Abstract
FLOWER FLAVONOID TRANSPORTER (FFT) encodes a multidrug and toxin efflux family transporter in Arabidopsis thaliana. FFT (AtDTX35) is highly transcribed in floral tissues, the transcript being localized to epidermal guard cells, including those of the anthers, stigma, siliques and nectaries. Mutant analysis demonstrates that the absence of FFT transcript affects flavonoid levels in the plant and that the altered flavonoid metabolism has wide-ranging consequences. Root growth, seed development and germination, and pollen development, release and viability are all affected. Spectrometry of mutant versus wild-type flowers shows altered levels of a glycosylated flavonol whereas anthocyanin seems unlikely to be the substrate as previously speculated. Thus, as well as adding FFT to the incompletely described flavonoid transport network, it is found that correct reproductive development in Arabidopsis is perturbed when this particular transporter is missing.
Keywords: Anther, fertility, flavonoid, MATE transporter, nectary, pollen
Introduction
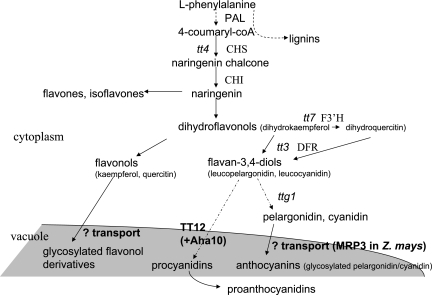
Flavonoids are phenolic secondary metabolites. Among the molecules sharing the same chemical backbone are three major classes, the pink and purple anthocyanins, the pale yellow flavonols, and the proanthocyanidins (PAs) of seedcoats (Fig. 1). Various ‘tt’ (transparent testa) mutants have provided a route for the study of flavonoid metabolic pathways, particularly the stages involving PAs. When these condensed tannins are perturbed in Arabidopsis thaliana, the seedcoat is paler than normal, giving rise to the name of the mutant series (Koornneef, 1990).
Fig. 1.
Flavonoid biosynthesis pathways and transport steps, in outline. Enzymes and mutants are marked if mentioned in the text. PAL, phenylalanine ammonia lyase; CHS, chalcone synthase; CHI, chalcone isomerase; F3'H, flavonoid 3’ hydroxylase; DFR, dihydroflavonol 4-reductase; tt, transparent testa.
The presence of anthocyanins in Arabidopsis vegetative tissues is easily observed since stress often results in purple-pigmented rosette leaves. The main flavonoids in Arabidopsis leaves, however, are normally the paler-coloured kaempferol and quercitin glycosides (Pelletier et al., 1999). Most of the aglycone flavonols become glycosylated in the C-3 and C-7 positions, and the principal sugar substitutions are glucose and rhamnose (Kerhoas et al., 2006). Literature reports identify kaempferol as the origin of most vegetative flavonol glycosides, and kaempferol-3-O-rhamnoside-7-O-rhamnoside as a major component (Routaboul et al., 2006, and references therein). Flowers have also been reported to contain predominantly kaempferol compounds, but quercitin derivatives are known to accumulate in stamens (Shirley et al., 1995; Burbulis et al., 1996; Routaboul et al., 2006). Among the quercitin glycosides identified in inflorescence tissue are quercetin–glucoside–rhamnoside, quercetin–glucoside–rhamnoside–rhamnoside, and quercetin–rhamnoside–rhamnoside (Jones et al., 2003). Numerous others, including flavonol dimers and oligomers, have been identified in Arabidopsis seeds (Routaboul et al., 2006).
The roles of flavonoids in plants range from protection from ultraviolet (UV) light to the pigmentation of flowers to attract pollinators (Shirley, 2006). Evidence is also accumulating that these compounds are involved in regulating auxin transport since the flavonols quercitin and kaempferol, in particular, can displace synthetic auxin transport inhibitors in vitro (Jacobs and Rubery, 1998). Indeed, some flavonoid mutants have phenotypes suggestive of altered auxin transport (Shirley et al., 1995; Peer et al., 2001, 2004; Peer and Murphy, 2007). For example, the Arabidopsis chalcone synthase (CHS) tt4 mutant has delayed gravitropism and, although flavonoids are classically thought of as localized in the vacuole, the root tissue flavonols involved here may be cytosolic (Buer and Muday, 2004). Meanwhile, control of bud outgrowth is affected in the Arabidopsis max1 mutant, in which this regulator of flavonoid pathway genes is missing and which consequently has altered levels of auxin transporters (Lazar and Goodman, 2006).
Given the many roles of flavonoids, their transport from the site of synthesis (primarily the cytosol) to the correct cell compartment, and between tissues (Buer et al., 2007), is obviously important and complex. Recently, Kitamura (2006) and Marinova et al. (2007) have summarized the known steps for flavonoid synthesis and transport in Arabidopsis, clearly showing the gaps in our knowledge (Fig. 1): whereas flavonoid biosynthesis is well understood, the proteins transporting anthocyanins and glycosylated quercitins and kaempferols remain little known. Characterization of tt mutants, however, has provided some insight into flavonoid transport. The product of TT12 (At3g59030) in Arabidopsis is a vacuolar flavonoid/H+ antiporter expressed in seeds (Debeaujon et al., 2001, 2003; Marinova et al., 2007). TT12 is a member of the multidrug and toxin efflux (MATE) transporter family, a group of proteins thought likely to fill some of the other missing steps (Kitamura, 2006; Fig. 1). These eukaryotic and prokaryotic transporters are one of the five groups that together comprise the multidrug transporter superfamily (MTS). MATE proteins typically contain 12 transmembrane helices, and are characterized by the absence of the signature sequences found in their four sister groups—the ATP-binding cassette, major facilitator, multidrug resistance (MRP), and resistance–nodulation–cell division families (Omote et al., 2006). Arabidopsis MATE proteins have been found in the tonoplast (Debeaujon et al., 2001, 2003; Jaquinod et al., 2007; Marinova et al., 2007), as was an acylated anthocyanin transporter in recent work in grape (Gomez et al., 2009), but one family member from lupin was identified in the plasma membrane (Uhde-Stone et al., 2005). The sequestration of anthocyanin and other flavonoids in the vacuole is already linked to other MTS members: Goodman et al. (2004), using antisense technology to investigate a Zea mays (maize) MRP (ZmMRP3), found perturbed anthocyanin accumulation. Arabidopsis MRP may also be responsible for vacuolar uptake of glutathione–anthocyanin conjugates (Kitamura, 2006).
In Arabidopsis, there is a large group of >50 MATE proteins (Kitamura, 2006) of which only a few have been characterized. There are two subgroups, the smaller one being most similar to the bacterial MATE proteins. One of these has been studied: FRD3 (ferric reductase defective 3, At3g08040) is involved in root iron metabolism (Rogers and Guerinot, 2002; Green and Rogers, 2004). The second subgroup contains the majority of A. thaliana members, is similar to human and yeast MATE proteins, and includes the protein described here, a flower flavonoid transporter (FFT). Three proteins from this group have been studied: the product of TT12, mentioned above; ALF5 (aberrant lateral root formation 5, At3g23560; Diener et al., 2001), thought to be involved in formation of lateral roots and toxin sensitivity; and EDS5 (enhanced disease susceptibility 5, At4g39030; Nawrath et al., 2002).
The MATE family member under study here, FFT, is the product of At4g25640 [named, amongst a family of so-called detoxifying efflux carriers, as AtDTX35 by Li et al. (2002)] in A. thaliana. It is most similar to putative Solanum lycopersicum (tomato), Vitis vinifera (grape) and Oryza sativa (rice) MATE proteins, and four other Arabidopsis proteins (Supplementary Fig. S1 available at JXB online). Its existence was noted previously in work on tomato describing a MYB overexpression line with altered anthocyanin regulatory pathways (Mathews et al., 2003). Because of the sequence similarity, these authors speculated that FFT might be involved in anthocyanin sequestration in vegetative tissues. Now identified as a possible vacuolar membrane protein (in a mass spectrometry study of vacuoles isolated from Arabidopsis cell culture; Jaquinod et al., 2007), loss of FFT could have profound effects.
It was found that FFT expression was widespread but particularly high in inflorescence tissues, especially in floral epidermal guard cells and those of the anther and nectary. Mutant analysis confirmed that abolishing FFT expression affects flavonoid levels in the plant, also altering root growth, seed development and germination, and pollen development and release. The FFT substrate is not established here, but the data suggest it is more likely to be a glycosylated flavonol than an anthocyanin as previously speculated. FFT can be added to the incomplete flavonoid transport network, and the results also show that correct reproductive development in Arabidopsis requires this putative transporter.
Materials and methods
Plants and growth conditions
Wild-type (WT) and mutant lines were grown in compost in greenhouses maintained at 20 °C. For growth assays and transformed plants, half-strength Murashige and Skoog (1/2MS) agar (pH 5.7; Duchefa, Haarlem, The Netherlands) was used, and plates or pots were incubated in a growth room at 20 °C with light of 10 μmol m−2 s−1 (16 h light cycles). For high-intensity light assays when testing photosynthetic parameters, a Fitotron growth chamber (Weiss-Gallenkamp, Loughborough, Leicestershire, UK) was used. N552331 and N604224 T-DNA insertion lines were purchased from the Nottingham Arabidopsis Stock Centre (NASC; Nottingham, UK) and segregated until homozygous. The presence and location of single T-DNA insertions were confirmed by Southern analysis (according to Church and Gilbert, 1984) and the absence of transcript by RT-PCR. Growth assays of fft-1 were conducted on plates in which one half of each plate was sowed with mutant and the other half with WT seed, with 6–11 seeds on each half for germination counts.
Molecular biology
DNA manipulation was performed essentially according to Sambrook et al. (1989). Primers for cloning the FFT full-length cDNA were 5′-CCCTGCAGATGGATCCGACGGCGCCGTT-3′ and 5′-CCGAATTCTCACGCAAGTATATCCTTGGATGTC-3′. Primers for the FFT promoter were 5′-CGAGCTCTCTATTTTATCCTCCCGAACA-3′ and 5′-CATCTTCTGAAAATGATTAACC-3′. For transcript analysis via RT-PCR, primers were used that would generate product across introns to differentiate any genomic contamination in template from transcript. To investigate transcript levels in specific tissues, RT-PCR was carried out using primers to actin8 (5′-AGAAAGATGCGTATGTTGGTGA-3′ and 5′-CTGCTGGAAAGTGCTGAGGGAA-3′), and the number of cycles of amplification determined that would produce the exponential phase of product amplification. The amount of template cDNA required to give this phase of PCR at the same cycle number in all tissue samples was also determined, and then the appropriate template concentration was used for RT-PCR with primers to FFT (5′-GGATGATAGCTGCTCCTGTG-3′ and 5′- CCGAATTCTCACGCAAGTATATCCTTGGATGTC-3′).
Binary vectors and plant transformation
Vectors for β-glucuronidase (GUS)–promoter lines and complemented mutant were made using pGreenII0029-based constructs (John Innes Centre, Norwich, UK) and Agrobacterium tumefaciens-mediated transformation via floral dipping (Gilmartin and Bowler, 2002). Transformed plants (T1) were grown on 1/2MS plates as above, supplemented with antibiotic as appropriate, and antibiotic-resistant plants grown on in compost.
Complemented mutant
The fft-1 mutant was complemented to ensure a return of WT characteristics by transforming it with a WT copy of the transcript under the control of the cauliflower mosaic virus (CaMV) 35S promoter. Effects on growth, seed and floral phenotypes were checked in three independent complemented transformant lines.
GUS staining
For promoter–GUS localization, tissues were stained according to Jefferson et al. (1987). Tissues fixed in 90% (v/v) ice-cold acetone for 20 min were rinsed twice in GUS working solution [0.1 M sodium phosphate buffer, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 10 mM Na2EDTA, 0.1% (v/v) Triton X-100; pH 7.0] and stained by adding 40 mg ml−1 stock 5-bromo-4-chloro-3-indolyl β-D-glucuronide cyclohexamine salt at 12.5 μl ml−1 buffer, vacuum infiltrating tissue, and incubating for 12–48 h as required at 37 °C. Tissues were cleared with 70% (v/v) ethanol. Three independent lines were examined to ensure reproducibility.
Microscopy of inflorescence tissues
Pollen and inflorescence tissues were examined using cryo-scanning electron microscopy (SEM) (Oxford Instruments CT 1500, Abingdon, Oxfordshire, UK). The Alexander stain was carried out essentially according to Pline et al. (2002). For a general screen of phenylpropanoid and flavonoid compounds in the fft-1 mutant, the method of Hutzler et al. (1998) was followed, in which secondary fluorescence was observed following UV excitation of tissues stained with 0.1% (w/v) DPBA [diphenylboric acid 2-aminoethylester; in 0.1 M potassium phosphate buffer pH 6.8, 1% NaCl (w/v) (Hutzler et al., 1998), since alkaline conditions induce fluorescence in flavonoids (Pfundel et al., 2006)]. For both epifluorescence and confocal microscopy, tissues were excited at the peak of flavonoid absorbance, at ∼350 nm; specifically, this was 330–380 nm (Nikon Eclipse 50i, epifluorescence attachment visualized using the UV-2A filter block) or, by confocal microscopy [Leica (Milton Keynes, UK) using TCS-SP1 equipped with Coherent Enterprise (Santa Paula, CA, USA) UV laser], selected areas were irradiated with the 351 nm laser line, and lambda scans were collected over 400–730 nm. Samples were examined with a ×63 oil immersion objective lens with a numerical aperture of 1.3. Fluorescence intensity was measured in 10 nm windows and displayed in the TCS (Leica) software as spectra.
Liquid chromatography–mass spectrometry (LC-MS)
Samples were run on a Thermo Finnigan (Thermo Fisher, Waltham, MA, USA) DecaXP+ ion trap LC-MS and an Agilent (Santa Clara, CA, USA) 1100 single quadrupole LC-MS system. Tentative quantification by mass spectrometry was checked against UV absorbance to help confirm identities. Flavonols were measured using the Agilent system as follows: analytes were separated on a 100×2 mm 3 μm Luna C18(2) column (Phenomenex, Torrance, CA, USA) at 23 °C using the following gradient of solvent A [0.1% (v/v) formic acid in water] versus solvent B (methanol) at 230 μl min−1 (0 min, 2% B; 30 min, 40% B; 40 min, 70% B; 42 min, 70% B; 43 min, 2% B; 51 min, 2% B; all v/v). Detection was by UV absorbance at 254 nm and 325 nm, and by positive electrospray mass spectrometry from 100 to 1500 amu. Spray chamber conditions were 172 Pa nebulizer pressure, 11.0 l min−1 drying gas at 350 °C, 4000 V capillary voltage, and 70 V fragmentor. Selected peaks in extracted ion chromatograms were integrated automatically using Chemstation software. Anthocyanins were analysed on a Surveyor LC system (Thermo Fisher) attached to a DecaXPplus ion trap mass spectrometer. Samples were acidified by addition of HCl to 0.1 M and separated on a 100×2 mm 3 μm Luna C18(2) column at 30 °C using the following gradient of solvent A [1% (v/v) formic acid in water] versus solvent B (methanol) at 230 μl min−1 (0 min, 2% B; 40 min, 70% B; 41 min, 2% B; 50 min, 2% B; all v/v). Anthocyanins were detected by UV absorbance at 520 nm and by positive electrospray mass spectrometry with spray chamber conditions of 350 °C capillary temperature, 50 units of sheath gas, 5 units of auxiliary gas, and a source voltage of 5.2 kV. Identification of anthocyanins was confirmed from MS2 and MS3 spectra collected at an isolation width of 4 amu and 35% collision energy. For ion-trap analysis of flavonols and glucosinolates, the identification of flavonols was confirmed by analysis in a Surveyor LC system attached to a DecaXP+ ion trap mass spectrometer. The gradient, column, and solvents were as used in the Agilent system, and spray chamber conditions were identical to those used for anthocyanins. Glucosinolates were identified using the same gradient, and negative electrospray ionization, with the same spray chamber conditions except that the source voltage was 5.0 kV.
Photosynthesis
Pulse amplitude modulation fluorimetry was used to measure the quantum yield of photosystem II (Blankenship, 2002) according to the manufacturer's instructions (Hansatech, King's Lynn, Norfolk, UK). Estimation of chlorophyll content was carried out according to Hipkins and Baker (1986) and Porra et al. (1989), extracting pigment with TRIS-Cl (pH 8.0)-buffered 80% (v/v) acetone.
Results
FFT encodes a MATE protein
The 488 amino acid protein (AtDTX35; estimated mol. wt, 53.2 kDa) is classified as a MATE protein (see TAIR; www.arabidopsis.org/abrc/). The most similar proteins to FFT in BLAST searches are an anthocyanin permease in tomato (Mathews et al., 2003; 72% identity at the protein level), an unknown protein in grape (70% identity), a putative ripening-regulated protein in rice (DDTFR18, Os08g0562800; 61% identity), and four other Arabidopsis MATE proteins (48–67% identity; Supplementary Fig. S1 at JXB online). As expected, hydropathy plots (TMPred; www.cbs.dtu.dk/services/TMHMM-2.0/) predict 12 transmembrane spans for the protein encoded by At4g25640. There is no recognizable signal peptide to predict targeting of the translated protein (SignalP, PredoTar; urgi.versailles.inra.fr/predotar/predotar.html). Glutamate residues may be important for activity in MATE proteins, and one such, found in almost every eukaryotic and prokaryotic MATE protein's seventh transmembrane domain, is also seen in FFT (Supplementary Fig. S1). Site-directed mutagenesis of this amino acid in the human MATE1 protein severely affected expression and prevented uptake of tetraethylammonium substrate in human embryonic kidney-293 cells (Matsumoto et al., 2008). Consistent with a likely role in flavonoid metabolism, a search for cis regulatory elements (PLACE database; Higo et al., 1999) finds MYB recognition sequences upstream of At4g25640 including two MYB-binding sites commonly found in flavonoid pathway genes such as CHS (consensus MACCWAMC; Sablowski et al., 1994), situated at −99 nt and −23 nt relative to the start codon.
FFT is most strongly expressed in floral tissues and guard cells
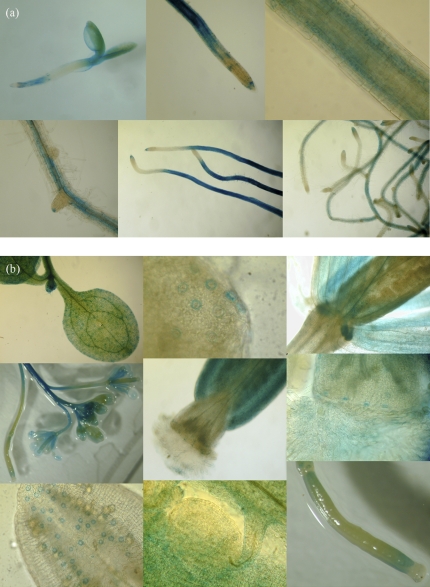
RT-PCR indicates that FFT is highly transcribed in floral organs and is present to some degree in most tissues, including young seedlings, senescent leaves and roots (Supplementary Fig. S2 at JXB online). To investigate further the spatial and temporal expression of FFT, a 1.4 kb genomic fragment from immediately upstream of the coding regions was amplified and fused to the uidA reporter gene. Floral tissues of plants transformed with this construct again showed the strongest activity (Fig. 2). Notably, GUS expression directed by the FFT promoter was almost exclusively found in guard cells in aerial tissues, including the specialized guard cells of nectaries and the hydathodes of leaves. Anther, stigma and silique guard cells were strongly stained, whereas the activity in mature leaf guard cells was reduced compared with those of the inflorescence and of the cotyledon (Fig. 2b). In newly germinated seedlings, as well as strong staining in leaf guard cells, there was GUS activity in the roots (Fig. 2a). By day 4 post-stratification (ps), on 1/2MS plates (t=0 at transfer of plants to growth chamber), staining was found in two discrete areas of the roots, namely at the root apex (meristematic zone) and in the elongation zone through to the fully expanded cells of the differentiation zone. The transition zone showed little staining. GUS activity in the elongation–differentiation zone was strongest internal to the epidermis, apparently in the endodermis and cortex. The tips of lateral roots were also stained blue (Fig. 2a).
Fig. 2.
X-gluc staining of GUS–FFT–promoter-transformed plants (each row, left–right). (a) Seedling roots: day 4 post-stratification; day 7 root tip, elongation zone, and developing lateral root; day 12; day 17. (b) Aerial tissues: (row 1) cotyledon guard cells (GC); mature leaf hydathode GC; nectaries; (row 2) inflorescence apex; silique apex; close-up of papillae and stigma GC; (row 3) anther GC; developing seed in siliques; developing silique.
Isolation of the fft-1 null mutant
Seeds of two mutants with T-DNA insertions in the At4g25640 open reading frame (ORF) were obtained from the NASC. Both were segregated to homozygosity. The insertion in line N552331 was situated ∼200 bp into the 3′-untranslated region: this mutant (fft-2) was not null so was not studied further. Transcript was abolished in line N604224 (fft-1), in which the T-DNA was inserted into the third intron (Supplementary Fig. S3 at JXB online). Having established that only one T-DNA was present in this line (Supplementary Fig. S3), phenotypic analyses were conducted on the homozygous mutant fft-1.
Several differences were observed between fft-1 and WT (Col0) plants in terms of growth and fertility. To ensure these phenotypic effects were the consequence of loss of function of FFT, genetic rescue was carried out, re-introducing a WT cDNA into the mutant line. The ectopic copy of the WT gene restored normal growth and fertility: root growth, seed and pollen development phenotypes were tested, and it was found that the FFT cDNA complemented all aspects of the mutation (Fig. 3–5; Supplementary Figs S4, S5 at JXB online).
Fig. 3.
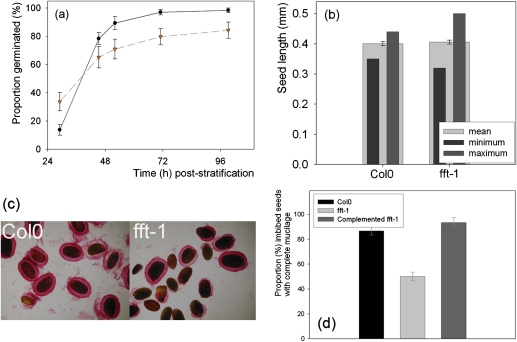
(a) Germination of the fft-1 (- -) mutant versus Col0 (—). (b) Seed size in Col0 and the mutant. (c) Ruthenium red staining of imbibing seed mucilage. (d) Proportion of imbibing Col0, fft-1 and complemented fft-1 seeds showing complete mucilage when stained with ruthenium red. (This figure is available in colour at JXB online.)
Fig. 4.
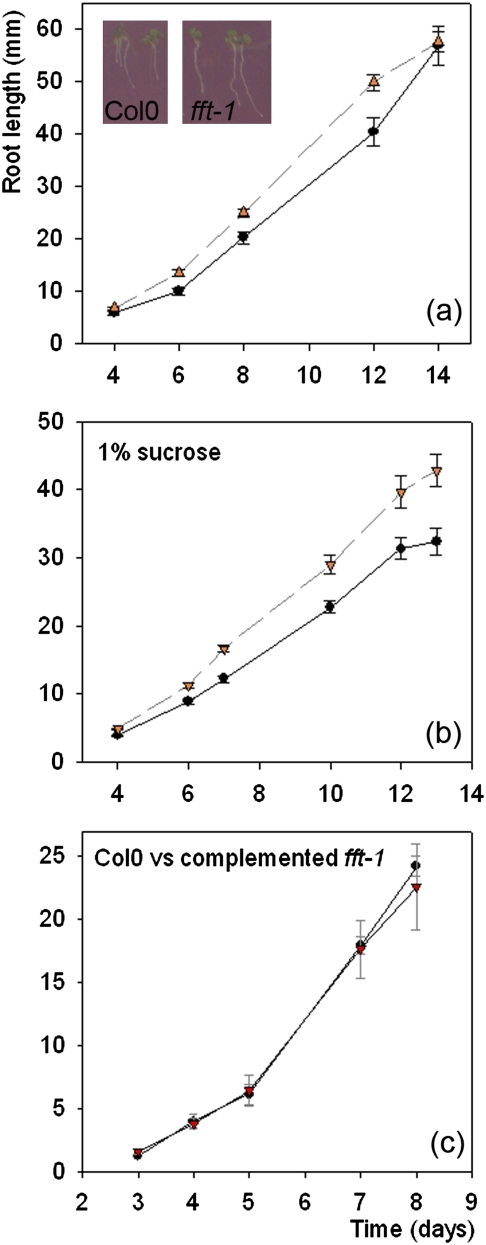
Mutant fft-1 (- -) roots grow more quickly than Col0 (—) up to ∼2 weeks. (a) No sucrose: day 6–12, P=0.002–0.003 (n=70–81 for each time point). Inset: Col0 and fft-1 seedlings at day 4 post-stratification. (b) 1% sucrose: day 6–12, P <0.0001–0.001 (n=60–97 for each time point). (c) The fft-1 mutant complemented with 35S-At4g25640 returns to wild-type growth characteristics. (This figure is available in colour at JXB online.)
Fig. 5.
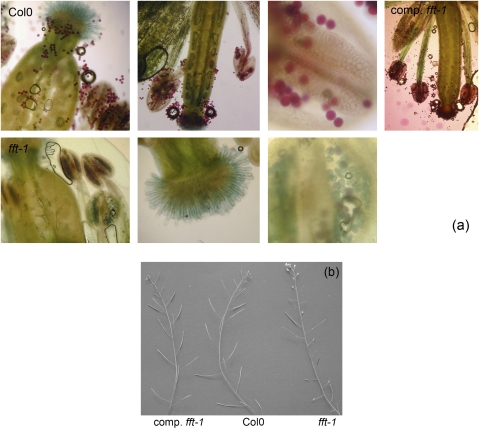
(a) Alexander's staining showing (left–right) lack of dehiscence, no pollen on the stigma, and reduced proportion of viable pollen in anthers in fft-1 (second row) versus Col0 (top row). Top right: pollen production and dehiscence in complemented fft-1. (b) Variable success of silique production in fft-1 versus Col0 and complemented fft-1.
Germination and seed morphology is perturbed in fft-1
Of plate-grown plants, >30% of the fft-1 mutant seeds [standard error of the mean (SE), 6%] had germinated by 29 h ps whereas in the WT this was <14% (SE, 4%; t-test P = 0.01, 14 df; Fig. 3a]. Fewer mutant seeds in total, however, germinated successfully in the mutant by 99 h [WT, 100% (SE, 2%); fft-1, 84% (SE, 6%)]. Because flavonoid mutants often show altered seedcoats and, because germination in fft-1 was variable, seeds were examined for physical abnormalities. The seedcoat of fft-1 was brown pigmented like that of the WT, but ruthenium red (RR) staining showed altered seed mucilage. RR stains acidic polysaccharides such as pectic polysaccharides: with imbibing Arabidopsis seeds, RR densely stains an inner layer of mucilage plus an outer, more diffuse (and easily disrupted) layer (Macquet et al., 2007). This staining of imbibing fft-1 seeds showed that the surrounding mucilage layer was absent or disrupted in the mutant. In the WT, 87% (SE, 3%) had complete mucilage compared with only 50% (SE, 4%) in the mutant (t-test, P <0.0001, 44 df; Fig. 3c, d). Seed mucilage contributes to seed dormancy and, thus, heterogeneous mucilage release in fft-1 mutant seeds is consistent with the abnormal germination rates. In addition, seed size was significantly more variable in the mutant (F-test, P <0.005), although mean seed size was not significantly different from that of the WT (Fig. 3b).
FFT affects growth rates
It was noticed that mutant seedlings appeared to grow and mature more quickly than the WT, so assays of root growth were carried out using plants on 1/2MS with and without sucrose (the former to promote anthocyanin production, in case of any effect). The mutant showed significantly faster root growth in all cases (Fig. 4a, b; for no sucrose: t-test, P <0.001 day 8, 64 df). Although the different germination characteristics meant that fewer fft-1 seeds were viable than WT seeds, the difference in mean rate of root growth developed after day 3–4 ps (when all viable seeds had successfully germinated) and continued to develop, suggesting that faster germination was not the primary cause of the root growth phenotype. The growth rate of fft-1 up to 2 weeks was greater under all conditions tested, including cold (4 °C) and higher intensity light (250 μmol m−2 s−1).
Fertility of fft-1 is reduced
When examined using cryo-SEM, a proportion of mutant pollen appeared shrunken and irregular, and/or had an aberrant surface structure (Supplementary Fig. S6 at JXB online). To examine this further, Alexander's stain was used to differentiate viable from non-viable pollen. Acid fuchsin stains the protoplasm of viable pollen purple, and malachite green (MG) is a background counterstain. This can give an overestimate of viability, since non-viable pollen can contain protoplasm (Pline et al., 2002), but here a distinct difference between WT and fft-1 flowers was shown (Fig. 5a). Series of flowers at identical stages and positions on the primary inflorescence were sampled and stained. In contrast to WT anthers, which were plump, full of purple-stained pollen, and showed many pollen grains released upon dehiscence, the mutant anthers appeared thinner and had a lower proportion of (but did contain some) round, purple pollen grains. Much of the pollen in mutant anthers stained only blue-green, the cell walls coloured only by MG (Fig. 5a).
Many fft-1 anthers fail to dehisce, with no visible pollen outside each (Fig. 5a, Supplementary Fig. S6 at JXB online). This was also obvious in epifluorescence microscopy. DPBA was used to enhance cytoplasmic flavonol fluorescence: it gives high emission levels when bound to flavonols rather than other flavonoids, for example fluorescing at 520 nm and 543 nm when bound to the flavonols kaempferol and quercitin, respectively (Saslowsky and Winkel-Shirley, 2001). WT flowers treated with DPBA showed bright yellow pollen: in contrast, only a few, non-fluorescent, pollen grains were present on mutant flowers (data not shown).
Arabidopsis siliques elongate as seeds develop following successful fertilization: reduced silique length is a visible demonstration of diminished fertility. In the fft-1 mutant plants, silique length is very variable (Fig. 5b) and seed number per silique was lower than in the WT [WT mean, 51 (SE, 11); fft-1 mean, 25 (SE, 5); t-test P=0.0001, 19 df].
Flavonoid metabolism is perturbed in mutant plants
In fft-1 mutant seedlings up to 1 week of age, extracted anthocyanins were slightly reduced in level compared with the WT (not shown), but the pigment was visible even in young tissues and, in quantity, in mature fft-1 and WT plants grown on high (5% w/v) sucrose, which were comparable, dark purple, colours.
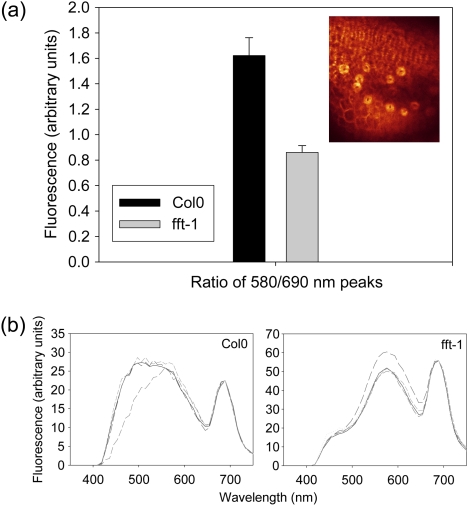
Following epifluorescent imaging of the WT and fft-1, it was hypothesized that confocal microscopy would reveal differences in flavonoid content of the plants. Confocal laser scanning microscopy was therefore used essentially to perform fluorescence spectroscopy, and thus indicate any changes to flavonoid levels (a technique previously used by Hideg et al., 2002, and discussed in Pfündel et al., 2006). Since GUS staining had occurred strongly in floral guard cells, inflorescence tissues treated with DPBA were excited with UV light (364 nm) and an emission spectrum was recorded (400–730 nm). Spectra were recorded from anther guard cells in WT or mutant flowers (inset, Fig. 6a) from the same position on an inflorescence (the first open flower below the unopened bud cluster), in which flavonoids should be comparable. A peak of emission from anther guard cells expected to result from flavonoid excitation could be visualized at 500–580 nm in WT tissues (Fig. 6b). Notably, a component of the fluorescence spectrum at ∼520 nm was absent in fft-1 (Fig. 6b), coinciding with the expected peak from flavonol–DPBA secondary fluorescence (Saslowsky and Winkel-Shirley, 2001). To quantify and test statistically the difference in replicate flowers, the well-defined flavonoid fluorescence peak at 580 nm and the peak from chlorophyll fluorescence were used to produce a ratio of flavonoid:chlorophyll emission. The ratios were significantly different between WT and fft-1, suggesting a lower flavonoid content in the mutant [WT, 1.60 (SE, 0.14) versus fft-1, 0.86 (SE, 0.05); t-test P=0.0016, 28 df; Fig. 6a]. Chlorophyll content was confirmed to be the same in WT and fft-1 mutant plants to make certain that levels of that pigment were not responsible for the differences observed, nor for any other aspect of the mutant phenotype (other photosynthetic parameters were also unaffected; see Supplementary Note S7 at JXB online). The products of the reactions of DPBA with flavonoids have not been comprehensively identified, however, so this can only be an indication of a difference in the flavonoid content in fft-1.
Fig. 6.
(a) Col0 versus fft-1 mean ratio of flavonoid (580 nm) and chlorophyll fluorescence (690 nm; Bolhar-Nordenkampf et al., 1989). Inset: example of the anther guard cell fluorescence scanned for analysis as viewed with the confocal microscope. (b) Examples of replicate fluorescence emission spectra from guard cells in Col0 and fft-1 anthers (background subtracted; normalized to 690 nm). (This figure is available in colour at JXB online.)
To investigate specific changes in flavonoid composition resulting from loss of the FFT-encoded protein, LC-MS was carried out on fft-1 mutant versus WT plants. A wide range of tissues was initially tested, guided by the position of GUS staining, to identify any differences between WT and fft-1 samples at the same stages of development. More plants were then grown so that the tissues where fft-1 apparently had different levels of particular metabolites could be sampled and analysed in replicate assays. Consistent differences were noted in plants from the separate sowings: in particular, mutant buds and siliques contained an altered spectrum of flavonol glycosides, with several peaks present at ∼40–70% the level in the WT, namely quercitin–rhamnoside–glucoside, kaempferol–glucoside–glucoside (kGG), and kaempferol–rhamnoside–glucoside. In contrast, there were almost doubled levels of quercitin–rhamnoside–glucoside–rhamnoside in buds (Table 1), and high levels of this flavonol were consistently found in mutant seedlings, suggesting that flavonoid synthesis and/or transport is perturbed in the fft-1 mutant. The difference in the levels of kGG was significant (t-test P=0.018, 5 df; see also Supplementary Fig. S8 at JXB online).
Table 1.
Relative level of flavonol glycosides in mutant versus wild-type (WT) floral tissues (further data in Supplementary Fig. S8 at JXB online)
| Peak and tissue type | Mean content in fft-1 versus WT (%) |
| kGG bud* | 40 |
| kRG bud | 70 |
| kRG immature silique | 73 |
| qRGR bud | 180 |
| qRG bud | 65 |
| qRG immature silique | 49 |
P=0.018, 5 df.
q, quercitin; R, rhamnoside; G, glucoside; IS, immature silique; k, kaempferol.
Discussion
The lack of detail known about the transport of flavonoids and about some of their roles in plants contrasts with our knowledge of their biosynthetic pathways. Here, it is shown that a MATE protein in Arabidopsis, FFT, is necessary for correct accumulation of flavonols in the plant. The effects of removing this protein support a role for flavonols in reproduction in A. thaliana and suggest an effect on growth, in line with current literature documenting an influence of flavonoids on auxin transport.
Flavonoids are necessary for full fertility
A GUS–promoter construct showed that guard cells are the principal location of the FFT transcript in aerial tissues. This is not surprising in the context of flavonoid pathways: a large proportion of a plant's phenylpropanoid and flavonoid compounds are found in the vacuoles of guard cells, whose location and physiological importance places them at the forefront of UV protection and, indeed, other responses to the environment (Hutzler et al., 1998; Gitz and Liu-Gitz, 2003). A possible role in flavonoid pathways makes the location of FFT in the vacuolar membrane likely and, by coincidence during the course of this work, it was one of the 30 most abundant proteins identified as likely to be located on the A. thaliana tonoplast membrane (Jaquinod et al., 2007).
Of particular note is the presence of FFT transcript in specialized guard cells of the nectaries and those of anthers and hydathodes. There is sparse published literature describing the roles of guard cells in these three locations. The nectaries develop as a ring at the base of the stamens in the Brassicaceae, which is among the families of plants that exude nectar through modified stomata (Davis et al., 1998). Flavonoids including quercitin and kaempferol and other phenolics are found in nectar, and CHS, the enzyme at the root of the flavonoid biosynthesis pathway (Fig. 1), is up-regulated during nectary development in tobacco (Nicolson et al., 2007). The presence of FFT transcript in these specific guard cells suggests a direct role in transporting flavonols for the production of nectar. Nectar release and nectar fall coincide with the start and end of anthesis (Durkee, 1983; Davis et al., 1998; Nicolson et al., 2007), so it is interesting that FFT transcription is also directed to the anthers.
As a self-fertile plant, nectar composition is unlikely to affect reproductive success in A. thaliana, but male sterility can result from defects in anther or pollen development. fft-1 mutant anthers contained a proportion of non-viable pollen, and dehiscence of anthers was perturbed. As there was no GUS staining in the pollen of FFT–promoter–GUS-transformed plants (in contrast to anther guard cells), the altered flavonoid profiles in anthers are the probable cause of the fft-1 pollen and fertility phenotypes. This was seen to an even great degree in the white anther petunia mutant which lacks CHS and is functionally male sterile (Napoli et al., 1999). Other mutants besides fft-1 are defective in both anther and pollen development, e.g. coi1 (coronatine insensitive 1) or fad (fatty acid desaturase 6) mutants, both related to jasmonic acid signalling in the regulation of anther dehiscence (Xie et al., 1998; Sanders et al., 1999). As was seen here, other mutations also reduce rather than abolish fertility. Altered exine development in the ms2 (male sterility 2) mutant (Aarts et al., 1997), for example, caused a range of effects, many similar to those seen in the fft-1 mutant, including reduced seed set, shrivelled anthers, and the majority of the seeds being produced from apical flowers of the mature inflorescence.
An effect of altered flavonoid content could be perturbed lignification, as seen after disrupting the function of the first enzyme of the phenylpropanoid pathway, phenylalanine ammonia lyase (PAL; Fig. 1; Rohde et al., 2004). Anther development and dehiscence follow defined pathways of maturation and cell death (Sanders et al., 1999), lignified secondary walls in the anther being important in its dehiscence (and also of relevance here, in breaking open mature siliques; Mitsuda et al., 2005). If anthers dehisce late, stigmatic papillae may no longer be receptive to pollination (in a self-fertile plant such as Arabidopsis). The guard cell location of the FFT transcript, however, suggests a connection in particular with appropriate dehydration of pollen and/or anther (Keijzer et al., 1996; Taylor and Heppler, 1997; Scott et al., 2004; Mitsuda et al., 2005; Jung et al., 2006), which would be expected to require proper function of the guard cells on the epidermis of the anther theca (Fig. 2b). Flavonoids in anther guard cells may also act within regulatory pathways that control co-ordinated development and maturation of pollen and anther (see comments on auxin, below). A role for FFT protein in dehiscence or dehydration of anthers and thus pollen coat formation and pollen viability seems most likely, but a direct chemical effect is also possible. Pollen's highly sculpted outer exine layer is mostly made up of sporopollenin, derived from long-chain fatty acids, but also contains some phenylpropanoids (Morant et al., 2007). Preston et al. (2004) speculated that altered flux along phenylpropanoid pathways was the cause of male sterility in myb4 and myb32 pollen mutants. Likewise, altered flavonoid composition in fft-1 could disturb the pollen coat and reduce pollination success, since exine is involved in recognition and adhesion.
The fft-1 mutant phenotype certainly indicates that perturbing flavonoid metabolism in Arabidopsis affects this plant's fertility. Although this is known to be the case in other plants, e.g. petunia (Napoli et al., 1999), the Arabidopsis CHS mutant, tt4, is fertile. The requirement for particular flavonoids for viable pollen and pollination in Arabidopsis has been discounted, therefore (Burbulis et al., 1996), although these authors do point out that the high levels of flavonols in pollen argue for an indirect role, at least, in fertility of the male gametophyte. In addition, a PAL double mutant (lesions in two of four PAL genes) did become sterile, with the siliques remaining small, and empty ovule sacs initially enlarging but then becoming shrivelled, suggesting male sterility (Rohde et al., 2004).
As with fft-1, variability was seen in the fertility phenotype of the mpk6 (mitogen-activated protein kinase 6) mutant (Bush and Krysan, 2007). Neither the present nor the latter authors could pinpoint specific environmental conditions that determined the severity of the effects. These mutations are clearly important but can (later in development) be overcome for reproduction in Arabidopsis, probably by developmental homeostasis. The variable success of fertilization in fft-1 does suggest that there may be redundancy in the putative flavonoid transporters of Arabidopsis. If a threshold level of a, or some, flavonoids must be achieved for some fertilization, low levels of transport by other transporters might eventually compensate for the missing FFT. Interestingly, the TT12 MATE transporter was able to transport glycosylated anthocyanidins in in vitro transport assays although in vivo these do not appear to be the normal substrate (Marinova et al., 2007).
FFT is necessary for correct seed development, germination, and growth
Numerous mutants with seedcoat phenotypes are defective in components of flavonoid biosynthesis or of its transcriptional regulation, e.g. tt8, ttg1 and ttg2 (Walker et al., 1999; Broun, 2005; Ishida et al., 2007). Changing the levels of flavonoids can have a range of effects on seed: in the ttg1 mutant, seedcoat anthocyanin levels, dormancy, and seed mucilage are all affected (Walker et al., 1999). In fft-1, accumulation of PAs and tannins is apparently not affected, since seeds are not paler than the WT. Fewer seed are set in the fft-1 mutant, however, and varied seed sizes and heterogeneous mucilage release upon imbibition are seen amongst those that are produced. There is no evidence, from GUS staining, of FFT promoter activity in seeds, so it seems likely that the seeds’ heterogeneity is a result of faulty flavonoid transport in the carpel and/or developing silique, in which RT-PCR did reveal high levels of transcript, and where there was strong GUS activity in FFT–promoter–GUS plants. Since, for example, oxidation of PAs contributes to seed dormancy (and protection) by strengthening the seedcoat (Debeaujon et al., 2000), reduced dormancy in the fft-1 mutant does also imply some change in seedcoat composition.
The GUS staining seen in FFT–promoter–GUS-transformed seedlings corresponds to the fluorescence from flavonoids seen previously in Arabidopsis roots using epifluorescent microscopy (Buer and Muday, 2004) and, notably, is consistent with possible accumulation of flavonoids in discrete locations of the root. FFT-directed GUS staining was notable in the cortex in the elongation zone and in the root tip, close to the locations of CHS and chalcone isomerase (Saslowsky and Winkel-Shirley, 2001). The faster root growth of seedlings is intriguing, since flavonoid mutants can have altered auxin transport (Buer and Muday, 2004; Lazar and Goodman, 2005). After studies using several tt mutants, quite a body of literature (see Peer and Murphy, 2009) now documents a reduction or elevation in polar auxin transport because of increased or absent flavonols (respectively), and thus altered accumulation of auxin in apical tissues. Cecchetti and colleagues (2008) also recently discussed how auxin regulates anther dehiscence and pollen maturation: flavonol–auxin effects on fertility cannot be ruled out in fft-1 and present an interesting avenue for future work.
MATE proteins transport-specific subsets of flavonoids
fft-1 has variable fertility, not complete infertility. Another flavonoid mutant's altered gravitational response was eventually corrected (Buer and Muday, 2004). These results suggest the existence of redundant mechanisms that allow roots or aerial tissues to recover. The existence of several Arabidopsis MATE proteins similar to FFT may explain redundancy of function, although some degree of specificity would correspond to the TT12 MATE transporter's limited substrates. Although it transports cyanidin-3-O-glucoside (acting as an H+-antiporter) in yeast assays in vitro, work on the tt12 mutant suggests that anthocyanins and glycosylated flavan-3-ols are its principal substrates in vivo, where it acts at the vacuolar membrane of PA-synthesizing cells of the seedcoat (Marinova et al., 2007). This work has recently been extended, where complementation of the tt12 mutant with a vacuolar Medicago truncata orthologue corroborates the identity of its substrate, epicatechin-3′-O-glucoside (Zhao and Dixon, 2009). In the present case, data from a tomato orthologue (Mathews et al., 2003) led to speculation that Arabidopsis FFT is a transporter of anthocyanin in vegetative tissues. The present work does not support this suggestion. Spectroscopic measurement of anthocyanin extracts does show a small reduction in young seedlings, and RT-PCR reveals FFT transcript in senescent leaves (which have high levels of anthocyanin) but, conversely, anthocyanin visibly accumulates in the mutant. It is hypothezised that the small changes in anthocyanin levels are a result of other perturbations in the flavonoid biosynthetic pathways since FFT–promoter–GUS staining was most significant in non-anthocyanin-containing tissues (inflorescence guard cells and roots). In addition, LC-MS measurements did not show any significant change in peaks likely to be flavonoids arising from samples of vegetative tissues (data not shown).
Fluorescence spectroscopy with DPBA did suggest altered flavonoid content in mutant versus WT anther guard cells, and LC-MS data revealed reductions in the fft-1 mutant in several flavonol glycosides in buds and immature siliques. Work to date suggests that a lack of FFT most affects levels of kaempferol (the more predominant of the two flavonols in whole Arabidopsis flowers; Shirley et al., 1995), specifically a diglucoside and probably therefore kaempferol 3,7-O-diglucoside (see www.plantcyc.org:1555/ARA/NEW-IMAGE?type=PATHWAY&object=PWY1F-FLAVSYN for detailed pathways and published contents of flavonoids and enzymes), but further work is required to confirm whether this is a transporter for that compound. Numerous novel dimers and oligomers of flavonols have been identified in Arabidopsis seeds (Routaboul et al., 2006), totalling 26 different flavonoids, so perturbing this pathway in fft-1 plants could clearly affect various aspects of reproductive development.
In summary, the characteristics of FFT support the case that MATE proteins are a component of flavonoid biosynthetic pathways. In addition, this protein appears to be necessary for proper reproductive development in Arabidopsis. Although chs mutants argue against an essential role in reproduction, transport of certain key flavonoids does seem to be essential for full male fertility.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. CLUSTALW alignment (N- and C-termini omitted to save space) and phylogram of FFT and the three most similar Arabidopsis MATE proteins with tomato putative anthocyanin permease (SlAnthP) and rice ripening protein (OsXP). The human MATE1 sequence (Q96FL8) is included to show an invariant glutamate (boxed) in the seventh transmembrane domain (Matsumoto et al., 2008). Twelve putative transmembrane domains (TMPred) are underlined.
Figure S2. Tissue or stress-induced transcription of FFT. RT-PCR was conducted as described in the Materials and methods. Sil, silique (green, >1 cm); YS, young silique (green, <0.5 cm); IS, immature silique (green, <1 cm); ISP, IS with dehydrating petals attached; Pet, petals; Bud, unopened bud; BS, bolt stem; CL, cauline leaf; ML, mature rosette leaf; SL, senescent anthocyanin-pigmented leaf; d2/d5, day 2 or day 5 seedling; LS, rosette leaf of plant grown with 1% sucrose; Pt, petioles; Fl, plates flooded (25 ml of sterile water for 36 h); H2, 1 mM H2O2 added to root tips for 3 h; 4C, plates incubated at 4 °C with light for 24 h; Pseudomonas syringae was infiltrated into the leaf as follows: M4, P.s. maculicola M4 (pathogenic); RW, P.s. phaseolica RW60 (non-pathogenic); Hrp, P.s. DC300 HrpA− (non-pathogenic).
Figure S3. Southern blot with a kanamycin probe showing one T-DNA band in genomic DNA from the fft-1 (N604224) mutant and the position of the T-DNA insertion.
Figure S4. RT-PCR in inflorescence tissues (top panel, FFT transcript; lower panel, actin8) from seven transformed mutants (lines A–G). Lines D, F, and G were investigated fully.
Figure S5. Cryo-SEM of inflorescence tissue from the WT (top panel, Col0-1), fft-1 mutant (middle panel, Ab-1), and complemented mutant (lower panel, F-1). The mutant has non-dehisced anthers but receptive papillae on the stigma, whereas the WT and complemented mutant have dehisced anthers and pollen germinating on stigma papillae.
Figure S6. Cryo-SEM of WT (Col0) and fft-1 mutant pollen and anthers, showing defective pollen and anthers in the mutant.
Figure S7. Photosynthetic parameters.
Figure S8. (a) Example of LC-MS runs for kGG in buds and immature siliques. Mutant, left (red); wild-type, right (green). (b) Mean values from which Table 1 proportions are derived. k, kaempferol; R, rhamnoside; G, glucoside; IS, immature silique; q, quercitin.
Supplementary Material
Acknowledgments
We thank Cathie Martin, Lionel Hill (LC-MS), Tony Burgess and Jeremy Skepper (SEM), and Sheila-Ann Johnson (pollen staining) for their help and advice. This work was funded by the BBSRC (grant no. C17526).
References
- Aarts MG, Hodge R, Kalantidis K, Florack D, Wilson ZA, Mulligan BJ, Stiekema WJ, Scott R, Pereira A. The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. The Plant Journal. 1997;12:615–623. doi: 10.1046/j.1365-313x.1997.00615.x. [DOI] [PubMed] [Google Scholar]
- Blankenship RE. Molecular mechanisms of photosynthesis. Oxford: Blackwell Science; 2002. [Google Scholar]
- Bolhar-Nordenkampf HR, Long SP, Baker NR, Oquist U, Schreiber U, Lechner EG. Chlorophyll fluorescence as a probe of the photosynthetic competence of leaves in the field: a review of current instrumentation. Functional Ecology. 1989;3:497–514. [Google Scholar]
- Broun P. Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Current Opinion in Plant Biology. 2005;8:272–279. doi: 10.1016/j.pbi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Buer CS, Muday GK. The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. The Plant Cell. 2004;16:1191–1205. doi: 10.1105/tpc.020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer CS, Muday GK, Djordjevic MA. Flavonoids are differentially taken up and transported long distances in Arabidopsis. Plant Physiology. 2007;145:478–490. doi: 10.1104/pp.107.101824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulis IE, Iacobucci M, Shirley BW. A null mutation in the first enzyme of flavonoid biosynthesis does not affect male fertility in Arabidpsis. The Plant Cell. 1996;8:1013–1025. doi: 10.1105/tpc.8.6.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush SM, Krysan PJ. Mutational evidence that the Arabidopsis MAP kinase MPK6 is involved in anther, inflorescence, and embryo development. Journal of Experimental Botany. 2007;58:2181–2191. doi: 10.1093/jxb/erm092. [DOI] [PubMed] [Google Scholar]
- Cecchetti V, Altamura MM, Falasca G, Costantino P, Cardarelli M. Auxin regulates Arabidopsis anther dehiscence, pollen maturation, and filament elongation. The Plant Cell. 2008;20:1760–1774. doi: 10.1105/tpc.107.057570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proceedings of the National Academy of Sciences, USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AD, Pylatuik JD, Paradis JC, Low NH. Nectar-carbohydrate production and composition vary in relation to nectary anatomy and location within individual flowers of several species of Brassicaceae. Planta. 1998;205:305–318. doi: 10.1007/s004250050325. [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Leon-Kloosterziel KM, Koornneef M. Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiology. 2000;122:403–413. doi: 10.1104/pp.122.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Nesi N, Perez P, Devic M, Grandjean O, Caboche M, Lepiniec L. Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. The Plant Cell. 2003;15:2514–2531. doi: 10.1105/tpc.014043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Peeters AJM, Léon-Kloosterziel KM, Koornneef M. The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. The Plant Cell. 2001;13:853–871. doi: 10.1105/tpc.13.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener AC, Gaxiola RA, Fink GR. Arabidopsis ALF5, a multidrug efflux transporter gene family member, confers resistance to toxins. The Plant Cell. 2001;13:1625–1637. doi: 10.1105/TPC.010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkee LT. Ultrastructure of nectaries. In: Bentley B, Thomas E, editors. The biology of nectaries. New York: Columbia University Press; 1983. pp. 1–29. [Google Scholar]
- Gilmartin PM, Bowler C. Molecular plant biology. Oxford: Oxford University Press; 2002. [Google Scholar]
- Gitz DC, Liu-Gitz L. How do UV photomorphogenic responses confer water stress tolerance? Photochemistry and Photobiology. 2003;78:529–534. doi: 10.1562/0031-8655(2003)078<0529:hduprc>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Gomez C, Terrier N, Torregrosa L, et al. Grapevine MATE-type proteins act as vacuolar H+-dependent acylated anthocyanin transporters. Plant Physiology. 2009;150:402–415. doi: 10.1104/pp.109.135624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CD, Casati P, Walbot V. A multidrug resistance-associated protein involved in anthocyanin transport in Zea mays. The Plant Cell. 2004;16:1812–1826. doi: 10.1105/tpc.022574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green LS, Rogers EE. FRD3 controls iron localization in Arabidopsis. Plant Physiology. 2004;136:2523–2531. doi: 10.1104/pp.104.045633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideg E, Juhász M, Bornman JF, Asada K. The distribution and possible origin of blue–green fluorescence in control and stressed barley leaves. Photochemical and Photobiological Sciences. 2002;1:934–941. doi: 10.1039/b201916g. [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acid Research. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipkins MF, Baker NR. Spectroscopy. In: Hipkins MF, Baker NR, editors. Photosynthesis energy transduction. a practical approach. 1986. IRL Press, Oxford, 51–101. [Google Scholar]
- Hutzler P, Fischbach R, Heller W, Jungblut T, Reuber S, Schmitz R, Veit M, Weissenbock G, Schnitzler J. Tissue localization of phenolic compounds in plants by confocal laser scanning microscopy. Journal of Experimental Botany. 1998;49:953–965. [Google Scholar]
- Ishida T, Hattori S, Sano R, et al. Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. The Plant Cell. 2007;19:2531–2543. doi: 10.1105/tpc.107.052274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs M, Rubery PH. Naturally occurring auxin transport regulators. Science. 1988;241:346–349. doi: 10.1126/science.241.4863.346. [DOI] [PubMed] [Google Scholar]
- Jaquinod M, Villiers F, Kieffer-Jaquinod S, Hugouvieux V, Bruley C, Garin J, Bourguignon J. A proteomics dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Molecular and Cellular Proteomics. 2007;6:394–412. doi: 10.1074/mcp.M600250-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Messner B, Nakajima J-I, Schäffner A-R, Saito K. UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. Journal of Biological Chemistry. 2003;278:43910–43918. doi: 10.1074/jbc.M303523200. [DOI] [PubMed] [Google Scholar]
- Jung K-H, Han M-J, Lee D-Y, et al. Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. The Plant Cell. 2006;18:3015–3032. doi: 10.1105/tpc.106.042044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keijzer CJ, Leferink-Ten Klooster HB, Reinders MC. The mechanisms of the grass flower: anther dehiscence and pollen shedding in maize. Annals of Botany. 1996;78:15–21. [Google Scholar]
- Kerhoas L, Aouak D, Cingöz A, Routaboul JM, Lepiniec L, Einhorn J, Birlirakis N. Structural characterization of the major flavonoid glycosides from Arabidopsis thaliana seeds. Journal of Agricultural and Food Chemistry. 2006;54:6603–6612. doi: 10.1021/jf061043n. [DOI] [PubMed] [Google Scholar]
- Kitamura S. Transport of flavonoids. In: Grotewold E, editor. The science of flavonoids. New York: Springer; 2006. pp. 123–146. [Google Scholar]
- Koornneef M. Mutations affecting the testa colour in Arabidopsis. Arabidopsis Information Service. 1990;28:1–4. [Google Scholar]
- Lazar G, Goodman HM. MAX1, a regulator of the flavonoid pathway, controls vegetative axillary bud outgrowth in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2006;103:472–476. doi: 10.1073/pnas.0509463102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, He Z, Pandey GK, Tsuchiya T, Luan S. Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification. Journal of Biological Chemistry. 2002;277:5360–5368. doi: 10.1074/jbc.M108777200. [DOI] [PubMed] [Google Scholar]
- Macquet A, Ralet M-C, Kronenberger J, Marion-Poll A, North HM. In situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant and Cell Physiology. 2007;48:984–999. doi: 10.1093/pcp/pcm068. [DOI] [PubMed] [Google Scholar]
- Marinova K, Pourcel L, Weder B, Schwarz M, Barron D, Routaboul JM, Debeaujon I, Klein M. The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+-antiporter active in proanthocyanidin-accumulating cells of the seed coat. The Plant Cell. 2007;19:2023–2038. doi: 10.1105/tpc.106.046029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews H, Clendennen SK, Caldwell CG, et al. Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. The Plant Cell. 2003;15:1689–1703. doi: 10.1105/tpc.012963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Kanamoto T, Otsuka M, Omote H, Moriyama Y. Role of glutamate residues in substrate recognition by human MATE1 polyspecific H+/organic cation exporter. American Journal of Physiology Cell Physiology. 2008;294:1074–1078. doi: 10.1152/ajpcell.00504.2007. [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Seki M, Shinazaki K, Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. The Plant Cell. 2005;17:2993–3006. doi: 10.1105/tpc.105.036004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morant M, Jørgensen K, Schaller H, Pinot F, Møller BL, Werck-Reichhart D, Bak S. CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. The Plant Cell. 2007;19:1473–1487. doi: 10.1105/tpc.106.045948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli CA, Fahy D, Wang H-Y, Taylor LP. white anther: a petunia mutant that abolishes pollen flavonol accumulation, induces male sterility, and is complemented by a chalcone synthase transgene. Plant Physiology. 1999;120:615–622. doi: 10.1104/pp.120.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Heck S, Parinthawong N, Métraux JP. EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. The Plant Cell. 2002;14:275–286. doi: 10.1105/tpc.010376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolson SW, Nepi M, Pacini E. Nectaries and nectar. Dordrecht: Springer; 2007. [Google Scholar]
- Omote H, Hiasa M, Matsumoto T, Otsuka M, Moriyama Y. The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends in Pharmacological Science. 2006;27:587–593. doi: 10.1016/j.tips.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Peer WA, Bandyopadhyay A, Blakeslee JJ, Makam SN, Chen RJ, Masson PH, Murphy AS. Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. The Plant Cell. 2004;16:1898–1911. doi: 10.1105/tpc.021501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Brown DE, Tague BW, Muday GK, Taiz L, Murphy AS. Flavonoid accumulation patterns of transparent testa mutants of Arabidopsis. Plant Physiology. 2001;126:536–548. doi: 10.1104/pp.126.2.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer WA, Murphy AS. Flavonoids and auxin transport: modulators or regulators? Trends in Plant Science. 2007;12:556–563. doi: 10.1016/j.tplants.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Pelletier MK, Burbulis IE, Shirley BW. Disruption of specific flavonoid genes enhances the accumulation of flavonoid enzymes and end-products in Arabidopsis seedlings. Plant Molecular Biology. 1999;40:45–54. doi: 10.1023/a:1026414301100. [DOI] [PubMed] [Google Scholar]
- Pfündel EE, Agati G, Cerovic ZG. Optical properties of plant surfaces. Annual Plant Reviews. 2006;23:216–249. [Google Scholar]
- Pline WA, Edmisten KL, Oliver T, Wilcut JW, Wells R, Allen NS. Use of digital image analysis, viability stains, and germination assays to estimate conventional and glyphosate resistant cotton pollen viability. Crop Science. 2002;42:2193–2200. [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica et Biophysica Acta. 1989;975:384–394. [Google Scholar]
- Preston J, Wheeler J, Heazlewood Li SF, Parish RW. At Myb32 is required for normal pollen development in Arabidopsis thaliana. The Plant Journal. 2004;40:979–995. doi: 10.1111/j.1365-313X.2004.02280.x. [DOI] [PubMed] [Google Scholar]
- Rogers EE, Guerinot ML. FRD3, a member of the multidrug and toxin efflux family, controls iron deficiency responses in Arabidopsis. The Plant Cell. 2002;14:1787–1799. doi: 10.1105/tpc.001495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde A, Morreel K, Ralph J, et al. Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. The Plant Cell. 2004;16:2749–2771. doi: 10.1105/tpc.104.023705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routaboul J-M, Kerhoas L, Debeaujon I, Pourcel L, Caboche M, Einhorn J, Lepiniec L. Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta. 2006;224:96–107. doi: 10.1007/s00425-005-0197-5. [DOI] [PubMed] [Google Scholar]
- Sablowski RWM, Moyano E, Culianez-Macia A, Schuch W, Martin C, Bevan M. A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO Journal. 1994;13:128–137. doi: 10.1002/j.1460-2075.1994.tb06242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. A laboratory manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu Y-C, Lee PY, Truong MT, Beals TP, Goldberg RB. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sexual Plant Reproduction. 1999;11:297–322. [Google Scholar]
- Saslowsky D, Winkel-Shirley B. Localization of flavonoid enzymes in Arabidopsis roots. The Plant Journal. 2001;27:37–48. doi: 10.1046/j.1365-313x.2001.01073.x. [DOI] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Dickinson HG. Stamen structure and function. The Plant Cell. 2004;16(Suppl):S46–S60. doi: 10.1105/tpc.017012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley BW. Flavonoid biosynthesis: ‘new’ functions for an ‘old’ pathway. Trends in Plant Science. 2006;1:377–382. [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM. Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. The Plant Journal. 1995;8:659–671. doi: 10.1046/j.1365-313x.1995.08050659.x. [DOI] [PubMed] [Google Scholar]
- Taylor LP, Hepler PK. Pollen germination and tube growth. Annual Review of Plant Physiology and Plant Molecular Biology. 1997;48:461–491. doi: 10.1146/annurev.arplant.48.1.461. [DOI] [PubMed] [Google Scholar]
- Uhde-Stone C, Liu J, Zinn KE, Allan DL, Vance CP. Transgenic proteoid roots of white lupin: a vehicle for characterizing and silencing root genes involved in adaptation to P stress. The Plant Journal. 2005;44:840–853. doi: 10.1111/j.1365-313X.2005.02573.x. [DOI] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, et al. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. The Plant Cell. 1999;11:1337–1349. doi: 10.1105/tpc.11.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D-X, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- Zhao J, Dixon RA. MATE transporters facilitate vacuolar uptake of epicatechin 3′-O-glucoside for proanthocyanidin biosynthesis in Medicago trunculata and Arabidopsis. The Plant Cell. 2009 doi: 10.1105/tpc.109.067819. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.