Abstract
The genome and its nucleotide precursor pool are under sustained attack by radiation, reactive oxygen and nitrogen species, chemical carcinogens, hydrolytic reactions, and certain drugs. As a result, a large and heterogeneous population of damaged nucleotides forms in all cells. Some of the lesions are repaired, but for those that remain, there can be serious biological consequences. For example, lesions that form in DNA can lead to altered gene expression, mutation, and death. This perspective examines systems developed over the past 20 years to study the biological properties of single DNA lesions.
1. Introduction
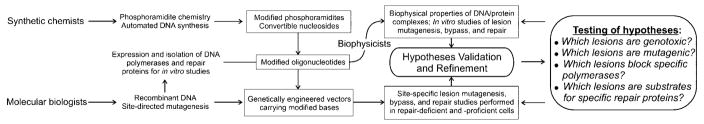
Twenty years ago, the field of site-specific mutagenesis by DNA lesions was at the transition between its infancy and early adolescence. At that time, the Editor of this journal asked one of us to write a perspective (1) that became the first article published in Chemical Research in Toxicology. Now that the field has reached maturity, the Editor has once again asked us to take stock of it. He has asked us to look back and assess the accomplishments of the entire field and then to look forward to see what might be expected as we gaze down the path ahead. The perspective is not meant to be a comprehensive review because, indeed, the field is now of such size that it is best reviewed in a fragmented way (see, for example, ref 2). Rather, we look back with 20/20 hindsight to identify past milestones that, in our opinion, have pushed the field ahead technologically or conceptually. As shown in Figure 1, advancements made by synthetic chemists, molecular biologists, and biophysicists have made possible the testing of hypotheses regarding lesion genotoxicity, mutagenicity, and repair. We apologize in advance to those who have made important contributions that did not fit into the boxed areas in Figure 1, which we describe below, or were not mentioned. We have been very selective and, at times, seemingly arbitrary in our choice of topics. Lastly, we look ahead and make guesses as to the new areas in which this field could contribute in the future.
Figure 1.
Evolution for understanding polymerase and repair contributions to lesion mutagenesis and bypass.
Site-specific lesion mutagenesis, bypass, or repair experiments use site-specifically modified oligonucleotides or genomes to monitor the fates of a chemical lesion situated at a known position within DNA or RNA. The fates studied are those evident after the lesion has been encountered by a polymerase (e.g., mutation and bypass) or repair protein (e.g., suppressed mutation and toxicity) in vivo. This field began as a logical offshoot of the fields of carcinogenesis and drug development, fields in which DNA damage is often a central concern. Most carcinogens and many drugs are or generate electrophilic intermediates that damage many sites in DNA. The resulting DNA lesion or adduct population is often so vast that it is difficult to determine with certainty which specific chemical modification of DNA was responsible for mutagenic or lethal events. Consequently, about a quarter of a century ago, workers started engineering the genomes of viruses and plasmids to contain, at specific genome sites, the DNA adducts caused by genotoxins. The genomes were replicated in cells, and the output progeny was analyzed to determine the type, amount, and genetic requirements for mutagenesis by the specific lesions. The objectives of the work were 2-fold: first, to rank order the toxicity and mutagenicity of DNA lesions and, second, to pinpoint the specific lesions that gave rise to the mutational spectra observed in the wake of DNA damage to tissues. In this perspective, we describe the advances made by groups concerning the synthesis of site-specifically modified oligonucleotides, as well as the methodology to track specific lesions of interest in vitro and in vivo.
2. Synthesis of Oligonucleotides Containing Site-Specifically Modified Bases
2.1. Synthesis of Modified Phosphoramidites
The current state of the art in DNA synthesis allows one to envision building an entire genome containing the damage of interest through solid-phase chemical synthesis (3, 4). However, because machine synthesis has not yet achieved the speed and accuracy that natural DNA replication can provide, from a technical and purity standpoint, it is easier to tap into an existing genome by splicing in a short piece of purified and characterized DNA containing the damage of interest located at a unique site. The first oligodeoxynucleotide containing site-specific damage contained O6-methyldeoxyguanosine in a tetramer made by manual solution-phase phosphotriester chemical synthesis (5). Today, thanks to the chemistry of the Caruthers’ group (6–8) and the instrumentation of Ogilvie (9) and Hood (10), automated solid-phase DNA synthesis using phosphoramidites has become the industry standard, and many phosphoramidites containing biologically interesting base modifications are now commercially available. The discovery of new chemical reactions, such as the Buchwald–Hartwig palladium-catalyzed cross-coupling aryl amination reaction (11–14), provides a route for synthetic chemists in the field of chemical toxicology to make DNA adducts stemming from industrial and dietary mutagens. Much pioneering work in the field was and continues to be performed by synthetic chemists, who work out conditions whereby the modified base in the phosphoramidite containing the customized damage is stable to not only the reagents involved in chemical synthesis but also to those involved in deprotection of the oligonucleotide. A short list of notable chemists in the modified DNA phosphoramidite field, who have made oligonucleotides with an emphasis on solving biological problems, is presented below.
Francis Johnson has made DNA containing 8-oxo purines (15), 8-aminoguanosine (16), ethano- and ethenocytidine (17, 18), ethenoguanosine (19), synthetic abasic sites (with Arthur Grollman) (20), guanosine adducts of acrolein (21), amino-naphthalene and aminofluorene (22), benzo[a]pyrene (23), the dietary mutagen PhIP (24), and tamoxifen (with Shinya Shibutani) (25). Shinya Shibutani has made DNA using guanine phosphoramidites bearing methyl, phenyl, ethyl, and tamoxifen adducts (25–29). Lawrence Marnett has made the deoxyguanosine malondialdehyde adduct (30). Lawrence Sowers has made DNA with pyrimidines oxidized at the C5-methyl or C5 ring positions (31–34). Jean Cadet has made DNA containing the 5′,8-cyclopurines (35), cyanuric acid (36), and the thymidine oxidation products 5-hydroxy-5-methylhydantoin (37), 5-carboxyuridine (38), and thymidine glycol (39). Shigenori Iwai synthesized oligonucleotides containing both stereoisomers of thymine glycol (40–42), as well as pyrimidine (6-4) pyrimidone, Dewar, and cyclobutane pyrimidine dimers (43–46). The incorporation of radical precursors into DNA by Bernd Giese at the C4′ sugar position (47–50) and by Marc Greenberg at the C1′ and C4′ sugar positions (51–54) and at the corresponding C5-methyl, C5, and C6 pyrimidine ring positions (55–62) has furthered our understanding of the products and mechanism of how site-specific radical formation may lead to DNA strand breaks, sugar and base damage, tandem lesions, and cross-links, which may themselves be used for further biological studies. Marc Greenberg has also made DNA containing bona fide formamidopyrimidine (FAPY) guanine and adenine lesions (63, 64), oxidized abasic sites (65, 66), and dihydro-5-hydroxythymidine (67). Thomas Harris has made the DNA acrolein adducts of guanine (68) and epoxybutene adducts at N1 of inosine (69). Carmelo Rizzo has made deoxyguanosine lesions containing etheno and hydroxyethano damage (70), as well as the C8 and N2 derivatives from the dietary mutagen IQ (71, 72). John-Steven Taylor has made the cis- and trans-syn photoproduct lesion between vicinal and nonadjacent thymidines (73–76). We have made DNA containing 5-hydroxy derivatives of cytidine and uridine (77) and the complex psoralen-thymidine monoadduct (78). Paul Hopkins has used a phosphoramidite strategy for making DNA interstrand cross-links between guanosines joined together by a common exocyclic nitrogen (79). Donald Jerina has made DNA bearing polycyclic aromatic hydrocarbon adducts of adenosine and guanosine (80–84) and, with the late Anthony Dipple, studied the effect of sequence context and stereochemistry on mutagenicity (85–92). The late Bea Singer was successful in making ethano-deoxyadenosine (93) and hydroxymethyl-etheno-deoxycytidine (94), and she, along with Dezider Grunberger, contributed enormously to the scholarly foundation of the field of alkylation chemistry of nucleic acids (95).
2.2. Postsynthetic Modification of DNA Bearing a Convertible Nucleoside
In contrast with the preceding section dealing with phosphoramidites that are fully equipped with their lesion, Mark Matteucci (96), Gregory Verdine (97–100), Peter Swann (101–103), Constance and Thomas Harris (104–107), and Roger Jones (108) pioneered the convertible nucleoside approach to site-specifically replace the exocyclic groups of DNA bases postsynthetically with a variety of N-, O-, and S-substitutions. The phosphoramidites containing postsynthetically displaceable leaving groups are commercially available, and researchers in the field can now, in many cases, easily gain access to their modified base of interest by simply displacing the leaving group in the postoligomerized oligonucleotide with an amine, alcohol, or thiol, rather than having to do a total synthesis of each phosphoramidite of interest (e.g., displacement of the O6-guanine leaving group with methanol will give the carcinogenic lesion O6-methyldeoxyguanosine) (102). Skilled synthetic chemists are still needed for the construction of complex nucleophiles containing specific stereochemistry, and the Harris’ have employed this umpolung technique to make several aromatic and aliphatic DNA lesions (109–114). Indeed, they made such diverse purine adducts as the polycyclic aromatic hydrocarbon benz[a]anthracene (105), styrene oxide (104), butadiene diol epoxides (109), and interstrand cross-links (115) in quantities large enough for Michael Stone to obtain NMR structures (116–120). This postsynthetic route was perhaps the only way to obtain DNA containing the malondialdehyde adduct of adenosine, which Carmelo Rizzo has made in addition to the guanosine adducts of malondialdehyde and trans-4-hydroxynonenal (121, 122). We have used this technique to make a furfuryl-N2-deoxyguanosine adduct (123) and, using a nonconventional convertible nucleoside, 5-guanidino-4-nitroimidazole (124). Isao Saito and Jean Cadet have used one-electron oxidation chemistry on DNA to postsynthetically convert 8-methoxyguanosine to 2-aminoimidazolone (125) and 5-hydroxyuracil to isodialuric acid (126), respectively. Using radical precursors, Jean Cadet and Yinsheng Wang have made C8-guanosine-pyrimidine DNA intrastrand cross-links (127, 128). Traditional convertible nucleosides have also been used to isotopically label the exocyclic groups for NMR studies (129, 130).
2.3. Postsynthetic Modification of DNA Bearing 8-Oxo-guanosine
While not a convertible nucleoside in the umpolung sense, one can view 8-oxoguanosine (and possibly even guanosine) as an “Achilles heel” convertible nucleoside, in that its low oxidation potential will attract damage to its position when an oligonucleotide is exposed to oxidizing agents. This potential liability is sometimes exploited in that one can choose the oxidant that will form the largest percentage of a sought-after specific lesion and then fish out their lesion of interest by HPLC or capillary gel electrophoresis purification. Cynthia Burrows was able to gain control over the DNA product distribution of 8-oxoguanosine conversion to spiroiminodihydantoin and guanidinohydantoin by an iridium oxidant using temperature and pH (131). Steven Tannenbaum modulated the distribution of DNA products from 8-oxoguanosine conversion by varying the concentration of peroxynitrite oxidant (132), and this site-specific oxidation technique was used to make oligonucleotides containing oxaluric acid, oxazolone, and cyanuric acid (133). Using photochemical oxidation, Nicholas Geacintov converted a DNA oligonucleotide containing a single guanine to one containing 8-nitroguanine (134). Conversion of a normal base within an oligonucleotide is not limited to guanine and its analogues, since site-specific transformations can be performed using base-specific chemistry, given that the base or reaction site is unique within the oligonucleotide; for example, we have converted an oligonucleotide containing a single thymine to one containing thymine glycol using permanganate oxidation (135), and John-Steven Taylor has converted vicinal thymidines to their (6-4) and Dewar photoproducts using 254 nm irradiation (136).
2.4. Postsynthetic Modification of DNA by Direct Reaction with Electrophile
In certain cases, one can circumvent the need for modified phosphoramidites by allowing an oligonucleotide to react directly with the electrophilic carcinogen or model compound to obtain the desired adduct, which has been performed in high yield with some polycyclic aromatic hydrocarbons. Nicholas Geacintov took this approach to make adenosine adducts of benzo[c]phenanthrene, and guanosine lesions of methylchrysene (137, 138) in quantities large enough for NMR solution structures to be obtained by Dinshaw Patel, with molecular modeling by Suse Broyde (139, 140). Likewise, the preparation of the relatively stable exo-8,9-epoxide of aflatoxin B1 using dimethyldioxirane, devised by Thomas Harris (141), allowed him to make enough of the N7-guanosine adduct (142) for Michael Stone to solve the NMR solution structure (143). Robert Fuchs used the direct reaction technique to synthesize C8-acetylaminofluorene adducts of guanosine, as well as an intrastrand cisplatin ApG adduct (144, 145); we and Stephen Lippard made the cisplatin GpG adduct (146). Using DNA containing only one guanosine or adenosine, Shinya Shibutani has made tamoxifen adducts of guanosine and quinone-derived estrogen adducts of guanosine and adenosine (147–151). Ashis Basu used DNA containing a single guanosine to make C8-aminopyrene and nitropyrene lesions (152, 153) and targeted adduction of mitomycin C to a specific location (154). Edward Loechler made a nitrogen mustard interstrand cross-link (155), and Paul Hopkins made interstrand cross-links from the anticancer agent BCNU, cisplatin, and nitrous acid (156–158).
2.5. Utility of DNA Bearing Isotopically Labeled and Atomically Mutated Bases
Oligonucleotides containing modifications not directly related to natural or environmental damage can also have a deep impact on the field of biological chemistry in general. Although undamaged, the phosphoramidites made by Roger Jones containing specifically placed multiple heavy isotopes have great utility in solving NMR solution structures of DNA (159–161), in investigating the mechanism of DNA base transformation by oxidative damage (162), and in acting as internal standards for mass spectrometry (163). Natalia Tretyakova has used these products to determine the reactivity (hotspot formation) of bases in varying sequence contexts toward DNA alkylating agents (164–169), as well as to quantify the amount of inter- vs intrastrand DNA cross-links formed by bifunctional alkylating agents (170). Thomas Spratt has made phosphoramidites containing atomically mutated bases to study the mechanism of DNA repair by alkyltransferases (171, 172) as well as DNA replication (173, 174), while Eric Kool has replaced individual atoms of bases to gain insight into the steric vs hydrogen-bonding requirements of biological processes, such as DNA replication (175–180), DNA recognition by mismatch repair complexes (with Peggy Hsieh and Thomas Kunkel) (181, 182), and DNA repair by Fpg and MutY (with Shelia David) (183).
3. In Vitro Studies Using Site-Specifically Modified Oligonucleotides
3.1. DNA Purification, Characterization, and Curvature Assay
Once the site-specifically modified oligonucleotides have been made, they are purified by one or several methods. Oligonucleotides are usually resolved based on charge using anion-exchange HPLC and an increasing ammonium acetate salt gradient or based on hydrophobicity using reversed-phase HPLC and an ion-pairing agent, such as triethylammonium acetate, with an increasing acetonitrile organic gradient. Denaturing high-percentage polyacrylamide gel electrophoresis resolves oligonucleotides based on their mass-to-charge ratio, but practically speaking, smaller failure sequences migrate faster, as is also seen for anion-exchange HPLC. After desalting and characterization by mass spectrometry and/or HPLC of the nucleosides or bases generated from an enzymatic or acid chemical hydrolyzate, respectively, the oligonucleotides can be used directly for biophysical studies, such as NMR, crystallographic, or melting temperature analyses to extract thermodynamic parameters on the helical stability of a lesion. Donald Crothers pioneered yet another biophysical method, which uses the ligation of properly phased DNA duplexes to measure DNA curvature, as bent DNA migrates more slowly through a nondenaturing gel (184, 185); he and Stephen Lippard used this to determine that cisplatin GpG intrastrand cross-links bend DNA 40° toward the major groove (186). While Nicholas Geacintov used the assay to show the stereochemical influence of benzo[a]pyrene-N2-guanine derivatives on DNA bending (187–189), the Stone, Hopkins, and Harris groups have used this assay to measure DNA bending induced by propanoguanine opposite a 2 bp deletion, mechlorethamine-N7-guanine interstrand cross-links, and alkane-N2-guanine intrastrand cross-links, respectively (190–192).
3.2. Cocrystal Structures of DNA Lesions with Replication and Repair Proteins
NMR solution studies of DNA lesions, such as the recently investigated ring-opened aflatoxin-N7-guanine FAPY adduct by Stone and the Harris’ (193), are informative in providing data on equilibrating isomers, which are valuable for the structural interpretation behind observed mutational spectra (194). However, the majority of structural studies using site-specifically modified oligonucleotides and DNA polymerases or repair proteins have been obtained using X-ray crystallography, which has shed light on mechanism. Tom Ellenberger’s cocrystal structures of DNA containing 8-oxoguanine or a thymine dimer with bacteriophage T7 DNA polymerase reveal how a lesion can be accurately or inaccurately replicated or act as a lethal replication block (195–197); his structure of DNA containing 1-azaribose with AlkA suggests a large 66° induced DNA bending, with base flipping and SN1-type mechanisms during repair (198). Ellenberger’s structures of the human AlkA homologue AAG bound to 1-azaribose or ethenoadenine lesions maps out a route for base flipping and excision, providing insight into the mechanism of discrimination between damaged and normal bases (199, 200). Sylvie Doublié and Susan Wallace captured stalled complexes of bacteriophage RB69 replicative DNA polymerase with abasic sites in the active and exonuclease sites, and their structure with thymine glycol shows the C5-methyl destacking the 5′-base, providing a rationale for why the lesion is not highly mutagenic and why adenine can be inserted opposite the glycol but cannot be extended further (201–203). Samuel Wilson’s cocrystals of pol β with DNA containing 8-oxoguanine, abasic sites, and benzo[c]phenanthrene-N2-guanine in gapped structures provides mechanistic data on dNTP insertion and deoxyribose phosphate lyase repair (204–206). Lorena Beese’s cocrystal of DNA bearing benzo[a]pyrene-N2-guanine with the high-fidelity Bacillus DNA polymerase shows that the adduct situated in the minor groove greatly distorts the contacts between the DNA and the polymerase (207); additionally, her structural snapshots of O6-methylguanine with the polymerase reveal how an opposing C and T can evade proofreading, and her structures, obtained with Paul Modrich, of MutSα bound to O6-methylguanine:T, G:T, G:U, and a one base loop shows all lesions to be recognized similarly by the complex (208, 209). Gregory Verdine’s cocrystal structures of DNA containing 8-oxoguanine with hOGG1 or catalytically inactive MutM, of trapped DNA repair intermediates with MutM or EndoIII, and of engineered adenine-MutY and other DNA–protein disulfide cross-links provide a framework for understanding how lesions are recognized, the step by step process by which excision of the lesions occurs during repair, as well as how a repair enzyme can find a single lesion on a vast landscape of unmodified DNA (210–218). John Tainer’s cocrystal structures of DNA containing uracil and abasic sites with human uracil-DNA glycosylases show how the enzyme can flip the base out of the helix and into the active site, by insertion of an amino acid residue from the minor groove with phosphate backbone compression, and how base excision repair is initiated (219, 220). His structure of endonuclease IV complexed with an abasic site, obtained with Richard Cunningham, reveals a 90° DNA kink, with both the abasic site and its partner flipped out of the helix, thus promoting specific insertion and endonucleic cleavage of the abasic site within the active site pocket (221); a kinked DNA structure was also seen with the APE1 structure (222). John Hunt’s crystal structure of 5′-T(1-methyldeoxyadenosine)T-3′ with AlkB shows the alkylated base to be flipped out and also provides a rationale for binding and repair of 1,N6-ethenoadenine, due to a small packing void adjacent to the 1-methyl group (223).
3.3. In Vitro Lesion Replication Studies
In many cases, the well-characterized DNA or RNA can be used directly or is ligated into a larger construct for in vitro lesion replication or repair studies. Such replication studies (mutagenicity and bypass) are straightforward, since the extension of an annealed 5′ 32P-labeled primer past a template lesion in the presence of nucleotide triphosphates and the polymerase under investigation can be monitored by PAGE resolution of 32P-labeled extension products. This method can also be used to determine the ability of a polymerase to incorporate modified nucleotide triphosphates. Myron Goodman established a gel assay for determining the kinetic parameters for the efficiency of nucleotide insertion (Vmax/Km) opposite a base, which determines the preference of different DNA polymerases for normal or modified dNTPs opposite normal or modified template bases (224–227). This advance led he and Roger Woodgate to show the relative impact of pol III, pol IV, pol V, and accessory proteins in trans-lesion synthesis (TLS) past photodimers and abasic sites, which led to a model for SOS lesion-targeted mutagenesis (228, 229). Graham Walker and we used such an assay to determine that Escherichia coli pol IV and its mammalian orthlogue, pol κ, are more efficient in accurate TLS past the model nitrofurazone-derived lesion N2-furfuryl-dG than past an undamaged template base (123). Using a variety of lesions and DNA polymerases, the Guengerich laboratory has performed several presteady-state (rapid quench) experiments, in addition to steady-state measurements, kinetic model fitting, and obtaining crystallographic data on lesion-polymerase-dNTP ternary complexes, to determine how individual steps in polymerization contribute to dNTP incorporation opposite the lesion, such as nucleotide binding, conformational change, phosphodiester bond formation, the efficiency of base pair extension, and the contribution of accessory proteins (230–244). Grollman and Shibutani, in addition to determining the traditional steady-state kinetic parameters for nucleotide incorporation, devised an ingenious in vitro method allowing for PAGE resolution of base substitutions and frameshift deletions resulting from trans-lesion extension of a labeled primer by DNA polymerases in the presence of equimolar triphosphates, thus screening for mutation frequency and specificity in a single-tube reaction (245–257). Indeed, Shibutani’s two-phase 20% PAGE modification allowed him to find the miscoding specificities of oxidized and alkylated purines by a variety of E. coli and mammalian DNA polymerases (26–28, 147, 149, 151, 258–263).
3.4. In Vitro Lesion Repair Studies
Repair studies performed in vitro are straightforward when a base excision mechanism is involved, since a cleavage event (either direct or induced chemically from the newly generated abasic site) can be followed easily by denaturing PAGE resolution of bands from a 5′ 32P-labeled substrate. However, lesions repaired by a direct reversal mechanism, such as O6-methylguanine by Ada or Ogt and 1-methyladenine by AlkB, will not produce cleavable substrates. In this case, the amount of alkylated and dealkylated oligonucleotide can be quantified by integration of peaks following reversed-phase HPLC resolution of the oligonucleotides and by mass spectrometry (using appropriate controls or standards). One can enzymatically induce cleavage of the repaired, dealkylated DNA if the lesion is situated in a restriction endonuclease site (264), which allows one to follow repair by PAGE resolution of 32P-labeled bands (265). Additionally, Anthony Pegg and F. Peter Guengerich showed that oligonucleotides containing or lacking O6-alkylguanine can be resolved by PAGE, provided that they are small enough (266).
Susan Wallace has melded in vitro bypass and repair techniques with in vivo analysis to determine the sequences (hotspots) surrounding 8-oxoguanosine that are most likely to be mutagenic and refractory to repair (267). The lesion is flanked by randomized bases, traversed by a DNA polymerase, and digested with MutM (weeding out nonmutagenic 8-oxoG:C pairings), and lesion-bearing strands that survived are PCR amplified and cloned into a vector to determine the nature of the mutation by sequencing; a similar study was done for abasic sites (268). Richard Wood, Peter Robins, and Tomas Lindahl devised an assay to monitor the repair and resynthesis of covalently closed DNA-damaged plasmids in vitro using mammalian cellular extracts and [α-32P]-dNTPs (269), which was used to measure site-specific repair and resynthesis of cisplatin intrastrand DNA cross-links, photoproducts, 2-acetylamino-fluoreneadducts,and5′,8-cyclodeoxynucleoside lesions(270–275). A comparable assay that achieves a similar goal was devised by Aziz Sancar and colleagues (276).
4. In Vivo Lesion Replication and Repair Studies
Lesion mutagenesis, bypass, and repair studies performed in vitro are unequivocal in that one knows exactly which polymerase or repair protein causes the observed effect. One issue, however, with this approach is that one can force a result that may not be biologically relevant; it is important to interpret results with the caveat that there are multiple DNA repair enzymes and multiple bypass polymerases within a cell, all competing for the same lesion. Accordingly, the most direct way to discern which protein may be the most relevant in lesion processing (and validating in vitro results) would be to place a site-specific lesion into a genome, which is allowed to be replicated and repaired in wild-type cells and, in parallel, in cells knocked out or down for, and/or induced for, specific polymerases or repair proteins. Methods for generating such site-specific genomes are described below.
4.1. Construction and Utility of Site-Specifically Modified Double-Stranded Vectors
Most studies on site-specific lesion replication and repair within cells involve the construction of double-stranded or single-stranded vectors bearing the lesion, the choice of which depends on the particular problem that one wishes to address. Double-stranded vectors are ideal for monitoring nucleotide excision repair (NER) or base excision events, since many repair systems require duplex DNA and one has control over what is placed opposite the lesion. However, numerous studies using duplex genomes have shown that many DNA lesions are blocks to DNA replication, strongly favoring the replication of the nonadducted strand and the reduction of mutation signal. Thomas Kunkel has advanced the fields of both protein engineering and site-specific lesion mutagenesis by devising a method for generating single-stranded M13 viral template strands containing uracil by growing phage in dUTPase and uracil-DNA-glycosylase-deficient E. coli (277). After priming with a mismatched (or lesion-bearing) oligonucleotide to program the mutation, extension occurs using T7 DNA polymerase to avoid displacement of the original primer (278), and T4 DNA ligase seals the nick prior to transformation into a uracil-DNA-glycosylase-proficient strain; replication in this strain generates abasic sites in the template strand, thus giving the newly synthesized mutagenic (or lesion-bearing) strand a competitive advantage in growth (279). Many groups have made double-stranded genomes bearing uracils in the nonlesion strand as a strategy for selective amplification of signal from the nonuracilated, lesion-bearing strand (280–290). Indeed, the Marnett laboratory showed that the uracilated (+) strand is 4 orders of magnitude less able to replicate than a nonuracilated (+) strand (284). For TLS and direct reversal of damage studies by methyltransferases, Gary Pauly and Robert Moschel ensured signal from the lesion strand by ligating into their vector a small duplex DNA in which several uracils oppose the lesion-bearing strand, with the generation of a gap after in vitro degradation of the uracils (287, 288). Starting with an unmodified duplex plasmid and one containing randomly incorporated uracils, Robert Fuchs constructed a heteroduplex with the lesion in the nonuracilated strand and then degraded the entire opposing strand to obtain single-stranded modified genomes (286).
The synthesis of double-stranded genomes containing lesions may seem straightforward at first glance. Indeed, why not just digest a double-stranded vector with two restriction endonucleases, remove the small excised piece, and ligate a site-specifically modified duplex containing compatible ends? In practice, however, problems plagued this “traditional cloning” approach. Site-specific genetic studies rely on material that is homogeneous, and the common problems one encounters with traditional cloning would be unacceptable; these problems include concatemerization of the ligation components and poor ligation efficiency due to small overhangs. In response to the need for high-quality site-specifically modified genomes, the gapped duplex method was developed and validated. We were able to ligate a 4-mer containing O6-methylguanine into a duplex vector by denaturing a mixture of two related duplex genomes that had been linearized at different locations and that differed only by the size and sequence of the complementary oligonucleotide to be inserted, followed by reannealing and ligating the tetramer into the 4-base gap (264). Dephosphorylation of the genome linearized distally from the insert site allows for the removal of the complementary wild-type strand and the analysis of single-stranded genomes, while omission of this step generates duplex vectors. We also used a duplexing strategy whereby a single-stranded circular DNA is annealed with the complementary strand from a related linearized duplex to form the gap, into which the modified oligonucleotide is ligated (291).
Robert Fuchs’ use of gapped duplexes to generate lesions in the leading or lagging strand of covalently closed duplex vectors showed that while C8-guanine-acetylaminofluorene (AAF) adducts are 20-fold more mutagenic in the lagging than in the leading strand, 8-oxoguanine exhibits no such preference (292, 293). By crippling the nonlesion strand by UV irradiation of one of the linearized plasmids prior to gapped duplex formation, Fuchs showed that the position of AAF affects −2 deletions within the NarI site (294) and −1 deletions within the SmaI site, where a delay of replication after accurate insertion of cytosine opposite the guanine lesion may allow time for the formation of a slipped intermediate (295). Fuchs’ strand segregation analysis assay uses genomes containing a 3–4 nucleotide insertion opposite the lesion, allowing one to distinguish (in mismatch repair-deficient cells) TLS from damage avoidance pathways, which circumvent lesion replication (296–299). Among other discoveries, Fuchs used this assay to show that TLS increases from 1 to 70% when AAF is deacetylated (296) and that AAF-induced −2 deletions in the sequence GCGAAFCX can vary 30–50-fold in E. coli, depending on X (298). Lawrence Marnett placed a C:C mismatch 3 kb away from malondialdehyde and cyclopropano guanine lesions and showed that the lesions are substrates for NER by tracking template strand utilization (300). Masaaki Moriya and Arthur Grollman used mismatches at multiple locations to delineate the various repair and replication processes for 1,N6-ethenoadenine and γ-hydroxypropanoguanine by linkage analysis (301, 302).
Another strategy for duplex genome construction uses primer extension on single-stranded circular viral templates. Lawrence Loeb and Bea Singer extended primers by one nucleotide using the dNTP of O4-methylthymine or N2,3-ethenoguanine, followed by further extension with normal dNTPs and ligation of the nick (281, 303). Thomas Kodadek and Howard Gamper used a primer bearing an internal psoralen lesion to obtain site-specifically modified duplexes (304).
4.2. Construction and Utility of Site-Specifically Modified Single-Stranded Vectors
Single-stranded genomes are ideal for measuring lesion traversal and mutagenesis by polymerases, since vector replication requires trans-lesion synthesis to occur. Such vectors are also ideal for studying lesion repair by proteins that use a direct reversal of base damage mechanism, such as Ada, Ogt, AlkB, and their eukaryotic counterparts, since repair prior to replication yields a normal base that is neither mutagenic nor genotoxic. Such vectors are not well-suited for addressing NER or base excision repair if the oligonucleotide or lesion is excised with equal efficiency regardless of the opposing base inserted by the DNA polymerase, since the mutation would have already been made permanent. However, repair data can be gained if (i) another enzyme can remove the improper base opposite the lesion, giving the polymerase a second chance for nonmutagenic dNTP insertion, and/or (ii) the lesion is excised only when paired with a nonmutagenic base. This scenario describes the MutM/MutY “GO” repair system that Jeffery Miller and Arthur Grollman discovered for 8-oxoguanine (305, 306); the paradigm may possibly exist for other lesions as well. The GO discovery is a classic example combining in vitro experiments using site-specifically modified oligonucleotides with an original in vivo observation of a mutator phenotype in repair-deficient cells (307, 308).
A simple method devised by Christopher Lawrence and J. Eugene LeClerc has gained wide acceptance for constructing site-specific lesions within single-stranded viral genomes. A single-stranded circular viral genome is linearized by cleavage within a hairpin using a restriction endonuclease, followed by annealing of a scaffold complementary to the cleaved termini, into which a phosphorylated oligonucleotide containing site-specific damage is ligated, with scaffold removal by heating in the presence of excess complement (309–311). Lawrence and co-workers have used these genomes to study TLS past photodimers and an abasic site in E. coli (309–315) and, with Roger Woodgate, explored the effect of Umu proteins and proofreading on TLS past these lesions (316–318). We adopted this method for the rapid construction of genomes containing highly unstable aflatoxin adducts using a uracilated scaffold, which is removed under gentle conditions by the combined activities of uracil-DNA-glycosylase and exonuclease III (319). Masaaki Moriya’s versatile shuttle phagemid pMS2 uses the methodology described above for genome construction (but with the scaffold annealed prior to hairpin cleavage) and allows one to study TLS in mammalian (COS-7 simian kidney) cells, as well as in E. coli, for a direct comparison of the cellular responses to a lesion in different environments using the same experimental system (320); the scaffold oligonucleotide is removed using the 3′ to 5′ exonuclease property of T4 DNA polymerase, which we have adopted into our protocols, as we found it to be more effective and to nick DNA less than exonuclease III. We also modified the hairpin cleavage construction technique by using two scaffolds, each of which bridges one end of the cleaved vector and modified oligonucleotide insert, but nothing opposes the lesion or its neighbors, thus providing identical ligation efficiencies regardless of lesion bulk or local sequence context, and assurance of signal from TLS events rather than from any residual scaffold (321).
While this perspective deals primarily with E. coli and mammalian studies, a method circumventing the need for genome construction altogether was introduced for studying lesion mutagenesis in yeast. On the basis of the discovery of the Sherman group (322, 323), the single-strand oligonucleotide-mediated yeast transformation method uses the electroporation of a single-stranded 26–30-mer oligonucleotide into yeast cells to restore a frameshift allele and has been used to study natural, unnatural, and oxidized abasic sites, uracil (which forms a natural abasic site in vivo upon glycosylase repair), a TT photoproduct, and unnatural lesions (324–327). Sequencing of revertants provides mutation data, and assuming that modified and unmodified control oligonucleotides integrate into the host chromosome equally, bypass data can be obtained by colony counting.
4.3. Assays for in Vivo Mutagenesis, Bypass, and Repair of Site-Specific Lesions
Once the genomes have been synthesized, they are introduced into cells by electroporation, calcium chloride, calcium phosphate, or lipofection methods. For lesion mutagenesis and repair studies, the goal is to get as many initial independent events as possible for statistical robustness, and it is our experience that electroporation is superior to the calcium method for the transformation of E. coli.
The in vivo mutagenicity of a lesion is measured by assessing the percentage of each type of base or frameshift mutation at the lesion site. The blocking power that a lesion has over the polymerase that tries to copy past it within the cell (trans-lesion synthesis/lesion bypass) is often scored as a decrease in survival, with genotoxicity measured as a decrease in the ability to form plaques or colonies. All site-specific lesion repair studies performed in vivo involve mutagenesis and bypass assays that indirectly monitor the event. Repair is inferred by a drop in the mutation frequency or an increase in trans-lesion synthesis when genomes assayed in repair-deficient cells are processed in their wild-type complement. A short list of groups making significant contributions to our understanding of how site-specific lesions are processed within cells is presented below.
Moriya and Grollman used a single-stranded vector bearing 8-oxoguanine to show a striking increase in G to T transversions in MutY-deficient E. coli, thus confirming elements of the “GO” system in vivo using site-specific mutagenesis (328). Their use of the pMS2/COS-7 system showed differential mutagenesis for propanoguanine, ethenocytosine, ethenoadenine, and (with Nicholas Geacintov) benzo[a]pyrene-N2-dG adducts as a function of the host cell and, for the latter, sequence context and chirality (329–332); differential oligonucleotide hybridization to individual colonies using probe sets specific for all outcomes provides mutation frequency and specificity, and positive clones can be further sequenced, if desired. Moriya and Grollman engineered yet another vector containing many desirable design elements, allowing for the study of lesions placed in the leading or lagging strand of plasmids replicated extrachromosomally in human cells, as well as in E. coli (333, 334). For mammalian analysis, plasmids from antibiotic-resistant cells are harvested, treated with DpnI to destroy genomes that have not replicated within the host, and can be treated with another restriction endonuclease to inactivate progeny from the strand complementary to the lesion prior to transformation into E. coli for mutational analysis. Because this region is mismatched, omission of the endonuclease will allow the amount of replication from each strand to be quantified, thus providing feedback on lesion-induced inhibition of replication. This versatile system has been used to measure the response of ethenoadenine, α- and γ-hydroxypropanoguanine (and related lesions), and a heptanone adduct of ethenocytosine (333–337).
Shibutani’s use of the pMS2/COS-7 system allowed him to determine the mutagenicity of α-tamoxifen-N2-guanine and estrogen-N2-guanine and -N6-adenine adducts (150, 338, 339). This system allowed him and Grollman to explore, within mammalian cells, such diverse lesions and topics as the effect of sequence context on the mutagenicity of N2-guanine adducts of (acetyl)aminofluorene and C8-adducts of the cooked food mutagen PhIP; the mutagenicity of N2-guanine adducts of acetylaminonaphthalene, benzo[a]pyrene, and 8-aminoguanine; and a head-to-head mutagenic comparison of 8-oxoguanine vs 8-oxoadenine (257, 340–347).
R. Stephen Lloyd evaluated the lethality (plaque forming or transforming ability) and mutagenicity (by differential oligonucleotide probe hybridization) of many alkylated lesions, mainly obtained by the Harris’, using the M13/E. coli technique of Lawrence (348–357) and the pMS2/COS-7 technique of Moriya (358–363). Some highlights found in E. coli were a dramatic effect of chirality and sequence context on the lethality of α-styrene oxide-N6-adenine, which is a block to pol III in vitro when in a 33-mer but much less so when in an intact M13; the β-isomers are neither toxic nor mutagenic (348, 350, 355). The S,S stereoisomer of butadiene-N2-guanine intrastrand cross-links is more mutagenic than the R,R isomer (354); the N1 deoxyinosine adduct resulting from reaction of 1,2-epoxy-3-butene with deoxyadenosine is 90% mutagenic, giving predominantly A to G mutations (356); DNA pol II appears responsible for the mutations caused by butadiene-N6-adenine intrastrand cross-links (357); and the butadiene-derived N3-deoxyuridine adduct is 97% mutagenic in COS-7 cells, giving C to T and C to A mutations (363).
Hiroyuki Kamiya and Eiko Ohtsuka performed studies on the mutagenicity of several lesions in the c-Ha-ras gene in mouse NIH3T3 cells (364–372), and later, Hiroyuki Kamiya and Hiroshi Kasai studied the mutagenicity of 2-hydroxyadenine, the (6-4) T-T photoproduct, and 5-formyluracil in simian COS-7 cells (373–375).
Edward Loechler (376) and Ashis Basu (135, 291, 377) performed pioneering work in the early days of site-specific mutagenesis while in this laboratory and have gone on to investigate other lesions and biological problems in their independent careers. Loechler has looked at the biological effects within E. coli of nitrogen mustard interstrand cross-links (378, 379) and of sequence context and stereochemistry of benzo[a]pyrene-N2-guanine adducts (380–389), combining observations from his single adduct studies with molecular modeling (390–396). Basu has investigated N2-guanine adducts of nitropyrene and mitomycin C and C8-guanine adducts of nitropyrene and ammonia (154, 397–401) in E. coli. He has also recently investigated N7-guanine adducts of mitomycin C, tandem 8-oxoguanine/abasic site damage, 8-oxopurines, bona fide ring-opened FAPY lesions, and C8-guanine adducts of nitropyrene in COS-7 monkey cells (402–405).
The plot of mutation type and frequency across a gene is called a mutational spectrum. Mutational spectra are usually nonuniform; that is, there are hot and cold spots (406). The reasons for this nonuniformity are not fully understood at the biochemical level, but several reasonable possibilities exist as follows: (i) At any given site, the formation of an adduct may be influenced by neighboring bases. (ii) Adducts in some contexts may be repaired better than in others. (iii) Polymerases may misreplicate adducts more or in a qualitiatively different manner in one context than in others. (iv) A mutation may be more easily selected or detected in some contexts than in others (perhaps because a particular mutant grows better than other clones). The tools of lesion-specific mutagenesis lend themselves well to help quantify the relative contributions of the four possibilities listed above, and one should be cognizant of the risk of generalizing a result obtained in only one sequence context.
Indeed, one of the more interesting applications of genomes containing specific DNA lesions has been the collection of studies aimed at determining the effect of local sequence context on the ways that DNA polymerases or repair proteins recognize lesions. While a comprehensive analysis on sequence context effects is beyond the scope of this perspective, we note a few studies of interest. Wallace generated consensus sequences for hotspot mutagenesis of 8-oxoguanine using randomized flanking bases and in vitro selection (267), and Grollman and Shibutani explicitly generated consensus sequences for acetylaminofluorene and aminofluorene mutagenesis in vivo by constructing individual genomes containing nearly half of the possible 16 nearest neighbors flanking the C8-guanine lesions, and transfecting these into mammalian COS-7 cells; progeny plasmids were treated with S1 nuclease to eliminate signal from unprocessed input DNA prior to their introduction into E. coli for mutant screening by 32P-probe hybridization (344). Our group was interested in finding out if consensus sequences existed in vivo for O6-methylguanine mutagenesis by the DNA polymerase or if such sequences existed that made the lesion refractory to repair by the methyltransferases Ada and Ogt. We took a nearest-neighbor approach in constructing the requisite 16 individual genomes but quickly realized that processing the data from five cell strains in duplicate would require the analysis of 160 samples. We concluded that a high-throughput method was needed that would allow analysis of the entire output population from each sample. Such a method would obviate the need to sequence over 16000 individual clones or count even more plaque hybridization spots requiring several probe sets to encompass all contexts for mutant identification (312, 348). The REAP procedure, which has become widely used in the field, was developed to address the need for such a high-throughput method. This method is described below.
4.4. REAP Assay for Determining the Mutation Frequency and Specificity of Lesions
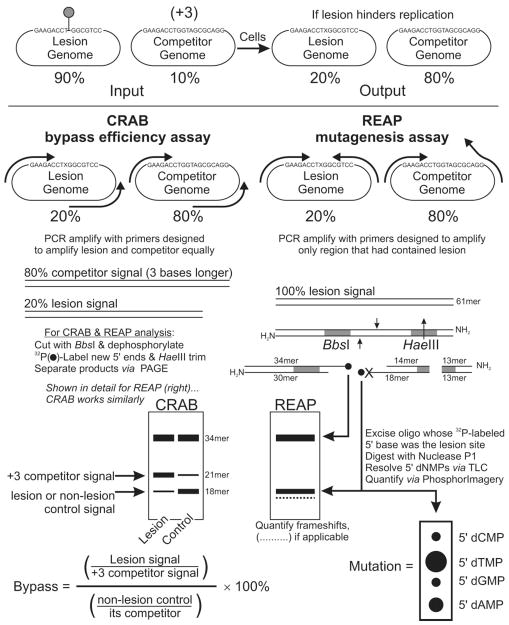
To overcome the obstacles described above, the approach that we took to obtain the base composition at the lesion site after biological processing involved incorporating our lesion of interest into the cleavage site for a type IIS restriction endonuclease, an enzyme that cleaves with surgical precision a fixed distance away from its recognition sequence. After restriction endonuclease cleavage of the progeny, the newly exposed 5′-phosphate associated with the site that had contained the lesion is postlabeled with 32P. The radiolabeled DNA is then trimmed down to a small size by another restriction endonuclease, gel purified, and digested to 5′-dNMPs, which are resolved by TLC and quantified by PhosphorImagery to obtain the mutation frequency and specificity. The Restriction Endonuclease And Postlabeling determination of mutation frequency (REAP) assay has evolved over the years to its present state, which is depicted in Figure 2 and described in detail with protocols in ref 407. Although Figure 2 depicts progeny from single-stranded DNA being analyzed, the use of PCR to amplify the region that had contained the lesion allows the assay to determine the mutation frequency and specificity of lesions from both single-stranded and double-stranded DNA genomes, which have been passaged through both E. coli or mammalian cells. Indeed, the REAP assay can extract data from lesions that have been absorbed intrachromosomally, and mutation data from RNA genomes containing RNA lesions can be obtained after reverse transcription of the lesion site. In addition to point mutations obtained by sampling from the entire progeny population, the nonphenotypic REAP procedure permits simultaneous statistically robust analysis of frameshifts.
Figure 2.
REAP assay for determining the mutation frequency and specificity of lesions (right) and CRAB assay for determining the replication blocking power of lesions (left).
Since its inception, the REAP assay has been used to define the coding specificities of over 50 lesions encompassing alkylative and oxidative damage, as well as unnatural atomically mutated bases; the lesions that have been published are shown in Figure 3 (321, 408–425). Using REAP, a number of large scale projects have been completed, and the results of those studies are highlighted as follows. First, REAP was used to define with high precision and accuracy the error frequency of the replicative DNA polymerase of E. coli as it attempts to copy O6-methylguanine (408); the polymerase nearly always reads the lesion as if it were an adenine regardless of sequence context. Repair, however, is highly sequence-dependent, with some sequences [e.g., 5′-A(O6-methylguanine)-3′] displaying conspicuously sluggish repair. Second, the high-throughput nature of the method allowed a host of guanine oxidation products to be analyzed in parallel, leading to the conclusion that these products are more highly mutagenic than 8-oxoguanine (409, 411, 413, 414). Similarly, the ability to study many lesions at the same time allowed characterization of the substrate requirements of the DNA repair enzyme AlkB, which was found to play a powerful protective role against certain alkylated bases, including etheno- and ethano-adenine lesions (412, 415, 417). Third, the use of REAP with a multiplexed set of cell strains in which various polymerases were expressed in combinations allowed a detailed picture to be constructed of the genetic requirements for lesion replication fidelity of a large array of oxidized DNA bases (419). Fourth, the method has been used to probe the fidelity of replication of a set of gradually expanded nonpolar thymine analogues (where both oxygens are simultaneously replaced by H, F, Cl, Br, or I); these studies provide basic data that help explain which structural features of normal DNA bases are critical determinants of replication fidelity and evolutionary adaptability (416).
Figure 3.
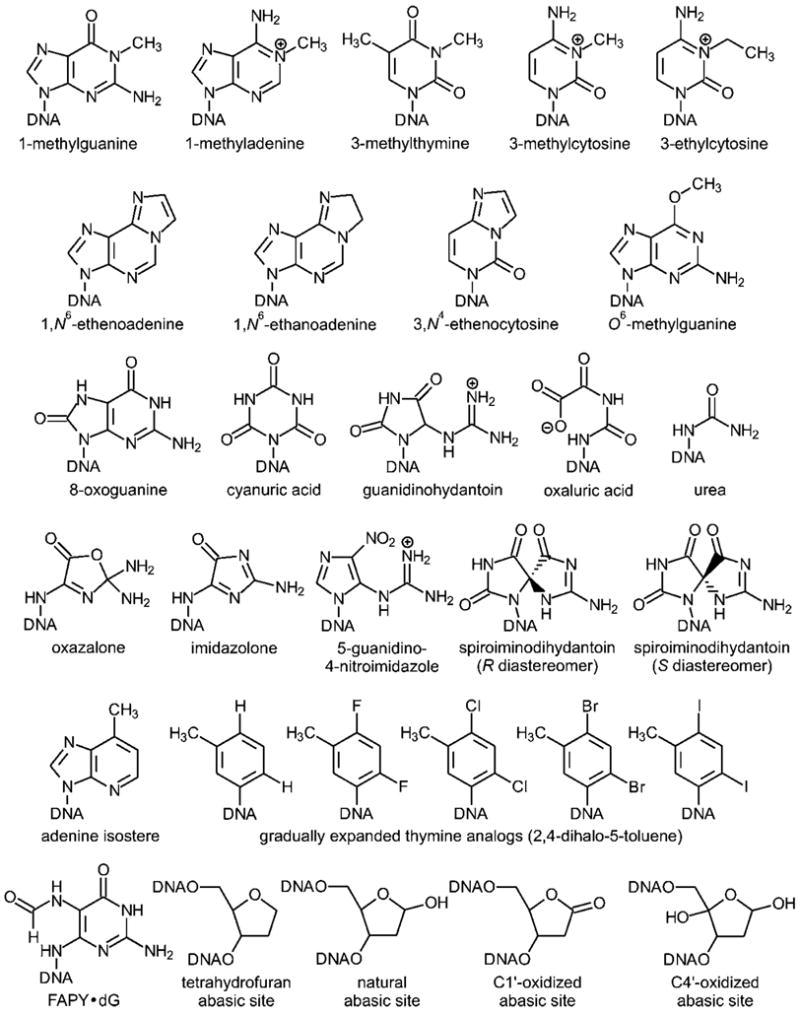
Lesions analyzed by the REAP and CRAB assays.
While designed primarily for the generation of in vivo results on lesion mutagenicity, an in vitro version of REAP can be a powerful tool to assess lesion integrity after genome construction or after exposure to specific chemical or enzymatic environments (412). After a scaffold is annealed opposite the lesion in a single-stranded genome (duplex genomes would need no such scaffold), the material is fed into the assay at the BbsI digestion step, with the ultimate resolution and quantification of the original modified 5′-dNMP from its 5′-dNMP conversion, degradation, or contamination products. As one example, REAP was used to study the Dimroth rearrangement of 1-methyladenine to 6-methyladenine (412). One can envision such an in vitro assay for monitoring the direct reversal repair of alkylated oligonucleotides, such as O6-methylguanine by Ada, Ogt, or O6-methylguanine-DNA-methyltransferase or 1-methyladenine by AlkB or its human homologues. By tracking the strand opposite the lesion, one may eventually obtain point and frameshift mutations induced by chosen DNA polymerases.
4.5. CRAB Assay for Determining the Polymerase Bypass Efficiency of Lesions
The ability to measure mutagenesis in vivo is only half the story. Indeed, lesions that are not mutagenic to the cell can still be extremely toxic (412, 417). Traditional experiments measure the ability of a lesion to hinder DNA replication by a reduction in plaque or colony formation, with respect to a nonlesion control, after immediate plating of transformed cells. Oftentimes, however, the transfection efficiency of cells, be they E. coli, mammalian, or yeast, is variable in practice. To address this problem, the Competitive Replication of Adduct Bypass (CRAB) assay was developed (Figure 2), in which a lesion-bearing genome is mixed with a nonlesion internal standard genome prior to transfection. In the initial version of the assay, plaques from lesion-induced point mutations would be dark or light blue, while the internal standard would be clear (412). A drop in blue plaques with respect to clears would indicate blocks to trans-lesion replication. The work also showed that such survival assays based on immediate plating of the transformation mixture can underestimate the blocking power of a DNA lesion; indeed, a lesion can be a block to replication, but if it is eventually traversed prior to host destruction of the vector, a colony (nonblocking “digital” signal) will form. Analysis of progeny after growth of the mixture in liquid culture, where both lesion and internal standard genomes can compete, gives a truer dynamic “analog” signal for lesion bypass determination. Two limitations of the phenotypic version of this assay were that many plaques had to be counted manually, and lesions that generate frameshifts would artificially decrease the bypass signal. To overcome these obstacles, the CRAB assay for determination of lesion bypass quickly evolved to its present state, depicted on the left-hand side of Figure 2 (407). Salient points are (i) that the proportion of signal from lesion and competitor genomes grown in liquid culture is obtained using REAP methodology but with primers that amplify lesion and competitor progeny equally; (ii) the stored progeny from one sample can be analyzed for both bypass and mutagenesis; (iii) the same input mixture containing lesion and competitor is used for an entire panel of repair- and polymerase-deficient and -proficient cell strains, instilling confidence that signal changes result from cellular variables and not from variations in the input formulation; (iv) the input asymmetry of lesion to competitor provides a reproducible, quantifiable signal for strongly blocking lesions, while an equal mixture would do so for nonblocking lesions.
A number of discoveries have been made using the CRAB assay for lesion bypass in vivo. As a first example, the replication of a series of bases possessing small alkyl functionalities provided a detailed picture of the features of lesion structure that allow polymerase bypass in vivo and indicate that some are well-repaired (412). These studies complemented the REAP results described above. In addition, replication of lesion-containing genomes in cells with known defects in repair (e.g., AlkB) or altered replication states (e.g., different SOS polymerases expressed) defined substrates for repair and for specific polymerases. It was found, for example, that certain replication-inhibiting etheno lesions, which form as a consequence of inflammation, are good substrates for AlkB in vivo (415); hence, the absence of AlkB results in severe genotoxicity. As a second example, it was demonstrated that the potent replication block of 1,N6-ethanoadenine, a major adduct formed by the antitumor agent BCNU, is relieved in AlkB-proficient E. coli (417). This discovery led to in vitro characterization of the mechanism of the “repair” reaction, which was found to be unique. The repair enzyme, using an oxidative route, converted the highly toxic cyclic adduct into a linear chain that was apparently readily bypassed in vivo.
In vitro biochemical studies, such as that of AlkB on ethanoadenine, nicely complement in vivo findings, especially for lesion repair. The unsaturated derivative of ethanoadenine, 1,N6-ethenoadenine, is a case in point. Using ESI-TOF mass spectrometry on site-specifically modified oligonucleotides treated with AlkB, it was possible to observe the accumulation of intermediates and product along the pathway of 1,N6-ethenoadenine repair by the enzyme. As was the case with ethanoadenine, it was possible to delineate the reaction path, which involved, in this case, direct reversal of base damage by epoxidation of the etheno double bond, followed by epoxide hydrolysis to form a glycol, which is removed as glyoxal, with restoration of the adenine within the oligonucleotide (415). Yinsheng Wang has recently adopted and adapted the CRAB and REAP assays, using a mass spectrometry platform for quantifying the genotoxicity and mutagenicity of a guanine-cytosine intrastrand cross-link in E. coli (128).
5. Future Directions
5.1. Intrachromosomal Probes for Mutagenesis
To look to the future, it behooves us to reflect on the past, picking up on areas that were pursued incompletely or neglected. Site-specific mutagenesis of DNA lesions has been performed mostly using an extrachromosomal approach, whereby an oligonucleotide is ligated into a single- or double-stranded vector containing an origin of replication, which is replicated independently of the host genome. Many important discoveries have been made using extrachromosomal probes; however, these vectors generally replicate more quickly than host chromosomes. They may, therefore, underestimate the effect of DNA repair on damage within a host chromosome, since the vector lesion site encounters a DNA polymerase more quickly, reducing time for repair, which is measured as an increase in lesion bypass (toxicity) and/or a decrease in mutation. On the other hand, mammalian chromosomes have a compacted nucleosome structure, which may shield the lesion from repair, and data from the more repair-accessible vector may overestimate the effect of DNA repair on damage within a host chromosome. Also, it may remain unknown if there are differences between a vector and a host chromosome in the interaction of a lesion with the DNA polymerase holoenzyme and its accessory proteins. For reasons detailed above, intrachromosomal probes may be the most biologically relevant system to study replication and repair of DNA lesions. We and the Guengerich laboratory performed such experiments by transfecting vectors containing site-specific O6-methylguanine, O4-methylthymine, or 1,N2-ethenoguanine lesions into Chinese hamster ovary cells deficient in methyltransferase or nucleotide excision repair, whereby the vector could not replicate within the host unless it integrated into the host chromosome (426–428). Intrachromosomal analysis of mutations should be a more widely used tool.
5.2. RNA Repair of Site-Specific Lesions in Vivo
The discovery that 1-methyladenine in RNA can be repaired by AlkB suggests an intriguing question and poses a significant research opportunity (429, 430). Some human transcripts are both long in size and long-lived temporally; examples include those coding for the muscle protein titin (34,350 amino acids) or for the β-and γ-globin transcripts, which have half-lives between 17 and 80 h, depending on induction (431, 432). After the large energy commitment to make such transcripts, it would be disadvantageous if chemical damage rendered an RNA species unreadable or misreadable. It seems reasonable to assume that the cell may use RNA repair as a defense against RNA adducts. An in vivo system in which RNA lesions site-specifically incorporated into biologically active RNA species would allow the hypothesis that RNA repair exists as a physiologically relevant process to be tested.
5.3. Kinetics of Site-Specific Lesion Replication and Repair in Vivo
The biological studies to this point on DNA lesions have defined the mutagenic and toxic properties of lesions. In the future, once the properties of the lesion are established, one could use the lesion as a probe to define the properties of biological systems, such as the kinetics of replication and repair, in vivo. For example, a lesion of predefined mutagenicity and toxicity could be introduced at several sites in a genome, and each site would be ever more distal to an origin of replication. Lesions distal to an origin or downstream from an impediment such as a second lesion or a secondary structural element might be expected to show lower mutation frequencies and lower toxicities than lesions that are more proximal to the origin, due to the increased time that they have for being repaired prior to being replicated. Using the highly quantitative methodology that now exists, the timing and hence the kinetics of replication and repair could be clocked in vivo by using mutagenic and toxicity end points. Such studies would help to establish a more detailed, granular image of the responses of cells to stress than exists at present.
5.4. Translational Aspects of Site-Specific Mutagenesis Studies
In any field, it is critical to identify where possible the translational elements that relate most directly to improvement of the human condition. Lesions of anticancer drugs kill cells, but they also can cause mutations that can engender second tumors in cancer patients treated with chemotherapy. Studies of lesion mutagenesis have helped and can continue to help the drug development process, as shown in studies by Yarema et al. (433, 434). Additionally, in work by Loeb and co-workers, lesions of established mutagenicity have been used to dope the nucleotide precursor pool of cells attempting to replicate HIV. The viral genomes accumulated mutations at an accelerated pace that apparently caused the virus to exceed the error threshold for viability. This work led to a novel antiviral therapy and opened a field termed “lethal mutagenesis”. Finally, lesion-specific mutagenesis has and will continue to be of central importance to field workers who need biomarkers predictive of carcinogenic risk. For example, the work of Thomas Kensler and John Groopman that established the aflatoxin-N7-guanine adduct in urine as an excellent biomarker for hepatocellular carcinoma risk in human populations benefited from and is mechanistically anchored to genetic studies showing that the aflatoxin adducts are indeed progenitors of the types of genetic change that occur in people with clinical disease (194, 435).
Twenty years ago, it would seem out of the question to process more than a few lesions of interest in vivo, yet alone in different sequence contexts and different cell strains. The progress described herein has increased the productivity of the field by several orders of magnitude, and fundamental discoveries on how cells respond to damage are being made at a staggering rate. It is clear that this field is robust and has a bright and dynamic future. It will be exciting to see what the next 20 years have in store.
Acknowledgments
We thank the National Institutes of Health (CA26731 and CA80024) for financial support. We dedicate this perspective to the memories of Drs. Bea Singer and Dezider Grunberger, who helped bring this field into existence and move it ahead with gentle nudges and, when necessary, timely shoves.
References
- 1.Basu AK, Essigmann JM. Site-specifically modified oligodeoxynucleotides as probes for the structural and biological effects of DNA-damaging agents. Chem Res Toxicol. 1988;1:1–18. doi: 10.1021/tx00001a001. [DOI] [PubMed] [Google Scholar]
- 2.Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 3.Smith HO, Hutchison CA, III, Pfannkoch C, Venter JC. Generating a synthetic genome by whole genome assembly: φX174 Bacteriophage from synthetic oligonucleotides. Proc Natl Acad Sci USA. 2003;100:15440–15445. doi: 10.1073/pnas.2237126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kodumal SJ, Patel KG, Reid R, Menzella HG, Welch M, Santi DV. Total synthesis of long DNA sequences: Synthesis of a contiguous 32-kb polyketide synthase gene cluster. Proc Natl Acad Sci USA. 2004;101:15573–15578. doi: 10.1073/pnas.0406911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fowler KW, Büchi G, Essigmann JM. Synthesis and characterization of an oligonucleotide containing a carcinogen-modified base: O6-methylguanine. J Am Chem Soc. 1982;104:1050–1054. [Google Scholar]
- 6.Matteucci MD, Caruthers MH. Synthesis of deoxyoligonucleotides on a polymer support. J Am Chem Soc. 1981;103:3185–3191. [PubMed] [Google Scholar]
- 7.Beaucage SL, Caruthers MH. Deoxynucleoside phosphoramidites. A new class of key intermediates for deoxypoly-nucleotide synthesis. Tetrahedron Lett. 1981;22:1859–1862. [Google Scholar]
- 8.McBride LJ, Caruthers MH. Nucleotide chemistry. X An investigation of several deoxynucleoside phosphoramidites useful for synthesizing deoxyoligonucleotides. Tetrahedron Lett. 1983;24:245–248. [Google Scholar]
- 9.Alvarado-Urbina G, Sathe GM, Liu WC, Gillen MF, Duck PD, Bender R, Ogilvie KK. Automated synthesis of gene fragments. Science. 1981;214:270–274. doi: 10.1126/science.6169150. [DOI] [PubMed] [Google Scholar]
- 10.Hunkapiller M, Kent S, Caruthers M, Dreyer W, Firca J, Giffin C, Horvath S, Hunkapiller T, Tempst P, Hood L. A microchemical facility for the analysis and synthesis of genes and proteins. Nature. 1984;310:105–111. doi: 10.1038/310105a0. [DOI] [PubMed] [Google Scholar]
- 11.Guram AS, Rennels RA, Buchwald SL. A simple catalytic method for the conversion of aryl bromides to arylamines. Angew Chem, Int Ed. 1995;34:1348–1350. [Google Scholar]
- 12.Louie J, Hartwig JF. Palladium-catalyzed synthesis of arylamines from aryl halides. Mechanistic studies lead to coupling in the absence of tin reagents. Tetrahedron Lett. 1995;36:3609–3612. [Google Scholar]
- 13.Wolfe JP, Wagaw S, Buchwald SL. An improved catalyst system for aromatic carbon–nitrogen bond formation: The possible involvement of bis(phosphine) palladium complexes as key intermediates. J Am Chem Soc. 1996;118:7215–7216. [Google Scholar]
- 14.Driver MS, Hartwig JF. A second-generation catalyst for aryl halide amination: Mixed secondary amines from aryl halides and primary amines catalyzed by (DPPF)PdCl2. J Am Chem Soc. 1996;118:7217–7218. [Google Scholar]
- 15.Bodepudi V, Shibutani S, Johnson F. Synthesis of 2′-deoxy-7,8-dihydro-8-oxoguanosine and 2′-deoxy-7,8-dihydro-8-oxoadenosine and their incorporation into oligomeric DNA. Chem Res Toxicol. 1992;5:608–617. doi: 10.1021/tx00029a004. [DOI] [PubMed] [Google Scholar]
- 16.Rieger RA, Iden CR, Gonikberg E, Johnson F. 8-Amino-2′-deoxyguanosine incorporation into oligomeric DNA. Nucleosides Nucleotides. 1999;18:73–88. doi: 10.1080/07328319908045595. [DOI] [PubMed] [Google Scholar]
- 17.Zhang W, Rieger R, Iden C, Johnson F. Synthesis of 3,N4-etheno, 3,N4-ethano, and 3-(2-hydroxyethyl) derivatives of 2′-deoxycytidine and their incorporation into oligomeric DNA. Chem Res Toxicol. 1995;8:148–156. doi: 10.1021/tx00043a020. [DOI] [PubMed] [Google Scholar]
- 18.Bonala RR, Rieger RA, Shibutani S, Grollman AP, Iden CR, Johnson F. 3,N4-ethano-2′-deoxycytidine: Chemistry of incorporation into oligomeric DNA and reassessment of miscoding potential. Nucleic Acids Res. 1999;27:4725–4733. doi: 10.1093/nar/27.24.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Y, Torres MC, Iden CR, Johnson F. Regioselective synthesis of 1,N2-etheno-2′-deoxyguanosine and its generation in oligomeric DNA. Chem Res Toxicol. 2003;16:708–714. doi: 10.1021/tx025659s. [DOI] [PubMed] [Google Scholar]
- 20.Takeshita M, Chang CN, Johnson F, Will S, Grollman AP. Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J Biol Chem. 1987;262:10171–10179. [PubMed] [Google Scholar]
- 21.Khullar S, Varaprasad CV, Johnson F. Postsynthetic generation of a major acrolein adduct of 2′-deoxyguanosine in oligomeric DNA. J Med Chem. 1999;42:947–950. doi: 10.1021/jm980605u. [DOI] [PubMed] [Google Scholar]
- 22.Bonala RR, Torres MC, Attaluri S, Iden CR, Johnson F. Incorporation of N2-deoxyguanosine metabolic adducts of 2-aminonaphthalene and 2-aminofluorene into oligomeric DNA. Chem Res Toxicol. 2005;18:457–465. doi: 10.1021/tx0497907. [DOI] [PubMed] [Google Scholar]
- 23.Johnson F, Bonala R, Tawde D, Torres MC, Iden CR. Efficient synthesis of the benzo[a]pyrene metabolic adducts of 2′-deoxyguanosine and 2′-deoxyadenosine and their direct incorporation into DNA. Chem Res Toxicol. 2002;15:1489–1494. doi: 10.1021/tx0256174. [DOI] [PubMed] [Google Scholar]
- 24.Bonala R, Torres MC, Iden CR, Johnson F. Synthesis of the PhIP adduct of 2′-deoxyguanosine and its incorporation into oligomeric DNA. Chem Res Toxicol. 2006;19:734–738. doi: 10.1021/tx0600191. [DOI] [PubMed] [Google Scholar]
- 25.Laxmi YRS, Suzuki N, Dasaradhi L, Johnson F, Shibutani S. Preparation of oligodeoxynucleotides containing a diaste-reoisomer of α-(N2-2′-deoxyguanosinyl)tamoxifen by phosphoramidite chemical synthesis. Chem Res Toxicol. 2002;15:218–225. doi: 10.1021/tx0101494. [DOI] [PubMed] [Google Scholar]
- 26.Kohda K, Tsunomoto H, Minoura Y, Tanabe K, Shibutani S. Synthesis, miscoding specificity, and thermodynamic stability of oligodeoxynucleotide containing 8-methyl-2′-deoxyguanosine. Chem Res Toxicol. 1996;9:1278–1284. doi: 10.1021/tx9601059. [DOI] [PubMed] [Google Scholar]
- 27.Kohda K, Tsunomoto H, Kasamatsu T, Sawamura F, Terashima I, Shibutani S. Synthesis and miscoding specificity of oligodeoxynucleotide containing 8-phenyl-2′-deoxyguanosine. Chem Res Toxicol. 1997;10:1351–1358. doi: 10.1021/tx970111k. [DOI] [PubMed] [Google Scholar]
- 28.Terashima I, Matsuda T, Fang TW, Suzuki N, Kobayashi J, Kohda K, Shibutani S. Miscoding potential of the N2-ethyl-2′-deoxyguanosine DNA adduct by the exonuclease-free Klenow fragment of Escherichia coli DNA polymerase I. Biochemistry. 2001;40:4106–4114. doi: 10.1021/bi002719p. [DOI] [PubMed] [Google Scholar]
- 29.Laxmi YRS, Suzuki N, Kim SY, Shibutani S. Synthesis of oligodeoxynucleotides containing a single diastereoisomer of α-(N2-2′-deoxyguanosinyl)-N-desmethyltamoxifen. Bioorg Med Chem Lett. 2004;14:4051–4054. doi: 10.1016/j.bmcl.2004.05.030. [DOI] [PubMed] [Google Scholar]
- 30.Schnetz-Boutaud NC, Mao H, Stone MP, Marnett LJ. Synthesis of oligonucleotides containing the alkali-labile pyrimidopurinone adduct, M1G. Chem Res Toxicol. 2000;13:90–95. doi: 10.1021/tx990141i. [DOI] [PubMed] [Google Scholar]
- 31.Sowers LC, Beardsley GP. Synthesis of oligonucleotides containing 5-(hydroxymethyl)-2′-deoxyuridine at defined sites. J Org Chem. 1993;58:1664–1665. doi: 10.1021/jo00059a011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujimoto J, Tran L, Sowers LC. Synthesis and cleavage of oligodeoxynucleotides containing a 5-hydroxyuracil residue at a defined site. Chem Res Toxicol. 1997;10:1254–1258. doi: 10.1021/tx970102b. [DOI] [PubMed] [Google Scholar]
- 33.Tardy-Planechaud S, Fujimoto J, Lin SS, Sowers LC. Solid phase synthesis and restriction endonuclease cleavage of oligodeoxynucleotides containing 5-(hydroxymethyl)-cytosine. Nucleic Acids Res. 1997;25:553–558. doi: 10.1093/nar/25.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang JI, Jr, Burdzy A, Liu P, Sowers LC. Synthesis and characterization of oligonucleotides containing 5-chlorocytosine. Chem Res Toxicol. 2004;17:1236–1244. doi: 10.1021/tx0498962. [DOI] [PubMed] [Google Scholar]
- 35.Romieu A, Gasparutto D, Cadet J. Synthesis and characterization of oligonucleotides containing 5′,8-cyclopurine 2′-deoxyribonucleosides: (5′R)-5′,8-cyclo-2′-deoxyadenosine, (5′S)-5′,8-cyclo-2′-deoxyguanosine, and (5′R)-5′,8-cyclo-2′-deoxyguanosine. Chem Res Toxicol. 1999;12:412–421. doi: 10.1021/tx9802668. [DOI] [PubMed] [Google Scholar]
- 36.Gasparutto D, Da Cruz S, Bourdat AG, Jaquinod M, Cadet J. Synthesis and biochemical properties of cyanuric acid nucleoside-containing DNA oligomers. Chem Res Toxicol. 1999;12:630–638. doi: 10.1021/tx980255e. [DOI] [PubMed] [Google Scholar]
- 37.Gasparutto D, Ait-Abbas M, Jaquinod M, Boiteux S, Cadet J. Repair and coding properties of 5-hydroxy-5-methylhydantoin nucleosides inserted into DNA oligomers. Chem Res Toxicol. 2000;13:575–584. doi: 10.1021/tx000005+. [DOI] [PubMed] [Google Scholar]
- 38.Guerniou V, Gasparutto D, Sauvaigo S, Favier A, Cadet J. New synthesis of 5-carboxy-2′-deoxyuridine and its incorporation into synthetic oligonucleotides. Nucleosides, Nucleotides Nucleic Acids. 2003;22:1073–1075. doi: 10.1081/NCN-120022739. [DOI] [PubMed] [Google Scholar]
- 39.Gasparutto D, Cognet S, Roussel S, Cadet J. Synthesis of a convenient thymidine glycol phosphoramidite monomer and its site-specific incorporation into DNA fragments. Nucleosides, Nucleotides Nucleic Acids. 2005;24:1831–1842. doi: 10.1080/15257770500267279. [DOI] [PubMed] [Google Scholar]
- 40.Iwai S. Synthesis of thymine glycol containing oligonucleotides from a building block with the oxidized base. Angew Chem, Int Ed. 2000;39:3874–3876. doi: 10.1002/1521-3773(20001103)39:21<3874::AID-ANIE3874>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 41.Iwai S. Synthesis and thermodynamic studies of oligonucleotides containing the two isomers of thymine glycol. Chem Eur J. 2001;7:4343–4351. doi: 10.1002/1521-3765(20011015)7:20<4343::aid-chem4343>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 42.Shimizu T, Manabe K, Yoshikawa S, Kawasaki Y, Iwai S. Preferential formation of (5S,6R)-thymine glycol for oligodeoxyribonucleotide synthesis and analysis of drug binding to thymine glycol-containing DNA. Nucleic Acids Res. 2006;34:313–321. doi: 10.1093/nar/gkj443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iwai S, Shimizu M, Kamiya H, Ohtsuka E. Synthesis of a phosphoramidite coupling unit of the pyrimidine (6–4) pyrimidone photoproduct and its incorporation into oligodeoxynucleotides. J Am Chem Soc. 1996;118:7642–7643. [Google Scholar]
- 44.Mizukoshi T, Hitomi K, Todo T, Iwai S. Studies on the chemical synthesis of oligonucleotides containing the (6–4) photoproduct of thymine-cytosine and its repair by (6–4) photolyase. J Am Chem Soc. 1998;120:10634–10642. [Google Scholar]
- 45.Yamamoto J, Hitomi K, Todo T, Iwai S. Chemical synthesis of oligodeoxyribonucleotides containing the Dewar valence isomer of the (6–4) photoproduct and their use in (6–4) photolyase studies. Nucleic Acids Res. 2006;34:4406–4415. doi: 10.1093/nar/gkl572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takasawa K, Masutani C, Hanaoka F, Iwai S. Chemical synthesis and translesion replication of a cis–syn cyclobutane thymine–uracil dimer. Nucleic Acids Res. 2004;32:1738–1745. doi: 10.1093/nar/gkh342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giese B, Erdmann P, Schaefer T, Schwitter U. Synthesis of a 4′-selenated 2-deoxyadenosine derivative: A novel precursor suitable for the preparation of modified oligonucleotides. Synthesis. 1994:1310–1312. [Google Scholar]
- 48.Giese B, Imwinkelried P, Petretta M. Synthesis of a modified thymidine and preparation of precursors of oligodeoxyri-bonucleotide radicals. Synlett. 1994:1003–1004. [Google Scholar]
- 49.Giese B, Beyrich-Graf X, Erdmann P, Giraud L, Imwinkelried P, Müeller SN, Schwitter U. Cleavage of single-stranded 4′-oligonucleotide radicals in the presence of O2. J Am Chem Soc. 1995;117:6146–6147. [Google Scholar]
- 50.Giese B, Beyrich-Graf X, Erdmann P, Petretta M, Schwitter U. The chemistry of single-stranded 4′-DNA radicals: Influence of the radical precursor on anaerobic and aerobic strand cleavage. Chem Biol. 1995;2:367–375. doi: 10.1016/1074-5521(95)90217-1. [DOI] [PubMed] [Google Scholar]
- 51.Tronche C, Goodman BK, Greenberg MM. DNA damage induced via independent generation of the radical resulting from formal hydrogen atom abstraction from the C1′-position of a nucleotide. Chem Biol. 1998;5:263–271. [PubMed] [Google Scholar]
- 52.Hwang JT, Greenberg MM. Kinetics and stereoselectivity of thiol trapping of deoxyuridin-1′-yl in biopolymers and their relationship to the formation of premutagenic α-deoxynucleotides. J Am Chem Soc. 1999;121:4311–4315. [Google Scholar]
- 53.Kim J, Gil JM, Greenberg MM. Synthesis and characterization of oligonucleotides containing the C4′-oxidized abasic site produced by bleomycin and other DNA damaging agents. Angew Chem, Int Ed. 2003;42:5882–5885. doi: 10.1002/anie.200352102. [DOI] [PubMed] [Google Scholar]
- 54.Kim J, Kreller CR, Greenberg MM. Preparation and analysis of oligonucleotides containing the C4′-oxidized abasic site and related mechanistic probes. J Org Chem. 2005;70:8122–8129. doi: 10.1021/jo0512249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greenberg MM, Barvian MR, Cook GP, Goodman BK, Matray TJ, Tronche C, Venkatesan H. DNA damage induced via 5,6-dihydrothymid-5-yl in single-stranded oligonucleotides. J Am Chem Soc. 1997;119:1828–1839. [Google Scholar]
- 56.Tallman KA, Greenberg MM. Oxygen-dependent DNA damage amplification involving 5,6-dihydrothymidin-5-yl in a structurally minimal system. J Am Chem Soc. 2001;123:5181–5187. doi: 10.1021/ja010180s. [DOI] [PubMed] [Google Scholar]
- 57.Hong IS, Carter KN, Greenberg MM. Evidence for glycosidic bond rotation in a nucleobase peroxyl radical and its effect on tandem lesion formation. J Org Chem. 2004;69:6974–6978. doi: 10.1021/jo0492158. [DOI] [PubMed] [Google Scholar]
- 58.Hong IS, Greenberg MM. DNA interstrand cross-link formation initiated by reaction between singlet oxygen and a modified nucleotide. J Am Chem Soc. 2005;127:10510–10511. doi: 10.1021/ja053493m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hong IS, Greenberg MM. Efficient DNA interstrand cross-link formation from a nucleotide radical. J Am Chem Soc. 2005;127:3692–3693. doi: 10.1021/ja042434q. [DOI] [PubMed] [Google Scholar]
- 60.Hong IS, Ding H, Greenberg MM. Oxygen independent DNA interstrand cross-link formation by a nucleotide radical. J Am Chem Soc. 2006;128:485–491. doi: 10.1021/ja0563657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hong IS, Carter KN, Sato K, Greenberg MM. Characterization and mechanism of formation of tandem lesions in DNA by a nucleobase peroxyl radical. J Am Chem Soc. 2007;129:4089–4098. doi: 10.1021/ja0692276. [DOI] [PubMed] [Google Scholar]
- 62.Ding H, Greenberg MM. γ-Radiolysis and hydroxyl radical produce interstrand cross-links in DNA involving thymidine. Chem Res Toxicol. 2007;20:1623–1628. doi: 10.1021/tx7002307. [DOI] [PubMed] [Google Scholar]
- 63.Haraguchi K, Delaney MO, Wiederholt CJ, Sambandam A, Hantosi Z, Greenberg MM. Synthesis and characterization of oligodeoxynucleotides containing formamidopyrimidine lesions and nonhydrolyzable analogs. J Am Chem Soc. 2002;124:3263–3269. doi: 10.1021/ja012135q. [DOI] [PubMed] [Google Scholar]
- 64.Jiang YL, Wiederholt CJ, Patro JN, Haraguchi K, Greenberg MM. Synthesis of oligonucleotides containing Fapy·dG (N6-(2-deoxy-α,β-D-erythropentofuranosyl)-2,6-diamino-4-hydroxy-5-formamidopyrimidine) using a 5′-dimethoxytrityl dinucleotide phosphoramidite. J Org Chem. 2005;70:141–149. doi: 10.1021/jo048253o. [DOI] [PubMed] [Google Scholar]
- 65.Hwang JT, Tallman KA, Greenberg MM. The reactivity of the 2-deoxyribonolactone lesion in single-stranded DNA and its implication in reaction mechanisms of DNA damage and repair. Nucleic Acids Res. 1999;27:3805–3810. doi: 10.1093/nar/27.19.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim J, Weledji YN, Greenberg MM. Independent generation and characterization of a C2′-oxidized abasic site in chemically synthesized oligonucleotides. J Org Chem. 2004;69:6100–6104. doi: 10.1021/jo049033d. [DOI] [PubMed] [Google Scholar]
- 67.Matray TJ, Greenberg MM. Site-specific incorporation of the alkaline labile, oxidative stress product (5R)-5,6-dihydro-5-hydroxythymidine in an oligonucleotide. J Am Chem Soc. 1994;116:6931–6932. [Google Scholar]
- 68.Nechev LV, Kozekov ID, Brock AK, Rizzo CJ, Harris TM. DNA adducts of acrolein: Site-specific synthesis of an oligodeoxynucleotide containing 6-hydroxy-5,6,7,8-tetrahydro-pyrimido[1,2-α]purin-10(3H)-one, an acrolein adduct of guanine. Chem Res Toxicol. 2002;15:607–613. doi: 10.1021/tx010181y. [DOI] [PubMed] [Google Scholar]
- 69.Kowalczyk A, Harris CM, Harris TM. Synthesis and characterization of oligodeoxynucleotides containing an N1 β-hydroxyalkyl adduct of 2′-deoxyinosine. Chem Res Toxicol. 2001;14:746–753. doi: 10.1021/tx010025r. [DOI] [PubMed] [Google Scholar]
- 70.Goodenough AK, Kozekov ID, Zang H, Choi JY, Guengerich FP, Harris TM, Rizzo CJ. Site specific synthesis and polymerase bypass of oligonucleotides containing a 6-hydroxy-3,5,6,7-tetrahydro-9H-imidazo[1,2-α]purin-9-one base, an intermediate in the formation of 1,N2-etheno-2′Gr 49-deoxyguanosine. Chem Res Toxicol. 2005;18:1701–1714. doi: 10.1021/tx050141k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Elmquist CE, Stover JS, Wang Z, Rizzo CJ. Site-specific synthesis and properties of oligonucleotides containing C8-deoxyguanosine adducts of the dietary mutagen IQ. J Am Chem Soc. 2004;126:11189–11201. doi: 10.1021/ja0487022. [DOI] [PubMed] [Google Scholar]
- 72.Stover JS, Rizzo CJ. Synthesis of oligonucleotides containing the N2-deoxyguanosine adduct of the dietary carcinogen 2-amino-3-methylimidazo[4,5-f]quinoline. Chem Res Toxicol. 2007 doi: 10.1021/tx7002867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Taylor JS, Brockie IR, O’Day CL. A building block for the sequence-specific introduction of cis-syn thymine dimers into oligonucleotides. Solid-phase synthesis of TpT[c,s]pTpT. J Am Chem Soc. 1987;109:6735–6742. [Google Scholar]
- 74.Taylor JS, Brockie IR. Synthesis of a trans-syn thymine dimer building block. Solid-phase synthesis of CG-TAT[t,s]TATGC. Nucleic Acids Res. 1988;16:5123–5136. doi: 10.1093/nar/16.11.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ordoukhanian P, Taylor JS. Solid phase-supported thymine dimers for the construction of dimer-containing DNA by combined chemical and enzymic synthesis: A potentially general method for the efficient incorporation of modified nucleotides into DNA. Nucleic Acids Res. 1997;25:3783–3786. doi: 10.1093/nar/25.19.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lingbeck JM, Taylor JS. Preparation and characterization of DNA containing a site-specific nonadjacent cyclobutane thymine dimer of the type implicated in UV-induced −1 frameshift mutagenesis. Biochemistry. 1999;38:13717–13724. doi: 10.1021/bi991035i. [DOI] [PubMed] [Google Scholar]
- 77.Morningstar ML, Kreutzer DA, Essigmann JM. Synthesis of oligonucleotides containing two putatively mutagenic DNA lesions: 5-Hydroxy-2′-deoxyuridine and 5-hydroxy-2′-deoxycytidine. Chem Res Toxicol. 1997;10:1345–1350. doi: 10.1021/tx970052a. [DOI] [PubMed] [Google Scholar]
- 78.Kobertz WR, Essigmann JM. Solid-phase synthesis of oligonucleotides containing a site-specific psoralen derivative. J Am Chem Soc. 1997;119:5960–5961. [Google Scholar]
- 79.Harwood EA, Sigurdsson ST, Edfeldt NBF, Reid BR, Hopkins PB. Chemical synthesis and preliminary structural characterization of a nitrous acid interstrand cross-linked duplex DNA. J Am Chem Soc. 1999;121:5081–5082. [Google Scholar]
- 80.Lakshman MK, Sayer JM, Jerina DM. Chemical synthesis of a bay-region polycyclic aromatic hydrocarbon tetrahydroepoxide-deoxyadenosine adduct and its site-specific incorporation into a DNA oligomer. J Am Chem Soc. 1991;113:6589–6594. [Google Scholar]
- 81.Lakshman MK, Sayer JM, Jerina DM. Synthesis and site-specific incorporation of a bay-region cis ring-opened tetrahydro epoxide-deoxyadenosine adduct into a DNA oligomer. J Org Chem. 1992;57:3438–3443. [Google Scholar]
- 82.Lakshman MK, Sayer JM, Yagi H, Jerina DM. Synthesis and duplex-forming properties of a nonanucleotide containing an N6-deoxyadenosine adduct of a bay-region diol epoxide. J Org Chem. 1992;57:4585–4590. [Google Scholar]
- 83.Kroth H, Yagi H, Sayer JM, Kumar S, Jerina DM. O6-Allyl protected deoxyguanosine adducts of polycyclic aromatic hydrocarbons as building blocks for the synthesis of oligonucleotides. Chem Res Toxicol. 2001;14:708–719. doi: 10.1021/tx0002637. [DOI] [PubMed] [Google Scholar]
- 84.Yagi H, Frank H, Seidel A, Jerina DM. Synthesis and absolute configuration of cis- and trans-opened cyclopenta[cd]-pyrene 3,4-oxide N2-deoxyguanosine adducts: Conversion to phosphoramidites for oligonucleotide synthesis. Chem Res Toxicol. 2007;20:650–661. doi: 10.1021/tx600286z. [DOI] [PubMed] [Google Scholar]
- 85.Page JE, Zajc B, Oh-hara T, Lakshman MK, Sayer JM, Jerina DM, Dipple A. Sequence context profoundly influences the mutagenic potency of trans-opened benzo[a]pyrene 7,8-diol 9,10-epoxide-purine nucleoside adducts in site-specific mutation studies. Biochemistry. 1998;37:9127–9137. doi: 10.1021/bi980273v. [DOI] [PubMed] [Google Scholar]
- 86.Page JE, Pilcher AS, Yagi H, Sayer JM, Jerina DM, Dipple A. Mutational consequences of replication of M13mp7L2 constructs containing cis-opened benzo[a]pyrene 7,8-diol 9,10-epoxide-deoxyadenosine adducts. Chem Res Toxicol. 1999;12:258–263. doi: 10.1021/tx980244l. [DOI] [PubMed] [Google Scholar]
- 87.Pontén I, Sayer JM, Pilcher AS, Yagi H, Kumar S, Jerina DM, Dipple A. Sequence context effects on mutational properties of cis-opened benzo[c]phenanthrene diol epoxide-deoxyadenosine adducts in site-specific mutation studies. Biochemistry. 1999;38:1144–1152. doi: 10.1021/bi982436l. [DOI] [PubMed] [Google Scholar]
- 88.Pontén I, Page JE, Dipple A, Kumar S, Sayer JM, Yagi H, Pilcher A, Jerina DM. Effect of structure and sequence on mutations induced by diol epoxide-DNA adducts in an E. coli-M13 vector system. Polycyclic Aromat Compd. 2000;21:63–72. [Google Scholar]
- 89.Pontén I, Sayer JM, Pilcher AS, Yagi H, Kumar S, Jerina DM, Dipple A. Factors determining mutagenic potential for individual cis and trans opened benzo[c]phenanthrene diol epoxide-deoxyadenosine adducts. Biochemistry. 2000;39:4136–4144. doi: 10.1021/bi991719q. [DOI] [PubMed] [Google Scholar]
- 90.Pontén I, Kroth H, Sayer JM, Dipple A, Jerina DM. Differences between the mutational consequences of replication of cis- and trans-opened benzo[a]pyrene 7,8-diol 9,10-epoxide-deoxyguanosine adducts in M13mp7L2 constructs. Chem Res Toxicol. 2001;14:720–726. doi: 10.1021/tx0002684. [DOI] [PubMed] [Google Scholar]
- 91.Ramos LA, Sayer JM, Yagi H, Shah JH, Dipple A, Jerina DM. Effect of benzo-ring hydroxyl groups on site-specific mutagenesis by tetrahydrobenzo[a]pyrene adducts at N6 of deoxyadenosine. Chem Res Toxicol. 2001;14:1082–1089. doi: 10.1021/tx010076o. [DOI] [PubMed] [Google Scholar]
- 92.Ramos LA, Pontén I, Dipple A, Kumar S, Yagi H, Sayer JM, Kroth H, Kalena G, Jerina DM. Site-specific mutagenesis in Escherichia coli by N2-deoxyguanosine adducts derived from the highly carcinogenic fjord-region benzo[c]phenanthrene 3,4-diol 1,2-epoxides. Chem Res Toxicol. 2002;15:1619–1626. doi: 10.1021/tx020073r. [DOI] [PubMed] [Google Scholar]
- 93.Maruenda H, Chenna A, Liem LK, Singer B. Synthesis of 1,N6-ethano-2′-deoxyadenosine, a metabolic product of 1,3-bis(2-chloroethyl)nitrosourea, and its incorporation into oligomeric DNA. J Org Chem. 1998;63:4385–4389. [Google Scholar]
- 94.Chenna A, Perry A, Singer B. Synthesis of 8-(hydroxymethyl)-3,N4-etheno-2′-deoxycytidine, a potential carcinogenic glycidaldehyde adduct, and its site-specific incorporation into DNA oligonucleotides. Chem Res Toxicol. 2000;13:208–213. doi: 10.1021/tx990181m. [DOI] [PubMed] [Google Scholar]
- 95.Singer B, Grunberger D. Molecular Biology of Mutagens and Carcinogens. Plenum; New York, NY: 1983. [Google Scholar]
- 96.Webb TR, Matteucci MD. Sequence-specific cross-linking of deoxyoligonucleotides via hybridization-triggered alkylation. J Am Chem Soc. 1986;108:2764–2765. [Google Scholar]
- 97.MacMillan AM, Verdine GL. Engineering tethered DNA molecules by the convertible nucleoside approach. Tetrahedron. 1991;47:2603–2616. [Google Scholar]
- 98.MacMillan AM, Verdine GL. Synthesis of functionally tethered oligodeoxynucleotides by the convertible nucleoside approach. J Org Chem. 1990;55:5931–5933. [Google Scholar]
- 99.Ferentz AE, Verdine GL. Aminolysis of 2′-deoxyinosine aryl ethers: Nucleoside model studies for the synthesis of functionally tethered oligonucleotides. Nucleosides Nucleotides. 1992;11:1749–1763. [Google Scholar]
- 100.Ferentz AE, Verdine GL. Disulfide-crosslinked oligonucleotides. J Am Chem Soc. 1991;113:4000–4002. [Google Scholar]
- 101.Xu YZ, Swann PF. Solid-phase synthesis of oligodeoxynucleotides containing O6-alkylguanine and O4-alkylthymine. Nucleosides Nucleotides. 1991;10:315–318. [Google Scholar]
- 102.Xu YZ, Zheng Q, Swann PF. Synthesis by post-synthetic substitution of oligomers containing guanine modified at the 6-position with sulfur, nitrogen, oxygen derivatives. Tetrahedron. 1992;48:1729–1740. [Google Scholar]
- 103.Xu YZ, Zheng Q, Swann PF. Synthesis of DNA containing modified bases by post-synthetic substitution. Synthesis of oligomers containing 4-substituted thymine: O4-alkylthymine, 5-methylcytosine, N4-dimethylamino-5-methylcytosine, and 4-thiothymine. J Org Chem. 1992;57:3839–3845. [Google Scholar]
- 104.Harris CM, Zhou L, Strand EA, Harris TM. New strategy for the synthesis of oligodeoxynucleotides bearing adducts at exocyclic amino sites of purine nucleosides. J Am Chem Soc. 1991;113:4328–4329. [Google Scholar]
- 105.Kim SJ, Stone MP, Harris CM, Harris TM. A postoligomerization synthesis of oligodeoxynucleotides containing polycyclic aromatic hydrocarbon adducts at the N6 position of deoxyadenosine. J Am Chem Soc. 1992;114:5480–5481. [Google Scholar]
- 106.DeCorte BL, Tsarouhtsis D, Kuchimanchi S, Cooper MD, Horton P, Harris CM, Harris TM. Improved strategies for postoligomerization synthesis of oligodeoxynucleotides bearing structurally defined adducts at the N2 position of deoxyguanosine. Chem Res Toxicol. 1996;9:630–637. doi: 10.1021/tx9501795. [DOI] [PubMed] [Google Scholar]
- 107.Kim HY, Nechev L, Zhou L, Tamura P, Harris CM, Harris TM. Synthesis and adduction of fully deprotected oligodeoxynucleotides containing 6-chloropurine. Tetrahedron Lett. 1998;39:6803–6806. [Google Scholar]
- 108.Gao H, Fathi R, Gaffney BL, Goswami B, Kung PP, Rhee Y, Jin R, Jones RA. 6-O-(Pentafluorophenyl)-2′-deoxyguanosine: A versatile synthon for nucleoside and oligonucleotide synthesis. J Org Chem. 1992;57:6954–6959. [Google Scholar]
- 109.Nechev LV, Zhang M, Tsarouhtsis D, Tamura PJ, Wilkinson AS, Harris CM, Harris TM. Synthesis and characterization of nucleosides and oligonucleotides bearing adducts of butadiene epoxides on adenine N6 and guanine N2. Chem Res Toxicol. 2001;14:379–388. doi: 10.1021/tx000241k. [DOI] [PubMed] [Google Scholar]
- 110.Dooley PA, Tsarouhtsis D, Korbel GA, Nechev LV, Shearer J, Zegar IS, Harris CM, Stone MP, Harris TM. Structural studies of an oligodeoxynucleotide containing a trimethylene interstrand cross-link in a 5′-(CpG) motif: Model of a malondialdehyde cross-link. J Am Chem Soc. 2001;123:1730–1739. doi: 10.1021/ja003163w. [DOI] [PubMed] [Google Scholar]
- 111.Nechev LV, Harris CM, Harris TM. Synthesis of nucleosides and oligonucleotides containing adducts of acrolein and vinyl chloride. Chem Res Toxicol. 2000;13:421–429. doi: 10.1021/tx990167+. [DOI] [PubMed] [Google Scholar]
- 112.Kim SJ, Jajoo HK, Kim HY, Zhou L, Horton P, Harris CM, Harris TM. An efficient route to N6 deoxyadenosine adducts of diol epoxides of carcinogenic polycyclic aromatic hydrocarbons. Bioorg Med Chem. 1995;3:811–822. doi: 10.1016/0968-0896(95)00065-o. [DOI] [PubMed] [Google Scholar]
- 113.Nechev LV, Kozekov I, Harris CM, Harris TM. Stereospecific synthesis of oligonucleotides containing crotonaldehyde adducts of deoxyguanosine. Chem Res Toxicol. 2001;14:1506–1512. doi: 10.1021/tx0100690. [DOI] [PubMed] [Google Scholar]
- 114.Kim HY, Cooper M, Nechev LV, Harris CM, Harris TM. Synthesis and characterization of nucleosides and oligonucleotides with a benzo[a]pyren-6-ylmethyl adduct at adenine N6 or guanine N2. Chem Res Toxicol. 2001;14:1306–1314. doi: 10.1021/tx010086p. [DOI] [PubMed] [Google Scholar]
- 115.Kozekov ID, Nechev LV, Moseley MS, Harris CM, Rizzo CJ, Stone MP, Harris TM. DNA Interchain cross-links formed by acrolein and crotonaldehyde. J Am Chem Soc. 2003;125:50–61. doi: 10.1021/ja020778f. [DOI] [PubMed] [Google Scholar]
- 116.Li Z, Tamura PJ, Wilkinson AS, Harris CM, Harris TM, Stone MP. Intercalation of the (1R,2S,3R,4S)-N6-[1-(1,2,3,4-tetrahydro-2,3,4-trihydroxybenz[a]anthracenyl)]-2′-deoxyadenosyl adduct in the N-ras codon 61 sequence: DNA sequence effects. Biochemistry. 2001;40:6743–6755. doi: 10.1021/bi002785r. [DOI] [PubMed] [Google Scholar]
- 117.Hennard C, Finneman J, Harris CM, Harris TM, Stone MP. The nonmutagenic (R)- and (S)-β-(N6-adenyl)styrene oxide adducts are oriented in the major groove and show little perturbation to DNA structure. Biochemistry. 2001;40:9780–9791. doi: 10.1021/bi010564v. [DOI] [PubMed] [Google Scholar]
- 118.Setayesh FR, DeCorte BL, Horton P, Harris CM, Harris TM, Stone MP. Styrene oxide adducts in an oligodeoxynucleotide containing the human N-ras codon 12: Minor groove structures of the R(12,1)- and S(12,1)-α-(N2-guanyl) stereoisomers determined by1H nuclear magnetic resonance. Chem Res Toxicol. 1998;11:766–777. doi: 10.1021/tx9800147. [DOI] [PubMed] [Google Scholar]
- 119.Scholdberg TA, Nechev LV, Merritt WK, Harris TM, Harris CM, Lloyd RS, Stone MP. Structure of a site specific major groove (2S,3S)-N6-(2,3,4-trihydroxybutyl)-2′-deoxyadenosyl DNA adduct of butadiene diol epoxide. Chem Res Toxicol. 2004;17:717–730. doi: 10.1021/tx034271+. [DOI] [PubMed] [Google Scholar]
- 120.Cho YJ, Wang H, Kozekov ID, Kurtz AJ, Jacob J, Voehler M, Smith J, Harris TM, Lloyd RS, Rizzo CJ, Stone MP. Stereospecific formation of interstrand carbinolamine DNA cross-links by crotonaldehyde- and acetaldehyde-derived α-CH3-γ-OH-1,N2-propano-2′-deoxyguanosine adducts in the 5′-CpG-3′ sequence. Chem Res Toxicol. 2006;19:195–208. doi: 10.1021/tx050239z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang H, Kozekov ID, Kozekova A, Tamura PJ, Marnett LJ, Harris TM, Rizzo CJ. Site-specific synthesis of oligonucleotides containing malondialdehyde adducts of deoxyguanosine and deoxyadenosine via a postsynthetic modification strategy. Chem Res Toxicol. 2006;19:1467–1474. doi: 10.1021/tx060137o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang H, Kozekov ID, Harris TM, Rizzo CJ. Site-specific synthesis and reactivity of oligonucleotides containing stereochemically defined 1,N2-deoxyguanosine adducts of the lipid peroxidation product trans-4-hydroxynonenal. J Am Chem Soc. 2003;125:5687–5700. doi: 10.1021/ja0288800. [DOI] [PubMed] [Google Scholar]
- 123.Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439:225–228. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- 124.Neeley WL, Henderson PT, Essigmann JM. Efficient synthesis of DNA containing the guanine oxidation-nitration product 5-guanidino-4-nitroimidazole: Generation by a postsynthetic substitution reaction. Org Lett. 2004;6:245–248. doi: 10.1021/ol036188j. [DOI] [PubMed] [Google Scholar]
- 125.Ikeda H, Saito I. 8-Methoxydeoxyguanosine as an effective precursor of 2-aminoimidazolone, a major guanine oxidation product in one-electron oxidation of DNA. J Am Chem Soc. 1999;121:10836–10837. [Google Scholar]
- 126.Simon P, Gasparutto D, Gambarelli S, Saint-Pierre C, Favier A, Cadet J. Formation of isodialuric acid lesion within DNA oligomers via one-electron oxidation of 5-hydroxyuracil: Characterization, stability and excision repair. Nucleic Acids Res. 2006;34:3660–3669. doi: 10.1093/nar/gkl496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bellon S, Gasparutto D, Saint-Pierre C, Cadet J. Guanine-thymine intrastrand cross-linked lesion containing oligonucleotides: From chemical synthesis to in vitro enzymatic replication. Org Biomol Chem. 2006;4:3831–3837. doi: 10.1039/b609460k. [DOI] [PubMed] [Google Scholar]
- 128.Hong H, Cao H, Wang Y. Formation and genotoxicity of a guanine-cytosine intrastrand cross-link lesion in vivo. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.MacMillan AM, Lee RJ, Verdine GL. Direct observation of a specific contact in the λ repressor-OL1 complex by isotope-edited NMR. J Am Chem Soc. 1993;115:4921–4922. [Google Scholar]
- 130.Acedo M, Fàbrega C, Aviño A, Goodman M, Fagan P, Wemmer D, Eritja R. A simple method for N-15 labeling of exocyclic amino groups in synthetic oligodeoxyribonucleotides. Nucleic Acids Res. 1994;22:2982–2989. doi: 10.1093/nar/22.15.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kornyushyna O, Berges AM, Muller JG, Burrows CJ. In vitro nucleotide misinsertion opposite the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin and DNA synthesis past the lesions using Escherichia coli DNA polymerase I (Klenow fragment) Biochemistry. 2002;41:15304–15314. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- 132.Tretyakova NY, Niles JC, Burney S, Wishnok JS, Tannenbaum SR. Peroxynitrite-induced reactions of synthetic oligonucleotides containing 8-oxoguanine. Chem Res Toxicol. 1999;12:459–466. doi: 10.1021/tx980235c. [DOI] [PubMed] [Google Scholar]
- 133.Tretyakova NY, Wishnok JS, Tannenbaum SR. Peroxynitrite-induced secondary oxidative lesions at guanine nucleo-bases: Chemical stability and recognition by the Fpg DNA repair enzyme. Chem Res Toxicol. 2000;13:658–664. doi: 10.1021/tx000083x. [DOI] [PubMed] [Google Scholar]
- 134.Shafirovich V, Mock S, Kolbanovskiy A, Geacintov NE. Photochemically catalyzed generation of site-specific 8-nitroguanine adducts in DNA by the reaction of long-lived neutral guanine radicals with nitrogen dioxide. Chem Res Toxicol. 2002;15:591–597. doi: 10.1021/tx015593l. [DOI] [PubMed] [Google Scholar]
- 135.Basu AK, Loechler EL, Leadon SA, Essigmann JM. Genetic effects of thymine glycol: Site-specific mutagenesis and molecular modeling studies. Proc Natl Acad Sci USA. 1989;86:7677–7681. doi: 10.1073/pnas.86.20.7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Smith CA, Taylor JS. Preparation and characterization of a set of deoxyoligonucleotide 49-mers containing site-specific cis-syn, trans-syn-I, (6–4), and Dewar photoproducts of thymidylyl(3′→5′)-thymidine. J Biol Chem. 1993;268:11143–11151. [PubMed] [Google Scholar]
- 137.Laryea A, Cosman M, Lin JM, Liu T, Agarwal R, Smirnov S, Amin S, Harvey RG, Dipple A, Geacintov NE. Direct synthesis and characterization of site-specific adenosyl adducts derived from the binding of a 3,4-dihydroxy-1,2-epoxybenzo[c]phenanthrene stereoisomer to an 11-mer oligodeoxyribonucleotide. Chem Res Toxicol. 1995;8:444–454. doi: 10.1021/tx00045a017. [DOI] [PubMed] [Google Scholar]
- 138.Xu R, Dwarakanath S, Cosman M, Amin S, Geacintov NE. Site-specific adducts derived from the binding of anti-5-methylchrysene diol epoxide enantiomers to DNA: Synthesis and characteristics. Carcinogenesis. 1996;17:2035–2042. doi: 10.1093/carcin/17.9.2035. [DOI] [PubMed] [Google Scholar]
- 139.Cosman M, Laryea A, Fiala R, Hingerty BE, Amin S, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (−)-trans-anti-benzo[c]phenanthrene-dA ([BPh] dA) adduct opposite dT in a DNA duplex: Intercalation of the covalently attached benzo[c]phenanthrenyl ring to the 3′-side of the adduct site and comparison with the (+)-trans-anti-[BPh]dA opposite dT stereoisomer. Biochemistry. 1995;34:1295–1307. doi: 10.1021/bi00004a024. [DOI] [PubMed] [Google Scholar]
- 140.Cosman M, Xu R, Hingerty BE, Amin S, Harvey RG, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (−)-trans-anti-5-methylchrysene-dG adduct opposite dC in a DNA duplex: DNA bending associated with wedging of the methyl group of 5-methylchrysene to the 3′-side of the modification site. Biochemistry. 1995;34:6247–6260. doi: 10.1021/bi00018a029. [DOI] [PubMed] [Google Scholar]
- 141.Baertschi SW, Raney KD, Stone MP, Harris TM. Preparation of the 8,9-epoxide of the mycotoxin aflatoxin B1: The ultimate carcinogenic species. J Am Chem Soc. 1988;110:7929–7931. [Google Scholar]
- 142.Gopalakrishnan S, Stone MP, Harris TM. Preparation and characterization of an aflatoxin B1 adduct with the oligodeoxynucleotide d(ATCGAT)2. J Am Chem Soc. 1989;111:7232–7239. [Google Scholar]
- 143.Gopalakrishnan S, Harris TM, Stone MP. Intercalation of aflatoxin B1 in two oligodeoxynucleotide adducts: Comparative1H NMR analysis of d(ATCAFBGAT)·d(ATCGAT) and d(ATAFBGCAT)2. Biochemistry. 1990;29:10438–10448. doi: 10.1021/bi00498a002. [DOI] [PubMed] [Google Scholar]
- 144.Koehl P, Burnouf D, Fuchs RP. Construction of plasmids containing a unique acetylaminofluorene adduct located within a mutation hot spot. A new probe for frameshift mutagenesis. J Mol Biol. 1989;207:355–364. doi: 10.1016/0022-2836(89)90259-3. [DOI] [PubMed] [Google Scholar]
- 145.Burnouf D, Gauthier C, Chottard JC, Fuchs RP. Single d(ApG)/cis-diamminedichloroplatinum(II) adduct-induced mutagenesis in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:6087–6091. doi: 10.1073/pnas.87.16.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Naser LJ, Pinto AL, Lippard SJ, Essigmann JM. Chemical and biological studies of the major DNA adduct of cis-diamminedichloroplatinum(II), cis-[Pt(NH3)2{d(GpG)}], built into a specific site in a viral genome. Biochemistry. 1988;27:4357–4367. doi: 10.1021/bi00412a024. [DOI] [PubMed] [Google Scholar]
- 147.Shibutani S, Dasaradhi L. Miscoding potential of tamoxifen-derived DNA adducts: α-(N2-deoxyguanosinyl)tamoxifen. Biochemistry. 1997;36:13010–13017. doi: 10.1021/bi970243c. [DOI] [PubMed] [Google Scholar]
- 148.Shibutani S, Itoh S, Yoshizawa I. Miscoding properties of model estrogen-DNA adducts in reactions catalyzed by mammalian and Escherichia coli DNA polymerases. Biochemistry. 1997;36:1755–1765. doi: 10.1021/bi962275q. [DOI] [PubMed] [Google Scholar]
- 149.Terashima I, Suzuki N, Dasaradhi L, Tan CK, Downey KM, Shibutani S. Translesional synthesis on DNA templates containing an estrogen quinone-derived adduct: N2-(2-hydroxyestron-6-yl)-2′-deoxyguanosine and N6-(2-hydroxyestron-6-yl)-2′-deoxyadenosine. Biochemistry. 1998;37:13807–13815. doi: 10.1021/bi981235e. [DOI] [PubMed] [Google Scholar]
- 150.Terashima I, Suzuki N, Shibutani S. Mutagenic properties of estrogen quinone-derived DNA adducts in simian kidney cells. Biochemistry. 2001;40:166–172. doi: 10.1021/bi002273c. [DOI] [PubMed] [Google Scholar]
- 151.Suzuki N, Itoh S, Poon K, Masutani C, Hanaoka F, Ohmori H, Yoshizawa I, Shibutani S. Translesion synthesis past estrogen-derived DNA adducts by human DNA polymerases η and κ. Biochemistry. 2004;43:6304–6311. doi: 10.1021/bi0360298. [DOI] [PubMed] [Google Scholar]
- 152.Vyas RR, Nolan SJ, Basu AK. Synthesis and characterization of oligodeoxynucleotides containing N-(deoxyguanosin-8-yl)-1-aminopyrene. Tetrahedron Lett. 1993;34:2247–2250. [Google Scholar]
- 153.Purohit V, Basu AK. Synthesis and characterization of oligodeoxynucleotides containing the major DNA adducts formed by 1,6- and 1,8-dinitropyrene. Org Lett. 2000;2:1871–1874. doi: 10.1021/ol000090c. [DOI] [PubMed] [Google Scholar]
- 154.Ramos LA, Lipman R, Tomasz M, Basu AK. The major mitomycin C-DNA monoadduct is cytotoxic but not mutagenic in Escherichia coli. Chem Res Toxicol. 1998;11:64–69. doi: 10.1021/tx970163+. [DOI] [PubMed] [Google Scholar]
- 155.Ojwang JO, Grueneberg DA, Loechler EL. Synthesis of a duplex oligonucleotide containing a nitrogen mustard interstrand DNA-DNA cross-link. Cancer Res. 1989;49:6529–6537. [PubMed] [Google Scholar]
- 156.Fischhaber PL, Gall AS, Duncan JA, Hopkins PB. Direct demonstration in synthetic oligonucleotides that N,N′-bis(2-chloroethyl)-nitrosourea cross-links N1 of deoxyguanosine to N3 of deoxycytidine on opposite strands of duplex DNA. Cancer Res. 1999;59:4363–4368. [PubMed] [Google Scholar]
- 157.Huang H, Woo J, Alley SC, Hopkins PB. DNA-DNA interstrand crosslinking by cis-diamminedichloroplatinum(II): N7(dG)-to-N7(dG) Crosslinking at 5′-d(GC) in synthetic oligonucleotides. Bioorg Med Chem. 1995;3:659–669. doi: 10.1016/0968-0896(95)00059-p. [DOI] [PubMed] [Google Scholar]
- 158.Kirchner JJ, Sigurdsson ST, Hopkins PB. Interstrand cross-linking of duplex DNA by nitrous acid: Covalent structure of the dG-to-dG cross-link at the sequence 5′-CG. J Am Chem Soc. 1992;114:4021–4027. [Google Scholar]
- 159.Zhao H, Pagano AR, Wang W, Shallop A, Gaffney BL, Jones RA. Use of a13C atom to differentiate two15N-labeled nucleosides. Syntheses of [15NH2]-adenosine, [1,NH2-15N2]-and [2-13C-1,NH2-15N2]-guanosine, and [1,7,NH2-15N3]- and [2-13C-1,7,NH2-15N3]-2′-deoxyguanosine. J Org Chem. 1997;62:7832–7835. [Google Scholar]
- 160.Pagano AR, Zhao H, Shallop A, Jones RA. Syntheses of [1,7-15N2]- and [1,7,NH2-15N3]adenosine and 2′-deoxyadenosine via an N1-alkoxy-mediated Dimroth rearrangement. J Org Chem. 1998;63:3213–3217. [Google Scholar]
- 161.Shallop AJ, Gaffney BL, Jones RA. Use of13C as an indirect tag in15N specifically labeled nucleosides. Syntheses of [8-13C-1,7,NH2-15N3]adenosine, -guanosine, and their deoxy analogues. J Org Chem. 2003;68:8657–8661. doi: 10.1021/jo0345446. [DOI] [PubMed] [Google Scholar]
- 162.Ye Y, Muller JG, Luo W, Mayne CL, Shallop AJ, Jones RA, Burrows CJ. Formation of13C-,15N-, and18O-labeled guanidinohydantoin from guanosine oxidation with singlet oxygen. Implications for structure and mechanism. J Am Chem Soc. 2003;125:13926–13927. doi: 10.1021/ja0378660. [DOI] [PubMed] [Google Scholar]
- 163.Gu F, Stillwell WG, Wishnok JS, Shallop AJ, Jones RA, Tannenbaum SR. Peroxynitrite-induced reactions of synthetic oligo 2′-deoxynucleotides and DNA containing guanine: Formation and stability of a 5-guanidino-4-nitroimidazole lesion. Biochemistry. 2002;41:7508–7518. doi: 10.1021/bi020148q. [DOI] [PubMed] [Google Scholar]
- 164.Tretyakova N, Matter B, Jones R, Shallop A. Formation of benzo[a]pyrene diol epoxide-DNA adducts at specific guanines within K-ras and p53 gene sequences: Stable isotope-labeling mass spectrometry approach. Biochemistry. 2002;41:9535–9544. doi: 10.1021/bi025540i. [DOI] [PubMed] [Google Scholar]
- 165.Ziegel R, Shallop A, Jones R, Tretyakova N. K-ras Gene sequence effects on the formation of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK)-DNA adducts. Chem Res Toxicol. 2003;16:541–550. doi: 10.1021/tx025619o. [DOI] [PubMed] [Google Scholar]
- 166.Matter B, Wang G, Jones R, Tretyakova N. Formation of diastereomeric benzo[a]pyrene diol epoxide-guanine adducts in p53 gene-derived DNA sequences. Chem Res Toxicol. 2004;17:731–741. doi: 10.1021/tx049974l. [DOI] [PubMed] [Google Scholar]
- 167.Ziegel R, Shallop A, Upadhyaya P, Jones R, Tretyakova N. Endogenous 5-methylcytosine protects neighboring guanines from N7 and O6-methylation and O6-pyridyloxobutylation by the tobacco carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Biochemistry. 2004;43:540–549. doi: 10.1021/bi035259j. [DOI] [PubMed] [Google Scholar]
- 168.Rajesh M, Wang G, Jones R, Tretyakova N. Stable isotope labeling-mass spectrometry analysis of methyl- and pyridyloxobutyl-guanine adducts of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in p53-derived DNA sequences. Biochemistry. 2005;44:2197–2207. doi: 10.1021/bi0480032. [DOI] [PubMed] [Google Scholar]
- 169.Matter B, Guza R, Zhao J, Li ZZ, Jones R, Tretyakova N. Sequence distribution of acetaldehyde-derived N2-ethyl-dG adducts along duplex DNA. Chem Res Toxicol. 2007;20:1379–1387. doi: 10.1021/tx7001146. [DOI] [PubMed] [Google Scholar]
- 170.Park S, Anderson C, Loeber R, Seetharaman M, Jones R, Tretyakova N. Interstrand and intrastrand DNA-DNA cross-linking by 1,2,3,4-diepoxybutane: Role of stereochemistry. J Am Chem Soc. 2005;127:14355–14365. doi: 10.1021/ja051979x. [DOI] [PubMed] [Google Scholar]
- 171.Spratt TE, Campbell CR. Synthesis of oligodeoxyribonucleotides containing analogs of O6-methylguanine and reaction with O6-alkylguanine-DNA alkyltransferase. Biochemistry. 1994;33:11364–11371. doi: 10.1021/bi00203a035. [DOI] [PubMed] [Google Scholar]
- 172.Spratt TE, Wu JD, Levy DE, Kanugula S, Pegg AE. Reaction and binding of oligodeoxynucleotides containing analogues of O6-methylguanine with wild-type and mutant human O6-alkylguanine-DNA alkyltransferase. Biochemistry. 1999;38:6801–6806. doi: 10.1021/bi982908w. [DOI] [PubMed] [Google Scholar]
- 173.Spratt TE. Klenow fragment-DNA interaction required for the incorporation of nucleotides opposite guanine and O6-methylguanine. Biochemistry. 1997;36:13292–13297. doi: 10.1021/bi971253g. [DOI] [PubMed] [Google Scholar]
- 174.Spratt TE. Identification of hydrogen bonds between Escherichia coli DNA polymerase I (Klenow fragment) and the minor groove of DNA by amino acid substitution of the polymerase and atomic substitution of the DNA. Biochemistry. 2001;40:2647–2652. doi: 10.1021/bi002641c. [DOI] [PubMed] [Google Scholar]
- 175.Moran S, Ren RXF, Rumney S, IV, Kool ET. Difluorotoluene, a nonpolar isostere for thymine, codes specifically and efficiently for adenine in DNA replication. J Am Chem Soc. 1997;119:2056–2057. doi: 10.1021/ja963718g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Morales JC, Kool ET. Minor groove interactions between polymerase and DNA: More essential to replication than Watson-Crick hydrogen bonds? J Am Chem Soc. 1999;121:2323–2324. doi: 10.1021/ja983502+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Morales JC, Kool ET. Functional hydrogen-bonding map of the minor groove binding tracks of six DNA polymerases. Biochemistry. 2000;39:12979–12988. doi: 10.1021/bi001578o. [DOI] [PubMed] [Google Scholar]
- 178.Kim TW, Kool ET. A series of nonpolar thymidine analogues of increasing size: DNA base pairing and stacking properties. J Org Chem. 2005;70:2048–2053. doi: 10.1021/jo048061t. [DOI] [PubMed] [Google Scholar]
- 179.Mizukami S, Kim TW, Helquist SA, Kool ET. Varying DNA base-pair size in subangstrom increments: Evidence for a loose, not large, active site in low-fidelity Dpo4 polymerase. Biochemistry. 2006;45:2772–2778. doi: 10.1021/bi051961z. [DOI] [PubMed] [Google Scholar]
- 180.Taniguchi Y, Kool ET. Nonpolar isosteres of damaged DNA bases: Effective mimicry of mutagenic properties of 8-oxopurines. J Am Chem Soc. 2007;129:8836–8844. doi: 10.1021/ja071970q. [DOI] [PubMed] [Google Scholar]
- 181.Schofield MJ, Brownewell FE, Nayak S, Du C, Kool ET, Hsieh P. The Phe-X-Glu DNA binding motif of MutS: The role of hydrogen bonding in mismatch recognition. J Biol Chem. 2001;276:45505–45508. doi: 10.1074/jbc.C100449200. [DOI] [PubMed] [Google Scholar]
- 182.Drotschmann K, Yang W, Brownewell FE, Kool ET, Kunkel TA. Asymmetric recognition of DNA local distortion: Structure-based functional studies of eukaryotic Msh2-Msh6. J Biol Chem. 2001;276:46225–46229. doi: 10.1074/jbc.C100450200. [DOI] [PubMed] [Google Scholar]
- 183.Francis AW, Helquist SA, Kool ET, David SS. Probing the requirements for recognition and catalysis in Fpg and MutY with nonpolar adenine isosteres. J Am Chem Soc. 2003;125:16235–16242. doi: 10.1021/ja0374426. [DOI] [PubMed] [Google Scholar]
- 184.Koo HS, Wu HM, Crothers DM. DNA bending at adenine·thymine tracts. Nature. 1986;320:501–516. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- 185.Koo HS, Crothers DM. Calibration of DNA curvature and a unified description of sequence-directed bending. Proc Natl Acad Sci USA. 1988;85:1763–1767. doi: 10.1073/pnas.85.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Rice JA, Crothers DM, Pinto AL, Lippard SJ. The major adduct of the antitumor drug cis-diamminedichloroplatinum(II) with DNA bends the duplex by ≈40° toward the major groove. Proc Natl Acad Sci USA. 1988;85:4158–4161. doi: 10.1073/pnas.85.12.4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Xu R, Mao B, Xu J, Li B, Birke S, Swenberg CE, Geacintov NE. Stereochemistry-dependent bending in oligonucleotide duplexes induced by site-specific covalent benzo[a]pyrene diol epoxide-guanine lesions. Nucleic Acids Res. 1995;23:2314–2319. doi: 10.1093/nar/23.12.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Tsao H, Mao B, Zhuang P, Xu R, Amin S, Geacintov NE. Sequence dependence and characteristics of bends induced by site-specific polynuclear aromatic carcinogen-deoxyguanosine lesions in oligonucleotides. Biochemistry. 1998;37:4993–5000. doi: 10.1021/bi980291c. [DOI] [PubMed] [Google Scholar]
- 189.Ruan Q, Zhuang P, Li S, Perlow R, Srinivasan AR, Lu XJ, Broyde S, Olson WK, Geacintov NE. Base sequence effects in bending induced by bulky carcinogen-DNA adducts: Experimental and computational analysis. Biochemistry. 2001;40:10458–10472. doi: 10.1021/bi002643x. [DOI] [PubMed] [Google Scholar]
- 190.Moe JG, Reddy GR, Marnett LJ, Stone MP. 1H NMR characterization of a duplex oligodeoxynucleotide containing propanodeoxyguanosine opposite a two-base deletion in the (CpG)3 frameshift hotspot of Salmonella typhimurium hisD3052. Chem Res Toxicol. 1994;7:319–328. doi: 10.1021/tx00039a008. [DOI] [PubMed] [Google Scholar]
- 191.Rink SM, Hopkins PB. A mechlorethamine-induced DNA interstrand cross-link bends duplex DNA. Biochemistry. 1995;34:1439–1445. doi: 10.1021/bi00004a039. [DOI] [PubMed] [Google Scholar]
- 192.Kowalczyk A, Carmical JR, Zou Y, Van Houten B, Lloyd RS, Harris CM, Harris TM. Intrastrand DNA cross-links as tools for studying DNA replication and repair: Two-, three-, and four-carbon tethers between the N2 positions of adjacent guanines. Biochemistry. 2002;41:3109–3118. doi: 10.1021/bi010450j. [DOI] [PubMed] [Google Scholar]
- 193.Brown KL, Deng JZ, Iyer RS, Iyer LG, Voehler MW, Stone MP, Harris CM, Harris TM. Unraveling the aflatoxin - FAPY conundrum: Structural basis for differential replicative processing of isomeric forms of the formamidopyrimidine-type DNA adduct of aflatoxin B1. J Am Chem Soc. 2006;128:15188–15199. doi: 10.1021/ja063781y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Smela ME, Hamm ML, Henderson PT, Harris CM, Harris TM, Essigmann JM. The aflatoxin B1 formamidopyrimidine adduct plays a major role in causing the types of mutations observed in human hepatocellular carcinoma. Proc Natl Acad Sci USA. 2002;99:6655–6660. doi: 10.1073/pnas.102167699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Li Y, Dutta S, Doublié S, Bdour HM, Taylor JS, Ellenberger T. Nucleotide insertion opposite a cis-syn thymine dimer by a replicative DNA polymerase from bacteriophage T7. Nat Struct Mol Biol. 2004;11:784–790. doi: 10.1038/nsmb792. [DOI] [PubMed] [Google Scholar]
- 196.Brieba LG, Eichman BF, Kokoska RJ, Doublié S, Kunkel TA, Ellenberger T. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J. 2004;23:3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Brieba LG, Kokoska RJ, Bebenek K, Kunkel TA, Ellenberger T. A lysine residue in the fingers subdomain of T7 DNA polymerase modulates the miscoding potential of 8-oxo-7,8-dihydroguanosine. Structure. 2005;13:1653–1659. doi: 10.1016/j.str.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 198.Hollis T, Ichikawa Y, Ellenberger T. DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase Escherichia coli AlkA. EMBO J. 2000;19:758–766. doi: 10.1093/emboj/19.4.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lau AY, Schärer OD, Samson L, Verdine GL, Ellenberger T. Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: Mechanisms for nucleotide flipping and base excision. Cell. 1998;95:249–258. doi: 10.1016/s0092-8674(00)81755-9. [DOI] [PubMed] [Google Scholar]
- 200.Lau AY, Wyatt MD, Glassner BJ, Samson LD, Ellenberger T. Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG. Proc Natl Acad Sci USA. 2000;97:13573–13578. doi: 10.1073/pnas.97.25.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Hogg M, Wallace SS, Doublié S. Crystallographic snapshots of a replicative DNA polymerase encountering an abasic site. EMBO J. 2004;23:1483–1493. doi: 10.1038/sj.emboj.7600150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Aller P, Rould MA, Hogg M, Wallace SS, Doublié S. A structural rationale for stalling of a replicative DNA polymerase at the most common oxidative thymine lesion, thymine glycol. Proc Natl Acad Sci USA. 2007;104:814–818. doi: 10.1073/pnas.0606648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zahn KE, Belrhali H, Wallace SS, Doublié S. Caught bending the A-rule: Crystal structures of translesion DNA synthesis with a non-natural nucleotide. Biochemistry. 2007;46:10551–10561. doi: 10.1021/bi7008807. [DOI] [PubMed] [Google Scholar]
- 204.Krahn JM, Beard WA, Miller H, Grollman AP, Wilson SH. Structure of DNA polymerase β with the mutagenic DNA lesion 8-oxodeoxyguanine reveals structural insights into its coding potential. Structure. 2003;11:121–127. doi: 10.1016/s0969-2126(02)00930-9. [DOI] [PubMed] [Google Scholar]
- 205.Batra VK, Shock DD, Prasad R, Beard WA, Hou EW, Pedersen LC, Sayer JM, Yagi H, Kumar S, Jerina DM, Wilson SH. Structure of DNA polymerase β with a benzo[c]phenanthrene diol epoxide-adducted template exhibits mutagenic features. Proc Natl Acad Sci USA. 2006;103:17231–17236. doi: 10.1073/pnas.0605069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Prasad R, Batra VK, Yang XP, Krahn JM, Pedersen LC, Beard WA, Wilson SH. Structural insight into the DNA polymerase β deoxyribose phosphate lyase mechanism. DNA Rep. 2005;4:1347–1357. doi: 10.1016/j.dnarep.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 207.Hsu GW, Huang X, Luneva NP, Geacintov NE, Beese LS. Structure of a high fidelity DNA polymerase bound to a benzo[a]pyrene adduct that blocks replication. J Biol Chem. 2005;280:3764–3770. doi: 10.1074/jbc.M411276200. [DOI] [PubMed] [Google Scholar]
- 208.Warren JJ, Forsberg LJ, Beese LS. The structural basis for the mutagenicity of O6-methyl-guanine lesions. Proc Natl Acad Sci USA. 2006;103:19701–19706. doi: 10.1073/pnas.0609580103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Modrich PL, Beese LS. Structure of the human MutSα DNA lesion recognition complex. Mol Cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 210.Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 211.Fromme JC, Verdine GL. Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM. Nat Struct Biol. 2002;9:544–552. doi: 10.1038/nsb809. [DOI] [PubMed] [Google Scholar]
- 212.Fromme JC, Verdine GL. Structure of a trapped endonuclease III-DNA covalent intermediate. EMBO J. 2003;22:3461–3471. doi: 10.1093/emboj/cdg311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Fromme JC, Verdine GL. DNA lesion recognition by the bacterial repair enzyme MutM. J Biol Chem. 2003;278:51543–51548. doi: 10.1074/jbc.M307768200. [DOI] [PubMed] [Google Scholar]
- 214.Fromme JC, Banerjee A, Huang SJ, Verdine GL. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature. 2004;427:652–656. doi: 10.1038/nature02306. [DOI] [PubMed] [Google Scholar]
- 215.Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 216.Banerjee A, Santos WL, Verdine GL. Structure of a DNA glycosylase searching for lesions. Science. 2006;311:1153–1157. doi: 10.1126/science.1120288. [DOI] [PubMed] [Google Scholar]
- 217.Banerjee A, Verdine GL. A nucleobase lesion remodels the interaction of its normal neighbor in a DNA glycosylase complex. Proc Natl Acad Sci USA. 2006;103:15020–15025. doi: 10.1073/pnas.0603644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Radom CT, Banerjee A, Verdine GL. Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations. J Biol Chem. 2007;282:9182–9194. doi: 10.1074/jbc.M608989200. [DOI] [PubMed] [Google Scholar]
- 219.Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- 220.Parikh SS, Mol CD, Slupphaug G, Bharati S, Krokan HE, Tainer JA. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J. 1998;17:5214–5226. doi: 10.1093/emboj/17.17.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Hosfield DJ, Guan Y, Haas BJ, Cunningham RP, Tainer JA. Structure of the DNA repair enzyme endonuclease IV and its DNA complex: Double-nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell. 1999;98:397–408. doi: 10.1016/s0092-8674(00)81968-6. [DOI] [PubMed] [Google Scholar]
- 222.Mol CD, Izumi T, Mitra S, Tainer JA. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 223.Yu B, Edstrom WC, Benach J, Hamuro Y, Weber PC, Gibney BR, Hunt JF. Crystal structures of catalytic complexes of the oxidative DNA/RNA repair enzyme AlkB. Nature. 2006;439:879–884. doi: 10.1038/nature04561. [DOI] [PubMed] [Google Scholar]
- 224.Randall SK, Eritja R, Kaplan BE, Petruska J, Goodman MF. Nucleotide insertion kinetics opposite abasic lesions in DNA. J Biol Chem. 1987;262:6864–6870. [PubMed] [Google Scholar]
- 225.Boosalis MS, Petruska J, Goodman MF. DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. J Biol Chem. 1987;262:14689–14696. [PubMed] [Google Scholar]
- 226.Singer B, Chavez F, Spengler SJ, Kušmierek JT, Mendelman L, Goodman MF. Comparison of polymerase insertion and extension kinetics of a series of O2-alkyldeoxythymidine triphosphates and O4-methyldeoxythymidine triphosphate. Biochemistry. 1989;28:1478–1483. doi: 10.1021/bi00430a008. [DOI] [PubMed] [Google Scholar]
- 227.Creighton S, Bloom LB, Goodman MF. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 1995;262:232–256. doi: 10.1016/0076-6879(95)62021-4. [DOI] [PubMed] [Google Scholar]
- 228.Tang M, Pham P, Shen X, Taylor JS, O’Donnell M, Woodgate R, Goodman MF. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature. 2000;404:1014–1018. doi: 10.1038/35010020. [DOI] [PubMed] [Google Scholar]
- 229.Pham P, Bertram JG, O’Donnell M, Woodgate R, Goodman MF. A model for SOS-lesion-targeted mutations in Escherichia coli. Nature. 2001;409:366–370. doi: 10.1038/35053116. [DOI] [PubMed] [Google Scholar]
- 230.Lowe LG, Guengerich FP. Steady-state and pre-steady-state kinetic analysis of dNTP insertion opposite 8-oxo-7,8-dihydroguanine by Escherichia coli polymerases I exo− and II exo−. Biochemistry. 1996;35:9840–9849. doi: 10.1021/bi960485x. [DOI] [PubMed] [Google Scholar]
- 231.Furge LL, Guengerich FP. Pre-steady-state kinetics of nucleotide insertion following 8-oxo-7,8-dihydroguanine base pair mismatches by bacteriophage T7 DNA polymerase exo−. Biochemistry. 1998;37:3567–3574. doi: 10.1021/bi9722094. [DOI] [PubMed] [Google Scholar]
- 232.Einolf HJ, Schnetz-Boutaud N, Guengerich FP. Steady-state and pre-steady-state kinetic analysis of 8-oxo-7,8-dihydroguanosine triphosphate incorporation and extension by replicative and repair DNA polymerases. Biochemistry. 1998;37:13300–13312. doi: 10.1021/bi981346d. [DOI] [PubMed] [Google Scholar]
- 233.Einolf HJ, Guengerich FP. Fidelity of nucleotide insertion at 8-oxo-7,8-dihydroguanine by mammalian DNA polymerase δ. Steady-state and pre-steady-state kinetic analysis. J Biol Chem. 2001;276:3764–3771. doi: 10.1074/jbc.M006696200. [DOI] [PubMed] [Google Scholar]
- 234.Woodside AM, Guengerich FP. Misincorporation and stalling at O6-methylguanine and O6-benzylguanine: Evidence for inactive polymerase complexes. Biochemistry. 2002;41:1039–1050. doi: 10.1021/bi011496f. [DOI] [PubMed] [Google Scholar]
- 235.Zang H, Harris TM, Guengerich FP. Kinetics of nucleotide incorporation opposite DNA bulky guanine N2 adducts by processive bacteriophage T7 DNA polymerase (exonuclease−) and HIV-1 reverse transcriptase. J Biol Chem. 2005;280:1165–1178. doi: 10.1074/jbc.M405996200. [DOI] [PubMed] [Google Scholar]
- 236.Zang H, Harris TM, Guengerich FP. Kinetics of nucleotide incorporation opposite polycyclic aromatic hydrocarbon-DNA adducts by processive bacteriophage T7 DNA polymerase. Chem Res Toxicol. 2005;18:389–400. doi: 10.1021/tx049683c. [DOI] [PubMed] [Google Scholar]
- 237.Choi JY, Guengerich FP. Adduct size limits efficient and error-free bypass across bulky N2-guanine DNA lesions by human DNA polymerase η. J Mol Biol. 2005;352:72–90. doi: 10.1016/j.jmb.2005.06.079. [DOI] [PubMed] [Google Scholar]
- 238.Choi JY, Guengerich FP. Kinetic evidence for inefficient and error-prone bypass across bulky N2-guanine DNA adducts by human DNA polymerase ι. J Biol Chem. 2006;281:12315–12324. doi: 10.1074/jbc.M600112200. [DOI] [PubMed] [Google Scholar]
- 239.Choi JY, Angel KC, Guengerich FP. Translesion synthesis across bulky N2-alkyl guanine DNA adducts by human DNA polymerase κ. J Biol Chem. 2006;281:21062–21072. doi: 10.1074/jbc.M602246200. [DOI] [PubMed] [Google Scholar]
- 240.Choi JY, Chowdhury G, Zang H, Angel KC, Vu CC, Peterson LA, Guengerich FP. Translesion synthesis across O6-alkylguanine DNA adducts by recombinant human DNA polymerases. J Biol Chem. 2006;281:38244–38256. doi: 10.1074/jbc.M608369200. [DOI] [PubMed] [Google Scholar]
- 241.Zang H, Irimia A, Choi JY, Angel KC, Loukachevitch LV, Egli M, Guengerich FP. Efficient and high fidelity incorporation of dCTP opposite 7,8-dihydro-8-oxodeoxyguanosine by Sulfolobus solfataricus DNA polymerase Dpo4. J Biol Chem. 2006;281:2358–2372. doi: 10.1074/jbc.M510889200. [DOI] [PubMed] [Google Scholar]
- 242.Eoff RL, Irimia A, Egli M, Guengerich FP. Sulfolobus solfataricus DNA polymerase Dpo4 is partially inhibited by ”wobble” pairing between O6-methylguanine and cytosine, but accurate bypass is preferred. J Biol Chem. 2007;282:1456–1467. doi: 10.1074/jbc.M609661200. [DOI] [PubMed] [Google Scholar]
- 243.Eoff RL, Angel KC, Egli M, Guengerich FP. Molecular basis of selectivity of nucleoside triphosphate incorporation opposite O6-benzylguanine by Sulfolobus solfataricus DNA polymerase Dpo4: Steady-state and pre-steady-state kinetics and X-ray crystallography of correct and incorrect pairing. J Biol Chem. 2007;282:13573–13584. doi: 10.1074/jbc.M700656200. [DOI] [PubMed] [Google Scholar]
- 244.Eoff RL, Irimia A, Angel KC, Egli M, Guengerich FP. Hydrogen bonding of 7,8-dihydro-8-oxodeoxyguanosine with a charged residue in the little finger domain determines miscoding events in Sulfolobus solfataricus DNA polymerase Dpo4. J Biol Chem. 2007;282:19831–19843. doi: 10.1074/jbc.M702290200. [DOI] [PubMed] [Google Scholar]
- 245.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 246.Shibutani S, Bodepudi V, Johnson F, Grollman AP. Translesional synthesis on DNA templates containing 8-oxo-7,8-dihydrodeoxyadenosine. Biochemistry. 1993;32:4615–4621. doi: 10.1021/bi00068a019. [DOI] [PubMed] [Google Scholar]
- 247.Shibutani S, Grollman AP. On the mechanism of frameshift (deletion) mutagenesis in vitro. J Biol Chem. 1993;268:11703–11710. [PubMed] [Google Scholar]
- 248.Shibutani S, Margulis LA, Geacintov NE, Grollman AP. Translesional synthesis on a DNA template containing a single stereoisomer of dG-(+)- or dG-(−)-anti-BPDE (7,8-dihydroxy-anti-9, 10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene) Biochemistry. 1993;32:7531–7541. doi: 10.1021/bi00080a027. [DOI] [PubMed] [Google Scholar]
- 249.Shibutani S, Grollman AP. Nucleotide misincorporation on DNA templates containing N-(deoxyguanosin-N2-yl)-2-(acetylamino)fluorene. Chem Res Toxicol. 1993;6:819–824. doi: 10.1021/tx00036a011. [DOI] [PubMed] [Google Scholar]
- 250.Shibutani S, Grollman AP. Miscoding during DNA synthesis on damaged DNA templates catalysed by mammalian cell extracts. Cancer Lett. 1994;83:315–322. doi: 10.1016/0304-3835(94)90335-2. [DOI] [PubMed] [Google Scholar]
- 251.Shibutani S, Suzuki N, Matsumoto Y, Grollman AP. Miscoding properties of 3,N4-etheno-2′-deoxycytidine in reactions catalyzed by mammalian DNA polymerases. Biochemistry. 1996;35:14992–14998. doi: 10.1021/bi961446o. [DOI] [PubMed] [Google Scholar]
- 252.Shibutani S, Takeshita M, Grollman AP. Translesional synthesis on DNA templates containing a single abasic site. A mechanistic study of the “A rule”. J Biol Chem. 1997;272:13916–13922. doi: 10.1074/jbc.272.21.13916. [DOI] [PubMed] [Google Scholar]
- 253.Shibutani S, Suzuki N, Grollman AP. Mechanism of frameshift (deletion) generated by acetylaminofluorene-derived DNA adducts in vitro. Biochemistry. 2004;43:15929–15935. doi: 10.1021/bi048087e. [DOI] [PubMed] [Google Scholar]
- 254.Zhang W, Johnson F, Grollman AP, Shibutani S. Miscoding by the exocyclic and related DNA adducts 3,N4-etheno-2′-deoxycytidine, 3,N4-ethano-2′-deoxycytidine, and 3-(2-hydroxyethyl)-2′-deoxyuridine. Chem Res Toxicol. 1995;8:157–163. doi: 10.1021/tx00043a021. [DOI] [PubMed] [Google Scholar]
- 255.Suzuki N, Ohashi E, Hayashi K, Ohmori H, Grollman AP, Shibutani S. Translesional synthesis past acetylaminofluorene-derived DNA adducts catalyzed by human DNA polymerase κ and Escherichia coli DNA polymerase IV. Biochemistry. 2001;40:15176–15183. doi: 10.1021/bi010702g. [DOI] [PubMed] [Google Scholar]
- 256.Suzuki N, Ohashi E, Kolbanovskiy A, Geacintov NE, Grollman AP, Ohmori H, Shibutani S. Translesion synthesis by human DNA polymerase κ on a DNA template containing a single stereoisomer of dG-(+)- or dG-(−)-anti-N2-BPDE (7,8-dihydroxy-anti-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene) Biochemistry. 2002;41:6100–6106. doi: 10.1021/bi020049c. [DOI] [PubMed] [Google Scholar]
- 257.Yasui M, Dong H, Bonala RR, Suzuki N, Ohmori H, Hanaoka F, Johnson F, Grollman AP, Shibutani S. Mutagenic properties of 3-(deoxyguanosin-N2-yl)-2-acetylaminofluorene, a persistent acetylaminofluorene-derived DNA adduct in mammalian cells. Biochemistry. 2004;43:15005–15013. doi: 10.1021/bi048279+. [DOI] [PubMed] [Google Scholar]
- 258.Shibutani S. Quantitation of base substitutions and deletions induced by chemical mutagens during DNA synthesis in vitro. Chem Res Toxicol. 1993;6:625–629. doi: 10.1021/tx00035a006. [DOI] [PubMed] [Google Scholar]
- 259.Yasui M, Matsui S, Ihara M, Laxmi YRS, Shibutani S, Matsuda T. Translesional synthesis on a DNA template containing N2-methyl-2′-deoxyguanosine catalyzed by the Klenow fragment of Escherichia coli DNA polymerase I. Nucleic Acids Res. 2001;29:1994–2001. doi: 10.1093/nar/29.9.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Yasui M, Suzuki N, Miller H, Matsuda T, Matsui S, Shibutani S. Translesion synthesis past 2′-deoxyxanthosine, a nitric oxide-derived DNA adduct, by mammalian DNA polymerases. J Mol Biol. 2004;344:665–674. doi: 10.1016/j.jmb.2004.09.064. [DOI] [PubMed] [Google Scholar]
- 261.Suzuki N, Yasui M, Geacintov NE, Shafirovich V, Shibutani S. Miscoding events during DNA synthesis past the nitration-damaged base 8-nitroguanine. Biochemistry. 2005;44:9238–9245. doi: 10.1021/bi050276p. [DOI] [PubMed] [Google Scholar]
- 262.Yasui M, Suzuki N, Laxmi YRS, Shibutani S. Translesion synthesis past tamoxifen-derived DNA adducts by human DNA polymerases η and κ. Biochemistry. 2006;45:12167–12174. doi: 10.1021/bi0608461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Yasui M, Suzuki N, Liu X, Okamoto Y, Kim SY, Laxmi YRS, Shibutani S. Mechanism of translesion synthesis past an equine estrogen-DNA adduct by Y-family DNA polymerases. J Mol Biol. 2007;371:1151–1162. doi: 10.1016/j.jmb.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Green CL, Loechler EL, Fowler KW, Essigmann JM. Construction and characterization of extrachromosomal probes for mutagenesis by carcinogens: Site-specific incorporation of O6-methylguanine into viral and plasmid genomes. Proc Natl Acad Sci USA. 1984;81:13–17. doi: 10.1073/pnas.81.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Lee DH, Jin SG, Cai S, Chen Y, Pfeifer GP, O’Connor TR. Repair of methylation damage in DNA and RNA by mammalian AlkB homologues. J Biol Chem. 2005;280:39448–39459. doi: 10.1074/jbc.M509881200. [DOI] [PubMed] [Google Scholar]
- 266.Zang H, Fang Q, Pegg AE, Guengerich FP. Kinetic analysis of steps in the repair of damaged DNA by human O6-alkylguanine-DNA alkyltransferase. J Biol Chem. 2005;280:30873–30881. doi: 10.1074/jbc.M505283200. [DOI] [PubMed] [Google Scholar]
- 267.Hatahet Z, Zhou M, Reha-Krantz LJ, Morrical SW, Wallace SS. In search of a mutational hotspot. Proc Natl Acad Sci USA. 1998;95:8556–8561. doi: 10.1073/pnas.95.15.8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Hatahet Z, Zhou M, Reha-Krantz LJ, Ide H, Morrical SW, Wallace SS. In vitro selection of sequence contexts which enhance bypass of abasic sites and tetrahydrofuran by T4 DNA polymerase holoenzyme. J Mol Biol. 1999;286:1045–1057. doi: 10.1006/jmbi.1998.2520. [DOI] [PubMed] [Google Scholar]
- 269.Wood RD, Robins P, Lindahl T. Complementation of the xeroderma pigmentosum DNA repair defect in cell-free extracts. Cell. 1988;53:97–106. doi: 10.1016/0092-8674(88)90491-6. [DOI] [PubMed] [Google Scholar]
- 270.Szymkowski DE, Yarema K, Essigmann JM, Lippard SJ, Wood RD. An intrastrand d(GpG) platinum crosslink in duplex M13 DNA is refractory to repair by human cell extracts. Proc Natl Acad Sci USA. 1992;89:10772–10776. doi: 10.1073/pnas.89.22.10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Szymkowski DE, Lawrence CW, Wood RD. Repair by human cell extracts of single (6–4) and cyclobutane thymine-thymine photoproducts in DNA. Proc Natl Acad Sci USA. 1993;90:9823–9827. doi: 10.1073/pnas.90.21.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Moggs JG, Szymkowski DE, Yamada M, Karran P, Wood RD. Differential human nucleotide excision repair of paired and mispaired cisplatin-DNA adducts. Nucleic Acids Res. 1997;25:480–491. doi: 10.1093/nar/25.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Kuraoka I, Bender C, Romieu A, Cadet J, Wood RD, Lindahl T. Removal of oxygen free-radical-induced 5′,8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc Natl Acad Sci USA. 2000;97:3832–3837. doi: 10.1073/pnas.070471597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Shivji MK, Moggs JG, Kuraoka I, Wood RD. Assaying for the dual incisions of nucleotide excision repair using DNA with a lesion at a specific site. Methods Mol Biol. 2006;314:435–456. doi: 10.1385/1-59259-973-7:435. [DOI] [PubMed] [Google Scholar]
- 275.Biggerstaff M, Wood RD. Repair synthesis assay for nucleotide excision repair activity using fractionated cell extracts and UV-damaged plasmid DNA. Methods Mol Biol. 2006;314:417–434. doi: 10.1385/1-59259-973-7:417. [DOI] [PubMed] [Google Scholar]
- 276.Sibghat-Ullah, Husain I, Carlton W, Sancar A. Human nucleotide excision repair in vitro: Repair of pyrimidine dimers, psoralen and cisplatin adducts by HeLa cell-free extract. Nucleic Acids Res. 1989;17:4471–4484. doi: 10.1093/nar/17.12.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Bebenek K, Kunkel TA. The use of native T7 DNA polymerase for site-directed mutagenesis. Nucleic Acids Res. 1989;17:5408. doi: 10.1093/nar/17.13.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Kunkel TA, Bebenek K, McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 280.Reid TM, Lee MS, King CM. Mutagenesis by site-specific arylamine adducts in plasmid DNA: Enhancing replication of the adducted strand alters mutation frequency. Biochemistry. 1990;29:6153–6161. doi: 10.1021/bi00478a007. [DOI] [PubMed] [Google Scholar]
- 281.Cheng KC, Preston BD, Cahill DS, Dosanjh MK, Singer B, Loeb LA. The vinyl chloride DNA derivative N2,3-ethenoguanine produces G→A transitions in Escherichia coli. Proc Natl Acad Sci USA. 1991;88:9974–9978. doi: 10.1073/pnas.88.22.9974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G→T and A→C substitutions. J Biol Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- 283.Benamira M, Singh U, Marnett LJ. Site-specific frameshift mutagenesis by a propanedeoxyguanosine adduct positioned in the (CpG)4 hot-spot of Salmonella typhimurium hisD3052 carried on an M13 vector. J Biol Chem. 1992;267:22392–22400. [PubMed] [Google Scholar]
- 284.Benamira M, Marnett LJ. Construction of a vector for site-specific frameshift mutagenesis containing the mutable hotspot of Salmonella typhimurium TA98 on an M13 bacteriophage. Chem Res Toxicol. 1993;6:317–327. doi: 10.1021/tx00033a011. [DOI] [PubMed] [Google Scholar]
- 285.Burcham PC, Marnett LJ. Site-specific mutagenesis by a propanodeoxyguanosine adduct carried on an M13 genome. J Biol Chem. 1994;269:28844–28850. [PubMed] [Google Scholar]
- 286.Napolitano RL, Fuchs RP. New strategy for the construction of single-stranded plasmids with single mutagenic lesions. Chem Res Toxicol. 1997;10:667–671. doi: 10.1021/tx970018w. [DOI] [PubMed] [Google Scholar]
- 287.Pauly GT, Hughes SH, Moschel RC. Mutagenesis in Escherichia coli by three O6-substituted guanines in double-stranded or gapped plasmids. Biochemistry. 1995;34:8924–8930. doi: 10.1021/bi00027a045. [DOI] [PubMed] [Google Scholar]
- 288.Pauly GT, Hughes SH, Moschel RC. Comparison of mutagenesis by O6-methyl- and O6-ethylguanine and O4-methylthymine in Escherichia coli using double-stranded and gapped plasmids. Carcinogenesis. 1998;19:457–461. doi: 10.1093/carcin/19.3.457. [DOI] [PubMed] [Google Scholar]
- 289.Pauly GT, Moschel RC. Mutagenesis by O6-methyl-, O6-ethyl-, and O6-benzylguanine and O4-methylthymine in human cells: Effects of O6-alkylguanine-DNA alkyltransferase and mismatch repair. Chem Res Toxicol. 2001;14:894–900. doi: 10.1021/tx010032f. [DOI] [PubMed] [Google Scholar]
- 290.Pauly GT, Peterson LA, Moschel RC. Mutagenesis by O6-[4-oxo-4-(3-pyridyl)butyl]guanine in Escherichia coli and human cells. Chem Res Toxicol. 2002;15:165–169. doi: 10.1021/tx0101245. [DOI] [PubMed] [Google Scholar]
- 291.Basu AK, Niedernhofer LJ, Essigmann JM. Deoxyhexanucleotide containing a vinyl chloride induced DNA lesion, 1,N6-ethenoadenine: Synthesis, physical characterization, and incorporation into a duplex bacteriophage M13 genome as part of an amber codon. Biochemistry. 1987;26:5626–5635. doi: 10.1021/bi00392a007. [DOI] [PubMed] [Google Scholar]
- 292.Veaute X, Fuchs RP. Greater susceptibility to mutations in lagging strand of DNA replication in Escherichia coli than in leading strand. Science. 1993;261:598–600. doi: 10.1126/science.8342022. [DOI] [PubMed] [Google Scholar]
- 293.Wagner J, Kamiya H, Fuchs RP. Leading versus lagging strand mutagenesis induced by 7,8-dihydro-8-oxo-2′-deoxyguanosine in Escherichia coli. J Mol Biol. 1997;265:302–309. doi: 10.1006/jmbi.1996.0740. [DOI] [PubMed] [Google Scholar]
- 294.Burnouf D, Koehl P, Fuchs RP. Single adduct mutagenesis: Strong effect of the position of a single acetylaminofluorene adduct within a mutation hot spot. Proc Natl Acad Sci USA. 1989;86:4147–4151. doi: 10.1073/pnas.86.11.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Lambert IB, Napolitano RL, Fuchs RP. Carcinogen-induced frameshift mutagenesis in repetitive sequences. Proc Natl Acad Sci USA. 1992;89:1310–1314. doi: 10.1073/pnas.89.4.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Koffel-Schwartz N, Coin F, Veaute X, Fuchs RP. Cellular strategies for accommodating replication-hindering adducts in DNA: Control by the SOS response in Escherichia coli. Proc Natl Acad Sci U.S.A. 1996;93:7805–7810. doi: 10.1073/pnas.93.15.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Baynton K, Bresson-Roy A, Fuchs RP. Analysis of damage tolerance pathways in Saccharomyces cerevisiae: A requirement for Rev3 DNA polymerase in translesion synthesis. Mol Cell Biol. 1998;18:960–966. doi: 10.1128/mcb.18.2.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Broschard TH, Koffel-Schwartz N, Fuchs RP. Sequence-dependent modulation of frameshift mutagenesis at NarI-derived mutation hot spots. J Mol Biol. 1999;288:191–199. doi: 10.1006/jmbi.1999.2667. [DOI] [PubMed] [Google Scholar]
- 299.Lenne-Samuel N, Janel-Bintz R, Kolbanovskiy A, Geacintov NE, Fuchs RP. The processing of a benzo[a]pyrene adduct into a frameshift or a base substitution mutation requires a different set of genes in Escherichia coli. Mol Microbiol. 2000;38:299–307. doi: 10.1046/j.1365-2958.2000.02116.x. [DOI] [PubMed] [Google Scholar]
- 300.Fink SP, Marnett LJ. The relative contribution of adduct blockage and DNA repair on template utilization during replication of 1,N2-propanodeoxyguanosine and pyrimido[1,2-α]purin-10(3H)-one-adducted M13MB102 genomes. Mutat Res. 2001;485:209–218. doi: 10.1016/s0921-8777(01)00064-7. [DOI] [PubMed] [Google Scholar]
- 301.Pandya GA, Yang IY, Grollman AP, Moriya M. Escherichia coli responses to a single DNA adduct. J Bacteriol. 2000;182:6598–6604. doi: 10.1128/jb.182.23.6598-6604.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Yang IY, Hossain M, Miller H, Khullar S, Johnson F, Grollman A, Moriya M. Responses to the major acrolein-derived deoxyguanosine adduct in Escherichia coli. J Biol Chem. 2001;276:9071–9076. doi: 10.1074/jbc.M008918200. [DOI] [PubMed] [Google Scholar]
- 303.Preston BD, Singer B, Loeb LA. Mutagenic potential of O4-methylthymine in vivo determined by an enzymatic approach to site-specific mutagenesis. Proc Natl Acad Sci USA. 1986;83:8501–8505. doi: 10.1073/pnas.83.22.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Kodadek T, Gamper H. Efficient synthesis of a supercoiled M13 DNA molecule containing a site specifically placed psoralen adduct and its use as a substrate for DNA replication. Biochemistry. 1988;27:3210–3215. doi: 10.1021/bi00409a013. [DOI] [PubMed] [Google Scholar]
- 305.Michaels ML, Cruz C, Grollman AP, Miller JH. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc Natl Acad Sci USA. 1992;89:7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Michaels ML, Tchou J, Grollman AP, Miller JH. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry. 1992;31:10964–10968. doi: 10.1021/bi00160a004. [DOI] [PubMed] [Google Scholar]
- 307.Nghiem Y, Cabrera M, Cupples CG, Miller JH. The mutY gene: A mutator locus in Escherichia coli that generates G·C→T·A transversions. Proc Natl Acad Sci USA. 1988;85:2709–2713. doi: 10.1073/pnas.85.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Cabrera M, Nghiem Y, Miller JH. mutM, a second mutator locus in Escherichia coli that generates G·C→T·A transversions. J Bacteriol. 1988;170:5405–5407. doi: 10.1128/jb.170.11.5405-5407.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Banerjee SK, Christensen RB, Lawrence CW, LeClerc JE. Frequency and spectrum of mutations produced by a single cis-syn thymine-thymine cyclobutane dimer in a single-stranded vector. Proc Natl Acad Sci USA. 1988;85:8141–8145. doi: 10.1073/pnas.85.21.8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.LeClerc JE, Borden A, Lawrence CW. The thymine-thymine pyrimidine-pyrimidone(6–4) ultraviolet light photoproduct is highly mutagenic and specifically induces 3′ thymine-to-cytosine transitions in Escherichia coli. Proc Natl Acad Sci USA. 1991;88:9685–9689. doi: 10.1073/pnas.88.21.9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Gibbs PE, Lawrence CW. U-U and T-T cyclobutane dimers have different mutational properties. Nucleic Acids Res. 1993;21:4059–4065. doi: 10.1093/nar/21.17.4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Lawrence CW, Borden A, Banerjee SK, LeClerc JE. Mutation frequency and spectrum resulting from a single abasic site in a single-stranded vector. Nucleic Acids Res. 1990;18:2153–2157. doi: 10.1093/nar/18.8.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Banerjee SK, Borden A, Christensen RB, LeClerc JE, Lawrence CW. SOS-dependent replication past a single trans-syn T-T cyclobutane dimer gives a different mutation spectrum and increased error rate compared with replication past this lesion in uninduced cells. J Bacteriol. 1990;172:2105–2112. doi: 10.1128/jb.172.4.2105-2112.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Lawrence CW, Banerjee SK, Borden A, LeClerc JE. T-T cyclobutane dimers are misinstructive, rather than non-instructive, mutagenic lesions. Mol Gen Genet. 1990;222:166–168. doi: 10.1007/BF00283040. [DOI] [PubMed] [Google Scholar]
- 315.Horsfall MJ, Lawrence CW. Accuracy of replication past the T-C (6–4) adduct. J Mol Biol. 1994;235:465–471. doi: 10.1006/jmbi.1994.1006. [DOI] [PubMed] [Google Scholar]
- 316.Lawrence CW, Borden A, Woodgate R. Analysis of the mutagenic properties of the UmuDC, MucAB and RumAB proteins, using a site-specific abasic lesion. Mol Gen Genet. 1996;251:493–498. doi: 10.1007/BF02172378. [DOI] [PubMed] [Google Scholar]
- 317.Vandewiele D, Borden A, O’Grady PI, Woodgate R, Lawrence CW. Efficient translesion replication in the absence of Escherichia coli Umu proteins and 3′–5′ exonuclease proofreading function. Proc Natl Acad Sci USA. 1998;95:15519–15524. doi: 10.1073/pnas.95.26.15519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Borden A, O’Grady PI, Vandewiele D, Fernández de Henestrosa AR, Lawrence CW, Woodgate R. Escherichia coli DNA polymerase III can replicate efficiently past a T-T cis-syn cyclobutane dimer if DNA polymerase V and the 3′ to 5′ exonuclease proofreading function encoded by dnaQ are inactivated. J Bacteriol. 2002;184:2674–2681. doi: 10.1128/JB.184.10.2674-2681.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Bailey EA, Iyer RS, Harris TM, Essigmann JM. A viral genome containing an unstable aflatoxin B1-N7-guanine DNA adduct situated at a unique site. Nucleic Acids Res. 1996;24:2821–2828. doi: 10.1093/nar/24.14.2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Moriya M. Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-Oxoguanine in DNA induces targeted G·C→T·A transversions in simian kidney cells. Proc Natl Acad Sci USA. 1993;90:1122–1126. doi: 10.1073/pnas.90.3.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Delaney JC, Essigmann JM. Context-dependent mutagenesis by DNA lesions. Chem Biol. 1999;6:743–753. doi: 10.1016/s1074-5521(00)80021-6. [DOI] [PubMed] [Google Scholar]
- 322.Moerschell RP, Tsunasawa S, Sherman F. Transformation of yeast with synthetic oligonucleotides. Proc Natl Acad Sci USA. 1988;85:524–528. doi: 10.1073/pnas.85.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Moerschell RP, Das G, Sherman F. Transformation of yeast directly with synthetic oligonucleotides. Methods Enzymol. 1991;194:362–369. doi: 10.1016/0076-6879(91)94027-a. [DOI] [PubMed] [Google Scholar]
- 324.Noskov V, Negishi K, Ono A, Matsuda A, Ono B, Hayatsu H. Mutagenicity of 5-bromouracil and N6-hydroxyadenine studied by yeast oligonucleotide transformation assay. Mutat Res. 1994;308:43–51. doi: 10.1016/0027-5107(94)90197-x. [DOI] [PubMed] [Google Scholar]
- 325.Otsuka C, Kobayashi K, Kawaguchi N, Kunitomi N, Moriyama K, Hata Y, Iwai S, Loakes D, Noskov VN, Pavlov Y, Negishi K. Use of yeast transformation by oligonucleotides to study DNA lesion bypass in vivo. Mutat Res. 2002;502:53–60. doi: 10.1016/s0027-5107(02)00023-4. [DOI] [PubMed] [Google Scholar]
- 326.Otsuka C, Sanadai S, Hata Y, Okuto H, Noskov VN, Loakes D, Negishi K. Difference between deoxyribose- and tetrahydrofuran-type abasic sites in the in vivo mutagenic responses in yeast. Nucleic Acids Res. 2002;30:5129–5135. doi: 10.1093/nar/gkf666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Kow YW, Bao G, Minesinger B, Jinks-Robertson S, Siede W, Jiang YL, Greenberg MM. Mutagenic effects of abasic and oxidized abasic lesions in Saccharomyces cerevisiae. Nucleic Acids Res. 2005;33:6196–6202. doi: 10.1093/nar/gki926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Moriya M, Grollman AP. Mutations in the mutY gene of Escherichia coli enhance the frequency of targeted G:C→T:A transversions induced by a single 8-oxoguanine residue in single-stranded DNA. Mol Gen Genet. 1993;239:72–76. doi: 10.1007/BF00281603. [DOI] [PubMed] [Google Scholar]
- 329.Moriya M, Zhang W, Johnson F, Grollman AP. Mutagenic potency of exocyclic DNA adducts: Marked differences between Escherichia coli and simian kidney cells. Proc Natl Acad Sci USA. 1994;91:11899–11903. doi: 10.1073/pnas.91.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Pandya GA, Moriya M. 1,N6-Ethenodeoxyadenosine, a DNA adduct highly mutagenic in mammalian cells. Biochemistry. 1996;35:11487–11492. doi: 10.1021/bi960170h. [DOI] [PubMed] [Google Scholar]
- 331.Moriya M, Spiegel S, Fernandes A, Amin S, Liu T, Geacintov N, Grollman AP. Fidelity of translesional synthesis past benzo[a]pyrene diol epoxide-2′-deoxyguanosine DNA adducts: Marked effects of host cell, sequence context, and chirality. Biochemistry. 1996;35:16646–16651. doi: 10.1021/bi9608875. [DOI] [PubMed] [Google Scholar]
- 332.Fernandes A, Liu T, Amin S, Geacintov NE, Grollman AP, Moriya M. Mutagenic potential of stereoisomeric bay region (+)- and (−)-cis-anti-benzo[a]pyrene diol epoxide-N2-2′-deoxyguanosine adducts in Escherichia coli and simian kidney cells. Biochemistry. 1998;37:10164–10172. doi: 10.1021/bi980401f. [DOI] [PubMed] [Google Scholar]
- 333.Levine RL, Yang IY, Hossain M, Pandya GA, Grollman AP, Moriya M. Mutagenesis induced by a single 1,N6-ethenodeoxyadenosine adduct in human cells. Cancer Res. 2000;60:4098–4104. [PubMed] [Google Scholar]
- 334.Yang IY, Johnson F, Grollman AP, Moriya M. Genotoxic mechanism for the major acrolein-derived deoxyguanosine adduct in human cells. Chem Res Toxicol. 2002;15:160–164. doi: 10.1021/tx010123c. [DOI] [PubMed] [Google Scholar]
- 335.Yang IY, Chan G, Miller H, Huang Y, Torres MC, Johnson F, Moriya M. Mutagenesis by acrolein-derived propanodeoxyguanosine adducts in human cells. Biochemistry. 2002;41:13826–13832. doi: 10.1021/bi0264723. [DOI] [PubMed] [Google Scholar]
- 336.Pollack M, Yang IY, Kim HY, Blair IA, Moriya M. Translesion DNA synthesis across the heptanone-etheno-2′-deoxycytidine adduct in cells. Chem Res Toxicol. 2006;19:1074–1079. doi: 10.1021/tx0600503. [DOI] [PubMed] [Google Scholar]
- 337.Stein S, Lao Y, Yang IY, Hecht SS, Moriya M. Genotoxicity of acetaldehyde- and crotonaldehyde-induced 1,N2-propanodeoxyguanosine DNA adducts in human cells. Mutat Res. 2006;608:1–7. doi: 10.1016/j.mrgentox.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 338.Terashima I, Suzuki N, Itoh S, Yoshizawa I, Shibutani S. Mutagenic specificity of model estrogen-DNA adducts in mammalian cells. Biochemistry. 1998;37:8803–8807. doi: 10.1021/bi980336+. [DOI] [PubMed] [Google Scholar]
- 339.Terashima I, Suzuki N, Shibutani S. Mutagenic potential of α-(N2-deoxyguanosinyl)tamoxifen lesions, the major DNA adducts detected in endometrial tissues of patients treated with tamoxifen. Cancer Res. 1999;59:2091–2095. [PubMed] [Google Scholar]
- 340.Shibutani S, Suzuki N, Grollman AP. Mutagenic specificity of (acetylamino)fluorene-derived DNA adducts in mammalian cells. Biochemistry. 1998;37:12034–12041. doi: 10.1021/bi981059+. [DOI] [PubMed] [Google Scholar]
- 341.Tan X, Suzuki N, Johnson F, Grollman AP, Shibutani S. Mutagenic properties of the 8-amino-2′-deoxyguanosine DNA adduct in mammalian cells. Nucleic Acids Res. 1999;27:2310–2314. doi: 10.1093/nar/27.11.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Shibutani S, Fernandes A, Suzuki N, Zhou L, Johnson F, Grollman AP. Mutagenesis of the N-(deoxyguanosin-8-yl)-2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine DNA adduct in mammalian cells. Sequence context effects. J Biol Chem. 1999;274:27433–27438. doi: 10.1074/jbc.274.39.27433. [DOI] [PubMed] [Google Scholar]
- 343.Tan X, Grollman AP, Shibutani S. Comparison of the mutagenic properties of 8-oxo-7,8-dihydro-2′-deoxyadenosine and 8-oxo-7,8-dihydro-2′-deoxyguanosine DNA lesions in mammalian cells. Carcinogenesis. 1999;20:2287–2292. doi: 10.1093/carcin/20.12.2287. [DOI] [PubMed] [Google Scholar]
- 344.Shibutani S, Suzuki N, Tan X, Johnson F, Grollman AP. Influence of flanking sequence context on the mutagenicity of acetylaminofluorene-derived DNA adducts in mammalian cells. Biochemistry. 2001;40:3717–3722. doi: 10.1021/bi0027581. [DOI] [PubMed] [Google Scholar]
- 345.Tan X, Suzuki N, Grollman AP, Shibutani S. Mutagenic events in Escherichia coli and mammalian cells generated in response to acetylaminofluorene-derived DNA adducts positioned in the Nar I restriction enzyme site. Biochemistry. 2002;41:14255–14262. doi: 10.1021/bi0202878. [DOI] [PubMed] [Google Scholar]
- 346.Dong H, Bonala RR, Suzuki N, Johnson F, Grollman AP, Shibutani S. Mutagenic potential of benzo[a]pyrene-derived DNA adducts positioned in codon 273 of the human p53 gene. Biochemistry. 2004;43:15922–15928. doi: 10.1021/bi0482194. [DOI] [PubMed] [Google Scholar]
- 347.Tan X, Bonala RR, Suzuki N, Johnson F, Grollman AP, Shibutani S. Mutagenic specificity of 2-acetylaminonaphthalene-derived DNA adduct in mammalian cells. Chem-Biol Interact. 2005;152:131–138. doi: 10.1016/j.cbi.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 348.Latham GJ, Zhou L, Harris CM, Harris TM, Lloyd RS. The replication fate of R- and S-styrene oxide adducts on adenine N6 is dependent on both the chirality of the lesion and the local sequence context. J Biol Chem. 1993;268:23427–23434. [PubMed] [Google Scholar]
- 349.Chary P, Latham GJ, Robberson DL, Kim SJ, Han S, Harris CM, Harris TM, Lloyd RS. In vivo and in vitro replication consequences of stereoisomeric benzo[a]pyrene-7,8-dihydrodiol 9,10-epoxide adducts on adenine N6 at the second position of N-ras codon 61. J Biol Chem. 1995;270:4990–5000. doi: 10.1074/jbc.270.10.4990. [DOI] [PubMed] [Google Scholar]
- 350.Latham GJ, McNees AG, DeCorte B, Harris CM, Harris TM, O’Donnell M, Lloyd RS. Comparison of the efficiency of synthesis past single bulky DNA adducts in vivo and in vitro by the polymerase III holoenzyme. Chem Res Toxicol. 1996;9:1167–1175. doi: 10.1021/tx9600558. [DOI] [PubMed] [Google Scholar]
- 351.McNees AG, O’Donnell M, Horton PH, Kim HY, Kim SJ, Harris CM, Harris TM, Lloyd RS. Lack of correlation between in vitro and in vivo replication of precisely defined benz[a]anthracene adducted DNAs. J Biol Chem. 1997;272:33211–33219. doi: 10.1074/jbc.272.52.33211. [DOI] [PubMed] [Google Scholar]
- 352.Carmical JR, Nechev LV, Harris CM, Harris TM, Lloyd RS. Mutagenic potential of adenine N6 adducts of monoepoxide and diolepoxide derivatives of butadiene. Environ Mol Mutagen. 2000;35:48–56. [PubMed] [Google Scholar]
- 353.Carmical JR, Zhang M, Nechev L, Harris CM, Harris TM, Lloyd RS. Mutagenic potential of guanine N2 adducts of butadiene mono- and diolepoxide. Chem Res Toxicol. 2000;13:18–25. doi: 10.1021/tx9901332. [DOI] [PubMed] [Google Scholar]
- 354.Carmical JR, Kowalczyk A, Zou Y, Van Houten B, Nechev LV, Harris CM, Harris TM, Lloyd RS. Butadiene-induced intrastrand DNA cross-links: A possible role in deletion mutagenesis. J Biol Chem. 2000;275:19482–19489. doi: 10.1074/jbc.M002037200. [DOI] [PubMed] [Google Scholar]
- 355.Kanuri M, Finneman J, Harris CM, Harris TM, Lloyd RS. Efficient nonmutagenic replication bypass of DNAs containing β-adducts of styrene oxide at adenine N6. Environ Mol Mutagen. 2001;38:357–360. doi: 10.1002/em.10030. [DOI] [PubMed] [Google Scholar]
- 356.Rodriguez DA, Kowalczyk A, Ward JB, Jr, Harris CM, Harris TM, Lloyd RS. Point mutations induced by 1,2-epoxy-3-butene N1 deoxyinosine adducts. Environ Mol Mutagen. 2001;38:292–296. doi: 10.1002/em.10026. [DOI] [PubMed] [Google Scholar]
- 357.Kanuri M, Nechev LV, Kiehna SE, Tamura PJ, Harris CM, Harris TM, Lloyd RS. Evidence for Escherichia coli polymerase II mutagenic bypass of intrastrand DNA crosslinks. DNA Rep. 2005;4:1374–1380. doi: 10.1016/j.dnarep.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 358.Kanuri M, Minko IG, Nechev LV, Harris TM, Harris CM, Lloyd RS. Error prone translesion synthesis past γ-hydroxypropano deoxyguanosine, the primary acrolein-derived adduct in mammalian cells. J Biol Chem. 2002;277:18257–18265. doi: 10.1074/jbc.M112419200. [DOI] [PubMed] [Google Scholar]
- 359.Kanuri M, Nechev LV, Tamura PJ, Harris CM, Harris TM, Lloyd RS. Mutagenic spectrum of butadiene-derived N1-deoxyinosine adducts and N6,N6-deoxyadenosine intrastrand cross-links in mammalian cells. Chem Res Toxicol. 2002;15:1572–1580. doi: 10.1021/tx025591g. [DOI] [PubMed] [Google Scholar]
- 360.Fernandes PH, Wang H, Rizzo CJ, Lloyd RS. Site-specific mutagenicity of stereochemically defined 1,N2-deoxyguanosine adducts of trans-4-hydroxynonenal in mammalian cells. Environ Mol Mutagen. 2003;42:68–74. doi: 10.1002/em.10174. [DOI] [PubMed] [Google Scholar]
- 361.Sanchez AM, Minko IG, Kurtz AJ, Kanuri M, Moriya M, Lloyd RS. Comparative evaluation of the bioreactivity and mutagenic spectra of acrolein-derived α-HOPdG and γ-HOPdG regioisomeric deoxyguanosine adducts. Chem Res Toxicol. 2003;16:1019–1028. doi: 10.1021/tx034066u. [DOI] [PubMed] [Google Scholar]
- 362.Fernandes PH, Kanuri M, Nechev LV, Harris TM, Lloyd RS. Mammalian cell mutagenesis of the DNA adducts of vinyl chloride and crotonaldehyde. Environ Mol Mutagen. 2005;45:455–459. doi: 10.1002/em.20117. [DOI] [PubMed] [Google Scholar]
- 363.Fernandes PH, Hackfeld LC, Kozekov ID, Hodge RP, Lloyd RS. Synthesis and mutagenesis of the butadiene-derived N3 2′-deoxyuridine adducts. Chem Res Toxicol. 2006;19:968–976. doi: 10.1021/tx060016o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Kamiya H, Miura K, Ohtomo N, Nishimura S, Ohtsuka E. Transforming activity of a synthetic c-Ha-ras gene containing O6-methylguanine in codon 12. Jpn J Cancer Res. 1991;82:997–1002. doi: 10.1111/j.1349-7006.1991.tb01934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Kamiya H, Miura H, Kato H, Nishimura S, Ohtsuka E. Induction of mutation of a synthetic c-Ha-ras gene containing hypoxanthine. Cancer Res. 1992;52:1836–1839. [PubMed] [Google Scholar]
- 366.Kamiya H, Shimizu M, Suzuki M, Inoue H, Ohtsuka E. Mutation induced by deoxyxanthosine in codon 12 of a synthetic c-Ha-ras gene. Nucleosides Nucleotides. 1992;11:247–260. [PubMed] [Google Scholar]
- 367.Kamiya H, Miura K, Ishikawa H, Inoue H, Nishimura S, Ohtsuka E. c-Ha-ras containing 8-hydroxyguanine at codon 12 induces point mutations at the modified and adjacent positions. Cancer Res. 1992;52:3483–3485. [PubMed] [Google Scholar]
- 368.Kamiya H, Suzuki M, Komatsu Y, Miura H, Kikuchi K, Sakaguchi T, Murata N, Masutani C, Hanaoka F, Ohtsuka E. An abasic site analogue activates a c-Ha-ras gene by a point mutation at modified and adjacent positions. Nucleic Acids Res. 1992;20:4409–4415. doi: 10.1093/nar/20.17.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Kamiya H, Murata N, Murata T, Iwai S, Matsukage A, Masutani C, Hanaoka F, Ohtsuka E. Cyclobutane thymine dimers in a ras proto-oncogene hot spot activate the gene by point mutation. Nucleic Acids Res. 1993;21:2355–2361. doi: 10.1093/nar/21.10.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Kamiya H, Suzuki M, Ohtsuka E. Mutation-spectrum of a true abasic site in codon 12 of a c-Ha-ras gene in mammalian cells. FEBS Lett. 1993;328:125–129. doi: 10.1016/0014-5793(93)80979-5. [DOI] [PubMed] [Google Scholar]
- 371.Kamiya H, Murata-Kamiya N, Koizume S, Inoue H, Nishimura S, Ohtsuka E. 8-Hydroxyguanine (7,8-dihydro-8-oxoguanine) in hot spots of the c-Ha-ras gene: Effects of sequence contexts on mutation spectra. Carcinogenesis. 1995;16:883–889. doi: 10.1093/carcin/16.4.883. [DOI] [PubMed] [Google Scholar]
- 372.Kamiya H, Miura H, Murata-Kamiya N, Ishikawa H, Sakaguchi T, Inoue H, Sasaki T, Masutani C, Hanaoka F, Nishimura S, Ohtsuka E. 8-Hydroxyadenine (7,8-dihydro-8-oxoadenine) induces misincorporation in in vitro DNA synthesis and mutations in NIH 3T3 cells. Nucleic Acids Res. 1995;23:2893–2899. doi: 10.1093/nar/23.15.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Kamiya H, Kasai H. Mutations induced by 2-hydroxyadenine on a shuttle vector during leading and lagging strand syntheses in mammalian cells. Biochemistry. 1997;36:11125–11130. doi: 10.1021/bi970871u. [DOI] [PubMed] [Google Scholar]
- 374.Kamiya H, Iwai S, Kasai H. The (6–4) photoproduct of thymine-thymine induces targeted substitution mutations in mammalian cells. Nucleic Acids Res. 1998;26:2611–2617. doi: 10.1093/nar/26.11.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Kamiya H, Murata-Kamiya N, Karino N, Ueno Y, Matsuda A, Kasai H. Induction of T→G and T→A transversions by 5-formyluracil in mammalian cells. Mutat Res. 2002;513:213–222. doi: 10.1016/s1383-5718(01)00312-6. [DOI] [PubMed] [Google Scholar]
- 376.Loechler EL, Green CL, Essigmann JM. In vivo mutagenesis by O6-methylguanine built into a unique site in a viral genome. Proc Natl Acad Sci USA. 1984;81:6271–6275. doi: 10.1073/pnas.81.20.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Basu AK, Wood ML, Niedernhofer LJ, Ramos LA, Essigmann JM. Mutagenic and genotoxic effects of three vinyl chloride-induced DNA lesions: 1,N6-ethenoadenine, 3,N4-ethenocytosine, and 4-amino-5-(imidazol-2-yl)imidazole. Biochemistry. 1993;32:12793–12801. doi: 10.1021/bi00210a031. [DOI] [PubMed] [Google Scholar]
- 378.Berardini M, Mackay W, Loechler EL. Evidence for a recombination-independent pathway for the repair of DNA interstrand cross-links based on a site-specific study with nitrogen mustard. Biochemistry. 1997;36:3506–3513. doi: 10.1021/bi962778w. [DOI] [PubMed] [Google Scholar]
- 379.Berardini M, Foster PL, Loechler EL. DNA polymerase II (polB) is involved in a new DNA repair pathway for DNA interstrand cross-links in Escherichia coli. J Bacteriol. 1999;181:2878–2882. doi: 10.1093/gao/9781884446054.article.t031385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Mackay W, Benasutti M, Drouin E, Loechler EL. Mutagenesis by (+)-anti-B[a]P-N2-Gua, the major adduct of activated benzo[a]pyrene, when studied in an Escherichia coli plasmid using site-directed methods. Carcinogenesis. 1992;13:1415–1425. doi: 10.1093/carcin/13.8.1415. [DOI] [PubMed] [Google Scholar]
- 381.Jelinsky SA, Liu T, Geacintov NE, Loechler EL. The major, N2-Gua adduct of the (+)-anti-benzo[a]pyrene diol epoxide is capable of inducing G→A and G→C, in addition to G→T, mutations. Biochemistry. 1995;34:13545–13553. doi: 10.1021/bi00041a034. [DOI] [PubMed] [Google Scholar]
- 382.Hanrahan CJ, Bacolod MD, Vyas RR, Liu T, Geacintov NE, Loechler EL, Basu AK. Sequence specific mutagenesis of the major (+)-anti-benzo[a]pyrene diol epoxide-DNA adduct at a mutational hot spot in vitro and in Escherichia coli cells. Chem Res Toxicol. 1997;10:369–377. doi: 10.1021/tx9601925. [DOI] [PubMed] [Google Scholar]
- 383.Shukla R, Liu T, Geacintov NE, Loechler EL. The major, N2-dG adduct of (+)-anti-B[a]PDE shows a dramatically different mutagenic specificity (predominantly, G→A) in a 5′-CGT-3′ sequence context. Biochemistry. 1997;36:10256–10261. doi: 10.1021/bi970541+. [DOI] [PubMed] [Google Scholar]
- 384.Shukla R, Jelinsky S, Liu T, Geacintov NE, Loechler EL. How stereochemistry affects mutagenesis by N2-deoxyguanosine adducts of 7,8-dihydroxy-9,10-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene: Configuration of the adduct bond is more important than those of the hydroxyl groups. Biochemistry. 1997;36:13263–13269. doi: 10.1021/bi971195z. [DOI] [PubMed] [Google Scholar]
- 385.Shukla R, Geacintov NE, Loechler EL. The major, N2-dG adduct of (+)-anti-B[a]PDE induces G→A mutations in a 5′-AGA-3′ sequence context. Carcinogenesis. 1999;20:261–268. doi: 10.1093/carcin/20.2.261. [DOI] [PubMed] [Google Scholar]
- 386.Yin J, Seo KY, Loechler EL. A role for DNA polymerase V in G→T mutations from the major benzo[a]pyrene N2-dG adduct when studied in a 5′-TGT sequence in E. coli. DNA Rep. 2004;3:323–334. doi: 10.1016/j.dnarep.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 387.Nagalingam A, Seo KY, Loechler EL. Mutagenesis studies of the major benzo[a]pyrene N2-dG adduct in a 5′-TG versus a 5′-UG sequence: Removal of the methyl group causes a modest decrease in the [G→T/G→A] mutational ratio. Mutagenesis. 2005;20:105–110. doi: 10.1093/mutage/gei014. [DOI] [PubMed] [Google Scholar]
- 388.Seo KY, Nagalingam A, Tiffany M, Loechler EL. Mutagenesis studies with four stereoisomeric N2-dG benzo[a]pyrene adducts in the identical 5′-CGC sequence used in NMR studies: G→T mutations dominate in each case. Mutagenesis. 2005;20:441–448. doi: 10.1093/mutage/gei061. [DOI] [PubMed] [Google Scholar]
- 389.Seo KY, Nagalingam A, Miri S, Yin J, Chandani S, Kolbanovskiy A, Shastry A, Loechler EL. Mirror image stereoisomers of the major benzo[a]pyrene N2-dG adduct are bypassed by different lesion-bypass DNA polymerases in E. coli. DNA Rep. 2006;5:515–522. doi: 10.1016/j.dnarep.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 390.Kozack RE, Loechler EL. Molecular modeling of the conformational complexity of (+)-anti-B[a]PDE-adducted DNA using simulated annealing. Carcinogenesis. 1997;18:1585–1593. doi: 10.1093/carcin/18.8.1585. [DOI] [PubMed] [Google Scholar]
- 391.Kozack RE, Shukla R, Loechler EL. A hypothesis for what conformation of the major adduct of (+)-anti-B[a]PDE (N2-dG) causes G→T versus G→A mutations based upon a correlation between mutagenesis and molecular modeling results. Carcinogenesis. 1999;20:95–102. doi: 10.1093/carcin/20.1.95. [DOI] [PubMed] [Google Scholar]
- 392.Kozack RE, Loechler EL. Molecular modeling of the major adduct of (+)-anti-B[a]PDE (N2-dG) in the eight conformations and the five DNA sequences most relevant to base substitution mutagenesis. Carcinogenesis. 1999;20:85–94. doi: 10.1093/carcin/20.1.85. [DOI] [PubMed] [Google Scholar]
- 393.Lee CH, Chandani S, Loechler EL. Molecular modeling of four stereoisomers of the major B[a]PDE adduct (at N2-dG) in five cases where the structure is known from NMR studies: Molecular modeling is consistent with NMR results. Chem Res Toxicol. 2002;15:1429–1444. doi: 10.1021/tx0200257. [DOI] [PubMed] [Google Scholar]
- 394.Lee CH, Loechler EL. Molecular modeling of the major benzo[a]pyrene N2-dG adduct in cases where mutagenesis results are known in double stranded DNA. Mutat Res. 2003;529:59–76. doi: 10.1016/s0027-5107(03)00107-6. [DOI] [PubMed] [Google Scholar]
- 395.Chandani S, Lee CH, Loechler EL. Free-energy perturbation methods to study structure and energetics of DNA adducts: Results for the major N2-dG adduct of benzo[a]pyrene in two conformations and different sequence contexts. Chem Res Toxicol. 2005;18:1108–1123. doi: 10.1021/tx049646l. [DOI] [PubMed] [Google Scholar]
- 396.Chandani S, Loechler EL. Molecular modeling benzo[a]pyrene N2-dG adducts in the two overlapping active sites of the Y-family DNA polymerase Dpo4. J Mol Graphics Modell. 2007;25:658–670. doi: 10.1016/j.jmgm.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 397.Malia SA, Vyas RR, Basu AK. Site-specific frame-shift mutagenesis by the 1-nitropyrene-DNA adduct N-(deoxyguanosin-8-y1)-1-aminopyrene located in the (CG)3 sequence: Effects of SOS, proofreading, and mismatch repair. Biochemistry. 1996;35:4568–4577. doi: 10.1021/bi9525132. [DOI] [PubMed] [Google Scholar]
- 398.Bacolod MD, Krishnasamy R, Basu AK. Mutagenicity of the 1-nitropyrene-DNA adduct N-(deoxyguanosin-8-yl)-1-aminopyrene in Escherichia coli located in a nonrepetitive CGC sequence. Chem Res Toxicol. 2000;13:523–528. doi: 10.1021/tx000023r. [DOI] [PubMed] [Google Scholar]
- 399.Venkatarangan L, Sivaprasad A, Johnson F, Basu AK. Site-specifically located 8-amino-2′-deoxyguanosine: Thermodynamic stability and mutagenic properties in Escherichia coli. Nucleic Acids Res. 2001;29:1458–1463. doi: 10.1093/nar/29.7.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Bacolod MD, Basu AK. Mutagenicity of a single 1-nitropyrene-DNA adduct N-(deoxyguanosin-8-yl)-1-aminopyrene in Escherichia coli located in a GGC sequence. Mutagenesis. 2001;16:461–465. doi: 10.1093/mutage/16.6.461. [DOI] [PubMed] [Google Scholar]
- 401.Hilario P, Yan S, Hingerty BE, Broyde S, Basu AK. Comparative mutagenesis of the C8-guanine adducts of 1-nitropyrene and 1,6- and 1,8-dinitropyrene in a CpG repeat sequence. A slipped frameshift intermediate model for dinucleotide deletion. J Biol Chem. 2002;277:45068–45074. doi: 10.1074/jbc.M208103200. [DOI] [PubMed] [Google Scholar]
- 402.Utzat CD, Clement CC, Ramos LA, Das A, Tomasz M, Basu AK. DNA adduct of the mitomycin C metabolite 2,7-diaminomitosene is a nontoxic and nonmutagenic DNA lesion in vitro and in vivo. Chem Res Toxicol. 2005;18:213–223. doi: 10.1021/tx049813h. [DOI] [PubMed] [Google Scholar]
- 403.Kalam MA, Basu AK. Mutagenesis of 8-oxoguanine adjacent to an abasic site in simian kidney cells: Tandem mutations and enhancement of G→T transversions. Chem Res Toxicol. 2005;18:1187–1192. doi: 10.1021/tx050119r. [DOI] [PubMed] [Google Scholar]
- 404.Kalam MA, Haraguchi K, Chandani S, Loechler EL, Moriya M, Greenberg MM, Basu AK. Genetic effects of oxidative DNA damages: Comparative mutagenesis of the imidazole ring-opened formamidopyrimidines (Fapy lesions) and 8-oxo-purines in simian kidney cells. Nucleic Acids Res. 2006;34:2305–2315. doi: 10.1093/nar/gkl099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Watt DL, Utzat CD, Hilario P, Basu AK. Mutagenicity of the 1-nitropyrene-DNA adduct N-(deoxyguanosin-8-yl)-1-aminopyrene in mammalian cells. Chem Res Toxicol. 2007;20:1658–1664. doi: 10.1021/tx700131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Miller EC. Some current perspectives on chemical carcino-genesis in humans and experimental animals: Presidential address. Cancer Res. 1978;38:1479–1496. [PubMed] [Google Scholar]
- 407.Delaney JC, Essigmann JM. Assays for determining lesion bypass efficiency and mutagenicity of site-specific DNA lesions in vivo. Methods Enzymol. 2006;408:1–15. doi: 10.1016/S0076-6879(06)08001-3. [DOI] [PubMed] [Google Scholar]
- 408.Delaney JC, Essigmann JM. Effect of sequence context on O6-methylguanine repair and replication in vivo. Biochemistry. 2001;40:14968–14975. doi: 10.1021/bi015578f. [DOI] [PubMed] [Google Scholar]
- 409.Henderson PT, Delaney JC, Gu F, Tannenbaum SR, Essigmann JM. Oxidation of 7,8-dihydro-8-oxoguanine affords lesions that are potent sources of replication errors in vivo. Biochemistry. 2002;41:914–921. doi: 10.1021/bi0156355. [DOI] [PubMed] [Google Scholar]
- 410.Delaney JC, Henderson PT, Helquist SA, Morales JC, Essigmann JM, Kool ET. High-fidelity in vivo replication of DNA base shape mimics without Watson-Crick hydrogen bonds. Proc Natl Acad Sci USA. 2003;100:4469–4473. doi: 10.1073/pnas.0837277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Henderson PT, Delaney JC, Muller JG, Neeley WL, Tannenbaum SR, Burrows CJ, Essigmann JM. The hydantoin lesions formed from oxidation of 7,8-dihydro-8-oxoguanine are potent sources of replication errors in vivo. Biochemistry. 2003;42:9257–9262. doi: 10.1021/bi0347252. [DOI] [PubMed] [Google Scholar]
- 412.Delaney JC, Essigmann JM. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc Natl Acad Sci USA. 2004;101:14051–14056. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Neeley WL, Delaney JC, Henderson PT, Essigmann JM. In vivo bypass efficiencies and mutational signatures of the guanine oxidation products 2-aminoimidazolone and 5-guanidino-4-nitroimidazole. J Biol Chem. 2004;279:43568–43573. doi: 10.1074/jbc.M407117200. [DOI] [PubMed] [Google Scholar]
- 414.Henderson PT, Neeley WL, Delaney JC, Gu F, Niles JC, Hah SS, Tannenbaum SR, Essigmann JM. Urea lesion formation in DNA as a consequence of 7,8-dihydro-8-oxoguanine oxidation and hydrolysis provides a potent source of point mutations. Chem Res Toxicol. 2005;18:12–18. doi: 10.1021/tx049757k. [DOI] [PubMed] [Google Scholar]
- 415.Delaney JC, Smeester L, Wong C, Frick LE, Taghizadeh K, Wishnok JS, Drennan CL, Samson LD, Essigmann JM. AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat Struct Mol Biol. 2005;12:855–860. doi: 10.1038/nsmb996. [DOI] [PubMed] [Google Scholar]
- 416.Kim TW, Delaney JC, Essigmann JM, Kool ET. Probing the active site tightness of DNA polymerase in subangstrom increments. Proc Natl Acad Sci USA. 2005;102:15803–15808. doi: 10.1073/pnas.0505113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Frick LE, Delaney JC, Wong C, Drennan CL, Essigmann JM. Alleviation of 1,N6-ethanoadenine genotoxicity by the Escherichia coli adaptive response protein AlkB. Proc Natl Acad Sci USA. 2007;104:755–760. doi: 10.1073/pnas.0607377104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Delaney S, Neeley WL, Delaney JC, Essigmann JM. The substrate specificity of MutY for hyperoxidized guanine lesions in vivo. Biochemistry. 2007;46:1448–1455. doi: 10.1021/bi061174h. [DOI] [PubMed] [Google Scholar]
- 419.Neeley WL, Delaney S, Alekseyev YO, Jarosz DF, Delaney JC, Walker GC, Essigmann JM. DNA polymerase V allows bypass of toxic guanine oxidation products in vivo. J Biol Chem. 2007;282:12741–12748. doi: 10.1074/jbc.M700575200. [DOI] [PubMed] [Google Scholar]
- 420.Delaney S, Delaney JC, Essigmann JM. Chemical-biological fingerprinting: Probing the properties of DNA lesions formed by peroxynitrite. Chem Res Toxicol. 2007;20:1718–1729. doi: 10.1021/tx700273u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Kroeger KM, Jiang YL, Kow YW, Goodman MF, Greenberg MM. Mutagenic effects of 2-deoxyribonolactone in Escherichia coli. An abasic lesion that disobeys the A-rule. Biochemistry. 2004;43:6723–6733. doi: 10.1021/bi049813g. [DOI] [PubMed] [Google Scholar]
- 422.Kroeger KM, Goodman MF, Greenberg MM. A comprehensive comparison of DNA replication past 2-deoxyribose and its tetrahydrofuran analog in Escherichia coli. Nucleic Acids Res. 2004;32:5480–5485. doi: 10.1093/nar/gkh873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Kroeger KM, Kim J, Goodman MF, Greenberg MM. Effects of the C4′-oxidized abasic site on replication in Escherichia coli. An unusually large deletion is induced by a small lesion. Biochemistry. 2004;43:13621–13627. doi: 10.1021/bi048337r. [DOI] [PubMed] [Google Scholar]
- 424.Kroeger KM, Kim J, Goodman MF, Greenberg MM. Replication of an oxidized abasic site in Escherichia coli by a dNTP-stabilized misalignment mechanism that reads upstream and downstream nucleotides. Biochemistry. 2006;45:5048–5056. doi: 10.1021/bi052276v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Patro JN, Wiederholt CJ, Jiang YL, Delaney JC, Essigmann JM, Greenberg MM. Studies on the replication of the ring opened formamidopyrimidine, Fapy·dG in Escherichia coli. Biochemistry. 2007;46:10202–10212. doi: 10.1021/bi700628c. [DOI] [PubMed] [Google Scholar]
- 426.Ellison KS, Dogliotti E, Connors TD, Basu AK, Essigmann JM. Site-specific mutagenesis by O6-alkylguanines located in the chromosomes of mammalian cells: Influence of the mammalian O6-alkylguanine-DNA alkyltransferase. Proc Natl Acad Sci USA. 1989;86:8620–8624. doi: 10.1073/pnas.86.22.8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Altshuler KB, Hodes CS, Essigmann JM. Intrachromosomal probes for mutagenesis by alkylated DNA bases replicated in mammalian cells: A comparison of the mutagenicities of O4-methylthymine and O6-methylguanine in cells with different DNA repair backgrounds. Chem Res Toxicol. 1996;9:980–987. doi: 10.1021/tx960062w. [DOI] [PubMed] [Google Scholar]
- 428.Akasaka S, Guengerich FP. Mutagenicity of site-specifically located 1,N2-ethenoguanine in Chinese hamster ovary cell chromosomal DNA. Chem Res Toxicol. 1999;12:501–507. doi: 10.1021/tx980259j. [DOI] [PubMed] [Google Scholar]
- 429.Aas PA, Otterlei M, Falnes PO, Vågbø CB, Skorpen F, Akbari M, Sundheim O, Bjørås M, Slupphaug G, Seeberg E, Krokan HE. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 430.Ougland R, Zhang CM, Liiv A, Johansen RF, Seeberg E, Hou YM, Remme J, Falnes PO. AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol Cell. 2004;16:107–116. doi: 10.1016/j.molcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 431.Kabnick KS, Housman DE. Determinants that contribute to cytoplasmic stability of human c-fos and β-globin mRNAs are located at several sites in each mRNA. Mol Cell Biol. 1988;8:3244–3250. doi: 10.1128/mcb.8.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Morceau F, Dupont C, Palissot V, Borde-Chiché P, Trentesaux C, Dicato M, Diederich M. GTP-mediated differentiation of the human K562 cell line: Transient overexpression of GATA-1 and stabilization of the γ-globin mRNA. Leukemia. 2000;14:1589–1597. doi: 10.1038/sj.leu.2401890. [DOI] [PubMed] [Google Scholar]
- 433.Yarema KJ, Wilson JM, Lippard SJ, Essigmann JM. Effects of DNA adduct structure and distribution on the mutagenicity and genotoxicity of two platinum anticancer drugs. J Mol Biol. 1994;236:1034–1048. doi: 10.1016/0022-2836(94)90010-8. [DOI] [PubMed] [Google Scholar]
- 434.Yarema KJ, Lippard SJ, Essigmann JM. Mutagenic and genotoxic effects of DNA adducts formed by the anticancer drug cis-diamminedichloroplatinum(II) Nucleic Acids Res. 1995;23:4066–4072. doi: 10.1093/nar/23.20.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Bailey EA, Iyer RS, Stone MP, Harris TM, Essigmann JM. Mutational properties of the primary aflatoxin B1-DNA adduct. Proc Natl Acad Sci USA. 1996;93:1535–1539. doi: 10.1073/pnas.93.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]