 36.Kiss T, Fayet E, Jady BE, Richard P, Weber M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb. Symp. Quant. Biol. 2006;71:407–417. doi: 10.1101/sqb.2006.71.025. [DOI] [PubMed] [Google Scholar]
- 37.Filipowicz W, Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:319–327. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- 38.Semenov DV, Vratskih OV, Kuligina EV, Richter VA. Splicing by exon exclusion impaired by artificial box c/d RNA targeted to branch-point adenosine. Ann. NY Acad. Sci. 2008;1137:119–124. doi: 10.1196/annals.1448.037. [DOI] [PubMed] [Google Scholar]
- 39.Lukowiak AA, Narayanan A, Li ZH, Terns RM, Terns MP. The snoRNA domain of vertebrate telomerase RNA functions to localize the RNA within the nucleus. RNA. 2001;7:1833–1844. [PMC free article] [PubMed] [Google Scholar]
- 40.Nicholls RD, Knepper JL. Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annu. Rev. Genomics Hum. Genet. 2001;2:153–175. doi: 10.1146/annurev.genom.2.1.153. [DOI] [PubMed] [Google Scholar]
- 41.Cavaille J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Huttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl Acad. Sci. USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein RJ, Eddy SR. RSEARCH: finding homologs of single structured RNA sequences. BMC Bioinformatics. 2003;4:44. doi: 10.1186/1471-2105-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hertel J, Hofacker IL, Stadler PF. SnoReport: computational identification of snoRNAs with unknown targets. Bioinformatics. 2008;24:158–164. doi: 10.1093/bioinformatics/btm464. [DOI] [PubMed] [Google Scholar]
- 44.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng Q, Wang XJ. GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res. 2008;36:W358–W363. doi: 10.1093/nar/gkn276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang W, Zhang Y, Ma G, Zhao X, Chen Y, Zhu D. Identification of gene expression modifications in myostatin-stimulated myoblasts. Biochem. Biophys. Res. Commun. 2005;326:660–666. doi: 10.1016/j.bbrc.2004.11.096. [DOI] [PubMed] [Google Scholar]
- 47.Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, Wilkinson AC, Finn RD, Griffiths-Jones S, Eddy SR, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–D140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng Q, Zhang Y, Chen Y, Yang N, Wang XJ, Zhu D. Systematic identification of genes involved in divergent skeletal muscle growth rates of broiler and layer chickens. BMC Genomics. 2009;10:87. doi: 10.1186/1471-2164-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pang KC, Stephen S, Dinger ME, Engstrom PG, Lenhard B, Mattick JS. RNAdb 2.0–an expanded database of mammalian non-coding RNAs. Nucleic Acids Res. 2007;35:D178–D182. doi: 10.1093/nar/gkl926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34:D158–D162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Z, Pang AW, Gerstein M. Comparative analysis of genome tiling array data reveals many novel primate-specific functional RNAs in human. BMC Evol. Biol. 2007;7(Suppl. 1):S14. doi: 10.1186/1471-2148-7-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 54.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wegener G, Krause U. Different modes of activating phosphofructokinase, a key regulatory enzyme of glycolysis, in working vertebrate muscle. Biochem. Soc. Trans. 2002;30:264–270. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- 56.Mehler MF, Mattick JS. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol. Rev. 2007;87:799–823. doi: 10.1152/physrev.00036.2006. [DOI] [PubMed] [Google Scholar]
- 57.Peters J. Prader-Willi and snoRNAs. Nat. Genet. 2008;40:688–689. doi: 10.1038/ng0608-688. [DOI] [PubMed] [Google Scholar]
- 58.Pober BR, Johnson M, Urban Z. Mechanisms and treatment of cardiovascular disease in Williams-Beuren syndrome. J. Clin. Invest. 2008;118:1606–1615. doi: 10.1172/JCI35309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.