Abstract
An understanding of the mechanism(s) by which some individuals spontaneously control human immunodeficiency virus (HIV)/simian immunodeficiency virus replication may aid vaccine design. Approximately 50% of Indian rhesus macaques that express the major histocompatibility complex (MHC) class I allele Mamu-B*08 become elite controllers after infection with simian immunodeficiency virus SIVmac239. Mamu-B*08 has a binding motif that is very similar to that of HLA-B27, a human MHC class I allele associated with the elite control of HIV, suggesting that SIVmac239-infected Mamu-B*08-positive (Mamu-B*08+) animals may be a good model for the elite control of HIV. The association with MHC class I alleles implicates CD8+ T cells and/or natural killer cells in the control of viral replication. We therefore introduced point mutations into eight Mamu-B*08-restricted CD8+ T-cell epitopes to investigate the contribution of epitope-specific CD8+ T-cell responses to the development of the control of viral replication. Ten Mamu-B*08+ macaques were infected with this mutant virus, 8X-SIVmac239. We compared immune responses and viral loads of these animals to those of wild-type SIVmac239-infected Mamu-B*08+ macaques. The five most immunodominant Mamu-B*08-restricted CD8+ T-cell responses were barely detectable in 8X-SIVmac239-infected animals. By 48 weeks postinfection, 2 of 10 8X-SIVmac239-infected Mamu-B*08+ animals controlled viral replication to <20,000 viral RNA (vRNA) copy equivalents (eq)/ml plasma, while 10 of 15 wild-type-infected Mamu-B*08+ animals had viral loads of <20,000 vRNA copy eq/ml (P = 0.04). Our results suggest that these epitope-specific CD8+ T-cell responses may play a role in establishing the control of viral replication in Mamu-B*08+ macaques.
A few individuals spontaneously control the replication of human immunodeficiency virus (HIV) or simian immunodeficiency virus (SIV) to very low levels. The precise mechanisms underlying this control are of great interest, as a clear understanding of what constitutes a successful immune response may aid in developing an AIDS vaccine. Particularly pressing questions for vaccine design include which proteins to use as immunogens, the extent to which increasing the breadth and magnitude of responses is advantageous, how immunodomination affects T-cell responses, and if biasing the immune response toward particular effector profiles is beneficial. Characterization of immune responses made by elite controllers (ECs) may reveal patterns that can then be applied to vaccine formulation and evaluation.
HIV ECs are generally not infected with grossly unfit viruses (6, 42). Instead, elite control of immunodeficiency virus replication is correlated with the presence of particular major histocompatibility complex class I (MHC-I) alleles (11, 12, 18, 32, 41, 55). The association of MHC-I alleles with the control of viremia implicates CD8+ T cells as being mediators of this immune containment. Several lines of evidence support this hypothesis. These lines of evidence include the correlation between the appearance of CD8+ T-cell responses and the resolution of peak viremia during acute infection (7, 29), the finding that alleles associated with viral control restrict dominant acute-phase CD8+ T-cell responses (3), and the finding that responses directed against epitopes restricted by these alleles frequently select for viral escape variants (4, 27, 38). Perhaps most compelling is the observation that for a few HIV-infected individuals, the selection of escape variants by an immunodominant HLA-B27-restricted T-cell response temporally preceded substantial increases in viremia (17, 21, 53). While viruses exhibiting escape variants in epitopes restricted by protective alleles are often detectably less fit in vitro (10, 38, 43, 51), recent data have found normal, high levels of replication in vivo upon the transmission of some of these variants (15).
The association of control with MHC-I alleles does not, of course, implicate solely CD8+ T cells. MHC-I molecules are also ligands for killer immunoglobulin receptors (KIRs), which are predominantly expressed on natural killer (NK) cells. Genetic studies of HIV-infected humans suggest a model in which individuals with particular KIR/HLA combinations are predisposed to control HIV replication more readily than those with other KIR/HLA combinations (36, 37). These data were supported by functional studies of this KIR/HLA pairing in vitro, which demonstrated an inhibition of HIV replication by such NK cells (2). The relative contributions of NK and CD8+ T-cell responses to control have yet to be elucidated and may be closely intertwined.
Previously, the experimental depletion of circulating CD8+ cells from SIVmac239-infected ECs resulted in a sharp spike in viremia, which resolved as CD8+ cells repopulated the periphery (19). During the reestablishment of control of SIV replication, CD8+ T cells targeting multiple epitopes restricted by alleles associated with elite control expanded in frequency, providing strong circumstantial evidence for their role in maintaining elite control (19, 31). However, CD8 depletion antibodies used in macaques also remove NK cells, which, at least in vitro, also inhibit SIV replication (19). It was therefore difficult to make definitive conclusions regarding the separate contributions of these subsets to maintaining the control of SIV replication in vivo.
Here we investigate elite control in the rhesus macaque model for AIDS. We focused on the macaque MHC-I allele most tightly associated with the control of SIVmac239, Mamu-B*08. Approximately 50% of Mamu-B*08-positive (Mamu-B*08+) animals infected with SIVmac239 become ECs (32). Peptides presented by Mamu-B*08 share a binding motif with peptides presented by HLA-B27. Although these two MHC-I genes are dissimilar in domains that are important for peptide binding, each molecule can bind peptides that are presented by the other molecule (33). This striking similarity suggests that the elite control of SIVmac239 in Mamu-B*08+ animals is a good model for the elite control of HIV.
Seven SIVmac239 epitopes restricted by Mamu-B*08 accrue variation in Mamu-B*08+ rhesus macaques (30, 31). For an eighth Mamu-B*08-restricted epitope, which is also restricted by Mamu-B*03 (Mamu-B*03 differs from Mamu-B*08 by 2 amino acids in the α1 and α2 domains [9, 32]), escape has been documented only for SIV-infected Mamu-B*03+ macaques (16). Variation in these CD8+ T-cell epitopes accumulates with different kinetics, starting during acute infection for those targeted by high-magnitude responses.
In this study, we addressed the question of whether the elite control of SIVmac239 in Mamu-B*08+ animals is mediated by the known high-frequency CD8+ T-cell responses targeting Mamu-B*08-restricted epitopes. To this end, we introduced point mutations into eight epitopes, with the goal of reducing or abrogating immune responses directed against these epitopes during acute infection. We hypothesized that Mamu-B*08+ macaques would be unable to control SIV replication without these Mamu-B*08-restricted T-cell responses.
MATERIALS AND METHODS
Animals.
Indian rhesus macaques (Macacca mulatta) housed at the Wisconsin National Primate Research Center and cared for according to the Guide for the Care and Use of Laboratory Animals of the National Research Council (43a) were used in this study. Protocols were approved by the University of Wisconsin Institutional Animal Care and Use Committee. Animals used in this study were initially typed for the MHC-I alleles Mamu-A*01, Mamu-A*02, Mamu-A*08, Mamu-A*11, Mamu-B*01, Mamu-B*03, Mamu-B*04, Mamu-B*08, Mamu-B*17, and Mamu-B*29 using sequence-specific primers (26, 32). We excluded Mamu-B*17+ animals from this study because that allele is also enriched in ECs (55). Comprehensive typing of transcribed MHC-I sequences was determined by massively parallel sequencing of cDNAs (54). Due to the low frequency of Mamu-B*08 in the Wisconsin colony (32), many of the animals used in this study are closely related (Fig. 1).
FIG. 1.
Mamu-B*08+ macaques included in this study. Relationships are indicated by lines drawn between the animals’ identification numbers. Viral load outcomes are shown in parentheses next to an animal's number. Set points were calculated as the geometric mean of all of the viral load measurements from each animal between weeks 15 and 48 postinfection. EC indicates a set point viral load of <1,000 vRNA copy eq/ml plasma. C indicates a set point of <20,000 copies/ml. P indicates a set point of >20,000 copies/ml.
Immunological and virological data from four of the SIVmac239-infected animals, animals r91003, r01027, r00032, and r02019, were published as part of a prior study (30). Historical viral load data for five additional SIVmac239-infected, Mamu-B*17-negative, Mamu-B*08+ macaques were included in the viral load analysis. Animals r96104 and r96113 were controls from a previous vaccine study (1), while animal r98057 was a control in an unpublished vaccine study. Macaques r00078 and r01064 were part of a previously reported CD8 depletion experiment (19).
Primers for site-directed mutagenesis.
We introduced mutations sequentially into SIVmac239 proviral plasmids using site-directed mutagenesis (QuikChange PCR; Stratagene, La Jolla, CA) according to the manufacturer's protocol. The forward primers used to introduce each mutation are as follows (the altered codon is underlined): Vif172-179RL8-R2G (GG AAA CAG TGG AGA GGA GAC AAT AGG AGA GG), Vif123-131RL9-R2K (GCG GGA GAA GTG AGA AAG GCC ATC AGG GG), Nef8-16RL9a-P5Q (CGG TCC AGG CAG TCT GGA GATC), Nef246-254RL9b-L9P (GCA AGA GGC CTT CCT AAC ATG GCT GAC), Nef137-146RL10-A1P (CTG GAA GGG ATT TAT TAC AGT CCA AGA AGA CAT AGA ATC TTAG), Rev12-20KL9-I6L (CCGA AAA AGG CTA AGG CTA TTA CAT CTT CTG CAT CAAAC), Rev44-51RL8-L7I (G TGG CAA CAG ATC CTG GCC TTG GCA GAT AG), and Env573-581KL9-L9M (GAA TTG TTG CGA ATG ACC GTC TGG GG). The reverse complement of each primer was also used for mutagenesis PCR. Virus stocks were produced by transfecting Vero cells with proviral DNA by using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) as recommended by the manufacturer. CEMx174 cells were overlaid onto Vero cells 1 day posttransfection for expansion of the virus. We then transferred the CEMx174 cells to flasks 2 days later and maintained them until syncytia were widespread in the culture (about 1 week). Filtered supernatants from these cultures were stored in the vapor phase of liquid nitrogen until use. Using a commercial enzyme-linked immunosorbent assay kit (ZeptoMetrix Corporation, Buffalo, NY), we determined the Gag-p27 content of the viral stocks. Viral RNA (vRNA) was isolated and sequenced as described below to confirm that only the desired mutations were present. Animals were infected intravenously with 1 ng Gag-p27 equivalent of this virus resuspended in 0.5 ml phosphate-buffered saline (PBS). Animals newly infected with wild-type (WT) SIVmac239 as part of this study were also infected intravenously with 1 ng Gag-p27 equivalent resuspended in 0.5 ml PBS.
Cells.
EDTA-anticoagulated blood was centrifuged over Ficoll (GE Healthcare Systems, Piscataway, NJ) for 30 min at 1,860 × g to obtain peripheral blood mononuclear cells (PBMC). Plasma was removed and clarified by an additional centrifugation. Resulting PBMC were washed once in R10 medium (RPMI medium, 10% fetal calf serum [FCS], 1% antibiotic-antimycotic, 1% l-glutamine [all from HyClone Laboratories, Logan, UT]) and treated with ACK buffer to lyse remaining red blood cells. After washing, the cells were resuspended in R10 medium for use in immunological assays. Cells were cryopreserved using a Cryostor CS5 solution (BioLife Solutions, Inc., Bothell, WA). Some tetramer stainings were conducted on cryopreserved PBMC. These cells were thawed at 37°C, washed once with R10 medium, and used the same day in tetramer stains, as described above.
In vitro viral fitness assays.
For growth curves, PBMC were isolated from an SIV-negative macaque. We then depleted the PBMC of CD8α-positive cells using nonhuman primate CD8 Microbeads (Miltenyi Biotec, Inc., Auburn, CA) according to the manufacturer's protocol and activated them using phytohemagglutinin (5 μg/ml) for 18 to 24 h. One million activated cells were resuspended in 500 μl medium and infected with 100 pg, 10 pg, or 1 pg of virus. After 4 h, the cells were washed once and plated in duplicate (500,000 cells per well in a volume of 2 ml R15-50 medium [RPMI medium, 15% FCS, 1% antibiotic-antimycotic, 1% l-glutamine {all from HyClone Laboratories} containing 50 units of interleukin-2/ml [NIH AIDS Research and Reference Reagent Program, Germantown, MD]) in 24-well plates. At each time point, 500 μl of supernatant was removed and then replaced for vRNA quantitation.
For competition assays, SIVmac239 containing 10 synonymous mutations (referred to as Δ10s) in gag was constructed using QuikChange PCR. This mutated gag is not detected using our standard primers for quantifying SIV gag, allowing the specific, separate quantification of WT and Δ10s sequences from a mixed sample (data not shown), although the amino acid sequence of Gag is identical. The Δ10s virus shows no fitness defect in in vitro competition assays with WT SIVmac239.
Target cells were isolated and activated as described above for the growth curves. Activated cells were then cultured an additional day in R15-50 medium. For the competition assay, 1 million cells were resuspended in 100 μl medium and were infected with the competitor viruses at ratios of 10 pg:100 pg Gag-p27, 50 pg:50 pg Gag-p27, or 100 pg:10 pg Gag-p27. After 4 h, the cells were washed once and plated in duplicate (500,000 per well in a volume of 2 ml R15-50 medium in 24-well plates). Five hundred microliters of supernatant was removed and replaced for the quantitation of vRNAs.
We isolated vRNA using a Qiagen M48 Biorobot apparatus with the M48 virus minikit (Qiagen Corp., Valencia, CA). Quantitation of vRNA for the growth curve samples was performed as described above in “Detection of SIV RNA in plasma.” Separate quantitative reverse transcription-PCRs (RT-PCRs) were performed to quantify WT and Δ10s virus in each competition assay sample by using sequence-specific amplification primers. Each reaction mixture contained the following reagents: 10 μl SuperScript III Platinum one-step 2× buffer containing 5 mM MgSO4 and 0.2 mM of each deoxynucleoside triphosphate, 0.8 μl SuperScript III Platinum one-step enzyme mix (Invitrogen, Carlsbad, CA), forward and reverse primers with a final concentration of 500 nM, probe with a final concentration of 100 nM, 0.5% bovine serum albumin, 5 μl RNA template, and diethyl pyrocarbonate-treated water to 20 μl. The probe used for both the WT and Δ10s reactions and primers to amplify WT gag are described above in “Detection of SIV RNA in plasma.” Primers used to amplify the Δ10s sequence were LifMUT1F (5′-GTG TGC GTG ATC TGG TGT ATAC-3′) and LifMUT3R (5′-GAC TAG GTG ACT CTG GAC AAT CTG TTT AG-3′) (underlining indicates the differences from WT gag). Reactions were run using a Roche (Indianapolis, IN) LightCycler 480 apparatus under the following cycling conditions: cDNA synthesis at 50°C for 30 min, an initial denaturation step at 95°C for 2 min, and 45 amplification cycles of 95°C for 15 s and 68°C for 30 s (analysis mode, quantification; acquisition mode, single). A final cooling step was performed at 40°C for 30 s. Tenfold serial dilutions of an SIV gag in vitro transcript and a Δ10s gag in vitro transcript served as standard curves for each run.
Cell line recognition of mutant viruses.
CD8+ T-cell lines specific for Mamu-B*08-restricted epitopes were generated from SIV-infected Mamu-B*08+ animals by the repeated stimulation of PBMC with irradiated autologous B-lymphoblastoid cell lines pulsed with 1 μM peptide. We maintained the cell lines in R15-50 medium using an incubator with 5% CO2 at 37°C. CD4+ T-cell targets were generated, and virus was sucrose purified by centrifugation through 20% sucrose as described previously (49). Target cells were spinoculated (46) with the virus of interest in order to infect a high percentage of cells. Two days later, infected cells were mixed at a 1:1 ratio with cell lines specific for the epitope being tested and incubated at 37°C for 6 h, in the presence of 10 μg/ml brefeldin A (Sigma) for the last 4.5 h. Cells were stained for 30 min at room temperature with anti-CD4 and anti-CD8 monoclonal antibodies, washed twice with fluorescence-activated cell sorter (FACS) buffer (PBS containing 2% FCS and 1% bovine serum albumin), and fixed with 1% paraformaldehyde diluted in PBS for either 15 min or stored overnight at 4°C. Cells were then washed once with FACS buffer and once with FACS buffer containing 0.1% saponin to permeabilize cells. Anti-gamma interferon (IFN-γ) and anti-tumor necrosis factor alpha (TNF-α) antibodies were added to the cells, and the cells were incubated for 30 min at room temperature, washed twice with saponin-containing FACS buffer, and fixed with 1% paraformaldehyde diluted in PBS. Fixed cells were run on a FACSCalibur flow cytometer (Becton Dickinson) and analyzed using FlowJo software (Tree Star, Ashland, OR).
Detection of SIV RNA in plasma.
vRNA was isolated from EDTA-anticoagulated plasma as described previously, and viral loads were determined using a modification of the previously reported protocol (13). Briefly, vRNA was reverse transcribed and quantified using the SuperScript III Platinum one-step quantitative RT-PCR system (Invitrogen, Carlsbad, CA) with a LightCycler 2.0 apparatus (Roche, Indianapolis, IN). The final reaction mixtures (20-μl total volume) contained 0.2 mM (each) deoxynucleoside triphosphates, 3.5 mM MgSO4, 150 ng random hexamer primers (Promega, Madison, WI), 0.8 μl SuperScript III reverse transcriptase and Platinum Taq DNA polymerase in a single enzyme mix, 600 nM of each amplification primer (forward primer SIV1552 [5′-GTCTGCGTCATCTGGTGCATTC-3′] and reverse primer SIV1635 [5′-CACTAGCTGTCTCTGCACTATGTGTTTTG-3′]), and 100 nM probe (5′-6-carboxyfluorescein-CTTCCTCAGTGTGTTTCACTTTCTCTTCTGCG-BHQ1-3′). Cycling conditions were 37°C for 15 min, 50°C for 30 min, and 95°C for 2 min, followed by 50 amplification cycles of 95°C for 15 s and 62°C for 1 min with ramp times set to 3°C/s. Tenfold serial dilutions of an SIV gag in vitro transcript were quantified during each run and served as a standard curve. We determined copy numbers for samples by interpolation onto the standard curve by using LightCycler software, version 4.0. The lower limit of detection of this assay was 30 vRNA copy equivalents (eq)/ml plasma under standard assay conditions.
Tetramer and subset staining.
Tetramers conjugated to either phycoerythrin or allophycocyanin (APC) were produced by the Wisconsin National Primate Center Tetramer Core. Between 500,000 and 1,000,000 PBMC were stained with tetramers in a volume of 100 to 200 μl R10 medium for 1 h at 37°C. Surface stains for CD3 and CD8 were then added, and cells were incubated at room temperature for an additional 30 to 40 min, washed twice, and fixed with 1% paraformaldehyde diluted in PBS. Fixed cells were run on either a BD-LSRII or FACSCalibur flow cytometer (Becton Dickinson) and analyzed using FlowJo software, version 8.8.6 (Tree Star). We gated lymphocytes based on forward and side scatters. Tetramer frequencies are expressed as a percentage of CD3+ CD8+-gated events. Only tetramer staining frequencies of at least 0.05% were considered positive.
For determinations of CD4 counts, PBMC were stained with titrated anti-CD3, -CD4, -CD8, -CD28, and -CD95 antibodies for 30 to 40 min at room temperature, washed twice, and fixed with 1% paraformaldehyde in PBS. Subsets were determined as a percentage of the lymphocyte gate. We then calculated absolute counts using lymphocytes/μl blood determined by a complete blood count performed using a Pentra 60C+ hematology analyzer (ABX Diagnostics, Irvine, CA).
The following antibodies from BD Biosciences were used over the course of this study: anti-CD3-fluorescein isothiocyanate (clone SP34), APC (clone SP34-2) or Alexa700 (clone SP34-2), anti-CD28-phycoerythrin (clone CD28.2), anti-CD95-fluorescein isothiocyanate (clone DX2), anti-CD8a Pacific Blue (clone RPA-T8) or -peridinin chlorophyll protein (clone SK1), and anti-CD4-peridinin chlorophyll protein (clone L200). For assays measuring the cell line recognition of mutant viruses, anti-IFN-γ (clone 4S.B3), anti-TNF-α (clone MAb11), and anti-CD4-APC (clone M-T466 from Miltenyi Biotec) were used.
Sequencing of plasma vRNA.
vRNA was extracted from plasma using the Qiagen MinElute kit or by guanidium thiocyanate extraction. Amplicons containing the 14 Mamu-B*08-restricted epitopes were amplified for each vRNA sample using the Qiagen one-step RT-PCR kit and SIVmac239-specific primer pairs as previously described (45). In addition, we designed four new primer sets using Primer3 (http://frodo.wi.mit.edu/) to amplify target regions. These primers were SIV-5557-F (5′-AGG TAG TAC CCA GAA GAA AGG CTA-3′) and SIV-6349-R (5′-ATG TCC TTC CCC TAG ACA TCT ACA-3′), SIV-6465-F (5′-TAG TGG AGG TTC TGG AAG AAC TG-3′) and SIV-7021-R (5′-TGT TCC CCA AGT ATC CCT ATT CT-3′), SIV-8294-F (5′-AAT ATC ACC ATG AGT GCA GAG GT-3′) and SIV-8777-R (5′-ACT CTT GCC AAG TCT CATT GTT C-3′), and SIV-8903-F (5′-TTT GAC CTT GCT TCT TGG ATA AA-3′) and SIV-9575-R (5′-TGC TAA TTT TTC TCT CTC TTC AGC-3′). RT-PCR cycling conditions were 50°C for 30 min; 95°C for 15 min; 45 cycles of 94°C for 30 s, 53°C for 60 s, and 72°C for 150 s; and then 68°C for 20 min. Cycling ramp rates were 2°C per second. Unincorporated primers and nucleotides were enzymatically removed from the PCR products using ExoSAP-IT (USB Corporation, Cleveland, OH). Forward and reverse sequencing reactions for each amplicon were carried out the using DYEnamic ET Terminators sequencing kit (GE Healthcare, Piscataway, NJ) with the same primers as those used for RT-PCR. The cycling conditions for sequencing reactions were 30 cycles of 95°C for 20 s, 50°C for 15 s, and 60°C for 1 min. Products were then purified with the Agencourt CleanSEQ dye terminator removal kit (Agencourt Bioscience Corporation, Beverly, MA) and run on a 3730xl DNA analyzer (Applied Biosystems, Foster City, CA). Sequence assembly and base calling were done using CodonCode Aligner, version 2.0.6 (CodonCode Corporation, Dedham, MA). Bases were called as mixed whenever the secondary peak had an intensity that was greater than 40% compared to the dominant peak. DNA sequences were conceptually translated and aligned to WT SIVmac239 (GenBank accession no. M33262) in MacVector 10.6, trial version (MacVector, Inc., Cary, NC).
IFN-γ ELISPOT assay.
Enzyme-linked immunospot (ELISPOT) assays were performed using previously described methods (30). Briefly, PBMC were used directly in precoated ELISpotPLUS kits (Mabtech, Inc., Mariemont, OH) for the detection of IFN-γ-secreting cells according to the manufacturer's protocols. All tests were performed in duplicate. Wells were imaged and counted with an AID EliSpot reader, version 4.0, and analyzed as described previously (30).
Statistical analyses.
The “set-point” viral load for an animal was calculated as the geometric mean of all available viral loads from each animal between weeks 15 and 48 postinfection. The incidences of “control” (e.g., set point at least 1 log lower than the geometric mean chronic-phase viral load for SIVmac239-infected rhesus macaques) in the WT SIVmac239 (n = 15) and 8X-SIVmac239 (n = 10) groups were compared using the two-sided Fisher's exact test. Set points for each animal in the 8X- and WT-infected groups were log transformed, and the average set points for these groups were then compared using the t test with Welch correction. The nonparametric Wilcoxon signed-rank test was used to compare the magnitudes of epitope-specific CD8+ T-cell responses measured by MHC class I tetramer staining between WT SIVmac239-infected (n = 10) and 8X-SIVmac239-infected (n = 10) animals. Additional tests not shown (t test with Welch correction following log transformation) were performed and gave convergent results. Central memory CD4 counts of the groups were compared using the t test with Welch correction following log transformation. Statistical analyses were performed using SPSS v.12.0.
RESULTS
Construction and characterization of 8X-SIVmac239.
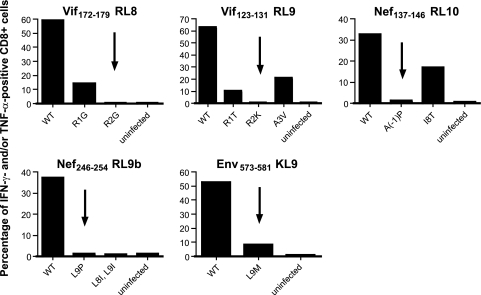
We introduced mutations into SIVmac239 with the goal of abrogating the immunogenicity of Mamu-B*08-restricted epitopes. We selected mutations for use in this study from mutations that we had observed previously in viral sequences from SIVmac239-infected Mamu-B*08/B*03+ animals (16, 30, 31). For some epitopes in which a variety of escape mutations had been observed, we screened viral variants for their effectiveness at reducing the recognition of virally infected cells by epitope-specific CD8+ T cells in vitro (Fig. 2). The L9M mutation introduced into the Env573-581KL9 epitope was also tested because viral escape has been observed only for SIV-infected Mamu-B*03+ macaques (16). We selected the mutations that were the most effective at reducing the recognition of virally infected cells by cell lines specific for the epitope for inclusion in the challenge virus. Thus, the selected mutations are not consensus escape sequences but rather those least likely to engender T-cell responses. There are 13 defined Mamu-B*08-restricted epitopes (33). Escape variation had not previously been observed in five of the subdominant epitopes.
FIG. 2.
In vitro recognition of escape variant viruses by epitope-specific CD8+ T-cell lines. We mixed polyclonal epitope-specific cell lines with CD4+ T cells infected with either WT SIVmac239 or SIVmac239 with the indicated mutation(s) in the epitope being tested. We then performed an intracellular cytokine staining assay to identify cells secreting IFN-γ and TNF-α. The percentages of CD4− CD8+ cells that were IFN-γ and/or TNF-α positive following stimulation are displayed. We intracellularly stained infected target cells for the expression of Gag p27 in parallel to ensure that comparable percentages were infected with the different viruses (data not shown). Results are representative of at least two independent experiments per epitope, using cell lines cultured from at least two different animals. Mutations that we selected for use in the challenge virus are indicated with arrows.
We altered eight epitopes using site-directed mutagenesis (Table 1). These included epitopes targeted by what are typically the four most immunodominant responses during acute SIVmac239 infection (Vif172-179RL8, Vif123-131RL9, Nef137-146RL10, and Nef246-254RL9b). We also mutated four epitopes that are typically targeted by lower-magnitude responses and/or in a lower frequency of animals. We changed the position 2 primary anchor residues for both epitopes in Vif. Env573-581KL9 and Nef246-254RL9b were changed at the C-terminal primary anchor residues; these changes also substantially reduce the binding of these peptides to Mamu-B*08. We introduced a proline immediately upstream of the defined minimal epitope for Nef137-146RL10. This mutation is a putative processing escape mutation, as prolines are rarely found at the N termini or immediately upstream of MHC-I epitopes (50). We mutated the remaining three epitopes, Nef8-16RL9a, Rev12-20KL9, and Rev44-51RL8, at positions 5, 6, and 7, respectively. These changes have relatively little effect on binding to Mamu-B*08 but may affect T-cell receptor (TCR) recognition (Table 1). The mutations that we introduced into the Rev epitopes are nonsynonymous in overlapping reading frames. The mutation in Rev KL9 resulted in an asparagine-to-isoleucine change at position 92 in Tat, while the Rev RL8 mutation resulted in a change at position 125 from alanine to aspartate in Tat and also in a change at position 760 from serine to arginine in the envelope protein.
TABLE 1.
Mamu-B*08-restricted CD8+ T-cell epitopes mutated in 8X-SIVmac239a
| Epitope | Amino acid sequenceb | Mamu-B*08 binding affinity (IC50 [nM])c | Fold reduction in binding to Mamu-B*08 |
|---|---|---|---|
| Vif172-179RL8 | RRDNRRGL | 125 | |
| .G...... | 594 | 4.8d | |
| Vif123-131RL9 | RRAIRGEQL | 7.5 | |
| .K....... | 2,901 | 387d | |
| Nef137-146RL10 | (A)RRHRILDIYL | 11 | |
| (P).......... | NT | NAe | |
| Nef246-254RL9b | RRLTARGLL | 3.2 | |
| ........P | 477 | 151d | |
| Env573-581KL9 | KRQQELLRL | 12 | |
| ........M | 397 | 33d | |
| Rev12-20KL9 | KRLRLIHLL | 3.2 | |
| .....L... | 1.6 | Nonef | |
| Nef8-16RL9a | RRSRPSGDL | 105 | |
| ....Q.... | 428 | 4.1g | |
| Rev44-51RL8 | RRRWQQLL | 20 | |
| ......I. | 59 | 2.9f |
Epitopes are listed in order of the typical immunodominance hierarchy of responses targeting the epitopes. Subscript numbers in the epitope name indicate amino acid positions of the epitope.
The mutations introduced are indicated beneath the sequence of the epitope. These changes are indicated in boldface type.
Peptides with IC50s of >500 nM are not considered binders and are indicated in italics. The binding of WT and 8X variant peptides to Mamu-B*08 was described previously (33). NT, not tested.
The putative mechanism of escape is a primary anchor residue mutation.
The putative mechanism of escape is a processing mutation. NA, not applicable, as the mutation introduced is outside the defined epitope.
The putative mechanism of escape is a TCR contact mutation.
The putative mechanism of escape is TCR contact mutation along with some reduction in Mamu-B*08 binding.
Five subdominant Mamu-B*08-restricted epitopes (Gag263-271YL9, Vpr62-70IF9, Env524-532KF9, Env717-725LF9, and Env868-876RL9) in which variation had not been seen in vivo remained intact. We did not alter these epitopes, as we were concerned that the introduction of mutations not known to be replication competent in vivo could result in a virus that had attenuated replication in animals.
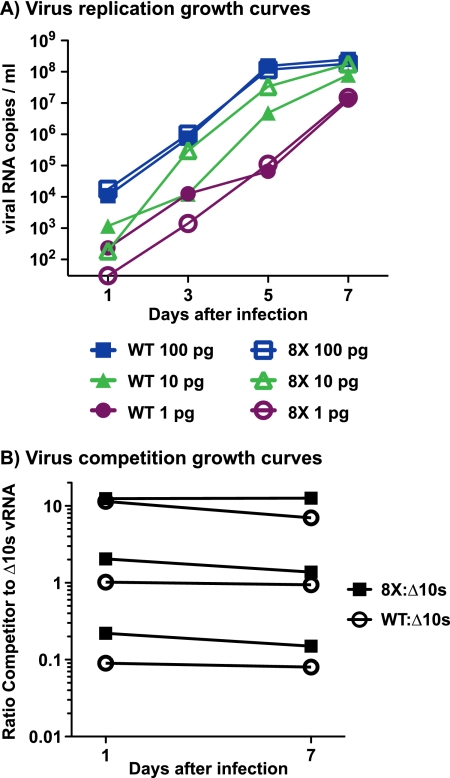
We first assessed whether this virus, 8X-SIVmac239 (referred to as 8X for the remainder of the manuscript), replicated normally in vitro. Growth curves did not reveal an obvious difference in replicative capacities between 8X and WT SIVmac239 (Fig. 3A). Therefore, we then assessed fitness in an in vitro coculture assay, which should reveal more subtle differences in fitness than simple growth curves (14). The competitor virus for this assay, SIVmac239-Δ10s (Δ10s), has 10 synonymous point mutations in gag. These mutations allow the differential amplification/quantification of Δ10s RNA and unchanged SIV gag RNA from mixed samples using separate sets of primers. We infected activated, CD8-depleted macaque PBMC with 8X and Δ10s at ratios of 1:1, 1:10, and 10:1 (Fig. 3B). A competition assay of WT SIVmac239 and Δ10s was done as a control. vRNA was isolated from culture supernatants, and the amount of Δ10s and WT SIV gag sequence was quantitated. We did not detect substantial changes in the ratios of 8X to the Δ10s virus over the course of a 7-day assay at any of the starting ratios. Therefore, we concluded that the 8X virus does not have an in vitro replicative defect. However, three of the mutations in 8X were located in Nef, a protein which is largely dispensable for replication in vitro (20).
FIG. 3.
In vitro viral fitness assays. (A) We infected 1 million cells with three different amounts of virus (100 pg, 10 pg, and 1 pg) and then plated cells in duplicate. The number of copies of SIVmac239 vRNA present in the supernatant was determined by quantitative PCR. Results displayed are averages of duplicates, except for the lowest infection amount, for which only one of the duplicate wells showed productive infection for both the 8X and WT viruses. (B) Cells were infected with three different starting ratios of SIVmac239-Δ10s (which has synonymous mutations in gag to allow for strain-specific PCR amplification [see Materials and Methods]) and a competitor virus, either WT SIVmac239 or 8X-SIVmac239. The numbers of copies of SIVmac239-Δ10s and competitor vRNA present in the supernatant were determined in separate quantitative PCRs. We calculated the ratio of competitor to SIVmac239-Δ10s by dividing the number of copies of competitor vRNA (e.g., 8X or WT) by the number of copies of SIVmac239-Δ10s vRNA. Results are representative of at least two independent assays.
Viral loads and central memory CD4 count outcomes.
We infected two Mamu-B*08-negative macaques with 8X virus in order to assess whether the virus replicated to normal levels in vivo (Fig. 4). The 8X virus peaked at >107 vRNA copy eq/ml plasma in both of these animals, indicating that the 8X virus was able to replicate to high levels in vivo. Outcomes for these two animals diverged, however. Animal r96141 became an EC, while animal rh2029 had a viral set point in excess of 106 vRNA copy eq/ml. Interestingly, animal r96141 expresses Mamu-B*0602 (see Table S1B in the supplemental material; our unpublished data). A similar allele, Mamu-B*06, is almost always carried on the same MHC-I haplotype as Mamu-B*08 (see Table S1 in the supplemental material) (47), and it has not been possible to independently evaluate its association with viral load outcomes.
FIG. 4.
Viral loads of two Mamu-B*08-negative rhesus macaques infected with 8X-SIVmac239. The gray bar indicates the lower limit of detection in the viral load assay (30 vRNA copy eq/ml).
We infected 10 Mamu-B*08+ macaques intravenously with 8X (Fig. 5A). Viral loads were determined weekly during acute infection and compared to the viral loads of six WT SIVmac239 animals infected here, along with nine Mamu-B*08+ animals previously infected with WT virus (Fig. 5B). It is important that animals also expressed a variety of MHC-I alleles in addition to Mamu-B*08. Therefore, comprehensive MHC-I genotypes of the 8X-infected animals and 10 of the WT virus-infected animals were determined using pyrosequencing (54) (see Table S1 in the supplemental material).
FIG. 5.
Viral loads of 8X- and WT-infected rhesus macaques. (A) Viral loads of the 10 8X-infected animals included in this study. The cross symbol indicates animal r03028, which was euthanized at 36 weeks postinfection due to AIDS-like symptoms. (B) Viral loads of the 15 WT SIVmac239-infected animals included in this study. The gray bar indicates the lower limit of detection in the viral load assay (30 vRNA copy eq/ml). (C) Set-point viral loads of WT- and 8X-infected Mamu-B*08+ animals. The viral set point was calculated as the geometric mean of all of each animal's viral loads between weeks 15 and 48 postinfection. Fewer animals in the 8X-infected group controlled viremia to <20,000 vRNA copy eq/ml than would be expected to control WT SIVmac239 infection (P = 0.04 by Fisher's exact t test, two sided). (D) Geometric mean viral loads of 8X-infected (orange line) and WT-infected (blue line) animals through 36 weeks postinfection, at which time an 8X-infected animal was euthanized. Due to the variable sampling of animals during the chronic phase of SIV infection, viral loads used in the geometric mean viral load calculation include some samples taken ±2 weeks from the indicated time point.
Seven of the 15 WT-infected macaques maintained a set-point viral load of <1,000 vRNA copy eq/ml plasma between weeks 15 and 48 postinfection. Three additional WT-infected animals controlled viral replication to <20,000 copy eq/ml plasma (Fig. 5B), which is a log below the chronic-phase geometric mean viral load of our cohort of 196 SIVmac239-infected Indian rhesus macaques (32). Of the 10 macaques infected with 8X-SIVmac239, 2 animals unexpectedly became ECs, maintaining viral loads of <1,000 vRNA copy eq/ml plasma (Fig. 5A). Interestingly, one of these 8X ECs, animal r04135, is a full sibling and MHC-I identical to animal r02040 (Fig. 1), a WT-infected animal with high viral loads. A detailed comparison of immune responses, viral loads, and central memory CD4 counts of these two animals is shown in Fig. S1 in the supplemental material.
We assessed whether fewer animals controlled 8X replication than would be expected if animals were infected with the WT virus. We calculated the set point as the geometric mean of all available viral loads from each animal between weeks 15 and 48 postinfection. Ten of 15 WT-infected Mamu-B*08+ animals had set points under 20,000 vRNA copy eq/ml, whereas 2 of the 10 8X-infected animals had set points under 20,000 copies/ml, a lower incidence of control (P = 0.04 by Fisher's exact t test, two sided) (Fig. 5C). Geometric mean viral loads for the 8X-infected group of animals were about 1 log higher than geometric mean viral loads for the WT-infected group during chronic infection (Fig. 5D). The difference in the average set-point viral loads of the groups was nearly significant (P = 0.06 by t test with Welch correction following log transformation).
The precise kinetics with which controllers’ viral loads diverge from progressors’ viral loads is unknown. To help focus on key time points for further immunological analysis, we examined viral loads to determine the time frame in which these groups diverged. We segregated viral loads from the 15 WT-infected animals and 10 8X-infected Mamu-B*08+ animals in this study based on whether animals ultimately became controllers (set point of <20,000 vRNA copy eq/ml plasma) or progressors (set point of >20,000 vRNA copy eq/ml plasma). We compared these groups’ viral loads at each week after infection. By 5 weeks postinfection, these two groups were largely distinct in terms of viral loads (see Fig. S2 in the supplemental material). Thus, immune responses present between 2 and 5 weeks postinfection may be important drivers of viral control.
In addition to viral loads, we monitored peripheral blood lymphocyte subsets. The CD4+ T-cell subset that was most depleted during acute infection was the central memory (CD28+ CD95+) subset. 8X-infected animals experienced levels of central memory CD4 loss during the first 2 weeks of infection that were comparable to the loss seen in WT-infected animals (see Fig. S3A and S3B in the supplemental material). Notably, controller animals infected with either 8X or WT virus experienced levels of central memory CD4 loss similar to that seen in progressor animals at 2 weeks postinfection (see Fig. S3C and S3D in the supplemental material). However, controllers’ central memory CD4 counts recovered to near-baseline levels by 24 weeks postinfection, while central memory CD4 counts in progressors failed to rebound.
Immunogenicity of 8X-SIVmac239.
We monitored acute-phase CD8+ T-cell responses in 8X-infected animals and in a subset of WT-infected animals using MHC-I tetramers for the 13 epitopes restricted by Mamu-B*08. These included the eight CD8+ T-cell epitopes which were mutated in 8X (Table 1) and five recently defined epitopes (33) in which viral variation had not been detected prior to this study and so were unchanged in 8X. These additional epitopes were Gag263-271YL9, Vpr62-70IF9, Env524-532KF9, Env717-725LF9, and Env868-876RL9.
Because tetramers were folded to WT peptides, we were concerned that they might underrepresent potential 8X variant-specific responses. For this reason, IFN-γ ELISPOT assays using both the WT and 8X variants of minimal epitopes were carried out in parallel to verify the tetramer results (see Fig. S4 in the supplemental material). Additionally, pools of SIVmac239-derived peptides known to bind Mamu-B*08 at biologically relevant affinities (50% inhibition concentration [IC50] of ≤500 nM) but which were not seen to be immunogenic in a screen of seven chronically infected animals (33) were tested in IFN-γ ELISPOT assays. In this manner, we sought to quantitate T-cell responses targeting Mamu-B*08-restricted epitopes in known SIV open reading frames as comprehensively as possible.
The five acute-phase T-cell responses against Mamu-B*08-restricted epitopes that are normally of the highest magnitude were either absent or severely impaired in 8X-infected animals (Fig. 6A). At week 3 postinfection, almost no responses to each of the Vif epitopes (which are typically the strongest responses seen in WT-infected animals) were detected by tetramer or IFN-γ ELISPOT assay. In contrast, WT-infected animals had mean responses of 4.5% and 1.3% of CD3+ CD8+ lymphocytes to Vif172-179RL8 and Vif123-131RL9, respectively. Small (<0.3%) responses directed against Nef137-146RL10, which had a processing mutation introduced, were detected in several 8X-infected animals, while WT-infected animals typically had robust cellular responses at this time (mean, 1.8% of CD3+ CD8+ lymphocytes). Likewise, responses to Nef246-254RL9b could be detected in several different 8X-infected animals at various times after infection. One 8X-infected EC animal, animal r04135, made a large (1.6% of CD3+ CD8+ lymphocytes at week 6 postinfection) response to this epitope, similar in magnitude to that seen with WT-infected animals. The response to Env573-581KL9, a response that contracts substantially between weeks 2 and 3 postinfection, was somewhat greater in WT-infected animals at the week 3 time point shown.
FIG. 6.
Tetramer responses to WT and 8X viruses in Mamu-B*08+ macaques at 3 weeks postinfection. (A) Responses to epitopes with mutations introduced into 8X, measured in 10 WT-infected and 10 8X-infected Mamu-B*08+ animals at 3 weeks postinfection. (B) Responses to Mamu-B*08-restricted epitopes that were unchanged in 8X. Tetramer responses are expressed as a percentage of CD3+ CD8+ lymphocytes. A 13th Mamu-B*08-restricted epitope, Env717-725LF9, is not shown here, as no animals responded to this epitope. Magnitudes of responses made by the groups of 8X- and WT-infected animals were compared using the Wilcoxon test. Responses from progressors (viral set point of >20,000 vRNA copy eq/ml) are in green, and responses from controllers (viral set point of <20,000 vRNA copy eq/ml) are in purple.
Of the three mutated epitopes with relatively unaltered binding to Mamu-B*08, two (Nef8-16RL9a and Rev44-51RL8) were rarely targeted by either WT- or 8X-infected animals, as measured by tetramer staining and ELISPOT assay. The third, Rev12-20KL9, had a slightly lower mean frequency of CD3+ CD8+ tetramer-positive lymphocytes in 8X-infected animals at week 3 postinfection, a difference that was not significantly different from the mean of WT-infected animals’ tetramer responses (Fig. 6A). Interestingly, in 8X-infected animals, ELISPOT responses to the 8X variant Rev12-20KL9 peptide (in which we had introduced a position 6 mutation, which still binds to Mamu-B*08 with an affinity equal to that of the WT peptide) were often stronger than responses to the WT peptide (see Fig. S4 in the supplemental material). This consistently increased responsiveness to the variant peptide, which was not observed for any of the other mutated epitopes, suggests that tetramer stains might underrepresent the Rev12-20KL9 response in 8X-infected animals.
In the 8X virus, five recently defined Mamu-B*08-restricted epitopes remained intact. T-cell responses targeting these epitopes are normally subdominant in WT-infected Mamu-B*08+ animals. Following infection with 8X, Mamu-B*08+ animals made significantly stronger CD8+ T-cell responses to two of these epitopes in the envelope protein (Fig. 6B). Another response, directed against the Vpr62-70IF9 epitope, was also somewhat greater in 8X-infected animals at 3 weeks postinfection. Of the remaining two Mamu-B*08-restricted epitopes, Gag263-271YL9 was rarely targeted by either 8X- or WT-infected Mamu-B*08+ animals (Fig. 6B), while no immune responses directed against Env717-725LF9 were detected in 8X- or WT-infected Mamu-B*08+ animals over the course of this study (data not shown). The overall differences between Mamu-B*08-restricted responses to WT and 8X viruses were maintained throughout acute infection. These results indicate that strong CD8+ T-cell responses dampened the magnitude of subdominant responses restricted by the same allele.
Pools of SIV-derived Mamu-B*08 binding peptides (IC50s of ≤500 nM) were also tested in IFN-γ ELISPOT assays to determine if we had missed any Mamu-B*08-restricted T-cell responses (data not shown). We identified one additional Mamu-B*08-restricted epitope using these peptides. This epitope overlaps with the above-defined Nef246-254RL9b epitope by 8 amino acids and is referred to as Nef245-253RL9c (see Fig. S5 in the supplemental material).
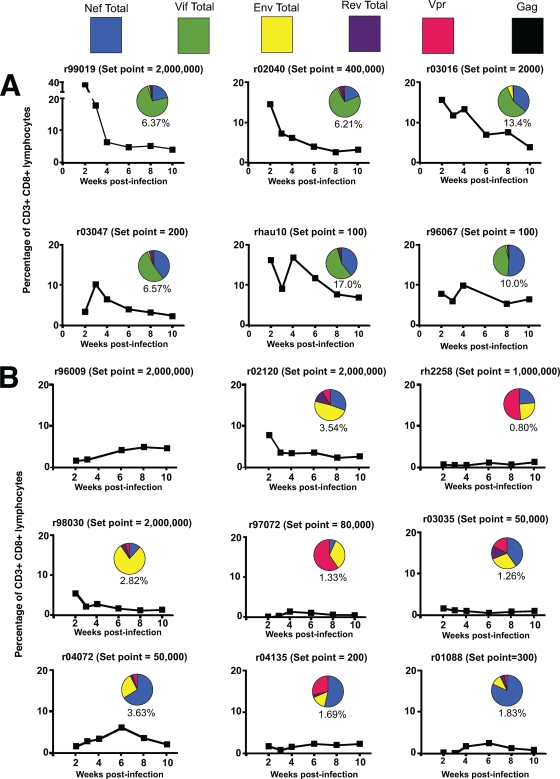
Although several typically subdominant CD8+ T-cell responses expanded to higher frequencies in 8X-infected animals than in WT-infected animals, these responses still did not approach the magnitude of the high-frequency Mamu-B*08-restricted responses seen in WT-infected animals. We calculated the total fraction of CD8+ T cells targeting Mamu-B*08-restricted epitopes in WT- and 8X-infected animals by summing all of their tetramer responses. The levels of Mamu-B*08-restricted responses in 8X-infected animals were lower than those of the WT-infected animals’ responses throughout the first 10 weeks of infection (Fig. 7). Additionally, the proteins targeted by these responses shifted. CD8+ T cells from WT-infected animals targeted predominately Vif and Nef. In contrast, CD8+ T cells from 8X-infected animals were directed against Env, Vpr, and/or Nef (Fig. 7).
FIG. 7.
Total Mamu-B*08-restricted responses during acute infection for individual macaques. (A) Responses from a subset of WT-infected animals. Graphs for WT-infected animals do not include the newly defined Nef245-253RL9c epitope. (B) Responses from 8X-infected animals. Tetramer responses to all of the Mamu-B*08 epitopes at the indicated weeks were summed and are expressed as a percentage of CD3+ CD8+ lymphocytes. These are plotted as line graphs. Inset pie charts represent the proportions of these tetramer responses targeting different SIV proteins at week 4 postinfection. Numbers underneath the pie charts indicate the total magnitudes of responses targeting Mamu-B*08-restricted epitopes at week 4 postinfection. No pie chart is presented for animal r96009, since samples from week 4 postinfection were not available.
8X variants are maintained in Mamu-B*08+ macaques.
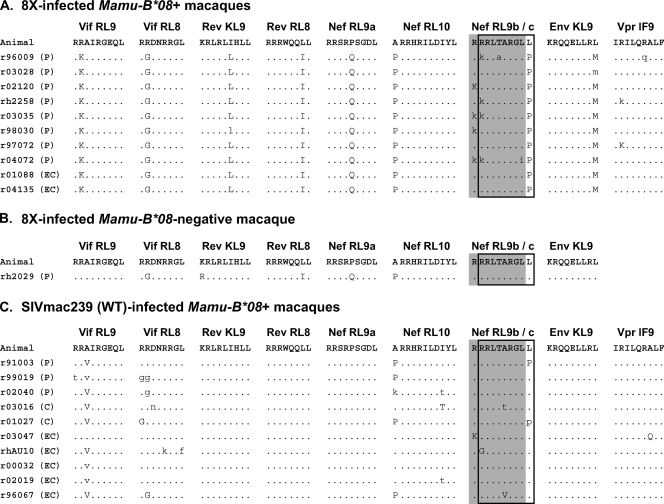
We next asked if viral sequence variation explained the differential viral load outcomes of 8X- and WT-infected animals. We first sequenced vRNA from weeks 18 to 20 postinfection in order to determine which Mamu-B*08-restricted epitopes had had variation selected for in these animals. In the 10 8X-infected animals, 8X mutations were maintained at 18 weeks postinfection (Fig. 8A). In a few instances, a mixed population was detected. In addition to maintaining the 8X mutations, we observed viral variation in 6 of 10 animals in the region containing the newly defined Nef245-253RL9c epitope. Interestingly, neither of the 8X-infected ECs had escape mutations in this region despite making CD8+ T-cell responses against it. However, Nef245-253RL9c also remained intact in one progressor macaque that made a CD8+ T-cell response against the epitope. Additionally, three 8X-infected animals had variation in the Vpr62-70IF9 epitope, an epitope which was targeted at a higher frequency by CD8+ T-cell responses in 8X-infected animals.
FIG. 8.
Sequence of Mamu-B*08-restricted epitopes in WT- and 8X-infected macaques. All known epitopes restricted by Mamu-B*08 were sequenced from plasma virus from Mamu-B*08+ animals between weeks 18 and 20 postinfection. The eight epitopes with mutations introduced into the 8X virus are shown, in addition to a ninth epitope in which we detected variation. We did not detect any variation in Env524-532KF9, Env868-876RL9, Env717-725LF9, or Gag263-271YL9 for either 8X- or WT-infected animals (data not shown). The Nef246-254RL9b epitope is indicated by an outlined box, and the Nef245-253RL9c epitope is indicated by a shaded box. (A) Epitope sequences in 8X-infected Mamu-B*08+ animals. (B) Sequence of the eight mutated epitopes at 46 weeks postinfection in one of the Mamu-B*08-negative animals infected with 8X. (C) Epitope sequences in WT-infected Mamu-B*08+ animals. Dots indicate the WT SIVmac239 sequence. Uppercase letters indicate differences from the WT sequence, and lowercase letters indicate the presence of a mixed base. Viral load outcomes are shown in parentheses next to an animal's identification number. EC indicates a set-point viral load of <1,000 vRNA copy eq/ml plasma. C indicates a set point of <20,000 copies/ml. P indicates a set point of >20,000 copies/ml.
The mutations introduced into the 8X virus did not cause a detectable fitness defect in vitro (Fig. 3). In vivo, however, four of these mutations (in Vif123-131RL9, Rev12-20KL9, Env573-581KL9, and Nef246-254RL9b) reverted in a Mamu-B*08-negative animal by 46 weeks postinfection (Fig. 8B), suggesting that some fitness cost is associated with these mutations.
In 10 WT-infected macaques, most animals—both ECs and progressors—had escape mutations in the two Vif epitopes and in two targeted Nef epitopes (Fig. 8C). Similar to previously reported observations (30), the kinetics with which these mutations accumulated did not differ between controllers and progressors (data not shown).
DISCUSSION
An understanding of the mechanism(s) by which some individuals spontaneously control HIV/SIV replication may aid in rational vaccine design. Here, we studied elite control of SIVmac239 in Mamu-B*08+ rhesus macaques. The association with MHC-I alleles implicates CD8+ T cells and/or natural killer cells in the control of viral replication. We attempted to break the ability of Mamu-B*08+ animals to control viremia by introducing point mutations into eight Mamu-B*08-restricted CD8+ T-cell epitopes, impairing the generation of epitope-specific CD8+ T-cell responses. Ten Mamu-B*08+ macaques were infected with this mutant virus, 8X. We compared the immune responses and viral loads of these animals to those of WT-infected Mamu-B*08+ macaques. The five typically strongest Mamu-B*08-restricted CD8+ T-cell responses were barely detectable in 8X-infected animals. Interestingly, these 8X-infected animals made unusually large responses to several unaltered, subdominant Mamu-B*08-restricted epitopes. By 48 weeks postinfection, 2 of 10 8X-infected Mamu-B*08+ animals controlled viral replication to <20,000 vRNA copy eq/ml plasma, while 10 of 15 WT-infected Mamu-B*08+ animals had viral loads of <20,000 vRNA copy eq/ml. These results suggest that high-frequency epitope-specific CD8+ T-cell responses restricted by Mamu-B*08 play an important role in establishing the control of viral replication.
Studies of chronically HIV-infected patients have failed to find correlations between the magnitude of T-cell responses directed against epitopes restricted by protective alleles and the control of viral replication (39, 41). Likewise, in recent work, no correlation between acute-phase T-cell responses and later set-point viral loads was seen in persons with a variety of HLA backgrounds (22). Similarly, in our study of acutely infected Mamu-B*08+ animals, there was no apparent relationship between the magnitude of responses and control of viremia. For instance, at week 2 postinfection, WT-infected animal r99019 had massive CD8+ T-cell responses against Mamu-B*08-restricted epitopes, in excess of 35% of its CD8+ T cells (Fig. 7A). This animal's viral set point was >1 million vRNA copy eq/ml plasma. Interestingly, animal r99019's Mamu-B*08-restricted CD8+ T-cell responses contracted dramatically between weeks 2 and 4 postinfection, which was not the case with WT-infected ECs’ responses, although they were not as high in frequency at week 2 (Fig. 7A). Additionally, there was no clear difference between which Mamu-B*08-restricted epitopes were targeted by WT-infected controllers and progressors. WT-infected animals generally made a consistent repertoire of responses, targeting multiple Mamu-B*08-restricted epitopes, albeit with various magnitudes. Thus, simply making immunodominant responses targeting Mamu-B*08 epitopes during the acute phase of infection is not sufficient to establish control in WT-infected animals.
The sequence of plasma virus did not clarify why some animals became ECs and others did not. Controllers’ and progressors’ viral loads diverged by 5 weeks postinfection (see Fig. S2 in the supplemental material). In 8X-infected animals, six of the eight progressors had variation in the region containing Nef246-254RL9b and Nef245-253RL9c, whereas neither of the two ECs had such variation despite making immune responses against this epitope (Fig. 8). However, the earliest viral variation detected in this epitope was at week 6 postinfection (data not shown), indicating that circulating virus in all animals should have been susceptible to T-cell responses targeting the Nef246-254RL9b and Nef245-253RL9c epitopes during at least the first 6 weeks of infection. Likewise, in the WT-infected animals, controllers and progressors had similar patterns and kinetics of escape (Fig. 8 and data not shown). However, a caveat to these conclusions is that our sequencing identifies only dominant quasispecies circulating in the plasma. It is possible that applying new deep-sequencing methodologies may reveal important dynamics in sequence variation that occur below our current level of detection (5, 52).
If simply making high-frequency CD8+ T-cell responses against Mamu-B*08-restricted epitopes is not sufficient for control, what other factors contribute to elite control in WT-infected animals? We have yet to extensively investigate the heterogeneity of T-cell responses against the same epitope in the setting of acute infection. For instance, the use of different T-cell receptor repertoires could render some T-cell responses more effective than others, as was shown previously in the setting of vaccination (48). Additionally, it is possible that different effector phenotypes of T cells are key. The responsiveness to antigen has been found to differ between cells from chronically infected progressors and controllers, with EC cells being able to secrete a wider array of cytokines and greater amounts of granzyme than cells from progressors (40). Perhaps most fundamentally, T-cell responses do not occur in isolation but act in concert with the many other components of the immune system. Qualitative differences in CD8+ T-cell responses may be determined by the immunological milieu in which the cells are initially primed. What sort of signals (numbers of T-cell receptors engaged, cytokines present, and levels of costimulatory engagement) are desirable during priming and whether such signals can be manipulated by vaccination could prove to be fruitful areas of future research.
We also investigated the effect of immunodomination (the phenomenon in which the presence of higher-frequency responses dampens the frequency of subdominant responses) on the magnitude of CD8+ T-cell responses. We observed that typically subdominant responses expanded to higher frequencies in the absence of immunodominant responses restricted by the same MHC allele. A recent vaccine study of macaques similarly found that the deletion of an immunodominant epitope from a simian-human immunodeficiency virus challenge virus resulted in a significantly more robust anamnestic expansion of a typically subdominant epitope (35). In our study, these subdominant responses did not, however, reach the levels of the high-frequency responses, which were “missing” (Fig. 6). Thus, immunodomination has a role in determining the magnitude of CD8+ T-cell responses, but the size of the responses is apparently also limited by other factors. This result, using outbred animals expressing a variety of other MHC-I alleles, agrees with data from previous work conducted largely using inbred mice as models (24, 25, 28, 44). Several groups have shown that the frequency of naïve precursor CD8+ T cells affects the magnitude of effector responses generated (23, 25, 28, 34, 44). Differences in precursor frequency could account for the failure of T cells directed against subdominant Mamu-B*08-restricted responses to expand to a greater extent in the absence of higher-frequency Mamu-B*08 responses. If TCR precursor frequency has a substantial role in dictating the magnitude of T-cell responses, altering immunodominance hierarchies by vaccination may prove to be difficult. However, it is also possible that the failure of these subdominant responses to expand to a greater extent could be due to immunodomination by T-cell responses restricted by MHC alleles other than Mamu-B*08—if this is the case, it is possible that dominance hierarchies are more malleable than our data indicate. The regulation of T-cell expansion and contraction during vaccination or chronic infection is currently poorly understood, and detailed studies of this topic would be very useful for vaccine design.
We had hypothesized that in the absence of the normal repertoire of Mamu-B*08-restricted responses, Mamu-B*08+ macaques would not become ECs. While the proportion of animals that controlled viremia was lower than would be expected for WT infection, 2 of 10 Mamu-B*08+ animals infected with the mutated virus still became ECs.
There are several hypotheses to explain why 2 of 10 Mamu-B*08+ macaques were still able to control 8X replication to very low levels. First, the mutant virus may be more easily controlled by an animal's immune system regardless of whether the animal expresses Mamu-B*08 or not. One of the two Mamu-B*08-negative animals infected with 8X became an EC, whereas the other had high viral loads (Fig. 4). This may indicate a replicative defect in the 8X virus in vivo. In the Mamu-B*08+ animals, at week 2 postinfection, the geometric mean viral load of those infected with 8X was as high as the geometric mean viral load of those infected with WT virus (Fig. 5). Therefore, no fitness defect in 8X was apparent during the first 2 weeks of infection. However, the reversion of four of the 8X mutations in vivo (Fig. 8B) suggests that mutations in 8X incur a replicative cost. Despite this, the majority of our 8X-infected Mamu-B*08+ macaques (which maintained these mutations) still had high levels of viral replication (Fig. 5A).
Another possibility is that the remaining responses against Mamu-B*08-restricted epitopes may in some cases be sufficient to control SIVmac239 replication to low levels. The sum of the Mamu-B*08-restricted responses made by the two 8X ECs was not notably different in magnitude than that of other 8X-infected animals (Fig. 7B). These two animals did have a somewhat skewed repertoire of Mamu-B*08-restricted responses compared to other 8X-infected animals, however, in that their CD8+ T cells targeted predominantly Nef (Fig. 7B). The strongest Mamu-B*08-restricted response of EC r01088 at week 4 was to Nef245-253RL9c, and the other EC, animal r04135, made an unusual response to the overlapping Nef246-254RL9b epitope, which had a primary anchor residue mutation introduced into 8X. The 8X-infected animals with higher levels of viremia tended to have their strongest Mamu-B*08 responses targeting Env or Vpr, although they generally made, and had escape in, Nef-directed responses also. The hypothesis that subdominant Mamu-B*08 responses mediated control could be tested by introducing point mutations into the additional Mamu-B*08-restricted epitopes that we found to accrue variation and by repeating the experiment reported here.
Third, it is possible that the surprising viral control by two 8X-infected macaques could be due to innate immune responses in Mamu-B*08+ animals tipping the balance to elite control. For instance, we know nothing about the different KIR alleles expressed by animals in this study. Based on human studies (36, 37), we could reasonably expect KIR to modulate the effect of Mamu-B*08 on viral replication. Information on KIR in rhesus macaques is limited, but a recent paper has defined a variety of KIR3DL polymorphisms, one of which was associated with higher viremia (8). An interesting future avenue of research with Mamu-B*08+ animals will be the characterization of the KIR genes (or other host factors) in animals that become ECs versus progressors.
In conclusion, we found that high-frequency CD8+ T-cell responses against Mamu-B*08-restricted epitopes may play an important role in establishing control of viral replication. In the case of 8X-infected animals, these responses were not strictly necessary to achieve control of viral replication. Two animals controlled the replication of the 8X virus in the absence of high-frequency Mamu-B*08-restricted responses. There are several possible explanations for this unexpected result, as discussed above. Additionally, the presence of high-magnitude responses was not always sufficient to establish elite control in Mamu-B*08+ macaques infected with WT virus. Thus, our results do not support a model in which the simple presence of a few particular T-cell responses can always determine the outcome of an infection. Our study suggests several lines of future investigation that may help further illuminate the components of a successful immune response against immunodeficiency viruses.
Supplementary Material
Acknowledgments
We are grateful to the staff at the Wisconsin National Primate Research Center (WNPRC) for the expert care of animals used in this study. We thank the MHC Typing Core at the WNPRC for PCR-SSP typing of these animals. We are grateful to Jason Reed for the production of the Mamu-B*08 tetramers used in this study. Kim Weisgrau and Jessica Furlott provided assistance with immunology assays, while Lyle Wallace and Richard Rudersdorf constructed MHC-I cDNA libraries from select animals in this study. We thank the AIDS Research and Reference Reagent Program for the interleukin-2 used in our assays.
This work was funded by NIH grants R01 AI052056, R01 AI049120, R24 RR015371, and R24 RR016038 in addition to NIAID contract number HHSN 266200400088C to D.I.W. Funding was also provided by NIH grant R24RR21745-01A1 to D.H.O. In addition, this work was supported by NCRR grant P51 RR000167 to the WNPRC, University of Wisconsin—Madison. This research was conducted in part at a facility constructed with support from Research Facilities Improvement Program grants RR15459 and RR020141.
Footnotes
Published ahead of print on 2 September 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Allen, T. M., P. Jing, B. Calore, H. Horton, D. H. O'Connor, T. Hanke, M. Piekarczyk, R. Ruddersdorf, B. R. Mothe, C. Emerson, N. Wilson, J. D. Lifson, I. M. Belyakov, J. A. Berzofsky, C. Wang, D. B. Allison, D. C. Montefiori, R. C. Desrosiers, S. Wolinsky, K. J. Kunstman, J. D. Altman, A. Sette, A. J. McMichael, and D. I. Watkins. 2002. Effects of cytotoxic T lymphocytes (CTL) directed against a single simian immunodeficiency virus (SIV) Gag CTL epitope on the course of SIVmac239 infection. J. Virol. 76:10507-10511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alter, G., M. P. Martin, N. Teigen, W. H. Carr, T. J. Suscovich, A. Schneidewind, H. Streeck, M. Waring, A. Meier, C. Brander, J. D. Lifson, T. M. Allen, M. Carrington, and M. Altfeld. 2007. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J. Exp. Med. 204:3027-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altfeld, M., E. T. Kalife, Y. Qi, H. Streeck, M. Lichterfeld, M. N. Johnston, N. Burgett, M. E. Swartz, A. Yang, G. Alter, X. G. Yu, A. Meier, J. K. Rockstroh, T. M. Allen, H. Jessen, E. S. Rosenberg, M. Carrington, and B. D. Walker. 2006. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8(+) T cell response against HIV-1. PLoS Med. 3:e403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey, J. R., T. M. Williams, R. F. Siliciano, and J. N. Blankson. 2006. Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations. J. Exp. Med. 203:1357-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bimber, B. N., B. J. Burwitz, S. O'Connor, A. Detmer, E. Gostick, S. M. Lank, D. A. Price, A. Hughes, and D. O'Connor. 2009. Ultradeep pyrosequencing detects complex patterns of CD8+ T-lymphocyte escape in simian immunodeficiency virus-infected macaques. J. Virol. 83:8247-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blankson, J. N., J. R. Bailey, S. Thayil, H. C. Yang, K. Lassen, J. Lai, S. K. Gandhi, J. D. Siliciano, T. M. Williams, and R. F. Siliciano. 2007. Isolation and characterization of replication-competent human immunodeficiency virus type 1 from a subset of elite suppressors. J. Virol. 81:2508-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68:6103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bostik, P., J. Kobkitjaroen, W. Tang, F. Villinger, L. E. Pereira, D. M. Little, S. T. Stephenson, M. Bouzyk, and A. A. Ansari. 2009. Decreased NK cell frequency and function is associated with increased risk of KIR3DL allele polymorphism in simian immunodeficiency virus-infected rhesus macaques with high viral loads. J. Immunol. 182:3638-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyson, J. E., C. Shufflebotham, L. F. Cadavid, J. A. Urvater, L. A. Knapp, A. L. Hughes, and D. I. Watkins. 1996. The MHC class I genes of the rhesus monkey. Different evolutionary histories of MHC class I and II genes in primates. J. Immunol. 156:4656-4665. [PubMed] [Google Scholar]
- 10.Brockman, M. A., A. Schneidewind, M. Lahaie, A. Schmidt, T. Miura, I. Desouza, F. Ryvkin, C. A. Derdeyn, S. Allen, E. Hunter, J. Mulenga, P. A. Goepfert, B. D. Walker, and T. M. Allen. 2007. Escape and compensation from early HLA-B57-mediated cytotoxic T-lymphocyte pressure on human immunodeficiency virus type 1 Gag alter capsid interactions with cyclophilin A. J. Virol. 81:12608-12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrington, M., G. W. Nelson, M. P. Martin, T. Kissner, D. Vlahov, J. J. Goedert, R. Kaslow, S. Buchbinder, K. Hoots, and S. J. O'Brien. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283:1748-1752. [DOI] [PubMed] [Google Scholar]
- 12.Carrington, M., and S. J. O'Brien. 2003. The influence of HLA genotype on AIDS. Annu. Rev. Med. 54:535-551. [DOI] [PubMed] [Google Scholar]
- 13.Cline, A. N., J. W. Bess, M. Piatak, Jr., and J. D. Lifson. 2005. Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J. Med. Primatol. 34:303-312. [DOI] [PubMed] [Google Scholar]
- 14.Collins, J. A., M. G. Thompson, E. Paintsil, M. Ricketts, J. Gedzior, and L. Alexander. 2004. Competitive fitness of nevirapine-resistant human immunodeficiency virus type 1 mutants. J. Virol. 78:603-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crawford, H., W. Lumm, A. Leslie, M. Schaefer, D. Boeras, J. G. Prado, J. Tang, P. Farmer, T. Ndung'u, S. Lakhi, J. Gilmour, P. Goepfert, B. D. Walker, R. Kaslow, J. Mulenga, S. Allen, P. J. Goulder, and E. Hunter. 2009. Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. J. Exp. Med. 206:909-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans, D. T., D. H. O'Connor, P. Jing, J. L. Dzuris, J. Sidney, J. da Silva, T. M. Allen, H. Horton, J. E. Venham, R. A. Rudersdorf, T. Vogel, C. D. Pauza, R. E. Bontrop, R. DeMars, A. Sette, A. L. Hughes, and D. I. Watkins. 1999. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat. Med. 5:1270-1276. [DOI] [PubMed] [Google Scholar]
- 17.Feeney, M. E., Y. Tang, K. A. Roosevelt, A. J. Leslie, K. McIntosh, N. Karthas, B. D. Walker, and P. J. Goulder. 2004. Immune escape precedes breakthrough human immunodeficiency virus type 1 viremia and broadening of the cytotoxic T-lymphocyte response in an HLA-B27-positive long-term-nonprogressing child. J. Virol. 78:8927-8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fellay, J., K. V. Shianna, D. Ge, S. Colombo, B. Ledergerber, M. Weale, K. Zhang, C. Gumbs, A. Castagna, A. Cossarizza, A. Cozzi-Lepri, A. De Luca, P. Easterbrook, P. Francioli, S. Mallal, J. Martinez-Picado, J. M. Miro, N. Obel, J. P. Smith, J. Wyniger, P. Descombes, S. E. Antonarakis, N. L. Letvin, A. J. McMichael, B. F. Haynes, A. Telenti, and D. B. Goldstein. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317:944-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedrich, T. C., L. E. Valentine, L. J. Yant, E. G. Rakasz, S. M. Piaskowski, J. R. Furlott, K. L. Weisgrau, B. Burwitz, G. E. May, E. J. Leon, T. Soma, G. Napoe, S. V. Capuano III, N. A. Wilson, and D. I. Watkins. 2007. Subdominant CD8+ T-cell responses are involved in durable control of AIDS virus replication. J. Virol. 81:3465-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibbs, J. S., D. A. Regier, and R. C. Desrosiers. 1994. Construction and in vitro properties of SIVmac mutants with deletions in “nonessential” genes. AIDS Res. Hum. Retrovir. 10:607-616. [DOI] [PubMed] [Google Scholar]
- 21.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 22.Gray, C. M., M. Mlotshwa, C. Riou, T. Mathebula, D. de Assis Rosa, T. Mashishi, C. Seoighe, N. Ngandu, F. van Loggerenberg, L. Morris, K. Mlisana, C. Williamson, and S. A. Karim. 2009. Human immunodeficiency virus-specific gamma interferon enzyme-linked immunospot assay responses targeting specific regions of the proteome during primary subtype C infection are poor predictors of the course of viremia and set point. J. Virol. 83:470-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haeryfar, S. M., H. D. Hickman, K. R. Irvine, D. C. Tscharke, J. R. Bennink, and J. W. Yewdell. 2008. Terminal deoxynucleotidyl transferase establishes and broadens antiviral CD8+ T cell immunodominance hierarchies. J. Immunol. 181:649-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holtappels, R., C. O. Simon, M. W. Munks, D. Thomas, P. Deegen, B. Kuhnapfel, T. Daubner, S. F. Emde, J. Podlech, N. K. Grzimek, S. A. Oehrlein-Karpi, A. B. Hill, and M. J. Reddehase. 2008. Subdominant CD8 T-cell epitopes account for protection against cytomegalovirus independent of immunodomination. J. Virol. 82:5781-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenkins, M. R., R. Webby, P. C. Doherty, and S. J. Turner. 2006. Addition of a prominent epitope affects influenza A virus-specific CD8+ T cell immunodominance hierarchies when antigen is limiting. J. Immunol. 177:2917-2925. [DOI] [PubMed] [Google Scholar]
- 26.Kaizu, M., G. J. Borchardt, C. E. Glidden, D. L. Fisk, J. T. Loffredo, D. I. Watkins, and W. M. Rehrauer. 2007. Molecular typing of major histocompatibility complex class I alleles in the Indian rhesus macaque which restrict SIV CD8+ T cell epitopes. Immunogenetics 59:693-703. [DOI] [PubMed] [Google Scholar]
- 27.Kelleher, A. D., C. Long, E. C. Holmes, R. L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, C. Brander, G. Ogg, J. S. Sullivan, W. Dyer, I. Jones, A. J. McMichael, S. Rowland-Jones, and R. E. Phillips. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kotturi, M. F., I. Scott, T. Wolfe, B. Peters, J. Sidney, H. Cheroutre, M. G. von Herrath, M. J. Buchmeier, H. Grey, and A. Sette. 2008. Naive precursor frequencies and MHC binding rather than the degree of epitope diversity shape CD8+ T cell immunodominance. J. Immunol. 181:2124-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loffredo, J. T., A. T. Bean, D. R. Beal, E. J. León, G. E. May, S. M. Piaskowski, J. R. Furlott, J. Reed, S. K. Musani, E. G. Rakasz, T. C. Friedrich, N. A. Wilson, D. B. Allison, and D. I. Watkins. 2008. Patterns of CD8+ immunodominance may influence the ability of Mamu-B*08-positive macaques to naturally control simian immunodeficiency virus SIVmac239 replication. J. Virol. 82:1723-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loffredo, J. T., T. C. Friedrich, E. J. Leon, J. J. Stephany, D. S. Rodrigues, S. P. Spencer, A. T. Bean, D. R. Beal, B. J. Burwitz, R. A. Rudersdorf, L. T. Wallace, S. M. Piaskowski, G. E. May, J. Sidney, E. Gostick, N. A. Wilson, D. A. Price, E. G. Kallas, H. Piontkivska, A. L. Hughes, A. Sette, and D. I. Watkins. 2007. CD8 T cells from SIV elite controller macaques recognize Mamu-B*08-bound epitopes and select for widespread viral variation. PLoS ONE 2:e1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loffredo, J. T., J. Maxwell, Y. Qi, C. E. Glidden, G. J. Borchardt, T. Soma, A. T. Bean, D. R. Beal, N. A. Wilson, W. M. Rehrauer, J. D. Lifson, M. Carrington, and D. I. Watkins. 2007. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J. Virol. 81:8827-8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loffredo, J. T., J. Sidney, A. T. Bean, D. R. Beal, W. Bardet, A. Wahl, O. E. Hawkins, S. Piaskowski, N. A. Wilson, W. H. Hildebrand, D. I. Watkins, and A. Sette. 2009. Two MHC class I molecules associated with elite control of immunodeficiency virus replication, Mamu-B*08 and HLA-B*2705, bind peptides with sequence similarity. J. Immunol. 182:7763-7775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manuel, E. R., W. A. Charini, P. Sen, F. W. Peyerl, M. J. Kuroda, J. E. Schmitz, P. Autissier, D. A. Sheeter, B. E. Torbett, and N. L. Letvin. 2006. Contribution of T-cell receptor repertoire breadth to the dominance of epitope-specific CD8+ T-lymphocyte responses. J. Virol. 80:12032-12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manuel, E. R., W. W. Yeh, M. S. Seaman, K. Furr, M. A. Lifton, S. L. Hulot, P. Autissier, and N. L. Letvin. 2009. Dominant CD8+ T-lymphocyte responses suppress expansion of vaccine-elicited subdominant T lymphocytes in rhesus monkeys challenged with pathogenic simian-human immunodeficiency virus. J. Virol. 83:10028-10035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin, M. P., X. Gao, J. H. Lee, G. W. Nelson, R. Detels, J. J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, J. Trowsdale, M. Wilson, S. J. O'Brien, and M. Carrington. 2002. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 31:429-434. [DOI] [PubMed] [Google Scholar]
- 37.Martin, M. P., Y. Qi, X. Gao, E. Yamada, J. N. Martin, F. Pereyra, S. Colombo, E. E. Brown, W. L. Shupert, J. Phair, J. J. Goedert, S. Buchbinder, G. D. Kirk, A. Telenti, M. Connors, S. J. O'Brien, B. D. Walker, P. Parham, S. G. Deeks, D. W. McVicar, and M. Carrington. 2007. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 39:733-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez-Picado, J., J. G. Prado, E. E. Fry, K. Pfafferott, A. Leslie, S. Chetty, C. Thobakgale, I. Honeyborne, H. Crawford, P. Matthews, T. Pillay, C. Rousseau, J. I. Mullins, C. Brander, B. D. Walker, D. I. Stuart, P. Kiepiela, and P. Goulder. 2006. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J. Virol. 80:3617-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Migueles, S. A., A. C. Laborico, H. Imamichi, W. L. Shupert, C. Royce, M. McLaughlin, L. Ehler, J. Metcalf, S. Liu, C. W. Hallahan, and M. Connors. 2003. The differential ability of HLA B*5701+ long-term nonprogressors and progressors to restrict human immunodeficiency virus replication is not caused by loss of recognition of autologous viral gag sequences. J. Virol. 77:6889-6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Migueles, S. A., C. M. Osborne, C. Royce, A. A. Compton, R. P. Joshi, K. A. Weeks, J. E. Rood, A. M. Berkley, J. B. Sacha, N. A. Cogliano-Shutta, M. Lloyd, G. Roby, R. Kwan, M. McLaughlin, S. Stallings, C. Rehm, M. A. O'Shea, J. Mican, B. Z. Packard, A. Komoriya, S. Palmer, A. P. Wiegand, F. Maldarelli, J. M. Coffin, J. W. Mellors, C. W. Hallahan, D. A. Follman, and M. Connors. 2008. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity 29:1009-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Migueles, S. A., M. S. Sabbaghian, W. L. Shupert, M. P. Bettinotti, F. M. Marincola, L. Martino, C. W. Hallahan, S. M. Selig, D. Schwartz, J. Sullivan, and M. Connors. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA 97:2709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miura, T., M. A. Brockman, C. J. Brumme, Z. L. Brumme, J. M. Carlson, F. Pereyra, A. Trocha, M. M. Addo, B. L. Block, A. C. Rothchild, B. M. Baker, T. Flynn, A. Schneidewind, B. Li, Y. E. Wang, D. Heckerman, T. M. Allen, and B. D. Walker. 2008. Genetic characterization of human immunodeficiency virus type 1 in elite controllers: lack of gross genetic defects or common amino acid changes. J. Virol. 82:8422-8430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miura, T., M. A. Brockman, A. Schneidewind, M. Lobritz, F. Pereyra, A. Rathod, B. L. Block, Z. L. Brumme, C. J. Brumme, B. Baker, A. C. Rothchild, B. Li, A. Trocha, E. Cutrell, N. Frahm, C. Brander, I. Toth, E. J. Arts, T. M. Allen, and B. D. Walker. 2009. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare Gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphocyte recognition. J. Virol. 83:2743-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43a.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC.
- 44.Obar, J. J., K. M. Khanna, and L. Lefrancois. 2008. Endogenous naive CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity 28:859-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Connor, D. H., A. B. McDermott, K. C. Krebs, E. J. Dodds, J. E. Miller, E. J. Gonzalez, T. J. Jacoby, L. Yant, H. Piontkivska, R. Pantophlet, D. R. Burton, W. M. Rehrauer, N. Wilson, A. L. Hughes, and D. I. Watkins. 2004. A dominant role for CD8+-T-lymphocyte selection in simian immunodeficiency virus sequence variation. J. Virol. 78:14012-14022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Doherty, U., W. J. Swiggard, and M. H. Malim. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otting, N., C. M. Heijmans, R. C. Noort, N. G. de Groot, G. G. Doxiadis, J. J. van Rood, D. I. Watkins, and R. E. Bontrop. 2005. Unparalleled complexity of the MHC class I region in rhesus macaques. Proc. Natl. Acad. Sci. USA 102:1626-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Price, D. A., T. E. Asher, N. A. Wilson, M. C. Nason, J. M. Brenchley, I. S. Metzler, V. Venturi, E. Gostick, P. K. Chattopadhyay, M. Roederer, M. P. Davenport, D. I. Watkins, and D. C. Douek. 2009. Public clonotype usage identifies protective Gag-specific CD8+ T cell responses in SIV infection. J. Exp. Med. 206:923-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sacha, J. B., C. Chung, E. G. Rakasz, S. P. Spencer, A. K. Jonas, A. T. Bean, W. Lee, B. J. Burwitz, J. J. Stephany, J. T. Loffredo, D. B. Allison, S. Adnan, A. Hoji, N. A. Wilson, T. C. Friedrich, J. D. Lifson, O. O. Yang, and D. I. Watkins. 2007. Gag-specific CD8+ T lymphocytes recognize infected cells before AIDS-virus integration and viral protein expression. J. Immunol. 178:2746-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schatz, M. M., B. Peters, N. Akkad, N. Ullrich, A. N. Martinez, O. Carroll, S. Bulik, H. G. Rammensee, P. van Endert, H. G. Holzhutter, S. Tenzer, and H. Schild. 2008. Characterizing the N-terminal processing motif of MHC class I ligands. J. Immunol. 180:3210-3217. [DOI] [PubMed] [Google Scholar]
- 51.Schneidewind, A., M. A. Brockman, R. Yang, R. I. Adam, B. Li, S. Le Gall, C. R. Rinaldo, S. L. Craggs, R. L. Allgaier, K. A. Power, T. Kuntzen, C. S. Tung, M. X. LaBute, S. M. Mueller, T. Harrer, A. J. McMichael, P. J. Goulder, C. Aiken, C. Brander, A. D. Kelleher, and T. M. Allen. 2007. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J. Virol. 81:12382-12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simen, B. B., J. F. Simons, K. H. Hullsiek, R. M. Novak, R. D. Macarthur, J. D. Baxter, C. Huang, C. Lubeski, G. S. Turenchalk, M. S. Braverman, B. Desany, J. M. Rothberg, M. Egholm, and M. J. Kozal. 2009. Low-abundance drug-resistant viral variants in chronically HIV-infected, antiretroviral treatment-naive patients significantly impact treatment outcomes. J. Infect. Dis. 199:693-701. [DOI] [PubMed] [Google Scholar]
- 53.Streeck, H., B. Li, A. F. Poon, A. Schneidewind, A. D. Gladden, K. A. Power, D. Daskalakis, S. Bazner, R. Zuniga, C. Brander, E. S. Rosenberg, S. D. Frost, M. Altfeld, and T. M. Allen. 2008. Immune-driven recombination and loss of control after HIV superinfection. J. Exp. Med. 205:1789-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiseman, R. W., J. A. Karl, B. N. Bimber, C. E. O'Leary, S. M. Lank, J. J. Tuscher, A. M. Detmer, P. Pouffard, N. Levenkova, C. L. Turcotte, E. Szekeres, Jr., C. Wright, T. Harkins, and D. H. O'Connor. MHC genotyping with massively parallel pyrosequencing. Nat. Med., in press. [DOI] [PMC free article] [PubMed]
- 55.Yant, L. J., T. C. Friedrich, R. C. Johnson, G. E. May, N. J. Maness, A. M. Enz, J. D. Lifson, D. H. O'Connor, M. Carrington, and D. I. Watkins. 2006. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 80:5074-5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.