Abstract
Prion diseases are neurodegenerative diseases associated with the accumulation of a pathogenic isoform of the host-encoded prion protein. The cellular responses to prion infection are not well defined. By performing microarray analysis on cultured neuronal cells infected with prion strain 22L, in the group of up-regulated genes we observed predominantly genes of the cholesterol pathway. Increased transcript levels of at least nine enzymes involved in cholesterol synthesis, including the gene for the rate-limiting hydroxymethylglutaryl-CoA reductase, were detected. Up-regulation of cholesterogenic genes was attributable to a prion-dependent increase in the amount and activity of the sterol regulatory element-binding protein Srebp2, resulting in elevated levels of total and free cellular cholesterol. The up-regulation of cholesterol biosynthesis appeared to be a characteristic response of neurons to prion challenge, as cholesterogenic transcripts were also elevated in persistently infected GT-1 cells and prion-exposed primary hippocampal neurons but not in microglial cells and primary astrocytes. These results convincingly demonstrate that prion propagation not only depends on the availability of cholesterol but that neuronal cells themselves respond to prions with specific up-regulation of cholesterol biosynthesis.
Introduction
The pathogenesis of prion diseases is typically associated with an abnormal accumulation of a misfolded protein (PrPSc),6 derived from the host-encoded prion protein (PrPC), in the nervous system of affected individuals (1–4). Although the pathology of the infected cells in the central nervous system is well documented (5), the host response to infection with prions and the changes within the cell caused by prions remain obscure. Transcriptome analysis of prion-infected animals and cell cultures suggested alterations in several cellular pathways (6–15), except for one study (16). However, no conclusive interpretation of the role of specific pathways altered by prion infection could be drawn. This might at least in part be due to the following: (a) different microarray platforms; (b) different prion strains; (c) different sampling time points during the course of infection; or (d) mixed cell populations. To better understand the neuronal response to prion infection, we performed a highly controlled genome-wide microarray analysis of paired populations of infected and mock-infected N2a cells, using the mouse-adapted scrapie strain 22L. Our statistical transcriptome analysis identified over 100 significantly and differentially expressed genes with putative function. Within the group of up-regulated genes, genes involved in cholesterol biosynthesis and uptake predominated in prion-infected cells. Transcription of genes associated with cholesterol biosynthesis and cholesterol uptake is tightly controlled by transcription factor Srebp2 that induces expression upon binding to the sterol regulatory element (SRE) in the promoter region of relevant genes (17, 18). Upon cholesterol depletion, Srebp2 is proteolytically cleaved to its active form and translocates to the nucleus where it binds to SRE in promoters of target genes participating in cholesterol metabolism. This way, Srebp2 activates cholesterol biosynthesis by increasing gene expression of individual target genes at every step of the pathway (17–19). Srebp2 is able to control its own synthesis by a positive feedback mechanism (20) and also regulates expression of the low density lipoprotein receptor (Ldlr), which is the main receptor for extracellular cholesterol acquisition in mammalian cells (17). Real time PCR and reporter gene assays confirmed up-regulation of cholesterogenic genes in prion-infected neuronal cells, including the gene for Srebp2, which was also reflected by increased cholesterol levels. Importantly, primary hippocampal neurons also exhibited elevated cholesterol biosynthesis transcripts upon exposure to prions. Up-regulation of cholesterol biosynthesis appeared to be a characteristic neuronal response, as a microglial cell line and primary astrocytes did not show increased levels of cholesterogenic transcripts in response to prion exposure. Thus, our results suggest that prions differentially alter cholesterol homeostasis in a cell type-specific manner.
EXPERIMENTAL PROCEDURES
Antibodies
Mouse antibody 4H11 directed against murine PrP has been described previously (21, 22). Rabbit polyclonal antibodies anti-Srebp2 (ab28482) and anti-Ldlr (ab30532) were purchased from Abcam plc (Cambridge, UK). Mouse monoclonal anti-GAPDH (MAB374) was from Millipore/Chemicon (Billerica, MA).
De Novo Infection of N2a Cells with Prion Strain 22L
Mouse neuroblastoma cell line N2a (ATCC, CCL-131) was maintained in Opti-MEM supplemented with 10% fetal bovine serum and penicillin/streptomycin (Invitrogen). The medium was exchanged every 48 h. Infection of cells was performed as described previously with small modifications (23). For exposure of N2a cell clones to prions, brain homogenates from 22L-infected terminally sick C57BL/6 mice or from uninfected mice were used. Brains were homogenized in phosphate-buffered saline using 0.9-mm gauge needles (Roth, Karlsruhe, Germany) and diluted to a final concentration of 10%. Cells were plated in 24-well plates at a density of 5 × 104 cells per well in 1 ml of medium. The following day the medium was replaced by 900 μl of fresh medium and 100 μl of brain homogenate (10%). 24 h post-infection, the brain homogenate was discarded, and cells were subsequently cultured. For Western blot analysis and RNA isolation, cells were always harvested when confluent.
Exposure of Microglia Cell Line BV-2 to Prions
Murine microglial cell line BV-2 was challenged with RML or 22L prions as described previously (24). Briefly, brain homogenates from terminally sick CD-1 mice (RML-infected) or C57BL/6 mice (22L infected) or normal mice were diluted to 1% in RPMI 1640 medium, 7.5% FCS (ultra low endotoxin), 50 μm mercaptoethanol, and penicillin/streptomycin. Cells were exposed to uninfected brain homogenate, 22L- or RML-infected brain homogenate for 24 h, and RNA was extracted.
Preparation of Primary Neurons and Astrocytes and Exposure to Prions
Primary neurons from hippocampi and astrocytes of C57B/L6 embryonic mice (gestation day 16) were prepared as described elsewhere (25) with some modifications. Briefly, hippocampi were excised, incubated with trypsin, and rinsed in Hanks' balanced salt solution (Invitrogen) complemented with 10% fetal calf serum. Single cell suspensions were obtained by mechanical dissociation. Approximately 1 × 106 viable cells were plated in poly-l-lysine-coated 6 wells in neurobasal medium supplemented with B-27 and antibiotics (Invitrogen). 10 μm of the antimitotics uridine and fluorodeoxyuridine (Sigma) were added to prevent astrocyte proliferation (26). Weekly, one-third of the medium was replaced with fresh medium. For isolation of primary astrocytes, meninges were removed, and hemispheres were rinsed with Hanks' balanced salt solution and dissected mechanically. Cells were grown in Dulbecco's modified Eagle's medium with penicillin/streptomycin and 10% FCS and rinsed thoroughly every other day with phosphate-buffered saline for 1 week. Established neuronal cultures showed typical neuronal growth and expressed neuron-specific marker protein neurofilaments (data not shown). Astrocyte cultures showed contact inhibition upon confluence and abundantly expressed glial fibrillary acidic protein (data not shown). Primary neurons were exposed to 0.1% brain homogenates (final concentration; mock and 22L) 1 day post-plating and astrocytes shortly before reaching confluence. The next day the medium of the neuronal populations was sterile-filtered to remove homogenate debris. Astrocytes were rinsed with phosphate-buffered saline 1 day after infection, and the medium was changed every 3 days. Cells were cultured for up to 3–4 weeks.
Western Blot Analysis
Cells were assayed for PrPSc content as described previously (22). All cell lysates were measured for protein content with the Bradford assay (Pierce). For normalization purposes, blots were stripped with stripping buffer (0.1 m glycine, 0.1% SDS, pH 2.2) and re-probed with mouse monoclonal antibody directed against GAPDH (MAB374; Millipore/Chemicon). For detection of Srebp2, cells were lysed (40 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 0.3% Nonidet P-40, 1 mm dithiothreitol) on ice for 10 min. Lysates were cleared of cell debris (20,000 × g, 1 min at room temperature). Samples were run on 7.5% gels, and Srebp2 or Ldlr and GAPDH were detected on the same Western blot. After densitometric analysis of the band intensities, Srebp2 and Ldlr levels were normalized to GAPDH levels on the same Western blot.
DNA Microarray (Chip) Design
cDNA microarrays were produced at the Helmholtz Zentrum München (German Research Center for Environment and Health) and contain the fully sequenced 20,000 cDNA mouse array-TAG library (Lion Bioscience, Heidelberg, Germany) and several hundred additional clones for genes not included in the commercial clone set. A full description of the probes on the microarrays has been submitted to the GEO data base (GPL3697). The RNA isolation, reverse transcription, fluorescent labeling, chip hybridization, and data analysis were done as described recently (27). The expression data have been submitted to the GEO data base (GSE7119).
Candidate genes were classified based on the relationship of their gene products to one or more functions. Classification was performed using the definitions in the LION and ENSEMBL data bases. BiblioSphere PathwayEdition (Genomatix, Munich, Germany) was used to calculate complete networks and rank pathway interactions by z scoring on the basis of Genomatix Knowledge Base Ontology. The over-representation of genes is calculated as a z score based on the expected number of genes within the filter category (Gene Ontology). Biological filtering was used to restrict the network to genes that are assigned to the biological function of the respective genes.
Quantitative Reverse Transcription-PCR
RNA samples were extracted with phenol/chloroform or the RNeasy mini kit (Qiagen, Hilden, Germany) and treated with RNase-free DNase I. RNA was reverse-transcribed using the SuperScript II reverse transcription kit (Invitrogen) and random hexamers. Quantitative real time PCR experiments for each gene were performed in triplicate using the LightCycler FastStart DNA Master SYBR Green I kit (Roche Applied Science). The 2−ΔΔCT method (28, 29) was used to calculate the relative expression levels. Each result was normalized to RNA Polymerase II (RPII). As the minimum detectable change, we used a 2-fold expression difference (30) and considered only expression differences equal to or higher than 2-fold as significant.
Luciferase Assay
Cells were plated at a density of 1.3 × 105 cells per 12 wells and co-transfected with the luciferase plasmid psynSRE-Luc harboring the sterol-responsive element (31) (1.6 μg) and transfection control pGL2 Renilla plasmid (160 ng; Promega) using FuGENE 6 (Roche Applied Science) transfection reagent. The dual luciferase reporter assay (E1910, Promega) was used to measure the bioluminescence 24 h post-transfection, and cell lysates were analyzed for firefly and Renilla luciferase activity. Relative bioluminescence generated by firefly luciferase was normalized to the bioluminescence generated by the Renilla luciferase.
Cholesterol Analysis
Cell pellets were resuspended in 3 m aqueous solution of guanidine hydrochloride to inactivate prion infectivity. Water was added; the samples were sonicated for 30 s, and the protein content was determined (32). After subsequent addition of 4 ml of methanol and 3 ml of chloroform/methanol/water (10:5:1, v/v), lipids were extracted for 24 h at 37 °C. The liquid phase was separated by filtration. The solvent was evaporated in a stream of nitrogen, and the residues were desalted by reversed phase chromatography on LiChroprep RP18 (Merck) columns (33). Lipids were applied to high performance thin layer chromatography silica gel 60 plates (Merck), which were pre-washed twice with chloroform/methanol (1:1, v/v) and air-dried for 30 min. Each lane of the TLC plate was loaded with the equivalent of 600 μg of protein. For quantification of free cholesterol, the TLC solvent system used was chloroform/methanol/glacial acetic acid (190:9:1, v/v). For the quantification of the total cholesterol content, cholesteryl esters were cleaved by alkaline hydrolysis with 2.5 ml of a 100 mm solution of sodium hydroxide in methanol for 2 h at 37 °C. After neutralization with acetic acid, the mixtures were desalted again on RP18 columns. Lipids were separated into acidic and neutral fractions by anion exchange chromatography on DEAE-cellulose columns (GE Healthcare) (34) with some modifications. The lipid mixture was solved in 1 ml of chloroform/methanol/water (3:7:1, v/v) and applied to the columns. Neutral lipids were eluted with 7 ml of the same solvent, and then acidic lipids were eluted with 8 ml of chloroform, methanol, 0.8 m ammonium acetate in water (3:7:1, v/v). Neutral lipids were applied to pre-washed high performance thin layer chromatography plates, loading each lane with the equivalent of 400 μg of protein. The TLC plates were developed in chloroform/methanol/glacial acetic acid (190:9:1, v/v). For quantitative analytical TLC determination, increasing amounts of standard cholesterol (Sigma) were applied to the TLC plates in addition to the lipid samples. After development, plates were dried under reduced pressure. For detection of lipid bands, the TLC plates were sprayed with a phosphoric acid/copper sulfate reagent (15.6 g of CuSO4(H2O)5 and 9.4 ml of H3PO4 (85%, w/v) in 100 ml of water) and charred at 180 °C for 10 min (35). Lipid bands were then quantified using a photo-densitometer (Shimadzu, Duisburg, Germany) at a wavelength of 595 nm.
Transient Knockdown of Srebp2 and Ldlr Expression by siRNA
Cells were transfected with siRNAs directed against the murine Srebp2 (SI01433131 and SI01433138, Qiagen, Hilden, Germany) or Ldlr (Sc35803, Santa Cruz Biotechnology, Santa Cruz, CA) or nonsilencing siRNA (AllStars Negative Control siRNA, 1027281, Qiagen, Hilden, Germany) and Lipofectamine 2000 (Invitrogen). Cells were subsequently cultured in serum-free Opti-MEM medium supplemented with antibiotics and 0.2 μm mevalonate. In additional experiments, cells were transfected with siRNA and subsequently cultured in the presence of 10% FCS or BSA (5% w/v) and cholesterol (0.1% v/v). BSA (A-4503, fraction V) and cholesterol (C3045, stock solution 35 mg/ml in absolute ethanol) were obtained from Sigma. After 48 h, cells were detached and split into two samples. Cells were lysed in two different buffers for detection of PrPSc, PrPC, Ldlr, and GAPDH (10 mm Tris-HCl, pH 7.5, 100 mm NaCl, 10 mm EDTA, 0.5% Triton X-100, 0.5% deoxycholate) or Srebp2 (40 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 0.3% Nonidet P-40, 1 mm dithiothreitol) and GAPDH.
Photodensitometric Analysis and Statistics
Photodensitometric analysis of signals on Western blots was performed using the ImageQuant TL software (GE Healthcare). Quantitative data are shown as mean ± S.D. For evaluation of statistical significance of the data, we used the paired t test. p values less than 0.05 were considered as significant.
RESULTS
Microarray Analysis Reveals Modulation of Cholesterol Biosynthesis upon 22L Prion Infection
To characterize the host cell response to 22L prion infection, N2a cells were cloned by limiting dilution, and the resultant cell clones were subsequently exposed to 1% 22L brain homogenate (supplemental Fig. 1A). Clone 5, which displayed a high PrPSc signal upon exposure to prions, was chosen for microarray experiments (supplemental Fig. 1B). Subclones of clone 5 were analyzed by Western blot for proteinase K-resistant PrP, and the infection rate was estimated to be ∼70% (supplemental Fig. 1C). The differential gene expression pattern of 22L prion-infected N2a cells (ScN2a) was compared with that of mock-infected cells using a genome-wide cDNA microarray. Mock-infected and 22L prion-infected cells were passaged 18 times under identical culture conditions to exclude any influences of residual brain homogenate or passage history on gene expression. A competitive hybridization was performed between RNA isolated from infected cells and RNA from mock-infected cells, including color flip experiments. In total, eight microarrays were hybridized. Statistical analysis revealed that in total 203 genes were significantly differentially regulated with a d(i) >4, a score assigned to each gene on the basis of changes in gene expression levels relative to the standard deviation. Reproducible differential expression was found for all microarray experiments. The number of false positives per 100 permutations was estimated as a false discovery rate of 0% for each data set. For 37 genes, increased expression levels were identified, although 166 genes were down-regulated. Differentially transcribed genes were classified according to the biological function of their gene products based on definitions in the LION and ENSEMBL data bases. Functional information was limited or absent for some of the candidate genes. For 27 up- and 94 down-regulated genes, a putative function is known or has been suggested (supplemental Table 1).
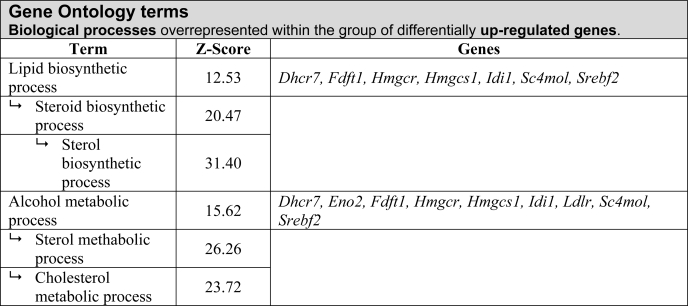
BiblioSphere software (Genomatix, Munich, Germany) was used to analyze and filter for Gene Ontology categories to integrate differentially regulated genes into pathways. BiblioSphere generated a list of biological process terms ranked by their probability of over- or under-representation in a specific gene list. z scores indicate how the results deviate from the distribution mean. Thus, it ranks the probability that the genes in this category are over- or under-represented in the gene list. The supplemental Table 2 lists biological processes over-represented within the group of down-regulated genes. Within the group of up-regulated genes, the lipid biosynthetic process was highly over-represented (Table 1), and genes of this GO term mainly belonged to the cholesterol biosynthetic pathway. Notably, transcripts for the sterol regulatory element-binding protein 2 (Srebp2) (Srebf2, 3.25) were increased. Furthermore, Ldlr transcripts were increased almost 4-fold in prion-infected N2a cells.
TABLE 1.
Classification of differentially expressed genes by biological processes
Microarray data was filtered for biological processes using the Gene Ontology filter structure of the BiblioSphere Pathway Edition. Gene Ontology filters consist of a hierarchy of terms and the corresponding annotations for the BiblioSphere analysis. Based on the number of observed and expected annotations for each term, a statistical analysis was performed. Annotation over- or underepresentation within the dataset of genes is indicated by the z score.
Prion Infection Induces Up-regulation of Srebp2-dependent Transcription in Neuronal Cell Lines
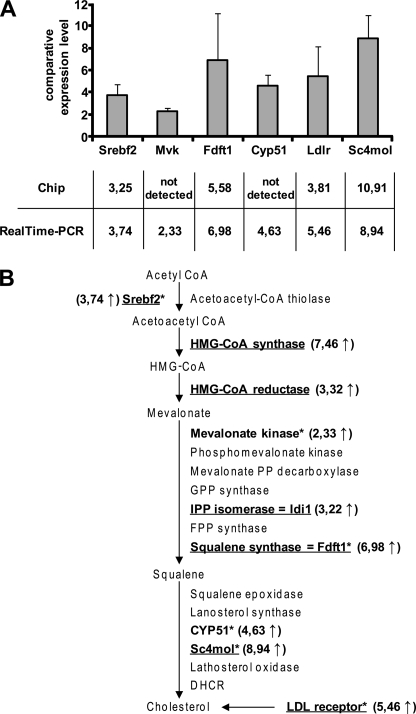
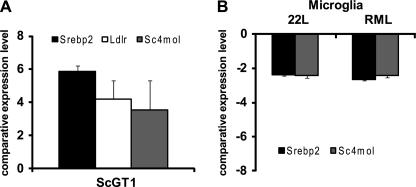
Microarray analysis is expected to underestimate the changes in expression of genes relative to real time PCR. For independent validation of microarray results, real time PCR analysis of cholesterogenic genes was performed. Srebp2 (gene Srebf2), as well as the genes for farnesyl-diphosphate farnesyltransferase 1 (Fdft1), C4-sterol methyl oxidase (Sc4mol), and Ldlr, was chosen based on substantial changes in expression levels as detected by microarray analysis. To elucidate if expression of further cholesterogenic genes was altered upon prion infection, relative expression levels of candidate genes mevalonate kinase (Mvk) and cytochrome p450 family 51 (Cyp51) were evaluated. Overall, the real time PCR results were highly consistent with the array results, demonstrating an up-regulation of all studied genes (Fig. 1A). In conclusion, we identified cholesterol biosynthesis as a main pathway up-regulated upon 22L prion infection of N2a cells (Fig. 1B). To study if prion infection leads to elevated expression of cholesterogenic genes in another neuronal cell line, the relative expression levels of Srebf2, Sc4mol, and Ldlr in RML prion strain-infected or uninfected GT-1 cells (12) were compared by real time PCR. Western blot analysis of RML-infected GT-1 cells prior to RNA isolation demonstrated that cells accumulated substantial amounts of PrPSc (data not shown). Transcript levels of all three genes were found increased in infected GT-1 cells compared with uninfected cells (Fig. 2A). Thus, these results strongly suggest that up-regulation of cholesterol metabolism-related genes is a general neuronal response to prion infection that might be independent of the prion strain.
FIGURE 1.
22L prion infection of N2a cells induces increased transcription of genes regulated by Srebp2. Differential gene expression of genes involved in cholesterol synthesis is shown. A, up-regulation of Srebf2 (sterol regulatory element-binding factor 2), Mvk (mevalonate kinase), Fdft1 (farnesyl-diphosphate farnesyltransferase 1), Cyp51 (cytochrome P450, family 51), Ldlr (low density lipoprotein receptor), and Sc4mol (C4-sterol methyl oxidase) was determined by real time PCR in the 22L-infected clone 5 (passage 18) compared with mock-infected clone 5 (passage 18). All results were normalized to the RPII (RNA polymerase 2) expression levels. For triplicate experiments, the standard deviation is shown for each gene. The y axis denotes the comparative gene expression levels. B, overview of important genes in the cholesterol pathway activated by Srbp2 (17). Genes in boldface were found up-regulated in prion-infected N2a cells compared with mock-infected N2a cells either in the chip experiment (underlined) or as detected by real time PCR (marked with asterisks). Srebp2 is the limiting transcription factor of cholesterol synthesis. HMG-CoA, hydroxymethylglutaryl-CoA; FPP, farnesyl diphosphate; IPP, isopentenyl diphosphate; GPP, geranyl diphosphate; DHCR, 7-dehydrocholesterol reductase.
FIGURE 2.
Up-regulation of cholesterol biosynthesis transcripts in neuronal cell lines. A, quantitative Srebf2, Ldlr, and Sc4mol expression levels in RML-infected and un-infected GT1 cells determined by real time PCR, normalized to the housekeeping gene RPII. B, BV-2 cells were exposed to 22L and RML prions (24). One day post-exposure, total RNA was extracted from cells, and relative mRNA concentrations were determined by real time PCR as above. Bars represent mean values ± S.D. (n = 3).
Microarray analysis of brains from prion-infected mice revealed down-regulation of cholesterogenic genes, in conflict with our data on neuronal cells (36). However, in the same study, a strong inflammatory reaction was evident, suggesting that the observed changes in gene expression reflect a profound microglial activation that could obscure potential transcriptional changes in neurons. Interestingly, microglial cells do not appear to become infected with prions in vivo (37). To test if prion exposure could impact cholesterol biosynthesis in microglial cells, relative transcript levels of Srebf2 and Sc4mol in murine BV-2 cells that had been exposed to 22L or RML prion strains or normal brain homogenate for 1 day (24) were analyzed by real time PCR (Fig. 2B). Immunofluorescence as well as Western blot analysis of BV-2 cells exposed to RML and 22L have previously shown that cells stained positive for PrPSc 1 day post-exposure (24). Surprisingly, both cholesterogenic genes were significantly down-regulated in microglial cell line BV-2 exposed to prions, suggesting that prions modulate cholesterol biosynthesis dependent on the cell type.
Increased Srebp2 Activity and Cholesterol Levels in Prion-infected N2a Cells
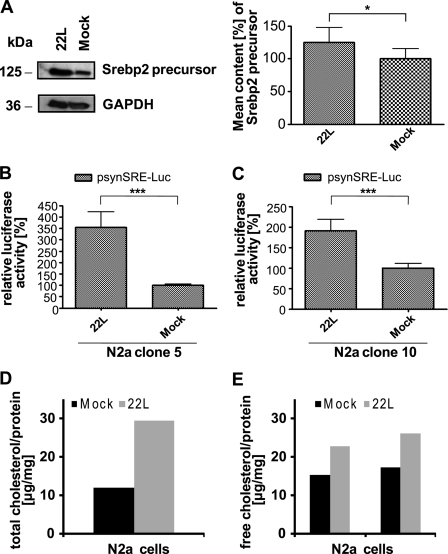
Srebp2 is synthesized as an inactive full-length protein that becomes inserted into the endoplasmic reticulum membrane. To study if increased Srebf2 transcript levels resulted in elevated Srebp2 protein levels, Srebp2 protein in whole lysates of 22L-infected and mock-infected N2a cells was detected (Fig. 3A). The amount of full-length Srebp2 was significantly increased in prion-infected cells compared with mock-infected N2a cells (p < 0.05). Upon cholesterol depletion, full-length Srebp2 is cleaved and translocates to the nucleus, where it binds to target genes harboring sterol regulatory elements. Activation (and thus cleavage) of Srebp2 can best be studied by reporter gene assays. To determine the Srebp2 transcriptional activity in prion-infected and uninfected cells, we used a plasmid vector coding for luciferase under the control of a synthetic promoter harboring two SRE motifs (31). Prion-infected and mock-infected N2a cells were co-transfected with psynSRE-firefly luciferase plasmid (reporter gene) and the pGL2-Renilla luciferase plasmid as a transfection efficiency control. A significantly higher luciferase activity (p ≤ 0.0005) was observed measuring bioluminescence in infected cells compared with mock-infected cells (Fig. 3B). When the luciferase reporter gene assay was performed with another N2a cell clone (clone 10), significantly increased luciferase activity (p ≤ 0.0005) was again detected in 22L prion-infected cells (Fig. 3C), arguing that clonal artifacts did not account for the observed differences in cholesterol metabolism between prion-infected and uninfected cells. High performance thin layer chromatography further revealed that both total cholesterol (including free and esterified cholesterol) and free cholesterol levels were elevated in prion-infected cells (Fig. 3, D and E). Thus, prion infection of neuronal cells induces up-regulation of cholesterol biosynthesis and leads to elevated total and free cholesterol levels.
FIGURE 3.
Increased Srebp2 transcriptional activity and cholesterol levels in prion-infected cells. A, Western blot analysis of full-length Srebp2 in cell lysates using rabbit polyclonal antibody ab28482. For photodensitometric analysis of the data, for each group (22L or mock) n = 6 samples were analyzed and normalized to the GAPDH signals detected on the same Western blot. Relative Srebp2 protein levels in mock-infected cells were set to 100%. p values were determined using a paired t test, and significant differences are indicated by asterisks. B and C, activity of Srebp2 in two independent sibling N2a clones (clones 5 and 10) was determined by a luciferase reporter gene assay, using the psynSRE-firefly luciferase construct and the pGL3-Renilla luciferase plasmid as a transfection control. The activity of Srebp2 was assessed by measuring the bioluminescence generated by the firefly luciferase normalized to the bioluminescence of Renilla luciferase in the same samples (n = 8). Relative firefly luciferase activities in the prion- and mock-infected cells clone 5 (B) and clone 10 (C). The activity in the mock-infected cells was set to 100%. p values were determined by the paired t test (*, p < 0.05; ***, p ≤ 0.0005). D and E, lipids from prion-infected and uninfected N2a cells were extracted and applied to high performance thin layer chromatography silica gel 60 plates. Free cholesterol and total cholesterol levels were determined as described under “Experimental Procedures.” For quantitative analytical TLC determination, lipid samples were compared with cholesterol standards. The amounts of total and free cholesterol per mg of protein are given. For free cholesterol, results of two independent samples are shown. Deviations were in the range of ± 1.7 μg/mg of protein.
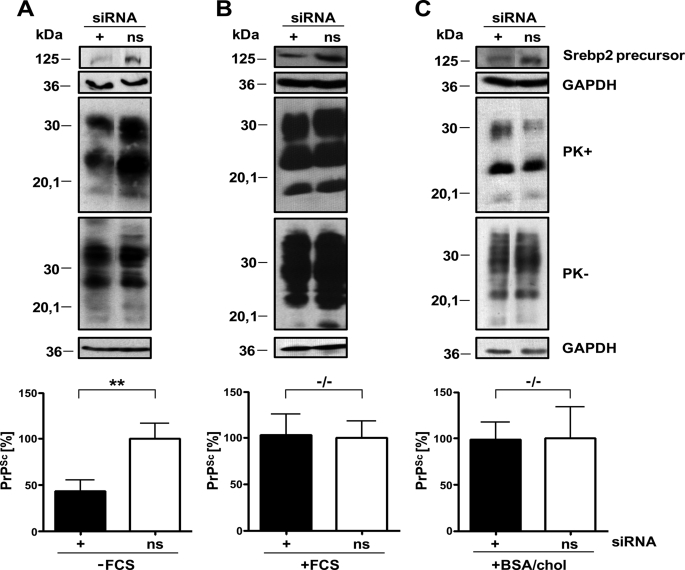
Transient Down-regulation of Srebp2 by siRNA Negatively Influences PrPSc Formation in N2a Cells in the Absence of Exogenous Cholesterol Sources
To determine whether reducing cholesterol biosynthesis by a transient down-regulation of Srebp2 has an effect on PrPSc accumulation, mock-infected and 22L prion-infected N2a cells (clone 5) were transfected either with siRNA targeting Srebf2 transcripts or with nonsilencing siRNA. Cells were kept in serum-free medium supplemented with 0.2 μm mevalonate for the course of the experiment. Cells transfected with siRNA targeting Srebf2 mRNA grew slightly slower than nonsilencing siRNA-transfected cells, but PrPC levels on cell membranes appeared unaltered as assessed by flow cytometry analysis (data not shown). Samples were adjusted to comparable protein contents and analyzed for total PrP and PrPSc (Fig. 4A). Interestingly, silencing of Srebf2 did not lead to a decrease in total PrP content but led to reduced levels of PrPSc compared with PrPSc levels in cells treated with nonsilencing siRNA. Similar results were obtained when cells were transfected with an alternative siRNA targeting Srebf2 transcripts (supplemental Fig. 2). We next tested if down-regulation of cholesterol biosynthesis affects PrPSc accumulation when cells are capable of replenishing their cholesterol from the medium. Silencing of Srebf2 expression in the presence of 10% FCS did not significantly influence PrPSc levels (Fig. 4B). To confirm that this effect depended on external cholesterol, the experiment was repeated with cells cultured with medium supplemented with 5% BSA and 0.1% cholesterol instead of FCS. Also, under these conditions PrPSc levels remained unaltered in cells transiently transfected with siRNA against Srebf2 compared with control cells treated with nonsilencing siRNA (Fig. 4C), arguing that external cholesterol rather than other compounds present in the FCS affected efficient PrPSc accumulation. Furthermore, silencing of the Ldlr necessary for extracellular cholesterol uptake significantly reduced the amount of PrPSc (supplemental Fig. 3). These results demonstrate that neuronal cholesterol uptake rather than cholesterol biosynthesis is required for efficient PrPSc accumulation. In conclusion, up-regulation of cholesterol biosynthesis is a direct response to prion infection, even in the presence of external cholesterol.
FIGURE 4.
Knockdown of Srebp2 only interferes with PrPSc accumulation in the absence of exogenous cholesterol. 22L-infected N2a cells (clone 5) were transfected with siRNAs against Srebp2 and nonsilencing siRNA (ns) either in the absence (A) or presence of FCS (B) or in medium supplemented with BSA/cholesterol (BSA/chol) (C) as an external source of cholesterol. Cells were lysed 48 h post-transfection and analyzed for total PrP (PK−) and PrPSc contents following proteinase K treatment (PK+). For PK− samples, only one-third of the sample was loaded for presentation purposes. GAPDH levels were determined for the PK− samples. Successful knockdown of Srebp2 was determined by using ab28482. Antibody 4H11 was used for PrP detection. Additional bands were excised for presentation purposes. Experiments were repeated four times, and results were normalized to GAPDH levels. PrPSc levels in Srebp2 siRNA-transfected cells were compared with PrPSc levels in nonsilencing siRNA-transfected cells (A–C, bottom panels). Significant changes are indicated by asterisks (**, p ≤ 0.005; paired t test). −/− denotes no significant differences.
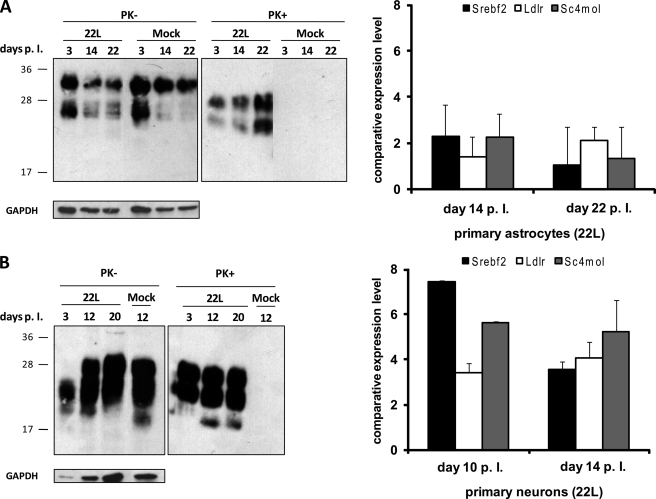
Primary Hippocampal Neurons Respond to Prion Exposure with Up-regulation of Cholesterogenic Genes
It has recently been reported that primary neurons and astrocytes are permissive to prions in vitro (38). To test if primary neuronal cultures also respond to prions by up-regulation of the cholesterol biosynthetic pathway, cultures of hippocampal neurons of C57BL/6 mice at gestation day 16 were prepared. As controls, primary astrocytes of these mice were also isolated. Cells were exposed to 0.1% brain homogenate from an uninfected mouse or to 22L brain homogenate. Cells were tested for PrPSc content 3–22 days post-exposure. Western blot analysis of astrocytic and neuronal cell lysates revealed that PrPSc was detectable in all lysates of cells exposed to 22L brain homogenate but not in lysates of cells exposed to mock brain homogenate (Fig. 5, A and B, left panels). Interestingly, in astrocytes the amount of PK-resistant PrP increased over time, suggesting that cells might generate PrPSc. Thus, although we cannot exclude that detected PrPSc originated from the inocula, our data clearly demonstrate that PrPSc was present throughout the course of the experiment. Transcript levels for cholesterogenic genes were assessed at different time points post-exposure. In neurons, cholesterogenic transcript levels were significantly increased (Fig. 5, A and B, right panels), although in astrocytes, transcript levels remained largely unaltered. In summary, the results presented in this study strongly argue that neurons up-regulate cholesterol biosynthesis in response to prions.
FIGURE 5.
Up-regulation of cholesterogenic genes in prion-exposed primary hippocampal neurons. Left panels, primary astrocytes (A) and neurons (B) were lysed at three different time points following inoculation with 0.1% 22L or normal brain homogenates, and cells were analyzed for total PrP (PK−) and PrPSc upon digestion with proteinase K (PK+). GAPDH levels in each lane are shown. Additional lanes were excised for presentation purposes. Right panels, after RNA extraction at two different time points per cell type Srebf2, Ldlr and Sc4mol expression levels were determined in prion-exposed cells compared with mock-treated cells by real time PCR. The experiments were performed in triplicate, and results were normalized to the housekeeping gene RPII. Bars represent mean values ± S.D.
DISCUSSION
Neuronal integrity is critically dependent on cholesterol, and cholesterol imbalance plays a pivotal role in several neurodegenerative diseases, including Alzheimer disease (39), Niemann Pick disease type C (40), and Huntington disease (41, 42). Up-regulation of cholesterol and lipid metabolism has been shown to cause neurological disease (43). Thus, imbalances in neuronal cholesterol homeostasis might also contribute to the pathology of prion diseases. The exact cellular location of prion conversion is unknown, but it appears to involve localization of PrPC to so-called lipid rafts, microdomains rich in cholesterol and sphingomyelin (44–47). Depletion of cholesterol from membranes abrogates correct PrPC cell surface localization and inhibits PrPSc formation (44, 48–50). Furthermore, drugs known to re-distribute cellular cholesterol (51) or to lower cholesterol biosynthesis (50) were shown to have anti-prion capacity in N2a cells or to increase incubation times in prion-infected mice (52, 53), suggesting that prion biogenesis is highly sensitive to changes in cholesterol homeostasis. In this study we now provide strong evidence that cholesterol homeostasis is directly altered upon prion infection of neuronal cells. Microarray analysis revealed that within the group of up-regulated genes, the predominant change observed was in the cholesterol biosynthetic pathway. Interestingly, several genes that are involved in cholesterol biosynthesis (Sc4mol and Cyp51) or harbor the SRE in their promoter region (e.g. Diazepam-binding inhibitor (Dbi) and stearoyl-coenzyme A desaturase 2 (Scd2)) were also found up-regulated in our previous gene expression studies utilizing N2a cells persistently infected with RML/Chandler mouse-adapted scrapie (12). Analysis of GT-1 cells persistently infected with RML scrapie confirmed increased levels of Srebf2 and Sc4mol transcripts. Thin layer chromatography analysis of N2a cells further revealed elevated total and free cholesterol levels upon prion infection. Intriguingly, up-regulation of cholesterogenic genes was not restricted to neuronal cell lines, as primary hippocampal neurons also strongly up-regulated cholesterol synthesis transcripts upon exposure to prions. Thus, our experiments suggest that induction of cholesterol biosynthesis and uptake might be a general consequence to a prion challenge of neuronal cells.
Of note, our data demonstrate that cholesterol biosynthesis was up-regulated as a consequence of prion infection even in the presence of external sources of FCS. We demonstrate by siRNA experiments that endogenous cholesterol biosynthesis is not crucial for PrPSc accumulation in case cholesterol can be replenished from external sources, indicating that prion-infected N2a cells are still capable of taking up exogenous cholesterol. However, as cholesterol deficiency is sensed in the endoplasmic reticulum, one possible explanation for our findings is that prion infection changes the cellular cholesterol distribution, leading to a decrease in endoplasmic reticulum cholesterol levels and thus to an activation of cholesterol biosynthesis. Interestingly, a recent study on neuronal cells now suggests an imbalance between free cholesterol and cholesterol esters in prion-infected cells (54).
It was recently reported that infection of N2a cells with RML scrapie did not lead to significant transcriptional changes (16), contrasting with microarray data on RML prion-infected GT1 and N2a cells (12) and data from this study. Although differences in experimental design, prion strains, and the use of different microarray platforms can certainly account for these results, the lack of differential gene expression of N2a cells reported by Julius et al. (16) could also be due to the specific cell clone PK1 that was used for the experiment. As this clone had been retrieved by several rounds of subcloning (as opposed to one round in our study) for drastically increased prion susceptibility (55), it is possible that this procedure led to the selection of a clone in which prion replication minimally interferes with normal cellular metabolism. Our findings are in line with microarray experiments of other groups that also showed a differential expression pattern of some genes belonging to the lipid or cholesterol pathway (9, 11, 13–15, 36, 56, 57). However, conflicting results exist as to whether prion infection leads to up-regulation (9, 11) or down-regulation (36) of cholesterol biosynthesis genes. Interestingly, up- or down-regulation appeared to be at least partially dependent on the time point of sampling during the infection process. In one study, the pre-clinical stage of prion infection had only marginal influence on cholesterogenic gene expression, whereas at terminal stages of disease, cholesterogenic transcripts decreased (36). In another study, pre-clinical animals infected with mouse-adapted ME7 scrapie prion strain displayed increased cholesterol biosynthesis gene expression in the central nervous system that decreased at terminal stages of the disease (11). Thus, cholesterol metabolism in the brain following prion infection appears to underlie dynamic changes that correlate with the disease state.
Brain cholesterol is almost exclusively synthesized locally, as the blood-brain barrier restricts import of plasma lipoproteins from peripheral circulation (58, 59). Cholesterol synthesis is mainly accomplished by astroglia, but neuronal cells also synthesize cholesterol at basal levels. Thus, the detected changes in cholesterol biosynthesis genes upon prion infection in vivo might reflect any of the following: (a) cell type-dependent differential expression; (b) disease progression-dependent differential expression; (c) neuronal cell loss-dependent differential expression; or (d) any combination thereof. Because microglial activation, mainly during clinical stages of disease, prominently influences transcript levels in mouse models of prion diseases (8, 10, 36, 56), it is possible that down-regulation of cholesterol biosynthesis during clinical prion disease reflects a glial response that masks neuronal up-regulation. This hypothesis is in agreement with our finding that exposure of microglial cells and primary astrocytes to prions in vitro either caused down-regulation of genes involved in cholesterol biosynthesis, e.g. Sc4mol and Srebf2, or left transcript levels relatively unaffected. Alternatively, neuronal cells in vivo might respond to prion infection dependent on the disease progression. Recent evidence shows that neuronal down-regulation of cholesterol biosynthesis can correlate with apoptosis (60). Neuronal loss is a hallmark of prion diseases, and several lines of evidence suggest that neurons infected with prions can undergo programmed cell death (61). Interestingly, recent studies on neuronal cultures demonstrated that de-regulation of intracellular cholesterol transport induced apoptotic cell death and was co-incident with decreased cholesterol biosynthesis transcripts. Notably, at earlier time points when no apoptosis was apparent, most cholesterol transcripts were increased, suggesting that damaged neurons might initially up-regulate cholesterol biosynthesis, potentially to compensate for cholesterol imbalances (60). Importantly, N2a cells can be persistently infected with prions and do not appear to undergo apoptosis, which could explain why N2a cells do not demonstrate decreased cholesterol biosynthesis transcripts upon prion infection. Similarly, primary hippocampal neurons exposed to 22L scrapie brain homogenate did not show decreased viability compared with primary neurons exposed to normal brain homogenate, at least not during the course of the experiment (data not shown). In summary, our results show that prions have the potential to alter the cholesterol homeostasis of cells in a cell type-specific manner. Our data provide new insights into the cellular and molecular biology and pathology of prion infection and might delineate new targets that can be used for selective interference in cellular prion propagation.
Supplementary Material
Acknowledgments
We are grateful to Dr. Osborne (Department of Molecular Biology and Biochemistry, University of California, Irvine) for providing the psynSRE plasmid and Dr. Martin Groschup (Friedrich-Loeffler-Institut, Germany) for providing 22L brain.
This work was supported by Deutsche Forschungsgemeinschaft Grants VO 1277/1-2, SFB 596 (Projects A8 and B14), by Bayerischer Forschungsverbund FORPRION (TUM 7), and by the European Commission Grant TSEUR LSHB-CT-2005-018805. This study was performed within the framework of EU FP6 Network of Excellence Neuroprion.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Tables 1 and 2.
- PrPSc
- abnormal prion protein associated with prion disease
- PrPC
- cellular prion protein
- FCS
- fetal calf serum
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- SRE
- sterol regulatory element
- BSA
- bovine serum albumin
- siRNA
- small interfering RNA
- Ldlr
- low density lipoprotein receptor.
REFERENCES
- 1.Prusiner S. B. (1991) Science 252, 1515–1522 [DOI] [PubMed] [Google Scholar]
- 2.Nunziante M., Gilch S., Schätzl H. M. (2003) ChemBioChem 4, 1268–1284 [DOI] [PubMed] [Google Scholar]
- 3.Wadsworth J. D., Hill A. F., Beck J. A., Collinge J. (2003) Br. Med. Bull. 66, 241–254 [DOI] [PubMed] [Google Scholar]
- 4.Aguzzi A., Polymenidou M. (2004) Cell 116, 313–327 [DOI] [PubMed] [Google Scholar]
- 5.Budka H. (2003) Br. Med. Bull. 66, 121–130 [DOI] [PubMed] [Google Scholar]
- 6.Doh-ura K., Perryman S., Race R., Chesebro B. (1995) Microb. Pathog. 18, 1–9 [PubMed] [Google Scholar]
- 7.Riemer C., Queck I., Simon D., Kurth R., Baier M. (2000) J. Virol. 74, 10245–10248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker C. A., Manuelidis L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 675–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riemer C., Neidhold S., Burwinkel M., Schwarz A., Schultz J., Krätzschmar J., Mönning U., Baier M. (2004) Biochem. Biophys. Res. Commun. 323, 556–564 [DOI] [PubMed] [Google Scholar]
- 10.Xiang W., Windl O., Wünsch G., Dugas M., Kohlmann A., Dierkes N., Westner I. M., Kretzschmar H. A. (2004) J. Virol. 78, 11051–11060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown A. R., Rebus S., McKimmie C. S., Robertson K., Williams A., Fazakerley J. K. (2005) Biochem. Biophys. Res. Commun. 334, 86–95 [DOI] [PubMed] [Google Scholar]
- 12.Greenwood A. D., Horsch M., Stengel A., Vorberg I., Lutzny G., Maas E., Schädler S., Erfle V., Beckers J., Schätzl H., Leib-Mösch C. (2005) J. Mol. Biol. 349, 487–500 [DOI] [PubMed] [Google Scholar]
- 13.Sorensen G., Medina S., Parchaliuk D., Phillipson C., Robertson C., Booth S. A. (2008) BMC Genomics 9, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hwang D., Lee I. Y., Yoo H., Gehlenborg N., Cho J. H., Petritis B., Baxter D., Pitstick R., Young R., Spicer D., Price N. D., Hohmann J. G., Dearmond S. J., Carlson G. A., Hood L. E. (2009) Mol. Syst. Biol. 5, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang Y., Xiang W., Hawkins S. A., Kretzschmar H. A., Windl O. (2009) J. Virol. 83, 9464–9473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Julius C., Hutter G., Wagner U., Seeger H., Kana V., Kranich J., Klöhn P. C., Klöhn P., Weissmann C., Miele G., Aguzzi A. (2008) J. Mol. Biol. 375, 1222–1233 [DOI] [PubMed] [Google Scholar]
- 17.Horton J. D., Shimomura I. (1999) Curr. Opin. Lipidol. 10, 143–150 [DOI] [PubMed] [Google Scholar]
- 18.Horton J. D., Shah N. A., Warrington J. A., Anderson N. N., Park S. W., Brown M. S., Goldstein J. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12027–12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakakura Y., Shimano H., Sone H., Takahashi A., Inoue N., Toyoshima H., Suzuki S., Yamada N., Inoue K. (2001) Biochem. Biophys. Res. Commun. 286, 176–183 [DOI] [PubMed] [Google Scholar]
- 20.Sato R., Inoue J., Kawabe Y., Kodama T., Takano T., Maeda M. (1996) J. Biol. Chem. 271, 26461–26464 [DOI] [PubMed] [Google Scholar]
- 21.Ertmer A., Gilch S., Yun S. W., Flechsig E., Klebl B., Stein-Gerlach M., Klein M. A., Schätzl H. M. (2004) J. Biol. Chem. 279, 41918–41927 [DOI] [PubMed] [Google Scholar]
- 22.Maas E., Geissen M., Groschup M. H., Rost R., Onodera T., Schätzl H., Vorberg I. M. (2007) J. Biol. Chem. 282, 18702–18710 [DOI] [PubMed] [Google Scholar]
- 23.Vorberg I., Raines A., Story B., Priola S. A. (2004) J. Infect. Dis. 189, 431–439 [DOI] [PubMed] [Google Scholar]
- 24.Gilch S., Schmitz F., Aguib Y., Kehler C., Bülow S., Bauer S., Kremmer E., Schätzl H. M. (2007) FEBS J. 274, 5834–5844 [DOI] [PubMed] [Google Scholar]
- 25.Lopes M. H., Hajj G. N., Muras A. G., Mancini G. L., Castro R. M., Ribeiro K. C., Brentani R. R., Linden R., Martins V. R. (2005) J. Neurosci. 25, 11330–11339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cronier S., Beringue V., Bellon A., Peyrin J. M., Laude H. (2007) J. Virol. 81, 13794–13800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horsch M., Schädler S., Gailus-Durner V., Fuchs H., Meyer H., de Angelis M. H., Beckers J. (2008) Proteomics 8, 1248–1256 [DOI] [PubMed] [Google Scholar]
- 28.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 29.Radonic A., Thulke S., Mackay I. M., Landt O., Siegert W., Nitsche A. (2004) Biochem. Biophys. Res. Commun. 313, 856–862 [DOI] [PubMed] [Google Scholar]
- 30.Bubner B., Gase K., Baldwin I. T. (2004) BMC Biotechnol. 4, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dooley K. A., Millinder S., Osborne T. F. (1998) J. Biol. Chem. 273, 1349–1356 [DOI] [PubMed] [Google Scholar]
- 32.Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. (1985) Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 33.Williams M. A., McCluer R. H. (1980) J. Neurochem. 35, 266–269 [DOI] [PubMed] [Google Scholar]
- 34.Momoi T., Ando S., Magai Y. (1976) Biochim. Biophys. Acta 441, 488–497 [PubMed] [Google Scholar]
- 35.Yao J. K., Rastetter G. M. (1985) Anal. Biochem. 150, 111–116 [DOI] [PubMed] [Google Scholar]
- 36.Xiang W., Hummel M., Mitteregger G., Pace C., Windl O., Mansmann U., Kretzschmar H. A. (2007) J. Neurochem. 102, 834–847 [DOI] [PubMed] [Google Scholar]
- 37.Priller J., Prinz M., Heikenwalder M., Zeller N., Schwarz P., Heppner F. L., Aguzzi A. (2006) J. Neurosci. 26, 11753–11762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cronier S., Laude H., Peyrin J. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12271–12276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burns M., Duff K. (2002) Ann. N. Y. Acad. Sci. 977, 367–375 [DOI] [PubMed] [Google Scholar]
- 40.Ohm T. G., Treiber-Held S., Distl R., Glöckner F., Schönheit B., Tamanai M., Meske V. (2003) Pharmacopsychiatry 36, Suppl. 2, S120–S126 [DOI] [PubMed] [Google Scholar]
- 41.Valenza M., Rigamonti D., Goffredo D., Zuccato C., Fenu S., Jamot L., Strand A., Tarditi A., Woodman B., Racchi M., Mariotti C., Di Donato S., Corsini A., Bates G., Pruss R., Olson J. M., Sipione S., Tartari M., Cattaneo E. (2005) J. Neurosci. 25, 9932–9939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valenza M., Cattaneo E. (2006) Prog. Neurobiol. 80, 165–176 [DOI] [PubMed] [Google Scholar]
- 43.Ntambi J. M., Miyazaki M. (2004) Prog. Lipid. Res. 43, 91–104 [DOI] [PubMed] [Google Scholar]
- 44.Taraboulos A., Scott M., Semenov A., Avrahami D., Laszlo L., Prusiner S. B., Avraham D. (1995) J. Cell Biol. 129, 121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vey M., Pilkuhn S., Wille H., Nixon R., DeArmond S. J., Smart E. J., Anderson R. G., Taraboulos A., Prusiner S. B. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 14945–14949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaneko K., Vey M., Scott M., Pilkuhn S., Cohen F. E., Prusiner S. B. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 2333–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marijanovic Z., Caputo A., Campana V., Zurzolo C. (2009) PLoS Pathog. 5, e1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gilch S., Kehler C., Schätzl H. M. (2006) Mol. Cell. Neurosci. 31, 346–353 [DOI] [PubMed] [Google Scholar]
- 49.Mangé A., Nishida N., Milhavet O., McMahon H. E., Casanova D., Lehmann S. (2000) J. Virol. 74, 3135–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bate C., Salmona M., Diomede L., Williams A. (2004) J. Biol. Chem. 279, 14983–14990 [DOI] [PubMed] [Google Scholar]
- 51.Klingenstein R., Löber S., Kujala P., Godsave S., Leliveld S. R., Gmeiner P., Peters P. J., Korth C. (2006) J. Neurochem. 98, 748–759 [DOI] [PubMed] [Google Scholar]
- 52.Kempster S., Bate C., Williams A. (2007) Neuroreport 18, 479–482 [DOI] [PubMed] [Google Scholar]
- 53.Mok S. W., Thelen K. M., Riemer C., Bamme T., Gültner S., Lütjohann D., Baier M. (2006) Biochem. Biophys. Res. Commun. 348, 697–702 [DOI] [PubMed] [Google Scholar]
- 54.Bate C., Tayebi M., Williams A. (2008) BMC Biol. 6, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klöhn P. C., Stoltze L., Flechsig E., Enari M., Weissmann C. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 11666–11671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Booth S., Bowman C., Baumgartner R., Sorensen G., Robertson C., Coulthart M., Phillipson C., Somorjai R. L. (2004) J. Gen. Virol. 85, 3459–3471 [DOI] [PubMed] [Google Scholar]
- 57.Skinner P. J., Abbassi H., Chesebro B., Race R. E., Reilly C., Haase A. T. (2006) BMC Genomics 7, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turley S. D., Burns D. K., Dietschy J. M. (1998) Am. J. Physiol. 274, E1099–E1105 [DOI] [PubMed] [Google Scholar]
- 59.Turley S. D., Burns D. K., Rosenfeld C. R., Dietschy J. M. (1996) J. Lipid Res. 37, 1953–1961 [PubMed] [Google Scholar]
- 60.Koh C. H., Peng Z. F., Ou K., Melendez A., Manikandan J., Qi R. Z., Cheung N. S. (2007) J. Cell. Physiol. 211, 63–87 [DOI] [PubMed] [Google Scholar]
- 61.Schätzl H. M., Laszlo L., Holtzman D. M., Tatzelt J., DeArmond S. J., Weiner R. I., Mobley W. C., Prusiner S. B. (1997) J. Virol. 71, 8821–8831 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.